Abstract
The gain in fitness during adaptation depends on the supply of beneficial mutations. Despite a good theoretical understanding of how evolution proceeds for a defined set of mutations, there is little understanding of constraints on net fitness - whether fitness will reach a limit despite ongoing selection and mutation, and if there is a limit, what determines it. Here, the dsDNA bacteriophage SP6, a virus of Salmonella, was adapted to Escherichia coli K-12. From an isolate capable of modest growth on E. coli, 4 lines were adapted for rapid growth by protocols differing in use of mutagen, propagation method, and duration, but using the same media, temperature, and a continual excess of the novel host. Nucleotide changes underlying those adaptations differed greatly in number and identity, but the four lines achieved similar absolute fitnesses at the end, an increase of more than 4000-fold phage descendants per hour. Thus the fitness landscape allows multiple genetic paths to the same approximate fitness limit. The existence and causes of fitness limits have ramifications to genome engineering, vaccine design, and ‘lethal mutagenesis’ treatments to cure viral infections.
Keywords: bacteriophage, experimental evolution, nucleotide, genome, DNA sequence
Introduction
Organisms are not infinitely adaptable. Constraints, typically in the form of trade-offs and ‘invariants,’ are widely accepted as influencing the evolution of individual phenotypes such as sex ratio and life history traits (Charnov 1982; Charnov 1993; Parker and Maynard Smith 1990; Smith and Fretwell 1974; Williams 1966). The easiest constraints to understand derive from universal laws of chemistry and physics, as they are not subject to species-specific biology. Thus, principles of mechanics apply to locomotion and skeletal performance, those of energy conservation apply to metabolism. Basic sex ratio theory assumes brood resources are constrained, independent of the proportion of males.
A special challenge is to understand evolutionary constraints on major fitness components. Just as an understanding of constraints in ‘minor’ phenotypes has proved useful in predicting evolutionary patterns, the understanding of constraints on major fitness components could potentially explain such fundamental properties as species distributions, abundances, population growth, and extinction. As one example, the basic reproductive number of a virus (R0) varies widely among species and has profound implications for epidemics and control (Anderson and May 1991). Knowing the evolutionary limit of R0 for a particular virus would have equally great implications for attempts at intervention, such as whether it is necessary to prevent zoonotic cases of H5N1 ‘bird’ flu in humans from spreading.
Constraints on major fitness components likely stem from complex interactions at multiple levels in an organism's biology so are not easily derived from first principles. The tools of molecular biology, systems biology and genomics may ultimately enable us to explain these high-dimensional constraints, but current progress comes mostly from identifying whether constraints exist and what form they take. For example, constraints in the form of observed allometries are used to make predictions about life history evolution and ecology, even when the bases of those allometries are unknown (Charnov 1993). Our study is offered in this spirit.
We identify constraints on the evolution of a major viral fitness component in a virus – the intrinsic rate of increase, or ‘growth’ rate when hosts are in excess. The virus is the dsDNA bacteriophage SP6, adapted to E coli, which is a novel host for this virus. By adapting the virus to an environment in which growth rate is virtually the sole determinant of fitness and by allowing the population to evolve for hundreds of generations in that environment, it is possible to observe fitness gains, limits, and the genetic changes involved. We address three questions. (1) Does a limit exist for this phage's growth rate? (2) Is the limit robust to different genetic pathways? (3) Is the limit similar for related genomes? The results affirm the first two questions but not the third.
Methods
Strains
The bacterial and phage strains are described in Table 1. Henceforth, we refer to IJ1133 as E. coli K-12 and to MS3849 as S. enterica LT2. The latter was a gift from M. M. Susskind; the other strains came from the collection of IJM; the origin of the E. coli K-12 strain is described in earlier work (Garcia and Molineux 1996). Adaptations of SP6 to E. coli were initiated with a derivative of wild-type SP6 (SP6*) because wild-type SP6 shows no detectable growth on E. coli (see below). SP6* carries five fixed substitutions differing from the ancestral SP6+; all substitutions are missense and four lie in the tail genes 32 (24264 A->G; gp32-D174G) and 33 (25027 G->A gp33-E182K; 25062 C->A gp33-S193R; 26833 A->C gp33-T784P).
Table 1.
Bacterial and phage strains
| Notation | Species/type | Genotype | Use |
|---|---|---|---|
| MS3849 | Salmonella enterica serovar Typhimurium | LT2 xyl-404 metA22 metE551 galE719 trpD2 ilv-452 hsdLT6 hsdSA29 hsdSB121 fla-66 rpsL120 H1-b H2-e nix | Adaptations of T7+ and SP6+ |
| IJ1133 | Escherichia coli | K-12 ΔlacX74 thi Δ(mcrC- mr r) 102 ::Tn 10 | 4 adaptations of SP6* |
| T7+ | phage | wild-type (39937 bases dsDNA) | Adaptation to MS3849 |
| SP6+ | phage | wild-type (43769 bases dsDNA) | Adaptation to MS3849 |
| SP6* | phage | A mutant of SP6 that can be propagated on E. coli K-12 | Adaptation to IJ1133 |
Media
All adaptations and fitness assays used LB broth (10g NaCl, 10g Bacto Tryptone, 5g Bacto yeast extract per liter). Plates used 15g Bacto agar per liter LB broth and, when growing phages, were overlaid with soft agar (7g Bacto agar per liter LB broth). The mutagen N-methyl-N’ nitro nitrosoguanidine (NG) was used in some adaptations (Table 2). NG was dissolved in 100% ethanol at 4 mg/mL and stored at -80° C. The concentrations used here (5 μg/mL and 2 μg/mL) are somewhat lower than that used in a study of T7 in which it was shown that 10 μg/mL induced 4 mutations/genome/infection; serial transfer of T7 led to the accumulation of an average of approximately 240 mutations per genome (Springman et al. 2010). Although the quantitative effect of the lower doses used here is not known, the lower doses used here were chosen heuristically on the basis of this prior study to elevate the mutation rate without creating an excessive fitness load.
Table 2.
Protocols of SP6* adaptations to E. coli K-12
| Strain | Propagation method | Duration hours/ generations | Mutagen | Outgrowth of final population |
|---|---|---|---|---|
| C | continuous culture | 307 / 1200 | none | 307-hour population was recombined with SP6*, subjected to serial transfer for 5 hr |
| Cm | continuous culture | 86 /350 | founding population was grown by serial transfer in NG (5 ug/mL) for 38 | 86-hour population was recombined with SP6*, subjected to serial transfer for 5 hr |
| S | serial transfer | 50 /200 | none | no additional outgrowth |
| Sm | serial transfer | 67 /250 | NG (2ug/mL) added to every fourth culture | for fitness determination, the 67-hr population was subjected to serial transfer for a further 10hr without mutagen; sequence was based on the 67-hr population |
Growth conditions and fitness assays
Four separate adaptations – one replicate each – of SP6* were conducted at 37°C using E. coli host K-12 in LB broth (Table 2). One line, referred to as ‘S,’ used the serial transfer method we have used in many other studies (Bull and Molineux 2008) and thus serves as a baseline for the other protocols. The other three lines used treatments expected to enhance adaptation beyond that obtained by the usual method: longer adaptation, mutagenesis or both. In addition to the four adaptations of SP6* on E. coli, one adaptation each of SP6+ and T7+ used S. enterica LT2 as host.
All protocols were designed to select rapid phage growth, but they varied in duration, culture environment, and exposure to mutagen. Two types of transfer protocols were used for adaptations of SP6* on E. coli, serial transfer and continuous culture (Table 2); the adaptations of T7+ and SP6+ on Salmonella used just continuous culture. For serial transfer, cells from frozen aliquots were inoculated into 10mL cultures in 125ml flasks, grown for 1hr at 37°C to a density of approximately 108 cells/mL. Typically, 105 phage were added, grown 30-60 minutes (until the phage density had reached at least 107/mL, but often to lysis), and then transferred directly to the next flask, usually resulting in a dilution of at least 1000-fold. Most phage generations in each culture thus experienced low levels of coinfection (phage/cell ratios < 0.1), but the final round of growth was at high levels, which allowed recombination between genomes. The transfer volume was adjusted to contain approximately 105 phage. After incubation, the last culture of the day was sterilized with chloroform and this phage stock was used to start the first transfer on the subsequent day.
Continuous culture conditions (referred to as phage ‘chemostats’) used a cell-only tube fed unidirectionally into a tube with phage (Bull et al. 1997; Wichman et al. 2005). Dilution was thus continuous and small during any short interval. The volume of liquid in the phage tube was maintained at 1-2 mL, with a flow through rate adjusted to maintain phage densities in the range of 106 – 108 (typically 20-26 mL/hr) and thus phage/cell ratios of 1 or less; higher phage densities can select traits other than rapid growth (Bull et al. 2006), but the range of phage densities employed was sufficient to permit some recombination during the adaptations. Both tubes were immersed in a heated water bath to maintain a temperature of 37° and aerated with filtered air bubbled into the liquid; the unidirectional air flow provided both the energy for media flow between tubes and for the exhaust. The system was replaced every two days, inoculating sterile tubes with fresh cells and phage from the most recent sample (at which a modest bottleneck was introduced, of perhaps a 10-fold reduction in population size). Samples from the phage tube were taken daily and filtered or treated with chloroform to kill cells. Two of the adaptations of SP6* to E. coli K-12 and the adaptations of T7+ and SP6+ to S. enterica LT2 used continuous culture; the adaptation of T7+ lasted 139hr, that of SP6+ for 166hr (Table 2). Generation time was estimated as 15min/hr for SP6*, based on lysis times (see Results). Hours of adaptation are thus multiplied by 4 to convert into the approximate number of phage generations.
For all phages, fitness was measured as a rate of growth in the serial transfer environment: the log2 change in phage density per hour; the fitness assay environment was the same as used for the S adaptation. Each individual measurement of fitness was based on two titers separated by at least one hour of growth. In addition, the initial titer was taken after 30-60 min of growth to dampen deviations from a stable age of infection distribution (Bull 2006). Typically, estimates of the mean growth rate were based on 3 replicate assays. In contrast to passage conditions, phage densities during fitness assays were kept at least 10-fold below cell densities to avoid exhaustion of hosts and thereby allow exponential phage growth across transfers; once fitness is known approximately, phage densities can be maintained arbitrarily low by choice of appropriate transfer intervals and volumes.
Fitness comparisons across different lines are meaningful only if measured in the same environment, but that environment should also match the adaptive environment. Here, the four adaptive environments were similar to each other but only one was strictly identical to the fitness assay environment. This complication was addressed by completing all adaptations in the serial transfer environment, i.e., the environment used for fitness assays. For each line propagated in the continuous culture environment (chemostat), equal multiplicities of the final population and the initial phage (SP6*) were coinfected to allow recombination and the mixture then grown by serial passage for 5 hours; fitness was estimated from the outgrown population. By starting the 5hr outgrowth with a recombinant pool, each mutation evolved in the chemostat would have been at approximately 50% frequency, and the 5 hr of subsequent serial transfer would purge any mutation detrimental to serial passage.
The 5hr recombinant outgrowth for chemostat lines was done as a precaution. Two subsequent observations suggested that the continuous culture environment is a suitable mimic of serial transfer. First, sequence comparisons from the C population at the end of the 307hr chemostat adaptation and those of its 5hr recombinant outgrowth revealed only minor differences, suggesting that the changes evolving in the chemostat are also beneficial in serial transfer (data not shown). Second, a fitness measurement of the C population at 102 hr (using no recombinant outgrowth) was indistinguishable from that of the 307 hr, recombinant outgrowth population.
For the SP6* line evolved in the periodic presence of mutagen (Sm), the 67 hr, evolved population was grown by serial transfer for a further 10hr in the absence of mutagen, and this population (which was not recombined against SP6*) was used for fitness assays. The logic for this protocol was that any mutation even moderately deleterious in the mutagen-free, serial transfer environment would not ascend to high frequency when three of every four transfers lacked mutagen – deleterious mutations would have remained polymorphic. So 10 hours of mutagen-free outgrowth should have been sufficient to purge deleterious mutations and minimize their impact on fitness.
In addition to the recombinant populations created between SP6* and an evolved population, a recombinant pool was created among all four evolved lines. This recombinant pool (designated as 4R) was outgrown for 10 hours by serial transfer and was then tested to determine whether higher fitness could be achieved by mixing substitutions from all independent lines.
Creating recombinant populations
T7-like phages replicate their genomes in a highly recombinogenic process. Co-infection by differentially marked T7 phages leads to ~ 0,01% recombination per base-pair between the two genomes (Molineux 2006; Studier 1969). Recombination frequencies between coinfecting SP6 genomes have not been measured directly, but are likely comparable because of the overall genetic similarity between T7 and SP6.
To obtain a population of varied recombinants, plates were overlaid with cells in soft agar and allowed to set. The surface was streaked with 2-5 μL of a phage suspension (at least 106 phage/mL), and this streak was then cross–streaked with a suspension of the other phage(s). The plate was incubated at 37°C for several hours, until the phage-induced clearings had expanded beyond the immediate zones of the streaks, ensuring high multiplicity of infection and thus recombination. The soft agar in which the two phage types had grown into each other were then scraped from the plate and diluted in broth. Resuspended phages were used directly as recombinant populations, which were typically propagated for five hours by serial transfer. Lysis was allowed before each transfer to allow additional recombination and to facilitate selection for beneficial mutations.
Adsorption assays
E. coli K-12 cells were grown for 1 hr to a density of approximately 108/mL and infected at low multiplicity. At five min, two samples were plated: (i) a sample of the suspension to estimate the total phage density (PT), and (ii) the supernatant of a centrifuged sample (12,000 rpm for 30 sec) to estimate the density of unadsorbed phages (PU). The adsorption rate coefficient k (mL/min) was calculated from PU = PT [exp(-5kC)], where C was the cell density per mL based on a titer made just prior to phage addition.
Lysis time assays
Lysis time was measured in liquid cultures by the decline in culture turbidity following a high multiplicity of infection (MOI). The MOI used for evolved phages was 10, that for SP6* was 100 because of its low adsorption rate. Turbidity was measured using a Klett-Summerson colorimeter and the data fit to a normal distribution function to estimate the mean lysis time (Heineman et al. 2005).
Sequence analysis
Nucleotide changes were determined with ‘454’ pyrosequencing technology. DNA of each phage population was subjected to sequencing, yielding frequency estimates of nucleotide changes but not genomic linkages beyond a few hundred nucleotides. Nucleotide changes whose frequencies were less than 40% were ignored, except for one change in the S line that appeared to be in complete linkage disequilibrium with another change. Discarding low- and moderate-frequency mutations eliminates common 454 sequencing errors; furthermore, in previous studies mutations achieving only moderate frequency by the end of an adaptation have not been found to be responsible for large fitness effects (Bull and Molineux 2008). Sequence files (sff format) have been deposited in Sequence Read Archive (SRP007575).
Results
Nature of selection in this host-range shift
Bacteriophage SP6, whose natural host is considered to be Salmonella enterica serovar Typhimurium, was adapted to a novel host, a strain of E. coli K-12. Although laboratory adaptation of SP6 to Salmonella would likely have resulted in measurable evolution, it seemed that adaptation to a novel host would increase the opportunity for evolution and possibly require multiple, sequential adaptive steps before any limit was reached. Like phage P22, SP6 virions display tailspikes containing endorhamnosidase activity that degrade the O-antigen of the host. Unlike P22, however, SP6 also grows on rough strains, which lack O-antigen (Scholl et al. 2004). SP6 also grows on Escherichia coli strains containing a S. enterica LT2 O-antigen cosmid but it fails to adsorb detectably to common laboratory E. coli strains (IJM, unpublished observations). Even spots of highly concentrated SP6 stocks do not elicit clearing on nascent lawns of E. coli, indicating a complete absence of infection. It was thus expected that successful adaptation of SP6 to E. coli would involve changes in adsorption, at a minimum affecting tail and/or tailspike genes. However, many other interactions with the novel host might also be subject to selection.
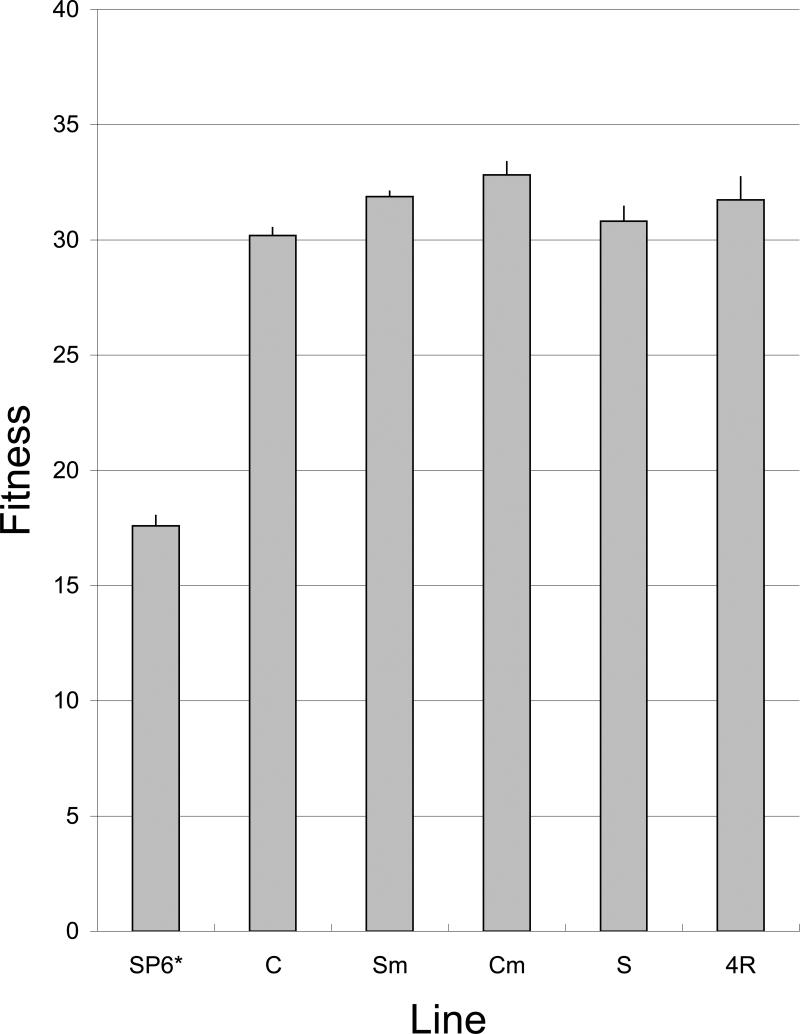
The adaptations performed here started with a mutant of SP6 (SP6*; isolation to be described elsewhere) that had expanded its host range from smooth and rough S. enterica LT2 to include rough E. coli K-12 strains. Its fitness was 17.6 doublings/hr on the E. coli K-12 host (Fig. 1). SP6* carries 4-5 mutations in the tail genes 32 and 33 (Table 3); at least three of the four are responsible for the host range expansion (IJM, unpublished data). Despite having overcome the initial hurdle of adsorption and genome penetration of the cell, it seemed that considerable opportunity for adaptive evolution of SP6* remained: the fitness of phage T7, a relative of SP6, can attain ~48 doublings/hr on this host (Heineman and Bull 2007).
Figure 1.
Fitnesses (doublings/hr) of the initial phage (SP6*), of endpoints from four lines adapted from it (C, Sm, Cm, and S), and of the adapted recombinant line created from a mixture of the four lines (4R). Fitness is measured as doubling/hr of the phage population under conditions of unlimited hosts (E. coli K-12) in broth at 37°C. Bars represent 1 std error.
Table 3.
Nucleotide change frequencies** of the evolved lines and SP6* (relative to SP6+, GenBank AY370673) the ‘weighted number of changes from SP6*’ is the sum of differences between SP6* and the evolved line, weighted by the frequency differences of those changes
| Position | Change (nuc) | Element (function) AA change | Phage line | ||||
|---|---|---|---|---|---|---|---|
| SP6* | C | S | Sm | Cm | |||
| 669-938 | Del | gene 0.2 | 1 | 0.84 | 0.86 | ||
| 1284-1911 | Del | partial gene 2 and partial gene 3 | 1 | 0.87 | |||
| 8428-9130 | Del | partial gene 9 and partial gene 10 | 1 | ||||
| 180 | Ins CCGTTC | just outside CJ | 0.99 | ||||
| 1129 | G -> A | gene 1 V12I | 0.85 | ||||
| 1901 | C -> T | gene 3 Q89X | 0.83 | ||||
| 2151 | Ins CCA | Potential RNase III site | 0.97 | ||||
| 2553 | G -> T | gene 6 R40L | 1 | 0.75 | |||
| 2630 | C -> A | gene 6 H66N | 0.47 | 0.74 | |||
| 2807 | C -> T | gene 6 P125S | 0.68 | ||||
| 3372 | G -> A | IG 6-7 | 0.8 | ||||
| 3696 | C -> T | gene 7 (RNAP) A102V | 1 | 1 | |||
| 6096 | C -> T | (between TE and a phage promoter) | 0.5 | ||||
| 6123 | A -> G | phage promoter | 1 | ||||
| 6178 | G -> A | (between two phage promoters) | 0.56 | ||||
| 6711 | G -> A | gene 9 (primase) K80K | 0.48 | ||||
| 8417 | G -> A | gene 9 G649D | 0.56 | ||||
| 8751 | G -> A | gene 10 A51T | 0.75 | ||||
| 11819 | G -> A | gene 13 E747E | 0.73 | ||||
| 12799 | C -> T | gene 15 R57R | 0.73 | ||||
| 13004 | Ins A | gene 15 (terminal codon) | 0.95 | ||||
| 13031 | G -> A | IG 15.5-16 | 0.75 | ||||
| 13485 | C -> T | gene 16 D90D | 0.62 | ||||
| 14732 | T -> G | IG (2 bases in front of putative terminator) | 0.99 | ||||
| 14735+ | C -> A | Putative terminator | 0.81 | ||||
| 14757 | G -> A | Putative terminator | 0.63 | ||||
| 14758 | G -> A | Putative terminator | 0.7 | ||||
| 14758+ | G -> T | Putative terminator | 0.26 | ||||
| 15326 | G -> A | gene 20 | 0.64 | ||||
| 15419 | G -> A | gene 20 G137D | 0.64 | ||||
| 16108 | G -> A | gene 21 G29D | 0.73 | ||||
| 16429 | G -> A | gene 21 (22) G136E (G2S) | 0.93 | ||||
| 16478 | A -> C | gene 22 E18A | 0.86 | ||||
| 17052 | C -> T | gene 22 Y209Y | 0.73 | ||||
| 19806 | C -> T | gene 28 A22V | 0.48 | ||||
| 19841 | A -> C | gene 28 T34P | 0.45 | ||||
| 19859 | G -> A | gene 28 D40N | 0.93 | ||||
| 20911 | C -> A | gene 29 (head- tail connector) A320E | 0.99 | ||||
| 20921 | C -> T | gene 29 I323I | 0.77 | ||||
| 20933 | G -> A | gene 29 Q327Q | 0.77 | ||||
| 21554 | T -> G | gene 30 A17A | 0.48 | ||||
| 23863 | T -> A | gene 32 (tail) G40G | 1 | ||||
| 24264 | A -> G | gene 32 D174G | 1 | 1 | 1 | 1 | 1 |
| 24689 | G -> A | gene 33 (tail) G69E | 0.78 | ||||
| 25027 | G -> A | gene 33 E182K | 1 | 1 | 1 | 1 | 1 |
| 25033 | C -> A | gene 33 H184N | 081 | ||||
| 25062 | C -> A | gene 33 S193R | 1 | 1 | 1 | 1 | 1 |
| 25093 | A -> C | gene 33 S204R | 1 | 0.62 | |||
| 25095 | T -> G | gene 33 R204R | 0.85 | ||||
| 25099 | G -> A | gene 33 D206N | 0.94 | ||||
| 25237 | G -> A | gene 33 A252T | 0.81 | ||||
| 25888 | G -> A | gene 33 V469I | 0.6 | ||||
| 26584 | A -> G | gene 33 R701G | 0.99 | 1 | 0.63 | ||
| 26833 | A -> C | gene 33 T784P | 1 | 0.98 | 1 | 1 | |
| 26833 | A -> T | gene 33 P784S | 1 | ||||
| 26890 | G -> A | gene 33 V803M | 0.93 | ||||
| 26996 | A -> G | gene 34 (internal virion) | 0.5 | ||||
| 27101 | A -> G | gene 34 D69G | 1 | 0.95 | |||
| 28014 | G -> A | gene 35 (internal capsid) | 0.95 | ||||
| 29164 | G -> A | gene 35 V517I | 0.94 | ||||
| 29250 | C -> T | gene 35 N545N | 0.94 | ||||
| 29425 | Del G | gene 35 (604th codon of 978) | 1 | ||||
| 30019 | G -> A | gene 35 D802N | 1 | ||||
| 30766 | G -> C | gene 36 (internal capsid) | 0.94 | ||||
| 33163 | A -> G | gene 36 N849D | 1 | ||||
| 33196 | C -> T | gene 36 R860C | 0.98 | 0.48 | |||
| 33509 | A -> G | gene 36 D964G | 0.46 | ||||
| 35056 | Del T | gene 36 | 0.4 | 0.41 | 0.4 | ||
| 36158 | C -> T | gene 40 P94S | 0.92 | ||||
| 36571 | G -> A | gene 40 G231G | 0.92 | ||||
| 37063 | C -> T | gene 40 D395D | 0.81 | ||||
| 37874 | Ins T | phage promoter | 0.49 | ||||
| 40864 | G -> A | gene 46 (tailspike) | 0.4 | ||||
| 41601 | G -> A | gene 46 E531K | 1 | 1 | 1 | 1 | |
| weighted number of changes from SP6* | 22.95 | 6.91 | 32.74 | 9.9 | |||
key: IG: intergenic, Del: deletion, Ins: insertion, AA: amino acid. Elements and functions listed here are taken from Scholl et al. (2004).
+ every sequence read fixed for one or the other mutation; frequencies do not add to 1.00 because not all reads spanned both sites
changes listed only if above 0.4 in frequency
Parallel evolution: assigning each of the observed base substitutions in a line to a position at random, the probability of observing evolution at the same site in all four lines is P<2×10-9; of observing two sites evolving in parallel across 3 lines is P<10-10; of observing 6 sites evolving in parallel across 2 lines is P<10-11.
To assess the fitness ‘limit’ of SP6* on E. coli as well as the possible impact of protocol on adaptation, we chose 4 different methods of evolving SP6* to higher fitness. All adaptations selected for rapid growth on an unlimited supply of naive E. coli K-12 cells in LB broth at 37°C, but they differed in the manner by which hosts were provided, the duration of adaptation, and the use of mutagen (Table 2). The expectation from this design was that, if our usual protocols of experimental phage evolution were inadequate for attaining maximal fitness, the S line (serial passage without mutagen, shortest duration) would attain the lowest fitness; higher fitness would be attained with any or all of the others.
Similar fitness endpoints despite different protocols
Final fitness, measured as growth rate in the serial transfer environment, was broadly similar across all four lines, ranging from 30-33 doublings/hr (Fig. 1). This represents a gain of at least 12 doublings/hr over the ancestral SP6*. In absolute numbers, the gain in fitness is minimally a 4,000-fold increase in phage descendants per hour. Fitness among adapted lines was marginally heterogeneous (F(3,7) = 4.5, P=0.046) but with no evidence of a difference among any pair of lines (pairwise t-tests among the 4 endpoint lines are not significant after correction for multiple comparisons). The overall similarities in final fitness are remarkable, given that durations and methods of adaptation were variable; in particular, protocols were not designed to ensure that maximum fitness had been reached. Although all adaptations were carried out at least as long as we have commonly employed in previous studies (Bull and Molineux 2008), some adaptations were extended for substantially longer times (e.g., C and Sm), and mutagen was used in two lines to increase the input of mutations.
The fact that similar fitnesses were reached using different protocols raises the possibility that ~32 doublings/hr is a practical fitness limit for SP6 under these conditions. It is of course likely that there are true differences in fitness among these lines that would be revealed with a larger set of fitness assays, but any such differences are necessarily small relative to the fitness improvement of 12 doublings/hr from the SP6* ancestor. Although continued adaptation of these lines would perhaps realize small fitness increases, note that for the C line, fitness at 102 hr was 30.7 ±0.32, indistinguishable from the 30.2 ±0.38 at 307 hr. The dominant picture, therefore, is that all four adaptations rapidly experienced major, similar fitness gains, relative to any remaining differences, and that in at least one line an approximate fitness plateau was reached long before the end of the adaptation protocol.
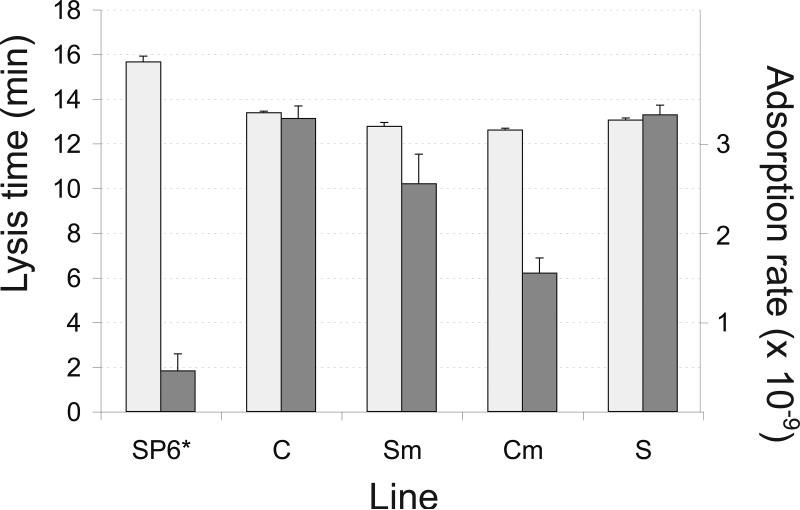
Phenotypic bases of fitness improvement were explored in the phenotypes of adsorption rate and lysis time (Fig. 2). Both traits showed significant improvements in the expected direction of higher adsorption rates and faster time to lysis.
Figure 2.
Lysis time (light bars, units of minutes) and adsorption rates (dark bars, units mL/min) of SP6* and of the four evolved lines. From a value of 4.5 × 10-10 (mL/min) for the SP6* ancestor, adsorption rate improved significantly in all four lines by a factor of 3-7, but also remained significantly heterogeneous among the four evolved lines (F(4,15) = 42, P< 10-4 for the 5 lines; F(3,12) = 16.2, P = 2 × 10-4 without SP6*). Lysis time declined 2-3 min from a value of 15.6 for the SP6* ancestor (F(4,5) = 63.4, P< 2 × 10-4 for the 5 lines); lysis times of the four evolved lines were not significantly heterogeneous. Bars represent 1 std error.
Big differences in nucleotide evolution among populations
In view of the similar endpoint fitnesses, the large differences in number of evolved changes among lines is striking (Table 3). The most changes were seen in the Sm line (37 new substitutions, not counting losses from SP6*, 32.7 differences from SP6* weighted by population frequency). The next most evolution was seen in the C line (24, 22.9) with substantially lower numbers of changes in the Cm (12, 9.9) and S (8, 6.9) lines. A few deletions and insertions were also observed. The number of evolved changes between the lowest and highest lines thus spans almost 5-fold. The number of changes correlates poorly with use of mutagen, chemostat versus serial transfer, or with estimated fitness (respective mean fitnesses in the order listed above are 31.9, 30.2 32.8 and 30.8, not significantly different, as noted earlier). Given the lack of replicates within each protocol, we cannot say whether individual protocols are prone to high or low numbers of changes.
Consistent with prior studies of experimental evolution in phages, the vast majority of observed changes in the SP6 lines are either missense mutations in coding sequences, indels, or substitutions in regulatory elements (Table 3). The near absence of silent substitutions in coding regions suggests that the large majority of changes are beneficial. Population sequences of the four lines reveal modest to high levels of parallel evolution of the same nucleotide, which were highly significant (P < 10-8, Table 3). The level of parallel evolution is especially high for the S line (4 of the 8 gains evolved in parallel with another line) and Cm line (6 of 12) but lower for the Sm line (6 of the 37) and C line (6 of 24). Thus parallel evolution was proportionately highest in the lines with fewest changes. Parallel evolution would have been reduced by any selective differences between the serial transfer and chemostat environments, but all six instances of parallelism between two lines involved a serial transfer line and a chemostat line. Any parallel changes could have been due to low frequency mutations present in the common SP6* lysate used to start the adaptations and thus may not have arisen independently, but regardless of their origins, the ascendance or fixation of parallel mutations in the different lines reveals similar selection across the different protocols.
Mutations affecting different nucleotides could also have the same phenotypic effect and be ‘phenotypically’ parallel (Pelosi et al. 2006). Phenotypic parallelism is difficult to demonstrate convincingly in the absence of biochemical and structural models of function, but other methods of inference are possible. One case of phenotypic parallelism was detected from strict linkage disequilibrium in the S line: all reads that spanned bases 14735-14758 revealed a change at either nucleotide 14735 or 14758 but never at both sites nor at neither site (Table 3). These changes likely affect the stability of a putative intrinsic transcriptional stem-loop terminator in the middle of the DNA metabolism genes (Scholl et al. 2004).
Recombination among lines does not yield higher fitness
Despite the similar final fitnesses, the lines might not have reached a maximum, but were still adapting by different pathways toward a common endpoint (a possibility that we ruled out for the C line). The four high-fitness lines were recombined into a common pool, and that population was subjected to serial passage for 10 hours. The ending fitness of 31.7 (± 0.82) was indistinguishable from that of the four founding lines (designated 4R in Fig. 1; a heterogeneity test of the 5 lines gives F(4,8) = 3.3, P = 0.07). This outcome further supports the conclusion of a robust fitness limit for SP6 under the conditions employed of 32 doublings/hr.
The genetic composition of the ending 4-way recombinant line was not an equal mix of the 4 input lines. It was dominated by changes of the C line but also carried a few changes from the Cm and Sm lines (data not shown). The four changes unique to the S line were not present and we cannot unequivocally determine the extent of its participation in creating the recombinant pool, The failure to evolve a balanced mix of changes from the different lines is expected if the mutations are highly epistatic – good only in specific backgrounds containing other mutations. Of course, epistasis is also evident by the lack of higher fitness in the recombinant population than in the progenitor lines.
Genomic basis of fitness limits: a higher limit for a relative of SP6
The approximate constancy of the SP6 fitness limit to variations in adaptation protocol suggests that the limit is robust, at least when the population can only explore the local landscape by point mutations and deletions and the environment is held constant. The question is how far this robustness extends. A previous study of phages suggested that fitness limits may be due to general properties of a genome (Bull et al. 2004). If so, phages closely related to SP6 may realize similar limits. A simple test of this possibility is afforded by T7, a more distant relative of SP6 that has been adapted to IJ1133 in many contexts (Bull and Molineux 2008).
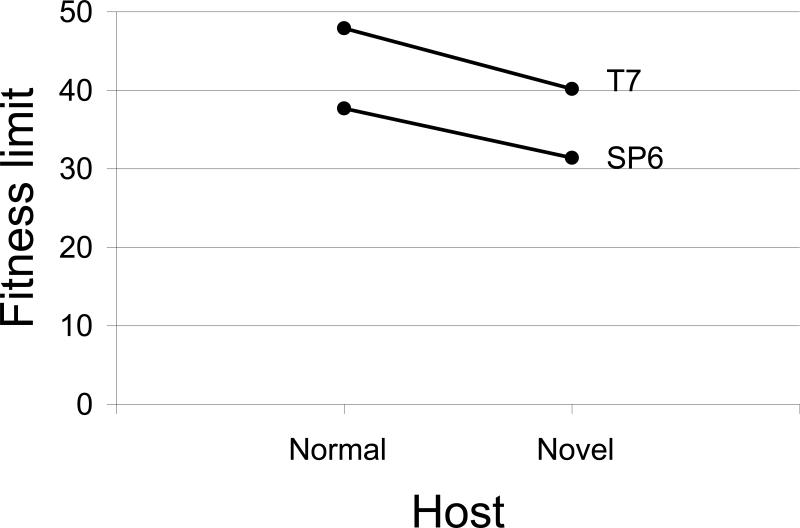
Extensive adaptation of T7 to growth on E. coli K-12 achieved a fitness of 47.9 doublings/hr (Heineman and Bull 2007). This value is much higher than the 32 of SP6*, corresponding to a more than 60,000-fold greater expansion per hour. However, it is premature to conclude that T7 is intrinsically superior to SP6 in its fitness limit, because E. coli is the normal host for T7 but a novel host for SP6. Perhaps the differential would be reversed when using the normal host for SP6.
To address the effect of host novelty, T7 and SP6 were both adapted to a S. enterica LT2 strain, the ancestral host for SP6 but a novel host for T7. Our S. enterica LT2 strain carries a galE mutation that prevents O-antigen biosynthesis, and it also carries several mutations inactivating various restriction systems (like some mutations in the E. coli K-12 strain used here). Both wild-type SP6 and wild-type T7 grow on this S. enterica LT2 strain, yielding plating efficiencies of unity relative to their defined preferred hosts. Fitnesses of T7 and SP6 adapted to S. enterica LT2 were far more similar than their fitnesses on E. coli (Fig. 3). When the fitnesses are plotted both by phage and by novelty of host (S. enterica is the novel host for T7, E. coli is the novel host for SP6), T7 is superior to SP6 on both hosts but there is a common effect of host novelty, indicated by the nearly parallel decline in fitness for each phage when growing on the novel host.
Figure 3.
Maximal fitnesses (doublings/hr) seen in T7 and SP6 adapted to the hosts E. coli and Salmonella enterica. The normal host for T7 is E. coli, for SP6 is Salmonella enterica. There is a strong and similar effect of normal versus novel host for both phages, indicated by the parallel declines in fitness on the novel host. There is also a superiority of T7 over SP6 when host type is held constant. T7 and SP6 are both members of the Podoviridae, but they belong to different genera within the subfamily Autographivirinae.
Discussion
A virus was adapted to a new host by several different protocols with the goal of studying the sensitivity of fitness and phenotypic outcomes to the genetic pathway. The virus used here was SP6, a member of the Podoviridae whose usual host is considered to be Salmonella. enterica sv. Typhimurium. The new host was Escherichia coli K-12. Viral host range evolution is often studied from the perspective of how a virus can evolve to use new hosts, but the emphasis here is merely to use a new host as a novel environment for viral adaptation under different protocols. Use of a new host offered the advantage that at least some of the initial impediments were known a priori, and there was reason to anticipate considerable fitness evolution.
Despite straightforward expectations that SP6 fitness would increase in our experimental environment, there was no clear expectation of whether fitness should reach a maximum in these brief adaptations. Observations from a century of selective breeding of domestic plants and animals reveals a plateau for some highly selected traits but continual progress for others (Hill 2008; Hill and Bunger 2003). Asexual bacterial lines evolved for tens of thousands of generations exhibit continual evolution and declining rates in fitness increases, as if a plateau is being approached but not obviously attained (de Visser and Lenski 2002). The novelty of our phage study is that the genome is small relative to bacteria and eukaryotes, and recombination occurs during adaptation. Selection is also largely free of a frequency-dependent component, similar to the breeding of domestic plants and animals. With this combination, there is clear potential for rapid evolution to any existing fitness maximum.
Four adaptations were conducted on E. coli K-12, all favoring rapid growth on a continual supply of naive hosts, but differing in the use of mutagen, the manner of continuous propagation and the duration of adaptation. The use of different protocols was in part a test of which properties of an adaptation would realize highest fitness. Surprisingly, final fitnesses of the four lines were broadly similar and all were substantially above the starting fitness. Theoretical models suggest that the large reduction in population size used in our standard serial transfer protocol will lose most beneficial mutations, so it was conceivable that 200 generations of serial transfer would be insufficient to attain a fitness limit (Hubbarde and Wahl 2008; Wahl and Krakauer 2000). Indeed, an adaptation using a 5th protocol and abandoned at 20hr – 80 generations – resulted in a fitness of 27.5, suggesting the limit had not been reached (data not shown). Adaptation should then be enhanced by raising the total input of beneficial mutations either from the occasional use of mutagen or from an increased duration of adaptation (in a chemostat). Furthermore, the chemostat environment does not employ extreme bottlenecks and fewer beneficial mutations should be lost than in serial transfer. However, it is also possible that, despite similarities, the chemostat and serial transfer environments are different enough that some mutations beneficial in serial transfer would not evolve in the chemostat. (We showed that the converse is not true for the C line.) There is therefore justification to consider that chemostat adaptation may have enhanced but also perhaps restricted opportunities to evolve high fitness as measured in the serial transfer environment.
The analyses and protocol variations we employed were neither as exhaustive nor as balanced as might have been attempted in a larger study, yet the observations permit several important conclusions about three empirical generalities developed from other work: (a) limits of adaptation, (b) parallel evolution, and (c) the importance of recombination in attaining high fitness.
(a) Limits of adaptation
Several aspects of the results suggest that a fitness of 32 doublings/hr is an approximate upper limit of fitness for growth of SP6 on E. coli K-12. There are undoubtedly real fitness differences among the evolved lines but they are small relative to the magnitude of the total fitness increase. It is also likely that continued evolution of each line would lead to further increases but we anticipate that they would be small. For these reasons, we consider the observed approximate limit to be a practical one, not an absolute limit. The surprising aspect of these results is that this approximate limit is robust to the different genetic pathways amid high levels of epistasis among the mutations.
One interesting implication of the robustness of the limit concerns the practice of lethal mutagenesis -- increasing the mutation rate of a virus to the point that its population dies out (Bull et al. 2007a; Vignuzzi et al. 2006). However, clinical mutagenic treatments often fail to clear the virus. The obvious concern from ‘failed’ lethal mutagenesis is an evolutionary saltation to a new fitness peak. The fact that fitness limits were similar between adaptive protocols with and without mutagenesis alleviates the concern, as did an earlier study addressing the evolution of enhanced ‘robustness’ during lethal mutagenesis (Martin et al. 2008).
In earlier work, it was observed that fitness limits were broadly similar within phages of the same type, hence it was suggested that fitness limits could be predicted from genome composition (Bull et al. 2004). Here that generality failed in two ways: fitness limits were lower on a novel host than on an ancestral host, and the limit of T7 was about 10 doublings/hr higher than that of SP6, even when correcting for host. SP6 and T7 are clearly related but sequence divergence across homologous proteins is substantial, and there are some differences in essential genes (Dobbins et al. 2004; Scholl et al. 2004), so the two genomes have many potential causes of differences in fitness limits. Furthermore, precedents already existed to suggest that fitness limits are not robustly determined by genome composition: the fitness limit of T3 (a much closer relative of T7 than is SP6) is 4 doublings/hr lower than that of T7, and the limit of T7 can itself be depressed by up to 20 doublings/hr by genome rearrangements (Bull et al. 2004; Springman et al. 2005).
(b) Parallel molecular evolution
Parallel nucleotide evolution across lines comprised a moderate to high fraction of total changes (50% in two lines), highly statistically significant. These levels are consistent with the similarity of selective conditions across the different environments, but of course, levels of parallelism could possibly have been even higher had the environments been more similar. There is in fact no consistent pattern of parallel molecular evolution in experimental adaptations of microbes. A low level has been observed in some prior adaptations of T7 and bacteria (Bull and Molineux 2008; Bull et al. 2007b; Ferris et al. 2007; Harcombe et al. 2009; Rokyta et al. 2008; Springman et al. 2005; Woods et al. 2006), but high levels were observed in experimental adaptations of the microviruses ϕX174, G4, and their relatives, as well as in some small-genome ssRNA phages (Bollback and Huelsenbeck 2009; Bull et al. 2000; Bull et al. 1997; Wichman et al. 2005; Wichman et al. 2000).
A biological perspective on SP6 provides a rationale for parallel evolution in these adaptations. Infection of a new host could require specific protein changes that would have few alternatives in a virus not well adapted, such as the missense substitutions in the tail proteins gp32 and gp33. These proteins constitute the bulk of the stubby SP6 tail, which interacts with a cell surface component - likely the lipopolysaccharide (LPS) inner core. The LPS sequence and structure differ between E. coli and S. enterica sv. Typhimurium LT2, and the rate of adsorption to E. coli of the adapted phages is much faster than of the parental SP6*. Mutations affecting the tail proteins were thus expected to enhance fitness, and indeed, some did exhibit parallel evolution.
A few instances of parallel evolution were also observed in the internal core proteins: gp34, gp35, and gp36. By analogy with T7, these proteins are ejected from the phage virion into the infected cell, creating a channel for phage DNA transport across the cell envelope and initiating DNA translocation into the infected cell (Chang et al. 2010; Kemp et al. 2005; Kemp et al. 2004) . As the cell envelopes of E. coli and S. enterica sv. Typhimurium LT2 are different, some substitutions in the internal core proteins of adapted phages may compensate for these differences, enhancing the rate of genome entry into the cell cytoplasm. In turn, this may explain the shorter latent period (and thus higher fitness) of the adapted phages relative to SP6*.
(c) Recombination and fitness improvement
Recombination is expected to enhance adaptation under many scenarios. Recombination enables each mutation to compete largely independently of the specific genetic background in which it arises, so that multiple, beneficial mutations can ascend in parallel and deleterious mutations cannot readily hitch-hike with beneficial mutations. However, it is also appreciated that recombination can thwart adaptation when mutations interact epistatically (Bell 1982; Crow and Kimura 1970; Maynard Smith 1978; Williams 1975). By allowing periodic co-infection of cells, our adaptations allowed extensive recombination to occur in each line. Indeed it would have been difficult to maintain recombination at low levels. Any effect of recombination on attainment or the magnitude of a fitness limit cannot be assessed from these adaptations.
There is, however, a second use of recombination commonly regarded to enhance adaptation that can be evaluated: recombination among moderately diverged lines. This type of gene and genome ‘shuffling’ has been an empirical cornerstone of ‘directed’ evolution (Crameri et al. 1998; Patnaik et al. 2002; Patten et al. 1997; Rowe et al. 2003; Voigt et al. 2000). However, recombination between the 4 adapted lines failed to enhance fitness. In part this reflects epistasis in the adaptive mutations in each line. Nevertheless, there may also be limits to the fitness improvements that genome shuffling can achieve, especially when the divergent lines are adapted before shuffling (as done here but not typically in other directed evolution studies).
The origins of fitness limits
It may be that simple characterizations, such as genome composition and other sequence-based metrics, will prove too superficial to be of much value in predicting fitness limits. However, the limits of some major fitness components should follow from well-understood principles in the intracellular dynamics of gene expression and protein synthesis. Although the specific parameters for any system are difficult to measure, appreciating the causes of fitness limits may be necessary before synthetic biology can realize its potential.
Acknowledgments
This work was supported by NIH GM57756 to JJB, GM 32095 to IJM; JJB also receives support as the Miescher Regents Professor. We thank two reviewers and S. Gandon for comments.
Literature Cited
- Anderson RM, May RM. Infectious diseases of humans : dynamics and control. Oxford University Press; Oxford ; New York: 1991. [Google Scholar]
- Bell G. The masterpiece of nature : the evolution and genetics of sexuality. Croom Helm; London: 1982. [Google Scholar]
- Bollback JP, Huelsenbeck JP. Parallel genetic evolution within and between bacteriophage species of varying degrees of divergence. Genetics. 2009;181:225–34. doi: 10.1534/genetics.107.085225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull JJ. Optimality models of phage life history and parallels in disease evolution. J Theor Biol. 2006;241:928–38. doi: 10.1016/j.jtbi.2006.01.027. [DOI] [PubMed] [Google Scholar]
- Bull JJ, Badgett MR, Springman R, Molineux IJ. Genome properties and the limits of adaptation in bacteriophages. Evolution. 2004;58:692–701. doi: 10.1111/j.0014-3820.2004.tb00402.x. [DOI] [PubMed] [Google Scholar]
- Bull JJ, Badgett MR, Wichman HA. Big-benefit mutations in a bacteriophage inhibited with heat. Mol Biol Evol. 2000;17:942–50. doi: 10.1093/oxfordjournals.molbev.a026375. [DOI] [PubMed] [Google Scholar]
- Bull JJ, Badgett MR, Wichman HA, Huelsenbeck JP, Hillis DM, Gulati A, Ho C, Molineux IJ. Exceptional convergent evolution in a virus. Genetics. 1997;147:1497–507. doi: 10.1093/genetics/147.4.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull JJ, Millstein J, Orcutt J, Wichman HA. Evolutionary feedback mediated through population density, illustrated with viruses in chemostats. The American Naturalist. 2006;167:E39–E51. doi: 10.1086/499374. [DOI] [PubMed] [Google Scholar]
- Bull JJ, Molineux IJ. Predicting evolution from genomics: experimental evolution of bacteriophage T7. Heredity. 2008;100:453–63. doi: 10.1038/sj.hdy.6801087. [DOI] [PubMed] [Google Scholar]
- Bull JJ, Sanjuan R, Wilke CO. Theory of lethal mutagenesis for viruses. J Virol. 2007a;81:2930–9. doi: 10.1128/JVI.01624-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull JJ, Springman R, Molineux IJ. Compensatory evolution in response to a novel RNA polymerase: orthologous replacement of a central network gene. Mol Biol Evol. 2007b;24:900–8. doi: 10.1093/molbev/msm006. [DOI] [PubMed] [Google Scholar]
- Chang CY, Kemp P, Molineux IJ. Gp15 and gp16 cooperate in translocating bacteriophage T7 DNA into the infected cell. Virology. 2010;398:176–86. doi: 10.1016/j.virol.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charnov EL. The theory of sex allocation. Princeton University Press; Princeton, NJ: 1982. [PubMed] [Google Scholar]
- Charnov EL. Life history invariants : some explorations of symmetry in evolutionary ecology. Oxford University Press; Oxford England ; New York: 1993. [Google Scholar]
- Crameri A, Raillard SA, Bermudez E, Stemmer WP. DNA shuffling of a family of genes from diverse species accelerates directed evolution. Nature. 1998;391:288–91. doi: 10.1038/34663. [DOI] [PubMed] [Google Scholar]
- Crow JF, Kimura M. An introduction to population genetics theory. Harper & Row, Publishers; New York, Evanston, London: 1970. [Google Scholar]
- de Visser JA, Lenski RE. Long-term experimental evolution in Escherichia coli. XI. Rejection of non-transitive interactions as cause of declining rate of adaptation. BMC Evol Biol. 2002;2:19. doi: 10.1186/1471-2148-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbins AT, George M, Jr., Basham DA, Ford ME, Houtz JM, Pedulla ML, Lawrence JG, Hatfull GF, Hendrix RW. Complete genomic sequence of the virulent Salmonella bacteriophage SP6. J Bacteriol. 2004;186:1933–44. doi: 10.1128/JB.186.7.1933-1944.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris MT, Joyce P, Burch CL. High frequency of mutations that expand the host range of an RNA virus. Genetics. 2007;176:1013–22. doi: 10.1534/genetics.106.064634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia LR, Molineux IJ. Transcription-independent DNA translocation of bacteriophage T7 DNA into Escherichia coli. J Bacteriol. 1996;178:6921–9. doi: 10.1128/jb.178.23.6921-6929.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcombe WR, Springman R, Bull JJ. Compensatory evolution for a gene deletion is not limited to its immediate functional network. BMC Evol Biol. 2009;9:106. doi: 10.1186/1471-2148-9-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heineman RH, Bull JJ. Testing optimality with experimental evolution: lysis time in a bacteriophage. Evolution Int J Org Evolution. 2007;61:1695–709. doi: 10.1111/j.1558-5646.2007.00132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heineman RH, Molineux IJ, Bull JJ. Evolutionary robustness of an optimal phenotype: re-evolution of lysis in a bacteriophage deleted for its lysin gene. J Mol Evol. 2005;61:181–91. doi: 10.1007/s00239-004-0304-4. [DOI] [PubMed] [Google Scholar]
- Hill WG. Estimation, effectiveness and opportunities of long term genetic improvement in animals and maize. Lohmann Information. 2008;43:3–20. [Google Scholar]
- Hill WG, Bunger L. Inferences on the Genetics of Quantitative Traits from Long-term Selection in Laboratory and Domestic Animals. Plant Breeding Reviews. 2003;24(part2):169–210. [Google Scholar]
- Hubbarde JE, Wahl LM. Estimating the optimal bottleneck ratio for experimental evolution: the burst-death model. Math Biosci. 2008;213:113–8. doi: 10.1016/j.mbs.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Kemp P, Garcia LR, Molineux IJ. Changes in bacteriophage T7 virion structure at the initiation of infection. Virology. 2005;340:307–17. doi: 10.1016/j.virol.2005.06.039. [DOI] [PubMed] [Google Scholar]
- Kemp P, Gupta M, Molineux IJ. Bacteriophage T7 DNA ejection into cells is initiated by an enzyme-like mechanism. Mol Microbiol. 2004;53:1251–65. doi: 10.1111/j.1365-2958.2004.04204.x. [DOI] [PubMed] [Google Scholar]
- Martin V, Grande-Perez A, Domingo E. No evidence of selection for mutational robustness during lethal mutagenesis of lymphocytic choriomeningitis virus. Virology. 2008;378:185–92. doi: 10.1016/j.virol.2008.05.016. [DOI] [PubMed] [Google Scholar]
- Maynard Smith J. The evolution of sex. Cambridge University Press; Cambridge Eng. ; New York: 1978. [Google Scholar]
- Molineux I. The T7 group. In: Calendar, editor. The Bacteriophages. Oxford Univ. Press; New York, NY, USA.: 2006. pp. 277–301. [Google Scholar]
- Parker GA, Maynard Smith J. Optimality theory in evolutionary biology. Nature. 1990;348:27–33. [Google Scholar]
- Patnaik R, Louie S, Gavrilovic V, Perry K, Stemmer WP, Ryan CM, del Cardayre S. Genome shuffling of Lactobacillus for improved acid tolerance. Nat Biotechnol. 2002;20:707–12. doi: 10.1038/nbt0702-707. [DOI] [PubMed] [Google Scholar]
- Patten PA, Howard RJ, Stemmer WP. Applications of DNA shuffling to pharmaceuticals and vaccines. Curr Opin Biotechnol. 1997;8:724–33. doi: 10.1016/s0958-1669(97)80127-9. [DOI] [PubMed] [Google Scholar]
- Pelosi L, Kuhn L, Guetta D, Garin J, Geiselmann J, Lenski RE, Schneider D. Parallel changes in global protein profiles during long-term experimental evolution in Escherichia coli. Genetics. 2006;173:1851–69. doi: 10.1534/genetics.105.049619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokyta DR, Beisel CJ, Joyce P, Ferris MT, Burch CL, Wichman HA. Beneficial fitness effects are not exponential for two viruses. J Mol Evol. 2008;67:368–76. doi: 10.1007/s00239-008-9153-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe LA, Geddie ML, Alexander OB, Matsumura I. A comparison of directed evolution approaches using the beta-glucuronidase model system. J Mol Biol. 2003;332:851–60. doi: 10.1016/s0022-2836(03)00972-0. [DOI] [PubMed] [Google Scholar]
- Scholl D, Kieleczawa J, Kemp P, Rush J, Richardson CC, Merril C, Adhya S, Molineux IJ. Genomic analysis of bacteriophages SP6 and K1-5, an estranged subgroup of the T7 supergroup. J Mol Biol. 2004;335:1151–71. doi: 10.1016/j.jmb.2003.11.035. [DOI] [PubMed] [Google Scholar]
- Smith CC, Fretwell SD. The optimal balance between size and number of offspring. The American Naturalist. 1974;108:499–506. [Google Scholar]
- Springman R, Badgett MR, Molineux IJ, Bull JJ. Gene order constrains adaptation in bacteriophage T7. Virology. 2005;341:141–52. doi: 10.1016/j.virol.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Springman R, Keller T, Molineux IJ, Bull JJ. Evolution at a high imposed mutation rate: adaptation obscures the load in phage T7. Genetics. 2010;184:221–32. doi: 10.1534/genetics.109.108803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW. The genetics and physiology of bacteriophage T7. Virology. 1969;39:562–74. doi: 10.1016/0042-6822(69)90104-4. [DOI] [PubMed] [Google Scholar]
- Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2006;439:344–8. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt CA, Kauffman S, Wang ZG. Rational evolutionary design: The theory of in vitro protein evolution. Advances in Protein Chemistry. 2000;55:79–160. doi: 10.1016/s0065-3233(01)55003-2. [DOI] [PubMed] [Google Scholar]
- Wahl LM, Krakauer DC. Models of experimental evolution: the role of genetic chance and selective necessity. Genetics. 2000;156:1437–48. doi: 10.1093/genetics/156.3.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichman HA, Millstein J, Bull JJ. Adaptive molecular evolution for 13,000 phage generations: a possible arms race. Genetics. 2005;170:19–31. doi: 10.1534/genetics.104.034488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichman HA, Scott LA, Yarber CD, Bull JJ. Experimental evolution recapitulates natural evolution. Philos Trans R Soc Lond B Biol Sci. 2000;355:1677–84. doi: 10.1098/rstb.2000.0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GC. Adaptation and natural selection. Princeton University Press; Princeton, NJ: 1966. [Google Scholar]
- Williams GC. Sex and evolution. Princeton University Press; Princeton, N.J.: 1975. [Google Scholar]
- Woods R, Schneider D, Winkworth CL, Riley MA, Lenski RE. Tests of parallel molecular evolution in a long-term experiment with Escherichia coli. Proc Natl Acad Sci U S A. 2006;103:9107–12. doi: 10.1073/pnas.0602917103. [DOI] [PMC free article] [PubMed] [Google Scholar]