Abstract
The diversity, ubiquity and prevalence in deep waters of the octocoral family Chrysogorgiidae Verrill, 1883 make it noteworthy as a model system to study radiation and diversification in the deep sea. Here we provide the first comprehensive phylogenetic analysis of the Chrysogorgiidae, and compare phylogeny and depth distribution. Phylogenetic relationships among 10 of 14 currently-described Chrysogorgiidae genera were inferred based on mitochondrial (mtMutS, cox1) and nuclear (18S) markers. Bathymetric distribution was estimated from multiple sources, including museum records, a literature review, and our own sampling records (985 stations, 2345 specimens). Genetic analyses suggest that the Chrysogorgiidae as currently described is a polyphyletic family. Shallow-water genera, and two of eight deep-water genera, appear more closely related to other octocoral families than to the remainder of the monophyletic, deep-water chrysogorgiid genera. Monophyletic chrysogorgiids are composed of strictly (Iridogorgia Verrill, 1883, Metallogorgia Versluys, 1902, Radicipes Stearns, 1883, Pseudochrysogorgia Pante & France, 2010) and predominantly (Chrysogorgia Duchassaing & Michelotti, 1864) deep-sea genera that diversified in situ. This group is sister to gold corals (Primnoidae Milne Edwards, 1857) and deep-sea bamboo corals (Keratoisidinae Gray, 1870), whose diversity also peaks in the deep sea. Nine species of Chrysogorgia that were described from depths shallower than 200 m, and mtMutS haplotypes sequenced from specimens sampled as shallow as 101 m, suggest a shallow-water emergence of some Chrysogorgia species.
Introduction
Corals of the family Chrysogorgiidae Verrill, 1883 are conspicuous members of deep benthic assemblages. They are found in all major oceans, as far north as Iceland [1] and as far south as Antarctica [2]. They have been described from a variety of habitats, including shallow-water reefs [3], soft sediments, and hard bottoms (e.g., [4]). They were recently described as predominant members of benthic communities on NW Atlantic seamounts [5], [6]. The Chrysogorgiidae are particularly diverse, with about one hundred described species. In his original description of the family, Verrill [7] presented the Chrysogorgiidae as including “some of the most beautiful and interesting of all the known Gorgonians.”
The family ranges between approximately 100 and 3375 m depth [8], most species (>75%) inhabiting deep waters. The family is an assemblage of deep-water specialists (e.g., Metallogorgia Versluys, 1902 and Iridogorgia Verrill, 1883), a shallow-water specialist (Stephanogorgia Bayer & Muzik 1976), and eurybathic genera (Chrysogorgia Duchassaing & Michelotti, 1864 and Radicipes Stearns, 1883). The variety and gradualism in the bathymetric range of the family makes it a noteworthy model system for the study of diversification and radiation in the deep sea. However, these advantages as a model system have little value if the family Chrysogorgiidae is an artificial assemblage of polyphyletic taxa.
Indeed, despite their ubiquity and relative abundance, little is known about the phylogeny of chrysogorgiid corals. The evolutionary history of the Chrysogorgiidae has, to date, not been appropriately studied. McFadden et al. [9], in their genus-level phylogenetic reconstruction of the subclass Octocorallia Haeckel, 1866, included four of the 12 genera described at the time, and retrieved a monophyletic group. However, the specimens used were all from deep waters and did not cover the morphological, ecological and biogeographic variation observed in the family.
In fact, most octocoral families – as they are currently known – are likely not monophyletic. McFadden et al. [9] included 28 of the 44 octocoral families in their analysis, 14 of which were represented by multiple genera. Only five of these 14 were monophyletic, but even these were based on limited taxonomic data: only a third or less of the genera described in each of these five families were analyzed. Under-representation was particularly important for the Isididae Lamouroux, 1812, for which only two out of 38 described genera (5%) were included, and these two both from only 1 of the 4 subfamilies.
In light of the prevalence of polyphyly among currently-described octocoral families, there is little guarantee that a phylogenetic analysis of the Chrysogorgiidae based on broader taxonomic sampling will recover a monophyletic group. In addition, none of the molecular phylogenies including chrysogorgiids produced to date [9]–[14] have assessed the monophyly of genera. The genus Chrysogorgia, in particular, is the most speciose (60+ species) and geographically the most wide-ranging of the Chrysogorgiidae [8]. As Chrysogorgia makes about 60% of the species diversity in the family, assessing its monophyly is of particular importance.
Our first goal was to test the monophyly of the family Chrysogorgiidae. Second, we compared depth distributions and evolutionary relationships among genera to evaluate the hypothesis that the two are correlated (i.e., shallow-water taxa are derived from deep-water ones, or vice-versa). We inferred phylogenetic relationships based on taxa from 10 of 14 currently-recognized genera, using mitochondrial (mtMutS and cox1) and nuclear (18S) markers. Bathymetric ranges were estimated based on our collections, museum records, and a literature review. The family Chrysogorgiidae is put in a broad evolutionary context by the inclusion of DNA sequences and distributional information from all other families of the suborder Calcaxonia (the Isididae, Primnoidae Milne Edwards, 1857, Ellisellidae Gray, 1859, and Ifalukellidae Bayer, 1955).
Results
Informativeness and Congruence among Genetic Markers
The information contents of mtMutS, cox1 and 18S were assessed by looking at 42 calcaxonian colonies (Table 1). The 5′ end of mtMutS was the most variable region, with 31% of sites being variable. The gene as a whole is slightly less variable (28%). 18S and cox11 were 50% less variable than mtMutS (Table 1). The same pattern was observed for parsimony-informative sites. A total of 34 haplotypes were differentiated by the first 781 bp of the mtMutS alignment. The entire gene sequence differentiated 37 haplotypes, while cox1 and 18S differentiated 28 and 40 haplotypes, respectively. All cox1 haplotypes were also differentiated by mtMutS. The additional richness observed at 18S is attributed to ambiguous positions. The molecular variation found at mtMutS and cox1, and the diagnostic potential of these markers for barcoding is further detailed in [15].
Table 1. Length and information content of mtMutS, cox1 and 18S alignments, alone and concatenated.
| Alignment | N. taxa | N. nt | Nt. min-max | N. var | N. pars | Model (AIC) | Model (BIC) | |
| 1 | mtMutS 5′ | 105 | 829 | 691–799 | 384 (46%) | 259 (31%) | TVM+G | TVM+G |
| 2 | mtMutS 5′ (Gblocks) | 105 | 688 | 679–688 | 316 (46%) | 227 (33%) | TVM+G | TVM+G |
| 3 | mtMutS | 46 | 3150 | 2889–2997 | 896 (28%) | 594 (19%) | TVM+I+G | TVM+G |
| 4 | mtMutS (Gblocks) | 46 | 2871 | 2832–2871 | 837 (29%) | 571 (20%) | TVM+I+G | TVM+G |
| 5 | cox1 | 64 | 786 | 786–786 | 133 (17%) | 92 (12%) | TVM+I+G | TPM1uf+G |
| 6 | 18S | 64 | 1315 | 1293–1307 | 215 (16%) | 181 (14%) | GTR+I+G | TIM2ef+I+G |
| 7 | mtMutS (5′), cox1, 18S | 64 | 2924 | 2773–2880 | 645 (22%) | 478 (16%) | GTR+I+G | GTR+I+G |
| 8 | mtMutS, cox1, 18S | 42 | 5236 | 4969–5078 | 1192 (23%) | 819 (16%) | GTR+I+G | TVM+I+G |
| 9 | mtMutS (5′) | 42 | 781 | 691–769 | 245 (31%) | 168 (22%) | ||
| 10 | 18S | 42 | 1300 | 1293–1297 | 189 (15%) | 160 (12%) | ||
| 11 | cox1 | 42 | 786 | 786–786 | 108 (14%) | 72 (9%) | ||
| 12 | mtMutS | 42 | 3150 | 2889–2997 | 895 (28%) | 587 (19%) |
Alignments 1–8 were used in phylogenetic analyses. Alignments 8–12 were used to compare levels of variation and information content, based on data from 42 individuals. Gblocks: alignment shortened using Gblocks. N. nt: alignment length in nucleotides. Nt. min-max: shortest and longest sequences (de-gapped). N. var and N. pars: number of variable and parsimony-informative sites (and percentage of total alignment length). Model (AIC) and Model (BIC): model of evolution that best described the data, based on jModelTest runs.
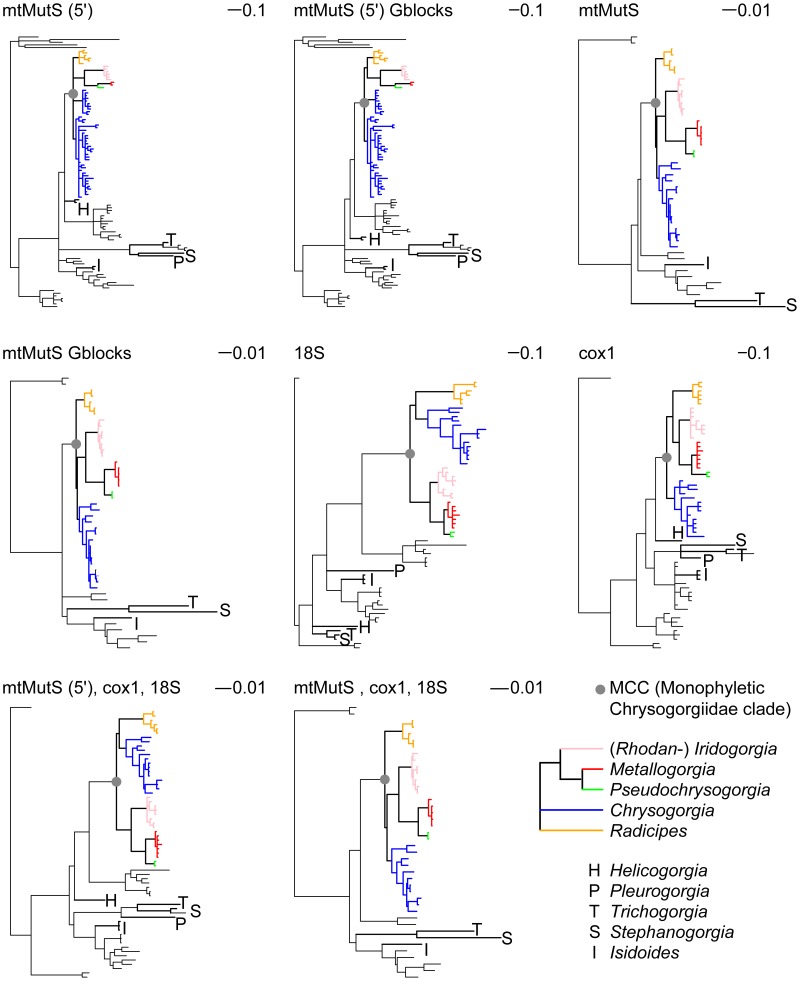
The phylogenetic signal contained in the three markers was congruent overall, and recovered the same clades at the genus and family levels (see below and Figure 1). Concatenating markers significantly improved clade support, with the notable exception of the relationship between Chrysogorgia, Radicipes, and the clade composed of Iridogorgia, Rhodaniridogorgia Watling, 2007, Metallogorgia, and Pseudochrysogorgia Pante & France, 2010 (see below). Removing complex indels of doubtful homology at mtMutS did not significantly improve taxonomic resolution. Indel removal, however, had the effect of influencing branch lengths (Figures 1 and S1).
Figure 1. Bayesian 50% majority rule consensus trees based on different markers (mtMutS, cox1 and 18S) and marker combinations.
For mtMutS, the effect of indels on phylogeny inference was tested by removing them with Gblocks. Chrysogorgiidae taxa are either color coded (MCC) or have bolded branches (nonMCC).The MCC clade is evidenced by a gray circle. All trees are rooted to the Pennatulacea, except trees using the entire mtMutS gene (rooted to the Ellisellidae). For sake of clarity, tip labels and node support values were removed, but can be consulted on Figure S1.
Family Monophyly
Genetic data strongly suggest that the family as currently described is not monophyletic. The following genera formed a strongly-supported phylogenetic clade: Iridogorgia, Rhodaniridogorgia, Metallogorgia, Pseudochrysogorgia, Radicipes and Chrysogorgia (Figures 2 and 3). We refer to this group as the “monophyletic Chrysogorgiidae clade” (MCC), as it is the only monophyletic group retrieved for genera within the family. In addition, the type species for the family, Chrysogorgia desbonni (Duchassaing & Michelotti, 1864), is from a genus in this clade. While the phylogenetic relationship between Radicipes and Chrysogorgia was difficult to retrieve with strong support, Iridogorgia, Metallogorgia and Pseudochrysogorgia consistently formed a very well-supported monophyletic clade.
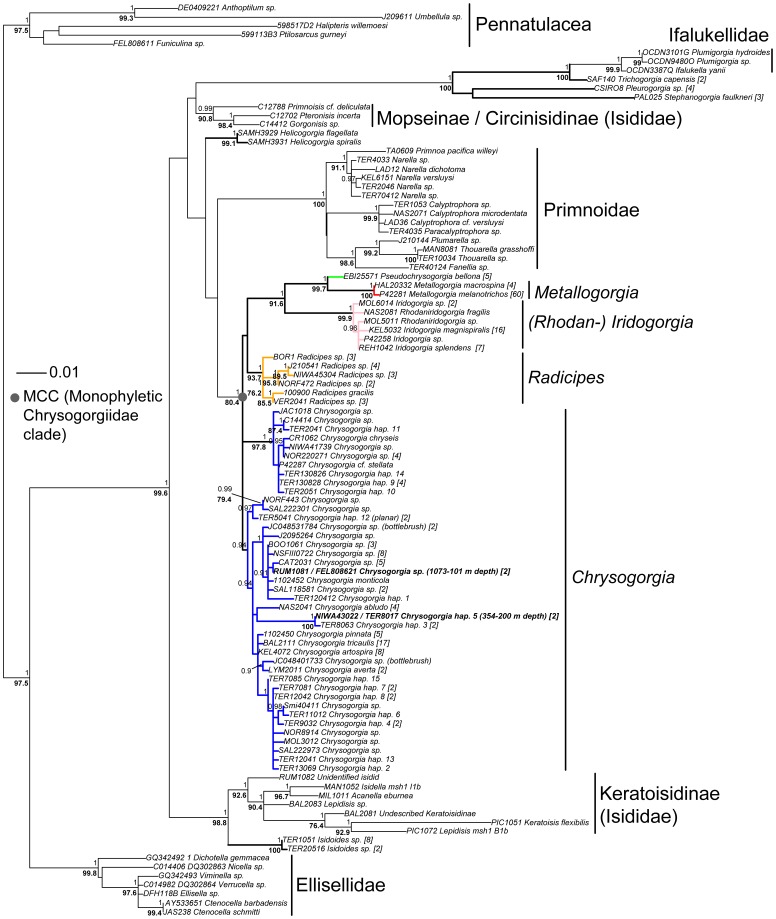
Figure 2. Maximum likelihood reconstruction of the suborder Calcaxonia (rooted to five sea pens; Order Pennatulacea) based on the 5′ end of mtMutS (102 taxa, 272 colonies, 829 bp; TVM+G model; 1000 bootstrap replicates).
All five families of the Calcaxonia are represented. Node support values from the ML analysis (bold, only values >70% shown) are indicated under each node, and node support values from the Bayesian analysis (>0.90) are above each node. Chrysogorgiidae taxa are either color coded (MCC) or have bolded branches (nonMCC). The collection depth of the shallowest Chrysogorgia specimens (≤200 m) for which a DNA sequence could be produced is indicated next to the tip label (bolded).
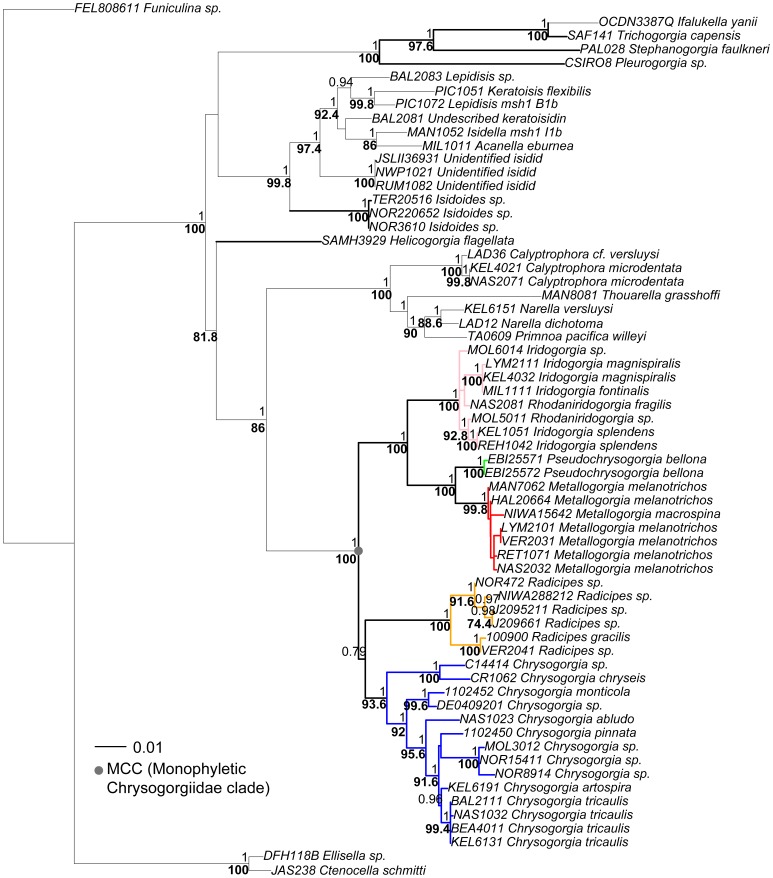
Figure 3. Maximum likelihood reconstruction of the suborder Calcaxonia (rooted to Funiculina, a sea pen) based on concatenated sequences of the 5′ end of mtMutS, cox1 and 18S (64 taxa, 2924 bp; GTR+I+G model; 500 bootstrap replicates).
All five families of the Calcaxonia are represented. Node support values from the ML analysis (>70%, bold) are indicated under each node, and node support values from the Bayesian analysis (>0.90) are above each node. Chrysogorgiidae taxa are either color coded (MCC) or have bolded branches (nonMCC).
All other genera for which we were able to retrieve DNA sequences fell outside of that clade. Stephanogorgia, Pleurogorgia Versluys, 1902 and Trichogorgia Hickson, 1904 formed a strongly-supported clade with the monophyletic ifalukellid genera Ifalukella Bayer, 1955 and Plumigorgia Nutting, 1910. Helicogorgia Bayer, 1981 systematically clustered outside the MCC, either sister to that clade (cox1 phylogeny), sister to the Primnoidae Milne Edwards, 1857/MCC clade (phylogenies based on mtMutS 5′ and concatenation of mtMutS 5′, cox1 and 18S) or sister to the clade composed of Stephanogorgia, Pleurogorgia and Trichogorgia (18S phylogeny). The entire mtMutS gene from Helicogorgia specimens was not sequenced. The genus Isidoides Nutting, 1910 was sister to the monophyletic sub-family Keratoisidinae (Isididae Lamouroux, 1812). This position is conserved across markers and combination of markers with strong statistical support. Only limited genetic data could be extracted from specimens of Chalcogorgia Bayer, 1949 (first 187 nt of mtMutS). This short sequence grouped with Helicogorgia with poor statistical support (56% node support on the ML phylogeny based on the 5′ end of mtMutS). The large amount of missing data negatively affected node support throughout the phylogeny; data from Chalcogorgia were therefore removed from all other analyses. Despite numerous attempts, no DNA could be amplified from specimens of Xenogorgia Bayer & Muzik, 1976. Specimens of Distichogorgia Bayer, 1979 could not be obtained.
Genus Monophyly
Within the MCC, Metallogorgia, Pseudochrysogorgia and Radicipes always formed monophyletic clades with strong statistical support. While Chrysogorgia formed two clades in the MCC based on the 5′ end of mtMutS (Figure 2), the genus formed a well-supported monophyletic clade based on the multiple-gene phylogeny (Figure 3). Genetic data cannot distinguish the genera Iridogorgia and Rhodaniridogorgia from each other, and there is very little divergence among haplotypes (uncorrected p distances at the 5′ end of mtMutS range from 0.1 to 0.7%; Figure 2 and 3). Outside the MCC, genera Stephanogorgia, Helicogorgia and Trichogorgia (n = 3, 5, 5 nominal species, respectively) are represented by multiple specimens (n = 3, 2, 2, respectively), but single haplotypes. Genus monophyly could therefore not be assessed. The two haplotypes sampled for the monotypic genus Isidoides formed a monophyletic clade.
Richness and Geographic Distribution of mtMutS Haplotypes
Chrysogorgia is the richest genus in terms of haplotypic diversity at mtMutS: we obtained 41 haplotypes, 31 of which were sampled in the Pacific and 11 from the Atlantic (Table 2). Only one out of 41 haplotypes was common between the Atlantic and the Pacific. Diversity was significantly lower (1–6 haplotypes) for the other genera of the MCC. For all other genera within the Chrysogorgiidae except Rhodaniridogorgia, more haplotypes were found in the Indo-Pacific compared to the Atlantic. Metallogorgia, Iridogorgia and Radicipes each had one haplotype in common between the Atlantic and the Pacific (Table 2). Relative to the number of colonies sampled, Metallogorgia was the least variable within the MCC. Among 64 colonies, two mtMutS haplotypes were sampled. The first, corresponding to Metallogorgia melanotrichos Versluys, 1902, was found 62 times across the N. Atlantic, SW and NE Pacific. Although M. melanotrichos was previously thought to be restricted to seamounts, we sampled colonies on a continental slope (Bahama Escarpment). The other haplotype, corresponding to Metallogorgia macrospina Kükenthal, 1919, was found only twice, on the Norfolk and the Kermadec ridges (vicinity of New Caledonia and New Zealand, respectively). The type of this species was originally described from West Sumatra, consistent with our observations of a SW Pacific distribution.
Table 2. Bathymetric range and biodiversity within the monophyletic, deep-sea Chrysogorgiidae (MCC).
| Genus | Data source | N. | Depth (m) | Total | Atl. | Ind. | Pac. | Ant. |
| Chrysogorgia | morphology | 615 (1589) | 31–4327 | 63 | 13 | 5 | 51 | 1 |
| mtMutS gene | 101 | 101–3860 | 41 | 11 | 0 | 31 | 0 | |
| Metallogorgia | morphology | 120 (159) | 570–2262 | 4 | 2 | 0 | 3 | 0 |
| mtMutS gene | 64 | 810–2262 | 2 | 1 | 0 | 2 | 0 | |
| Iridogorgia | morphology | 40 (49) | 567–2311 | 5 | 4 | 0 | 1 | 0 |
| mtMutS gene | 25 | 752–2311 | 4 | 2 | 0 | 3 | 0 | |
| Rhodaniridogorgia | morphology | 5 (6) | 568–2229 | 2 | 1 | 0 | 1 | 0 |
| mtMutS gene | 2 | 663–2229 | 2 | 1 | 0 | 1 | 0 | |
| Radicipes | morphology | 78 (208) | 196–3580 | 7 | 4 | 2 | 3 | 0 |
| mtMutS gene | 16 | 308–3000 | 6 | 3 | 0 | 4 | 0 | |
| Pseudochrysogorgia | morphology | 3 (5) | 861–1429 | 1 | 0 | 0 | 1 | 0 |
| mtMutS gene | 5 | 861–1429 | 1 | 0 | 0 | 1 | 0 |
Diversity: morphology estimate based on number of nominal species; genetic estimate based on number of mtMutS haplotypes. N: sample size (morphology: number of biogeographic records, and minimum number of colonies in parentheses), Atl: Atlantic Ocean, Ind: Indian Ocean, Pac: Pacific Ocean, Ant: Antarctic Ocean. Some depth estimates are based on depth ranges from trawling stations, in which case minimum and maximum depths were averaged (see notes in the Material and Methods section).
Bathymetric and Geographic Distribution
A total of 985 biogeographic records (2345 coral colonies) were gathered from the literature, museum collections, and our own collecting. Of these, 24 records had a depth range considered too large relative to the average depth, and were removed from the dataset (see Methods). In addition, two records were removed as being extremely shallow, and most probably errors (Metallogorgia sp. USNM 56792 and Iridogorgia pourtalesii Verrill, 1883 Blake station 259; see [16] and [5]). The final database used in the analysis contains 959 records representing 2302 colonies.
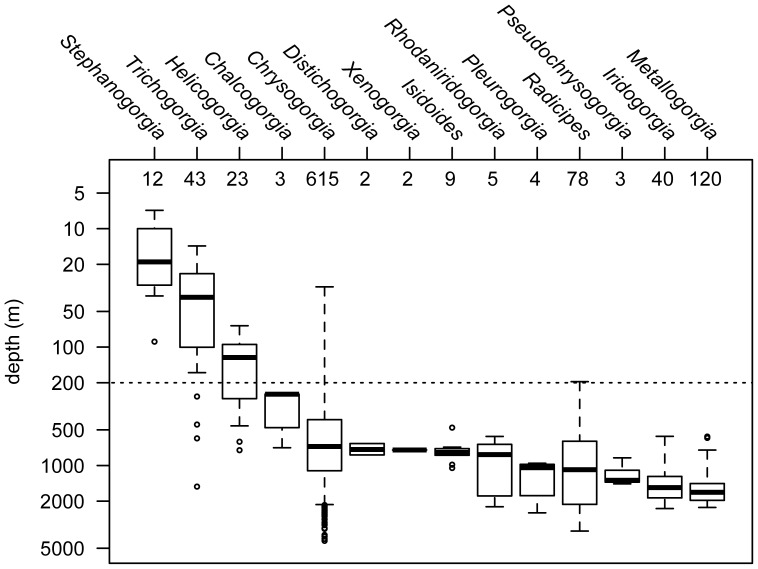
The genera Metallogorgia and Iridogorgia, relatively well sampled (n = 120 and 40, respectively), are found exclusively in deep waters between 567 and 2311 m. The genera Radicipes and Chrysogorgia, well sampled as well (n = 78 and 615), are more wide ranging, and Chrysogorgia in particular appears as a depth generalist, ranging from 31 to 4327 m (both extremes correspond to unidentified Chrysogorgia specimens held at the NMNH, Smithsonian Institution). Stephanogorgia is the only shallow-water specialist (n = 12, range 7–37 m, with one outlier, USNM 79628, sampled at 90 m and identified by Frederick Bayer), and Helicogorgia and Trichogorgia (n = 23 and 43), while being found predominantly in waters shallower than 200 m, were occasionally reported from about 1000 m deep (Figure 4). Many genera are only known from a few specimens and estimating their depth distribution is therefore inherently biased. Pseudochrysogorgia, Pleurogorgia, Isidoides, Xenogorgia, Distichogorgia and Chalcogorgia are all known from eight specimens or less. All are found in waters below 200 m (n = 24, 250–2509 m), and most have a narrow depth distribution (Figure 4). The depth distributions of haplotypes and nominal species were very similar (Tables 2 and S1).
Figure 4. Depth distribution of the 14 Chrysogorgiidae genera based on 959 depth records.
Depth records are summarized as box-and-whisker plots displaying the minimum, first quartile, median (bolded line), third quartile, and maximum values. Statistical outliers (>1.5x the inter-quartile range) are presented as open circles. Genera are sorted by increasing median depth (in meters, log scale) and sample size (number of biogeographic records) is provided on the top side of the plot. The 200 m isobath is represented (dashed line) as an arbitrary limit between deep and shallow waters.
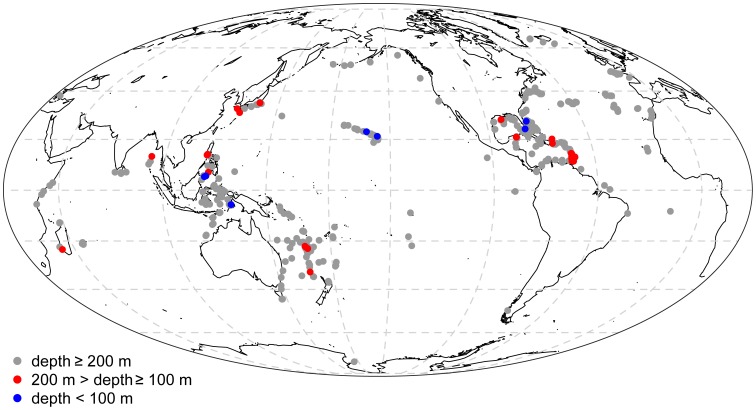
Among the genera belonging to the MCC, nine out of 89 species extend shallower than 200 m, all of them belonging to Chrysogorgia. Specimens reported in the taxonomic literature were collected from the NW Atlantic (C. desbonni Duchassaing & Michelotti, 1864, C. thyrsiformis Deichmann, 1936, C. fewkesii Verrill, 1883: [7], [8], [16], [17]), the NW Pacific (C. sphaerica Aurivillius, 1931, C. dichotoma Thomson & Henderson, 1906, C. axillaris (Wright & Studer, 1889), C. geniculata (Wright & Studer, 1889), C. cupressa (Wright & Studer, 1889): [18]–[21]), and the NW Indian Ocean (C. dichotoma Thomson & Henderson, 1906: [22]). With the exception of one specimen (C. cupressa, from the Banda Sea), all of these colonies were sampled in the northern hemisphere, south of 34°N (Figure 5). An additional species, held at the NMNH (USNM 91906) and identified by Charles Nutting as C. curvata Versluys, 1902 (but not included in his 1908 monograph [23]), was collected in the NE Pacific between 37–55 m depth. This specimen was confirmed by Dr. Stephen Cairns (Smithsonian Institution) as Chrysogorgia; this very shallow occurrence is therefore either real, or reflects a recording error of station data, or is the result of remnant specimens being retained in the net between dredges (Dr. Cairns notes that the preceding station for this collection was dredged from 1384–1829 m). One of these nine species, C. dichotoma, was exclusively found between 165 and 188 m, but is known from only three specimens collected in Japan and the Bay of Bengal [20], [22]. Other species extend much deeper, five of them being found below 1000 m.
Figure 5. Geographic distribution of Chrysogorgia based on our biogeographic database (all 634 records, Mollweide projection).
Records are displayed for three different depth ranges (in meters).
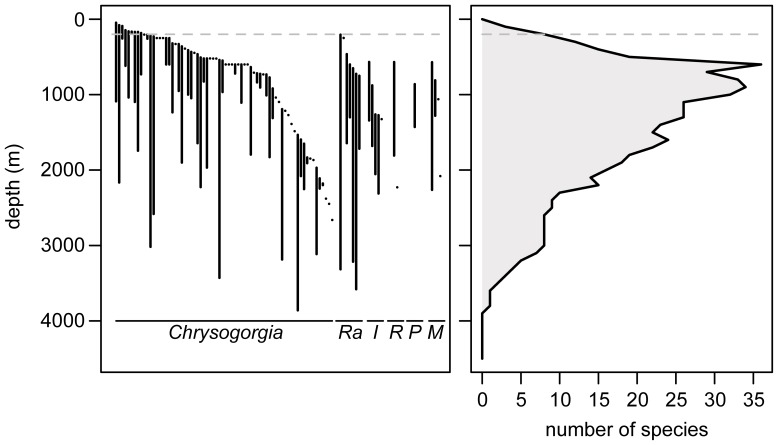
Among the genera belonging to the MCC, a diversity peak was observed at 600 m (36 species). Species diversity was maximum between 600 and 1000 m (29–36 species). All genera of the MCC were sampled within this depth interval. Diversity decreased progressively from 1000 m to 3860 m, the depth at which the last specimen identified to the species level was collected (Figure 6). This colony is a Chrysogorgia that was recently described by Pante and Watling [24], and was collected on Retriever Seamount in the NW Atlantic. Observations of a diversity gradient relative to depth are, of course, intimately linked to sampling effort in octocorals [25].
Figure 6. Depth range of 89 species (541 records) from the MCC (left) and resulting species diversity gradient (right).
On the left panel, each segment links the maximum and minimum collection depths for a particular species. Species within genera are sorted by increasing median depth (m). The 200 m isobath is represented as a dashed line. Ra: Radicipes, I: Iridogorgia, R: Rhodaniridogorgia, P: Pseudochrysogorgia, M: Metallogorgia.
Discussion
Family Monophyly and the Diagnosis of the Chrysogorgiidae
Genetic data strongly suggest that the Chrysogorgiidae is polyphyletic as currently described. In our analysis of mtMutS, chrysogorgiid genera appeared in three different clades: the “monophyletic Chrysogorgiidae clade” (MCC), the Pleurogorgia/Stephanogorgia/Trichogorgia/Ifalukellidae clade, and the Isidoides/Keratoisidinae clade. The position of Helicogorgia was labile among phylogenetic trees, being either sister to the MCC, or sister to the clade composed of the Primnoidae and the MCC. The very limited data available for Chalcogorgia suggests a close affinity with Helicogorgia.
The polyphyly of the family calls for a re-assessment of the diagnostic characters used to differentiate the Chrysogorgiidae. The family is currently diagnosed as (e.g., [8]): The axis is unjointed, solid (non-spicular), and made of concentrically-layered scleroproteinaceous material; these layers are not undulated; the axis surface is smooth and has a metallic or iridescent sheen; colonies are branched or unbranched; the holdfast is strongly calcified, and discoidal or rhizoidal; polyps are contractile, not retractile, and are arranged in rows, sometimes bi- or multiserially, never in whorls or on opposite sides of the branch; sclerites are mostly flat scales, plates, rods and needles; when exposed to polarized light, scales show a distinct circular (not cruciform) extinction pattern (for an example see p. F221 in [26]).
Trichogorgia and Stephanogorgia have in common biserially-arranged polyps, which contrasts with the organization in rows found in genera of the MCC. This suggests that polyp arrangement as a diagnostic character of the Chrysogorgiidae should be revised. Three of the four genera for which genetic information are lacking are characterized by bi- or multiserially-arranged polyps: Chalcogorgia, Distichogorgia and Helicogorgia. If indeed polyp arrangement is diagnostic of phylogenetic placement, then we can predict that these three genera should not belong to the MCC. Similarly, two genera (Pleurogorgia and Helicogorgia) have ornamented sclerites that are uncharacteristic of the Chrysogorgiidae. As Pleurogorgia does not belong to the MCC, it can be predicted that Helicogorgia is probably not a true chrysogorgiid. Our Pleurogorgia specimens are small whips with polyp and sclerite morphology, and biogeography, consistent with published records for this genus [19], [23]. Both described species of Pleurogorgia are branching, however, and our specimens might belong to undescribed species (Alderslade, pers. com) or colonies at an early pre-branching life stage.
On the other hand, some characters currently used in the diagnosis might not be good indicators of the uniqueness of chrysgorgiids. Nutting [27] initially placed the genus Isidoides in the Gorgonellidae (now Ellisellidae). He noted its unusual morphology and the resemblance of sclerites to those found in some isidids. Bayer and Stefani [28] later noted that the fine, smooth scales of Isidoides are typical of the Chrysogorgiidae. Although exactly when the genus Isidoides was placed in the family Chrysogorgiidae is unclear, the genus appears in a key of the family as early as 1979 [29], but without justification. Later, Bayer and Grasshoff [30] stated that Isidoides should belong to the Chrysogorgiidae rather than the Ellisellidae. Our phylogenetic data clearly show that Isidoides does not belong to the Chrysogorgiidae, and supports Nutting’s initial suggestion that Isidoides might be related to bamboo corals.
There is no published record that all nine characters diagnostic of the family have been scored for all extant genera. For example, Chrysogorgia is the only genus for which clear evidence was provided that, when exposed to polarized light, scales show a distinct circular extinction pattern [26]. For this study, we confirmed that specimens from all genera of the MCC show a circular extinction pattern. However, it is also the case for Pleurogorgia, Stephanogorgia, Helicogorgia, Xenogorgia, Isidoides, and even bamboo corals. Circular light extinction pattern is therefore clearly not suited as a diagnostic feature of the MCC. Similarly, the composition and arrangement of axial layers are rarely reported. These results imply that characters diagnostic of the Chrysogorgiidae need to be re-evaluated for these genera; the diagnosis of the family will have to be revised.
Trichogorgia, Stephanogorgia, Pleurogorgia, and the Ifalukellidae
Three of the 14 chrysogorgiid genera (Trichogorgia, Stephanogorgia, Pleurogorgia) formed a well-supported clade with the ifalukellid genera Plumigorgia and Ifalukella. The short genetic distance (mtMutS, uncorrected p: 0.72%) between the latter two genera supports the validity of the Ifalukellidae as a family (at mtMutS Plumigorgia and Ifalukella differ by only one amino acid, and have an identical indel structure, which is often quite variable among calcaxonians). Based on the taxa (Calcaxonia: Chrysogorgiidae, Primnoidae, Isididae) used in the study of McFadden et al. [15], the maximum intra-familial uncorrected p (at mtMutS) distance is 4.9%, while the minimum inter-familial distance is 3.8%. The distance between Trichogorgia and ifalukellids is <3.4%. Based on this criterion, we can legitimately suggest that Trichogorgia be considered as an ifalukellid. On the other hand, the genetic distances between Stephanogorgia, Pleurogorgia, and the Trichogorgia/Ifalukellidae group are all >6.7%, suggesting that two new families may need to be erected.
The phylogenetic grouping of Trichogorgia, Stephanogorgia, Pleurogorgia and the Ifalukellidae is only preliminary, and more comprehensive genetic and morphological analyses will have to be conducted. Only a single mtMutS haplotype could be used to represent each genus. More extensive taxon sampling will be required to comment further on the status of these genera, confirm their placement on the Calcaxonia tree, and rule-out spurious effects of long-branch attraction (e.g., [31]). There is nevertheless congruence among morphology, ecology and phylogeny. Morphologically, Trichogorgia, Stephanogorgia, and the Ifalukellidae share small, smooth sclerites in the form of scales and plates. In addition, both Trichogorgia and Ifalukella have relatively few sclerites in their tissues, to an extreme for T. capensis Kükenthal, 1919, which is completely lacking them. Also, the Chrysogorgiidae are defined as having an axis made of concentric layers that are not undulating. These layers are slightly undulating in the Ifalukellidae [26], [32]. However, the descriptions of Stephanogorgia, Trichogorgia and Pleurogorgia give no indication of how the layers are arranged. Examination of this character may inform us on the relationship of these taxa to ifalukellids. Ecologically, Trichogorgia and Stephanogorgia are two of the shallowest chrysogorgiid genera, and Stephanogorgia is a tropical lagoon-dwelling taxon. These observations are consistent with the fact that ifalukellids are strictly found above 50 m on coral reefs (e.g., [32], [33] and collection records from the Smithsonian Institution).
Genus Monophyly and Genetic Diversity
With the exception of Iridogorgia and Rhodaniridogorgia, all chrysogorgiid genera were monophyletic. Specimens of Iridogorgia and Rhodaniridogorgia have been separated by the morphology of their spiral (Iridogorgia: main stem coiled; Rhodaniridogorgia: main stem wavy) and the morphology and placement of sclerites (sclerites are larger in Rhodaniridogorgia, and consistently present in the branch coenenchyme) [5]. In contrast, genetic distances separating these genera were consistent with intra-generic variation. These genera might have diverged too recently for our markers to have detected their reciprocal monophyly. Therefore, more specimens and a fine-scale genetic analysis will be needed to decide whether these genera should be synonymized.
Both Radicipes and Chrysogorgia were supported as monophyletic genera, although the relationship between them and the clade composed of Metallogorgia, Pseudochrysogorgia, Iridogorgia and Rhodaniridogorgia, was difficult to recover. As predicted in the taxonomic literature (in particular [19]), Chrysogorgia appeared as a highly diversified genus: 73% of haplotypes within the MCC (41 out of 56) belong to Chrysogorgia. Pante and France [14] showed that the distribution of intra-generic and inter-generic genetic distances between Chrysogorgia and Radicipes are greatly overlapping, with some Chrysogorgia haplotypes being more divergent than Chrysogorgia-Radicipes pairs. In contrast to the relatively high level of divergence within Chrysogorgia, the very short genetic distances between most Chrysogorgia clades suggest a rapid diversification of the genus.
The available evidence does not allow us to determine whether the diversification of Chrysogorgia is the result of an adaptive radiation. Theory predicts that an adaptive radiation would be associated with a high diversity of filled ecological niches. Four main hypotheses should be evaluated to test for adaptive radiation: (1) all taxa share a common ancestor, (2) there is a correlation between the environmental conditions and phenotypes, (3) new traits are adaptive, and (4) speciation is rapid (reviewed in [34]). Although the first requirement is met, other hypotheses remain to be tested.
First, ecological information associated with individual Chrysogorgia haplotypes is scarce; most haplotypes are singletons (i.e., sampled only once), and correspond to colonies sampled using trawls. In the case of trawled specimens, ecological information can be inferred from the whole catch (residual substrate and associated species). Specimens collected using underwater vehicles have the advantage of being associated with more extensive ecological data, but are a minority at the moment. While ecological data is scarce, some observations are congruent with an adaptive radiation. Chrysogorgia is the most widely-distributed chrysogorgiid genus, both geographically and bathymetrically (Table 2). Specimens are found from soft and hard substrates (e.g., [35]), and in continental and oceanic environments (e.g., [8], [36]). Chrysogorgia colonies are characterized by rhizoidal and discoidal holdfasts, consistent with life in soft and hard substrates.
Second, phylogenetic relationships among Chrysogorgia haplotypes require more than sequencing of the 5′ end of mtMutS. Our comparative analysis shows that the relationships of some Chrysogorgia haplotypes could be resolved using mtMutS, cox1 and 18S together. Future research efforts will include a more comprehensive molecular analysis of Chrysogorgia, and the mapping of ecological and morphological characters on a better-resolved phylogeny. This exercise, however, might be complicated by the fact that morphological characters are notoriously plastic (e.g., [37]–[40]) and convergent [41], [42] in the Octocorallia. In addition, determining whether morphological traits confer an adaptive advantage will be technically very challenging.
Finally, studying the pace of speciation in the Octocorallia remains a daunting task, as informative markers that are variable at the intra- and inter-population levels are not yet readily available ([15] and references therein). Concepcion et al. [43] reported intra-specific variation at SRP54 (single-copy nuclear intron of the signal recognition particle 54) in the octocoral Carijoa, but preliminary results suggest this marker does not reveal more haplotypes than mtMutS in the Isididae (France, unpublished), and is very challenging to work with in the Chrysogorgiidae (Pante, unpublished).
Relationships of Evolutionary History, Depth, and Biogeography
The MCC formed a well-supported monophyletic clade within the Calcaxonia, sister to the well-supported family Primnoidae and sub-family Keratoisidinae (Isididae). The Primnoidae (“the quintessential deepwater octocoral family,” [44]) and Keratoisidinae are largely deep-water taxa. The phylogenetic position of the MCC within the Calcaxonia, and the fact that it mostly comprises deep-water taxa (80/89 species found strictly below 200 m) support the hypothesis that the MCC diversified in the deep sea from a deep-sea ancestor.
Metallogorgia, Iridogorgia, Rhodaniridogorgia and Pseudochrysogorgia formed a strong monophyletic group within the MCC, and specimens within this clade were exclusively sampled between 663 and 2311 m depth (Table 2). Observations from our biogeographic database confirmed that members of this group were exclusively confined to deep waters (567–2311 m). In contrast, specimens from the Radicipes and Chrysogorgia clades had a significantly wider depth range, extending between 101 and 3860 m (biogeography database: 31–4327 m). There is therefore a strong dichotomy between narrow-ranging (depth specialists) and wide-ranging (depth generalists) taxa within the MCC. Of all genera, Chrysogorgia was by far the widest-ranging taxon, both geographically and bathymetrically, and genotyped specimens were found as shallow as 100.3–101.7 m (average 101 m, Northern Gulf of Mexico). Phylogenetically, shallower specimens are nested within a clade of deep-water taxa (the MCC); we can therefore hypothesize that the shallowest Chrysogorgia have evolved from deep-water ancestors.
Lindner et al. [45] found evidence of four separate emergence events (three tropical, one temperate) leading to the colonization of deep stylasterid hydrocoral lineages into shallow waters (<50 m). As in their study, we found that the shallowest Chrysogorgia specimens occurred at tropical and subtropical latitudes (5°S - 34°N). Faunal exchange between deep and shallow water may occur where biophysical barriers are permeable (e.g., [46]). Emergence of deep-sea species may therefore occur at high latitude, where vertical movement of species adapted to cold-water may be facilitated (e.g., [47]–[49]). We have no evidence of Chrysogorgia from shallow waters at high latitudes (shallowest colony >40° latitude: C. flexilis, 219 m depth, coast of Chile; [18]), and the only species known from Antarctica was sampled between 445 and 448 m (C. antarctica [2]). Polar regions remain under-sampled, and additional sampling at high latitudes may reveal the presence of Chrysogorgia in shallow, cold waters. However, sampling efforts (>350 octocorals sampled) by us and colleagues in the Aleutian Islands (51–53°N) recovered no chrysogorgiids shallower than 1357 m, despite many other octocorals collected between 25–190 m [50], [51]. Faunal exchange between deep and shallow waters may have occurred at lower latitudes when deep waters were warmer than at present, between the Mesozoic and the early Cenozoic (180–50 mya, [48], [49]). This coincides with the period of opening of the Tethys Sea, and is consistent with the tethyan distribution of the shallowest Chrysogorgia specimens. Unfortunately, there are no published records of chrysogorgiid fossils (and no records on the Paleobiology Database) to support this hypothesis. In addition, our shallowest mtMutS haplotype was found between 100.3 and 101.7 m, and genetic evidence for colonization of Chrysogorgia above 50 m depth (as suggested by our biogeographic database) is yet unavailable. Unfortunately, available historical material from shallow depths was collected between 1868 and 1976, and we have had limited success extracting PCR-amplifiable DNA from such material. Future efforts might therefore depend on the collection of fresh tissue.
Materials and Methods
Collections and DNA Extraction
Most specimens were collected during a series of deep-sea coral expeditions to Hawaii (1996, 2009), Alaska (2004), the North Atlantic (2003–2005), New Caledonia (2008), and the Bahama Escarpment (2009). Additional specimens from museum collections (via formal loan requests) and colleagues were utilized (Table S1; we confirm that the museums and colleagues who provided these specimens gave us permission to use them). The species represented here are not protected and do not require collecting permits. Sampling in the North Atlantic was mostly in international waters, and when inside of the US EEZ, was outside of National Marine Sanctuaries and fishery-closed areas focused on coral conservation; no permits were required. Specimens from Alaska were collected under State of Alaska Fish Resource Permits CF-04-009 and CF-06-013. Specimens from Hawaii were either collected outside of state waters, or in the Marine National Monument under permit PMNM-2009-053. Collecting in the Bahamas was done with permission from the Department of Marine Resources, Ministry of Agriculture and Marine Resources of The Bahamas, as assigned to the University of Miami/RSMAS. Collecting in New Caledonia was done within the French EEZ, by a French boat, and no permit was required. Whole colonies or portions thereof were sampled using remotely-operated vehicles (ROV), human-occupied vehicles or scientific trawls. Fragments for genetic analyses were preserved in 80–100% ethanol, RNAlater (Ambion), or frozen at −80°C. DNA was extracted using a modified CTAB protocol [10] or using the MasterPure DNA purification kit (Epicenter).
DNA Amplification and Sequencing
Three gene regions were targeted. First, we attempted to PCR-amplify the 5′ end of the mitochondrial, protein-coding mtMutS for all available specimens. Although this gene has been most frequently referred to as msh1 in the octocoral systematics literature, we will refer to it as mtMutS throughout this paper as this term makes fewer assumptions about its evolutionary origins [52]. mtMutS was chosen because it is one of the most variable and informative markers available for octocorals to date (16S: [10]; mtMutS, nd3, nd4l: [53]; cox1: [54]; [55]; nd2, nd3, nd6: [56]; mtMutS, cox2-IGR-cox1: [15]; mtMutS, cox1, nd2, nd3, 16S, 28S, ITS2: [57]). All chrysogorgiids and outgroup taxa, except the Keratoisidinae, were amplified by priming in the flanking gene nd4l with upstream primer ND4L2475F [58] and extending into mtMutS with downstream primer MUT3458R [59]. This last primer was later replaced by MUTChry3458R (Table 3), a novel primer that is a perfect match to primnoids, isidids and chrysogorgiids tested to date. Mitochondrial gene order is not conserved across the sub-order Calcaxonia [58]. In the isidid sub-family Keratoisidinae, mtMutS is flanked by cox3 at its 5′ end. Thus, primers CO3BAM5647F and MUT3458R were paired to amplify mtMutS in this group of taxa. Internal primers were used when DNA was degraded (primers 1–16, Table 3).
Table 3. PCR primers used in the present study to amplify targeted gene regions.
| Name | Sequence (5′ ––3′) | Gene | Product size | Cycle | Reference | |
| 1 | CO3Bam5657f | gctgctagttggtattggcat | cox3 | 1–15: 1014 bp | 94:20,55:30,72:50 | [58] |
| 2 | ND4L2475F | tagttttactggcctctac | nad4L | [58] | ||
| 3 | ND42625F | tacgtggyacaattgctg | nad4L | 3–10: 476 bp | 94:20,50:30,72:30 | [58] |
| 4 | MSH2714F | cttaatggaggagaattattc | mtMutS | this publication | ||
| 5 | MSH2806F | taactcagcttgagagtatgc | mtMutS | [58] | ||
| 6 | msh2864r | gaggcaacttgttcaatgggaggtg | mtMutS | this publication | ||
| 7 | MSH3010F | ggataaaggttggactattatag | mtMutS | 7–16: 448 bp | 94:20,55:30,72:30 | [6] |
| 8 | MSHLA3034R | cctgagatactgcgcgttgtttaggccccg | mtMutS | [58] | ||
| 9 | MSH3055R | ggagaataaacctgagayac | mtMutS | [58] | ||
| 10 | MSH3101R | gatatcacataagataattccg | mtMutS | [59] | ||
| 11 | MSH3186F | gccatgartgggcatagtata | mtMutS | this publication | ||
| 12 | msh3208r | atcgagcyactttgtccckgtc | mtMutS | this publication | ||
| 13 | MSH3332F | cttattaattggttggaa | mtMutS | this publication | ||
| 14 | MSH3350F | gccatgartgggcatagtata | mtMutS | this publication | ||
| 15 | MUT3458R | tsgagcaaaagccactcc | mtMutS | 2–15: 940 bp | 94:20,50:30,72:50 | [59] |
| 16 | MUTChry3458R | tgaagyaaaagccactcc | mtMutS | 2–16: 940 bp | 94:20,50:30,72:50 | this publication |
| 17 | MSH3841F | ctgcgttatgaggagattgckac | mtMutS | this publication | ||
| 18 | MSH4094F | cagtcggacctcaattagaatcg | mtMutS | this publication | ||
| 19 | MSH4332R | gaaggcataaccctccttactg | mtMutS | 14–19: 920 bp | 94:20,50:30,72:50 | this publication |
| 20 | MSH4757R | gacttgcccgcaccatttactg | mtMutS | this publication | ||
| 21 | MSH4759F | tgtagctcatgatattag | mtMutS | [41] | ||
| 22 | MSH4915R | cgacctcaaaagtaccttgacc | mtMutS | 18–22: 830 bp | 94:20,50:30,72:50 | this publication |
| 23 | MSH5065F | gcaacaattgaaagattraca | mtMutS | this publication | ||
| 24 | MSH5075R | gagtagamagarcgaaactag | mtMutS | this publication | ||
| 25 | MSH5376R | agctccacatatttcacac | mtMutS | this publication | ||
| 26 | 16S5PR | tcacgtccttaccgatag | 16S | 21–26: 900 bp | [41] | |
| 27 | COII8068xF | ccataacaggrctwgcagcatc | cox2 | [15] | ||
| 28 | COIoctR | atcatagcatagaccatacc | cox1 | 27–28: 1080 bp | 94:20,50:30,72:60 | [54] |
| 29 | 18S-Af | aacctggttgatcctgccagt | 18S | mod. [76] | ||
| 30 | 18S-Lr | ccaactacgagctttttaactg | 18S | 29–30: 620 bp | 94:20,60:30,72:40 | [77] |
| 31 | 18S-Cf | cggtaattccagctccaatag | 18S | [77] | ||
| 32 | 18S-Yr | cagacaaatcgctccaccaac | 18S | 31–32: 710 bp | 94:20,60:30,72:40 | [77] |
| 33 | 18S-Of | aagggcaccaccaggagtggag | 18S | [77] | ||
| 34 | 18S-Br | tgatccttccgcaggttcacct | 18S | 33–34: 620 bp | 94:20,60:30,72:40 | mod. [76] |
Predicted fragment sizes (approximate, in bp) and PCR cycle profiles (temperature in °C: time in seconds) are given for the most commonly used primer pairs. Primer combinations are listed in the product size column, prior to the predicted fragment size, using the primer numbers defined in the first column. (mod.: modified from). Between 30 and 45 cycles were used for PCR.
For a set of taxa, additional gene sampling was performed. These taxa were chosen based on their position on the mtMutS phylogeny to achieve two goals: (1) to assess if additional sequencing would retrieve more variation from clades/taxa that show little or no variation at mtMutS (e.g., Metallogorgia melanotrichos), and (2) to further resolve phylogenetic relationships among the Chrysogorgiidae (e.g., relationship between Radicipes and Chrysogorgia). For one set (n = 64), cox2-IGR-cox1 and 18S were amplified (primers 27–28 and 29–34). Among these (n = 42), the nearly complete mtMutS was amplified (2889–2997 bp; primers 1–26).
PCR was performed in 25 µL total volume using 1x TaKaRa Ex Taq buffer (Mg2+-free, proprietary composition), 1.5 mM of MgCl2, 0.4 mM of dNTP mix, 0.5 U of Ex Taq polymerase (TaKaRa Bio USA Inc., now Clontech), 0.24 µM of each primer (Operon Biotechnologies, Inc., now Eurofins MWG Operon), and 40 ng of DNA template (quantified using BioRad VersaFluor fluorometer and/or Thermo Scientific Nanodrop ND-1000 spectrophotometer). 2.5 µg of acetylated BSA (Promega) was added for problematic samples. PCR amplification conditions were optimized for each primer pair (Table 3 provides cycling profiles for the most commonly-used pairs). PCR products were purified either by excising bands from low-melting point agarose gels followed by an agarase digestion (5 U; Sigma-Aldrich Co.; [54]) or by an Exo-SAP digestion (2 U of ExoI and 0.2 U of SAP/1 µL of DNA; Fermentas; [60]). The majority of purified PCR reactions were cycle-sequenced using the ABI BigDye Terminator v1.1 Cycle Sequencing Kit (1/4 reactions) and purified using either an EtOH/EDTA precipitation or Sephadex G-50 columns (Sigma-Aldrich). Purified products were electrophoresed on an ABI PRISM (R) 3100 or 3130xl Genetic Analyzer. A fraction of the PCR products were purified with AmPure XP beads, cycle-sequenced using the ABI BigDye Terminator v3.1 Cycle Sequencing Kit (1/32 reactions) and purified using an EtOH/Sodium Acetate precipitation. These products were electrophoresed on an ABI PRISM (R) 3730xl Genetic Analyzer. All sequence traces were edited using Sequencher (TM) v4.7 (Gene Codes). DNA sequences of specimens representing each haplotype, for each biogeographic region, were submitted to GenBank (Table S1).
Data Analysis
mtMutS sequences were translated to amino acids (Mold-Protozoan mitochondrial code) and aligned using MAFFT 6 (L-INS-i method, [61], [62]). TranslatorX was used to align nucleotides based on the amino-acid alignment [63]. There were no indels in the cox1 alignment. The 18S dataset contained only single-nucleotide indels, and could therefore be aligned by eye. Saturation plots and transition/transversion ratios were computed for each dataset using the ape 2.5–2 package [64] in R 2.12 [65]. The effect of complex indel motifs at mtMutS (particularly the 5′ region) on phylogenetic reconstruction was investigated by constructing trees with and without indels. Gblocks 0.91b [66], [67] was used to eliminate indel regions that are poorly conserved, while keeping the regions that contain informative sites. All alignments are available from the authors upon request. The phylogenetic information content of all gene regions (5′ end of mtMutS, whole mtMutS, partial cox1 and 18S) and these regions in combination was evaluated by calculating the number of variable and parsimony-informative sites in MEGA 5 [68] and constructing maximum-likelihood (ML) trees using PhyML 3.0 [69]. Node support was evaluated with 1000 bootstrap replicates (500 replicates for mtMutS, cox1 and 18S concatenated). The most-likely model of evolution was inferred using jModelTest 0.1.1 [70], and PhyML was parametrized accordingly. MrBayes 3.1 [71], [72] was used on the CIPRES Portal [73] to produce phylogenetic hypotheses based on Bayesian statistics (6 nucleotide substitution types, 4×4 substitution model with I+G among-site rate variation; 5 million generations, 2 runs, 4 chains, sampling every 1000 generations, burnin of 25% equaling 1250 samples). Convergence was assessed by checking that (1) standard deviations of split frequencies were <0.007, (2) potential scale reduction factors were close to one (they varied between 1 and 1.06), and (3) plots of log likelihood values did not show visible trends over time. Trees were rooted to the Pennatulacea (sea pens), which are the non-calcaxonian octocorals that are the most closely related to the Calcaxonia, based on the results of [9].
Biogeography Database
Biogeographic data were compiled from the taxonomic literature (37 published papers, one in press, one in preparation), collection information from museums (the Australian Museum (Sydney), the Florida Museum of Natural History (University of Florida, Gainesville), the Harvard Museum of Comparative Zoology, the Museum and Art Gallery of the Northern Territory (Darwin, Australia), the Muséum national d’Histoire naturelle (Paris, France), the National Institute of Water and Atmospheric Research (Wellington, New Zealand), the National Museum of Natural History (Smithsonian Institution), and the Yale Peabody Museum of Natural History (Yale University)), and our collections (including specimens provided by colleagues). Duplicate records (i.e., fragments from individual colonies held in multiple museums, use of material in different manuscripts) were removed. Depth distributions were compiled using sampling station information. Thirty five percent of the depth records (341 of 975) come from dredging or trawling, for which the minimum and maximum depth were reported. The average depth was computed for these records. However, in some cases the depth range was so large that it made the average meaningless. All records for which the depth range was more than half of the average depth were excluded from the dataset. A species-diversity profile across depth was computed for species belonging to the genera forming a monophyletic clade by counting the number of species found every 100 m between 0 and 4500 m. The biogeographic database is available from the authors upon request.
Supporting Information
Bayesian 50% majority rule consensus trees based on different markers ( mtMutS , cox1 and 18S) and marker combinations. For mtMutS, the effect of indels on phylogeny inference was tested by removing them with Gblocks. Chrysogorgiidae taxa are either color coded (MCC) or have bolded branches (nonMCC).All trees are rooted to the Pennatulacea, except trees using the entire mtMutS gene (rooted to the Ellisellidae).
(PDF)
Collection date, geographic coordinates, depth, and genetic markers sequenced for specimens used in this study. GenBank accession numbers representing each haplotype and biogeographic region are given in the final 3 columns.
(XLS)
Acknowledgments
We are grateful to our colleagues for kindly donating tissue and DNA sequences from rare specimens, and providing invaluable advice on octocoral systematics. A warm thank you to Phil Alderslade (CSIRO, Australia), Amy Baco-Taylor (Florida State University), Stephen Cairns (Smithsonian NMNH), Gary Williams (California Academy of Sciences), Laure Corbari (MNHN), Philippe Bouchet (MNHN), Bertrand Richer de Forges (MNHN/IRD), Ole Tendal (University of Copenhagen, Natural History Museum), Elly Beglinger (Zoological Museum of Amsterdam), Rob van Soest (Zoological Museum of Amsterdam), Jon Moore (Florida Atlantic University), Ron Etter (University of Massachusetts at Boston), Jess Adkins (CalTech), Bob Stone (NOAA, National Marine Fisheries Service), and Tina Molodsova (P.P. Shirshov Institute of Oceanology).
We thank Eric Lazo-Wasem (Yale Peabody Museum), Linda Ward (Smithsonian NMNH), Stephen Keable (American Museum), John Slapcinsky (Florida Museum), Gavin Dally (Museum and Art Gallery of the Northern Territory), and Paul Valentich-Scott (Santa Barbara Museum) for their help in retrieving museum collection records to build a dataset of biogeographic information on the Chrysogorgiidae. Students of Les Watling (University of Hawaii) were also instrumental in building this database. Boris Shoshitaishvili (Brown University) kindly translated Pasternak’s 1981 manuscript on Metallogorgia [74] from Russian to English.
We thank the following for their help with morphological, genetic and microscopy work: Aude Andouche, Philippe Maestrati, Sadie Mills, Kareen Schnabel and Peter Marriott (curatorial assistance), John Douzat, Kristina Samples, Tram Lam, Nguyen Loc, Ryan Picard, Michael Purpera and Tom Pesacreta (microscopy), Jana Thoma, Mercer Brugler, Esprit Heestand, Lance Renoux, Joris van der Ham (lab assistance, UL Lafayette), Annie Tillier, Céline Bonillo, and Josie Lambourdière (lab assistance, MNHN), and Magalie Castelin and Justine Thubaut (field work).
We thank the PIs, science parties, and crews of vessels (R/V Alis, R/V Atlantis, R/V Endeavor, R/V Ronald H. Brown, R/V F.G. Walton Smith), ROVs (Argus, Global Explorer and Hercules) and HOV Alvin, who were instrumental in the collection of the specimens involved in this study. More particularly, we thank all involved in the Mountains in the Sea, Deep Atlantic Stepping Stones, Bahamas Deep Coral 2009, Terrasses, and EN447. Thanks to the PIs of the “Tropical Deep-Sea Benthos” research program (formerly MUSORTSOM), spearheaded by the MNHN and the IRD [75].
Specimens from New Zealand were provided by the NIWA Invertebrate Collection, and were collected during the Voyages TAN0308 (“Norfolk Ridge/Lord Howe Rise Marine Biodiversity Discovery Survey” (NORFANZ), PI Malcom Clark, NIWA); TAN0413 (“Bay of Plenty & Hikurangi Plateau Seamounts: their importance to fisheries and marine ecosystems,” PI Malcolm Clark, NIWA); KAH0011 (“Bay of Plenty and southern Kermadec Arc Seamounts Biodiversity survey,” PI Malcolm Clark, NIWA); RENEWZ I – NEW ZEEPS (the first component of the project “Exploration of Chemosynthetic Habitats of the New Zealand Region”).
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Support for this research was provided by funding to SCF from National Science Foundation’s (NSF) Ocean Sciences Division-Biological Oceanography Program (OCE-0624601; http://www.nsf.gov/funding/pgm_summ.jsp?pims_id=11696), National Oceanic and Atmospheric Administration (NOAA)/National Marine Fisheries Service Auke Bay Laboratory (NFFS7400-5-00022; http://www.afsc.noaa.gov/abl/), NOAA’s National Undersea Research Program (NA05OAR4301001, NA66RU0186), NOAA-OER (NA05OAR4601061, NA03OAR4600116, NA08OAR4600756; http://explore.noaa.gov/), and NSF’s “Assembling the Cnidarian Tree of Life” (sub-contract on EF-0531570 to CSM and EF-0531779 to Paulyn Cartwright; http://www.nsf.gov/funding/pgm_summ.jsp?pims_id=5129&org=DEB&from=home); to Philippe Grandcolas from the French National Agency for Research (ANR Bioneocal; http://www.agence-nationale-recherche.fr/); to EGP from Sigma Xi (GIAR G20061021830514629; http://www.sigmaxi.org/programs/giar/), the American Museum of Natural History (Lerner Gray Fund; http://rggs.amnh.org/pages/academics_and_research/fellowship_and_grant_opportunities), and the University of Louisiana at Lafayette Graduate Student Organization. Specimens from New Zealand were provided by the National Institute of Water and Atmospheric (NIWA) Research Invertebrate Collection, and were collected during voyages undertaken by the NIWA (www.niwa.co.nz) with funding from the New Zealand Foundation for Research, Science and Technology (www.msi.govt.nz), with additional funding from the New Zealand Ministry of Fisheries (www.fish.govt.nz) and the Australian National Oceans Office (voyage TAN0308), as well as NOAA-OE (http://explore.noaa.gov/), Woods Hole Oceanographic Institution (www.whoi.edu), Scripps Institution of Oceanography (sio.ucsd.edu), and the University of Hawaii (www.hawaii.edu) (voyage RENEWZ I – NEW ZEEPS). This work was supported by the “Consortium National de Recherche en Génomique,” and the “Service de Systématique Moléculaire” of the Muséum national d’Histoire naturelle (MNHN) (CNRS UMS 2700; http://www.genoscope.cns.fr/spip/). It is part of the agreement n. 2005/67 between the Genoscope and the MNHN on the project “Macrophylogeny of life” directed by Guillaume Lecointre. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Madsen FJ. Octocorallia. The Danish Ingolf-Expedition. 1944;5:1–65. [Google Scholar]
- 2.Cairns SD. A new species of Chrysogorgia (Anthozoa : Octocorallia) from the Antarctic. Proceedings of the Biological Society of Washington. 2002;115:217–222. [Google Scholar]
- 3.Bayer F. A new species of Trichogorgia and records of two other octocorals new to the Palau Islands. Micronesica. 1974;10:257–271. [Google Scholar]
- 4.Grasshoff M. Outlines of Coelenterate evolution based on principles of constructional morphology. Proceedings of the 6th International Conference on Coelenterate Biology. 1995. pp. 195–208.
- 5.Watling L. A review of the genus Iridogorgia (Octocorallia : Chrysogorgiidae) and its relatives, chiey from the North Atlantic Ocean. Journal of the Marine Biological Association of the United Kingdom. 2007;87:393–402. [Google Scholar]
- 6.Thoma JN, Pante E, Brugler MR, France SC. Deep-sea octocorals and antipatharians show no evidence of seamount-scale endemism in the NW Atlantic. Marine Ecology Progress Series. 2009;397:25–35. [Google Scholar]
- 7.Verrill A. Report on the Anthozoa, and on some additional species dredged by the ‘Blake’ in 1877–79, and by the U. S. Fish Commission steamer ‘Fish Hawk’ in 1880–82. Bulletin of the Museum of Comparative Zoology, Harvard. 1883;11:72pp. [Google Scholar]
- 8.Cairns SD. Studies on western Atlantic Octocorallia (Coelenterata: Anthozoa). Part 1: The genus Chrysogorgia Duchassaing & Michelotti, 1864. Proceedings of the Biological Society of Washington. 2001;114:746–787. [Google Scholar]
- 9.McFadden CS, France SC, Sánchez JA, Alderslade P. A molecular phylogenetic analysis of the Octocorallia (Cnidaria: Anthozoa) based on mitochondrial protein-coding sequences. Molecular Phylogenetics and Evolution. 2006;41:513–527. doi: 10.1016/j.ympev.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 10.France SC, Rosel PE, Agenbroad JE, Mullineaux LS, Kocher TD. DNA sequence variation of mitochondrial large-subunit rRNA provides support for a two-subclass organization of the Anthozoa (Cnidaria). Molecular Marine Biology and Biotechnology. 1996;5:15–28. [PubMed] [Google Scholar]
- 11.Berntson EA, Bayer FM, McArthur AG, France SC. Phylogenetic relationships within the Octocorallia (Cnidaria : Anthozoa) based on nuclear 18s rRNA sequences. Marine Biology. 2001;138:235–246. [Google Scholar]
- 12.S_anchez JA, Lasker HR, Taylor DJ. Phylogenetic analyses among octocorals (Cnidaria): mitochondrial and nuclear DNA sequences (lsu-rRNA, 16S and ssu-rRNA, 18S) support two convergent clades of branching gorgonians. Molecular Phylogenetics and Evolution. 2003;29:31–42. doi: 10.1016/s1055-7903(03)00090-3. [DOI] [PubMed] [Google Scholar]
- 13.Strychar KB, Hamilton LC, Kenchington EL, Scott DB. Genetic circumscription of deepwater coral species in Canada using 18S rRNA. In: Freiwald A, Roberts J, editors, Cold-water corals and ecosystems. 2005. pp. 679–690.
- 14.Pante E, France SC. Pseudochrysogorgia bellona n. gen. n. sp.: a new genus and species of chrysogorgiid octocoral (Coelenterata: Anthozoa) from the Coral Sea. Zoosystema. 2010;32:595–612. [Google Scholar]
- 15.McFadden CS, Benayahu Y, Pante E, Thoma JN, Nevarez PA, et al. Limitations of mitochondrial gene barcoding in Octocorallia. Molecular Ecology Resources. 2011;11:19–31. doi: 10.1111/j.1755-0998.2010.02875.x. [DOI] [PubMed] [Google Scholar]
- 16.Deichmann E. The Alcyonaria of the western part of the Atlantic Ocean. Memoirs of the Museum of Comparative Zoology, Harvard. 1936;53:1–317. [Google Scholar]
- 17.de Pourtales LF. Contributions to the fauna of the Gulf Stream at great depths (2d series). Bulletin of the Museum of Comparative Zoology, Harvard. 1868;1:121–142. [Google Scholar]
- 18.Wright E, Studer T. Report on the Alcyonaria collected by the H.M.S. Challenger during the years 1873–76 - Report on the scientific results of the voyage of H.M.S. Challenger during the years 1872–76, volume Zoology 31. 1–314, pls 1–43 pp. 1889.
- 19.Versluys J. Die Gorgoniden der Siboga-Expedition. I. Die Chrysogorgiiden, volume 13. Monographie Siboga-Expeditie, 120 pp. 1902.
- 20.Nutting C. Description of the Alcyonaria collected by the U.S. fisheries steamer “Albatross,” mainly in Japanese waters, during 1906. Proceedings of the United States National Museum. 1912;43:1–104. [Google Scholar]
- 21.Aurivillius M. The Gorgonarians from Dr. Sixten Bock’s expedition to Japan and Bonin Islands 1914. Kungl Svenska Vetens-kapsakad Handlingar (3) 1931;9:1–337. [Google Scholar]
- 22.Thomson J, Henderson W. An account of the alcyonarians collected by the Royal Indian Marine Survey Ship Investigator in the Indian Ocean. Part 1. The Alcyonarians of the deep sea. Indian Museum, Calcutta, i-xvi +1–132, pls. 1–10 pp. 1906.
- 23.Nutting C. Descriptions of the Alcyonaria collected by the U.S. Bureau of Fisheries steamer Albatross in the vicinity of the Hawaiian Islands in 1902. Proceedings of the United States National Museum. 1908;34:543–601. [Google Scholar]
- 24.Pante E, Watling L. Chrysogorgia from the New England and Corner Seamounts: Atlantic – Pacific connections. Journal of the Marine Biological Association of the United Kingdom FirstView. 2011.
- 25.Watling L, France SC, Pante E, Simpson A. Biology of deep-water octocorals. Advances in Marine Biology. 2011;60:41–123. doi: 10.1016/B978-0-12-385529-9.00002-0. [DOI] [PubMed] [Google Scholar]
- 26.Bayer F. Octocorallia. In: Moore R, editor, Treatise on Invertebrate Paleontology. volume (F) Coelenterata, F166–F230. 1956.
- 27.Nutting C. The Gorgonacea of the Siboga Expedition I. The Chrysogorgiidae. Siboga-Expeditie. Monograph XIII. 1910.
- 28.Bayer F, Stefani J. A new species of Chrysogorgia (Octocorallia: Gorgonacea) from New Caledonia, with descriptions of some other species from the Western Pacific. Proceedings of the Biological Society of Washington. 1988;101:257–279. [Google Scholar]
- 29.Bayer F. Distichogorgia sconsa, a new genus and species of chrysogorgiid octocoral (Coelenterata: Anthozoa) from the Blake Plateau off Northern Florida. Proceedings of the Biological Society of Washington. 1979;92:876–882. [Google Scholar]
- 30.Bayer F, Grasshoff M. The genus group taxa of the family Ellisellidae, with clari_cation of the genera established by J. E. Gray (Cnidaria: Octocorallia). Senkenbergiana biologica. 1994;74:21–45. [Google Scholar]
- 31.Bergsten J. A review of long-branch attraction. Cladistics. 2005;21:163–193. doi: 10.1111/j.1096-0031.2005.00059.x. [DOI] [PubMed] [Google Scholar]
- 32.Bayer F. Contributions to the nomenclature, systematics, and morphology of the Octocorallia. Proceedings of the United States National Museum. 1955;105:207–220. [Google Scholar]
- 33.Fabricius K, Alderslade P. Soft Corals and Sea Fans: A comprehensive guide to the tropical shallow water genera of the Central West Pacific, the Indian Ocean and the Red Sea. Townsville, Australia: Australian Institute of Marine Science, 264 pp. 2001.
- 34.Schluter D. The ecology of adaptive radiation. Oxford: Oxford University Press. 2000.
- 35.Grasshoff M. Die Gorgonaria, Pennatularia und Antipatharia. In: Laubier L, Monniot C, editors, Peuplements profonds du Golfe de Gascogne, Ifremer. 1985. pp. 299–310.
- 36.Cairns SD. Calcaxonian octocorals (Cnidaria; Anthozoa) from Eastern Pacific Seamounts. Proceedings of the California Academy of Sciences. 2007;58:511–541. [Google Scholar]
- 37.West J, Harvell C, Walls A. Morphological plasticity in a gorgonian coral (Briareum as-bestinum) over a depth cline. Marine Ecology Progress Series. 1993;94:61–69. [Google Scholar]
- 38.West J. Plasticity in the sclerites of a gorgonian coral: Tests of water motion, light level, and damage cues. Biological Bulletin. 1997;192:279–289. doi: 10.2307/1542721. [DOI] [PubMed] [Google Scholar]
- 39.Sánchez JA, Aguilar C, Dorado D, Manrique N. Phenotypic plasticity and morphological integration in a marine modular invertebrate. BMC Evolutionary Biology. 2007;7:122. doi: 10.1186/1471-2148-7-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prada C, Schizas NV, Yoshioka PM. Phenotypic plasticity or speciation? a case from a clonal marine organism. BMC Evolutionary Biology. 2008;8:47. doi: 10.1186/1471-2148-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.France SC. Genetic analysis of bamboo corals (Cnidaria: Octocorallia: Isididae): does lack of colony branching distinguish Lepidisis from Keratoisis? Bulletin of Marine Science. 2007;81:323–333. [Google Scholar]
- 42.Due∼nas L, S_anchez J. Character lability in deep-sea bamboo corals (Octocorallia, Isididae, Keratoisidinae). Marine Ecology Progress Series. 2009;397:11–23. [Google Scholar]
- 43.Concepcion GT, Crepeau M, Toonen RJ. An alternative to ITS, a hypervariable, single-copy nuclear intron in corals, and its use in detecting cryptic species within the octocoral genus Carijoa. Coral Reefs. 2008;27:323–336. [Google Scholar]
- 44.Cairns SD, Bayer FM. A generic revision and phylogenetic analysis of the Primnoidae (Cnidaria: Octocorallia). Smithsonian Contributions to Zoology. 2009;629:1–79. [Google Scholar]
- 45.Lindner A, Cairns SD, Cunningham CW. From offshore to onshore: multiple origins of shallow-water corals from deep-sea ancestors. PLoS ONE. 2008;3:e2429. doi: 10.1371/journal.pone.0002429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Young C, Tyler P, Fenaux L. Potential for deep sea invasion by Mediterranean shallow water echinoids: Pressure and temperature as stage-specific dispersal barriers. Marine Ecology Progress Series. 1997;154:197–209. [Google Scholar]
- 47.Kussakin O. Peculiarities of the geographical and vertical distribution of marine isopods and the problem of deep-sea fauna origin. Marine Biology. 1973;23:19–34. [Google Scholar]
- 48.Menzies RH, George RY, Rowe GT. Abyssal environment and ecology of the world’s oceans. New York: Wiley. 1973.
- 49.Hessler R, Thistle D. On the place of origin of deep-sea isopods. Marine Biology. 1975;32:155–165. [Google Scholar]
- 50.Heifetz J, Wing B, Stone R, Malecha P, Courtney D. Corals of the Aleutian Islands. Fisheries Oceanography. 2005;14:131–138. [Google Scholar]
- 51.Stone R. Coral habitat in the Aleutian Islands of Alaska: depth distribution, fine-scale species associations, and fisheries interactions. Coral Reefs. 2006;25:229–238. [Google Scholar]
- 52.Bilewitch JP, Degnan SM. A unique horizontal gene transfer event has provided the octocoral mitochondrial genome with an active mismatch repair gene that has potential for an unusual selfcontained function. BMC Evolutionary Biology. 2011;11:228. doi: 10.1186/1471-2148-11-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.France SC, Hoover LL. Analysis of variation in mitochondrial DNA sequences (ND3, ND4L, MSH) among Octocorallia ( = Alcyonaria)(Cnidaria: Anthozoa). Bulletin of the Biological Society of Washington. 2001;10:110–118. [Google Scholar]
- 54.France SC, Hoover LL. DNA sequences of the mitochondrial COI gene have low levels of divergence among deep-sea octocorals (Cnidaria : Anthozoa). Hydrobiologia. 2002;471:149–155. [Google Scholar]
- 55.Calderon I, Garrabou J, Aurelle D. Evaluation of the utility of COI and ITS markers as tools for population genetic studies of temperate gorgonians. Journal of Experimental Marine Biology and Ecology. 2006;336:184–197. [Google Scholar]
- 56.McFadden CS, Tullis LD, Hutchinson MB, Winner K, Sohm JA. Variation in coding (NADH dehydrogenase subunits 2, 3, and 6) and noncoding intergenic spacer regions of the mitochondrial genome in Octocorallia (Cnidaria : Anthozoa). Marine Biotechnology. 2004;6:516–526. doi: 10.1007/s10126-002-0102-1. [DOI] [PubMed] [Google Scholar]
- 57.Herrera S, Baco A, Sánchez JA. Molecular systematics of the bubblegum coral genera (Paragorgiidae, Octocorallia) and description of a new deep-sea species. Molecular Phylogenetics and Evolution. 2010;55:123–135. doi: 10.1016/j.ympev.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 58.Brugler MR, France SC. The mitochondrial genome of a deep-sea bamboo coral (Cnidaria, Anthozoa, Octocorallia, Isididae): Genome structure and putative origins of replication are not conserved among octocorals. Journal of Molecular Evolution. 2008;67:125–136. doi: 10.1007/s00239-008-9116-2. [DOI] [PubMed] [Google Scholar]
- 59.Sánchez JA, McFadden CS, France SC, Lasker HR. Molecular phylogenetic analyses of shallow-water Caribbean octocorals. Marine Biology. 2003;142:975–987. [Google Scholar]
- 60.Werle E, Schneider C, Renner M, Volker M, Fiehn W. Convenient single-step, one tube purification of PCR products for direct sequencing. Nucleic Acids Research. 1994;22:4354–4355. doi: 10.1093/nar/22.20.4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Katoh K, Misawa K, Kuma Ki, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Briefings in Bioinformatics. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 63.Abascal F, Zardoya R, Telford MJ. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Research. 2010;38 doi: 10.1093/nar/gkq291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 65.R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. 2012. URL http://www.R-project.org. ISBN 3-900051-07-0.
- 66.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 67.Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Systematic Biology. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 68.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 70.Posada D. jModelTest: Phylogenetic model averaging. Molecular Biology and Evolution. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 71.Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 72.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 73.Miller M, Holder M, Vos R, Midford P, Liebowitz T, et al. The CIPRES Portals. CIPRES. 2009-08-04. 2011. URL http://www.phylo.org/sub sections/portal.
- 74.Pasternak F. Alcyonacea and Gorgonacea. In: Kuznetsov A, Mironov A, editors, Benthos of the submarine mountains Marcus-Necker and adjacent Pacific regions, Moscow: Akademiya Nauk. 1981. pp. 40–55.
- 75.Bouchet P, Héros V, Lozouet P, Maestrati P. A quarter-century of deep-sea malacological exploration in the South and West Pacific: Where do we stand? How far to go? In: Héros V, Cowie RH, Bouchet P, editors, Tropical Deep-Sea Benthos 25. volume 196, 9–40. 2008.
- 76.Medlin L, Elwood HJ, Stickel S, Sogin ML. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene. 1988;71:491–9. doi: 10.1016/0378-1119(88)90066-2. [DOI] [PubMed] [Google Scholar]
- 77.Apakupakul K, Siddall ME, Burreson EM. Higher level relationships of leeches (Annelida: Clitellata: Euhirudinea) based on morphology and gene sequences. Molecular Phylogenetics and Evolution. 1999;12:350–9. doi: 10.1006/mpev.1999.0639. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bayesian 50% majority rule consensus trees based on different markers ( mtMutS , cox1 and 18S) and marker combinations. For mtMutS, the effect of indels on phylogeny inference was tested by removing them with Gblocks. Chrysogorgiidae taxa are either color coded (MCC) or have bolded branches (nonMCC).All trees are rooted to the Pennatulacea, except trees using the entire mtMutS gene (rooted to the Ellisellidae).
(PDF)
Collection date, geographic coordinates, depth, and genetic markers sequenced for specimens used in this study. GenBank accession numbers representing each haplotype and biogeographic region are given in the final 3 columns.
(XLS)