Abstract
In recent years, better understanding of the molecular biology of non-small-cell lung carcinoma (nsclc) has led to a revolution in the work-up of these neoplasms. As a pathology diagnosis, “nsclc” without further attempt at subclassification is no longer accepted as a standard of care; separating squamous cell carcinoma from adenocarcinoma and large-cell carcinoma carries implications for prognosis and treatment decisions. Currently, detection of the presence in nsclc of mutations involving the epidermal growth factor receptor (EGFR) gene and fusion of the N-terminal portion of the protein encoded by EML4 (echinoderm microtubule-associated protein-like 4 gene) with the intracellular signaling portion of the receptor tyrosine kinase encoded by ALK (anaplastic lymphoma kinase gene)—that is, EML4–ALK—and variants has become routine in many centres because patients having tumours harbouring such alterations might benefit from tyrosine kinase inhibitors as part of their treatment regimen.
The purpose of the present review is to highlight important aspects of the screening for molecular derangements in nsclc and to briefly discuss the emergence of possible future biomarkers.
Keywords: Non-small-cell lung carcinoma, nsclc, epidermal growth factor receptor, EGFR, EML4–ALK, tyrosine kinase inhibitors, personalized medicine, biomarkers
1. INTRODUCTION
According to the American Cancer Society, approximately 157,300 deaths related to lung cancer (the highest among all cancers) were registered in the United States during 2010. Lung cancer is the leading cancer type for estimated new cancer cases; it also leads in deaths in men and women alike, at 29% and 26% respectively 1. Non-small-cell lung carcinoma (nsclc) is the most frequent subtype, representing approximately 85% of all cases, and most patients have locally advanced or distant metastatic disease (stage iii/iv) at the time of presentation 2.
Since the early 2000s, greater understanding of the molecular biology of lung cancer, particularly nsclc, has led to a revolution in the treatment of these neoplasms. Already, searching for specific mutations in individual cases so as to provide the most effective treatment with the least possible occurrence of side effects is a reality. Identification of mutations in the EGFR (epidermal growth factor receptor) and KRAS genes and, most recently, of rearrangements in the ALK gene has been incorporated into routine practice in several centres. These mutations and rearrangements are thought to alter the function or expression of several molecules that can be either located on the cell surface, acting as growth factor receptors, or participating in downstream intracellular pathways. Such derangement of the cellular apparatus ultimately leads to uncontrolled cell growth. The data provided by detection of such mutations can be used to generate prognostic information or to select patients for targeted therapies. In lung oncology, personalized medicine is slowly becoming the norm. Because of the immense amount of data constantly being generated and the rapid flow of information, arriving at a consensus on standardization of molecular testing creates a major challenge for pathologists, clinicians, and molecular biologists.
In this review, we briefly discuss the most common mutations and rearrangements associated with nsclc, the strategies potentially available to optimize their detection, and future trends in this now ever-changing field.
2. HISTOLOGIC ASPECTS AND COMMON MOLECULAR DERANGEMENTS
2.1. Histologic Aspects
Until recently, nearly all primary epithelial neoplasms of the lung would be classified either as small-cell carcinoma or nsclc, and that classification would suffice for institution by the treating clinician of the accepted standard-of-care treatment. However, three important and previously unrecognized aspects pertaining to treatment and prognosis in nsclc have been brought to light:
First, EGFR mutations are much more frequently detected in adenocarcinomas (adcs).
Second, bevacizumab, a monoclonal antibody that inhibits vascular endothelial growth factor A can induce life-threatening hemorrhage in patients with squamous cell carcinoma of the lung (thereby contraindicating its use in such patients) 3.
Third, pemetrexed, an inhibitor of purine and pyrimidine synthesis, has demonstrated superior efficacy in patients with adc or nsclc “not otherwise specified” than in those with squamous cell morphology 4.
Based on those findings, guidelines for good practice in handling pathology samples from lung neoplasms have begun recommending that small biopsies and cytology specimens be classified whenever possible as squamous cell carcinoma or adc, and that the use of immunohistochemistry and mucin stains be used in difficult cases, but kept at a minimum to spare tissue for molecular studies 5.
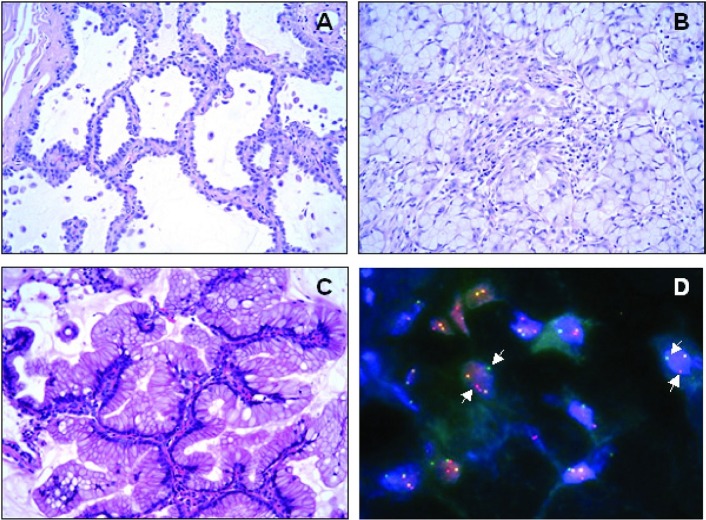
In terms of specific molecular abnormalities associated with predominant histology patterns, the presence of EGFR mutation is associated with adcs, particularly those containing a non-mucinous bronchioloalveolar (“lepidic”) pattern 6 [Figure 1(A)]. As for EML4–ALK, adcs are also the main histologic type, and the most striking correlation is with the presence of a signet-ring component 7 [Figure 1(B)]. Mutations in KRAS are also present almost exclusively in adcs, being strongly associated with the mucinous bronchioloalveolar pattern 8 [Figure 1(C)].
FIGURE 1.
Certain adenocarcinoma patterns are more frequently associated with specific mutations. Here, three patterns are shown: (A) lepidic (non-mucinous bronchioloalveolar), (B) signet-ring, and (C) mucinous bronchioloalveolar. These patterns correlate with abnormalities in the EGFR, ALK, and KRAS genes respectively. (D) A break-apart EML4–ALK fluorescence in-situ hybridization assay reveals split signal (arrows).
2.2. EGFR
The epidermal growth factor receptor (also known as her1 or ErbB1) is a member of the ErbB receptor tyrosine kinase family, comprising her1/ErbB1, her2/ErbB2, her3/ErbB3, and her4/ErbB4. It plays an important role in carcinogenesis and tumour progression through activation mechanisms including overexpression, mutation, and autocrine ligand production. Derangements in the EGFR gene are associated with cancer cell proliferation, cell growth, invasion, metastatic spread, apoptosis, and tumour angiogenesis. It accomplishes those actions through activation of the Ras/Raf/Mek/mapk and pi3k/Akt/mtor pathways 9.
The two most common EGFR mutations are short in-frame deletions of exon 19 and a point mutation (CTG to CGG) in exon 21 at nucleotide 2573 that results in substitution of leucine by arginine at codon 858 (L858R) 10.
Tumours with EGFR mutations occur at a higher frequency in East Asians than in non-Asians (30% vs. 8%), in women than in men (59% vs. 26%), in never-smokers than in ever-smokers (66% vs. 22%), and in adcs than in other nsclc histologies (49% vs. 2%) 11. In the United States, activating EGFR mutations are estimated to occur in 15% of patients with primary lung adc 12.
Gefitinib and erlotinib are the first generation of egfr tyrosine kinase inhibitors (tkis), which selectively target the intracellular tyrosine kinase domain of egfr, blocking the downstream signalling of the receptor 2. Recently, a multicentre phase iii trial demonstrated that patients with advanced-stage lung cancer containing EGFR mutations and treated with first-line gefitinib (compared with standard chemotherapy) showed improved progression-free survival. That finding led the authors to strongly recommend patient selection based on EGFR mutation status 13. In fact, the American Society of Clinical Oncology recommends EGFR mutation testing for patients with advanced nsclc of the lung who are being considered for first-line therapy with an egfr tki 12.
Unfortunately, all responders eventually develop resistance, most commonly because of the emergence of a secondary T790M mutation 14 or amplification of mesenchymal–epithelial transition factor (c-Met) 15.
Tissue samples are usually the specimen of choice. However, the approach of testing small cytology-based specimens, whether enriched for tumour cells by microdissection or not, has been published in various series with varying degrees of success. Tumour concentration and testing technique play an important role in test sensitivity 16–20. Direct sequencing is the first widely used method for the detection of EGFR mutation, but it lacks sensitivity and might miss 25% of positive cases 21. Several other more sensitive techniques have been applied, including polymerase chain reaction (pcr) single-strand conformation polymorphism 22, Taq-Man (Roche Molecular Diagnostics, Pleasanton, CA, U.S.A.) pcr 23, loop-hybrid mobility shift assay 24, Cycleave (Clontech Laboratories, Mountain View, CA, U.S.A.) pcr 25, pcr restriction fragment length polymorphism and length analysis 26, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry genotyping 27, peptide nucleic acid–locked nucleic acid pcr clamp 28, Scorpions (DxS Limited, Manchester, U.K.) amplified refractory mutation system 29, denaturing high-performance liquid chromatography 30, single-molecule sequencing 31, mutant-enriched pcr 32, and Smart Amplification Process (DNAFORM, Yokohama City, Japan) 33. These techniques not only vary in sensitivity, but also differ in terms of the detection of new and already-known mutations and the comprehensive detection of deletions and insertions 21.
2.3. ALK Gene Rearrangements
In generating a retroviral complementary dna (cdna) expression library from a lung adc sample, Soda et al. noted that one of the amplified cdnas had 3926 base pairs and contained an open reading frame for a protein of 1059 amino acids, with the amino-terminal portion of the protein being identical to that of human EML4, and the carboxy-terminal portion being identical to the intracellular domain of human ALK. That finding led to the conclusion that the cdna was derived from a fusion product of the genes EML4 and ALK. The EML4–ALK fusion was shown to result from a small inversion within chromosome 2p. The transforming potential of EML4–ALK was demonstrated using expression plasmids for EML4–ALK, which were introduced into mouse 3T3 fibroblasts, where transformed foci were readily identified. Additionally, transfected 3T3 cells in nude mice expressing EML4–ALK were able to form subcutaneous tumours. In this foundational work, EML4–ALK fusion transcript was detected in 5 of 75 nsclc patients (6.7%) 34.
Translocations involving the ALK gene have already been identified in a subset of T-cell lymphomas named anaplastic large-cell lymphomas and in an unusual mesenchymal neoplasm known as inflammatory myofibroblastic tumour or inflammatory pseudotumour 35–37. In these situations, ALK is associated with several other partner fusion genes, including, in anaplastic large-cell lymphoma, NPM, TPM3, CLTC, ATIC, and TFG 35,38–41, and in inflammatory myofibroblastic tumours, TPM3 and TPM4 42. Several EML4–ALK fusion variants—and also other rare fusion partners such as KIF5B—have been described in nsclc, all harbouring transforming activity 34,43,44.
Initially, it was thought that EML4–ALK fusions were restricted to nsclc. Reverse-transcriptase pcr (rt-pcr) was attempted, without success, to detect the fusion messenger rna in a variety of neoplasms, including acute myeloid leukemia, non-Hodgkin lymphoma, and gastric and colorectal carcinomas 34. However, in one study using rt-pcr screening of patient samples, EML4–ALK fusion was detected in 2.4% of breast and 2.4% colorectal carcinomas. In the same study, 11.3% of nsclcs tested showed EML4–ALK fusion 45.
The prevalence of EML4–ALK varies in different studies, with 4% being probably the most accurate figure. The presence of EML4–ALK appears to be mutually exclusive for EGFR and KRAS mutations 46,47. Patients are usually nonsmokers or light smokers and tend to be younger than the average patient with nsclc. They frequently present at an advanced clinical stage, and the tumours demonstrate adc with a solid pattern and signet-ring cells 48,49. In other studies looking at clinicopathologic correlations, the younger age of patients with lung adcs positive for EML4–ALK was emphasized. However, in a departure from previous reports, these patients were characterized by less differentiation and an acinarpredominant pattern 47.
Fluorescence in-situ hybridization (fish) is regarded as the preferred standard method for detecting ALK rearrangement [Figure 1(D)]. Recently, the U.S. Food and Drug Administration approved crizotinib (Xalkori: Pfizer, Mission, KS, U.S.A.) to treat certain patients with locally advanced or metastatic nsclc expressing the abnormal ALK gene. Crizotinib, an orally available small-molecule inhibitor of the Alk tyrosine kinase, is being approved with a companion diagnostic kit from Abbott Molecular (Abbott Park, IL, U.S.A.) 50,a.
The use of rt-pcr, immunohistochemistry, and chromogenic in-situ hybridization has also been described in clinical samples with variable sensitivity 51,52. Several studies have addressed the role of immunohistochemistry in the detection of ALK-positive cases. The sensitivity of the immunohistochemistry assay depends on several factors, including the antibody clone and the detection method used 47,48,52–54.
In 2010, only 3 years after the first description of EML4–ALK fusion in nsclc, the results of a clinical trial that enrolled 82 ALK-positive patients were made public. In that study, patients found to be ALK-positive by fish were evaluated for the therapeutic efficacy of crizotinib. The results were impressive, including an overall response rate of 57% (46 partial responses and 1 complete response) and 33% stable disease (27 patients). Of the 82 patients, 63 (77%) were continuing to receive crizotinib at the time of data cutoff, and the estimated probability of 6-month progression-free survival was 72%. Side effects were considered minor 55. The results of this trial have underscored the importance of establishing strategies for identifying ALK-positive patients.
2.4. KRAS
Kirsten rat sarcoma viral oncogene homolog (KRAS), a member of the ras gene family, encodes a small guanosine triphosphate gtpase that cycles between inactive gdp-bound and active gtp-bound conformations 56,57. An important downstream signalling target of egfr, kras has also been implicated in the development and prognosis of several cancers such adcs of colon, lung, and pancreas 58–60. Mutations that cause the loss of kras gtpase activity render the protein constitutively gtp-bound, resulting in sustained activation of downstream components and persistent proliferation signal 59,61.
In lung cancers, KRAS mutations occur primarily at codons 12 and 13; mutations at codons 10 and 61 are less frequently seen 56,62. The most common KRAS mutation in smoking patients with nsclc is a G-to-T transition (84%) resulting in substitution of cysteine (47%), valine (24%), aspartate (15%), or alanine (7%) for wild-type glycine 62,63.
Mutations in KRAS have been found in 15%–30% of patients with nsclc and are considered to be one of the more frequent mutations in these tumours 64–66. As with EGFR mutations, KRAS mutations are detected mainly in lung adcs and are less frequently observed in squamous cell carcinomas of the lung 67,68. By contrast with lung adcs harbouring EGFR mutations, tumours having KRAS mutations are seen at a higher frequency (20%–30%) in Caucasian patients than in East Asian patients (5%) 67,69. Also, compared with EGFR mutations, KRAS mutations are more common in current or former smokers than in never-smokers, although the absence of a history of tobacco use does not eliminate the possibility of such abnormalities 70–72.
Mutant KRAS plays an important role both in tumour development and in resistance to therapy. In colorectal cancer, it is well known that the presence of KRAS gene mutations is independently associated with poorer prognosis and is predictive of resistance to anti-egfr therapies 73–76. However, in nsclc, the value of KRAS status as a predictive biomarker for anti-egfr therapy is less clear. Although several studies have attempted to investigate the prognostic impact of KRAS mutations on survival and recurrence rates in nsclc, the results are difficult to interpret because of differences in histologic inclusion criteria (all nsclc vs. adc only), small sample size, and tumour stage 65,66. One group demonstrated that nsclc harbouring KRAS codon 12 mutation carries a strong and unfavourable prognostic factor and is associated with mortality and inferior disease-free survival 77. In addition, a meta-analysis of more than 53 studies showed that KRAS mutations are a factor correlated with poor survival in patients with nsclc 2,78. Interestingly, Ihle et al. recently found that nsclc patients whose tumours harbour either mutant KRAS-Gly12Cys or mutant KRAS-Gly12Val had worse progression-free survival outcomes than did those with all other mutant KRAS proteins or with wild-type KRAS 62.
The predictive value of KRAS status for response to conventional chemotherapy, adjuvant therapy, or targeted therapy is far from being established. When KRAS mutations were tested prospectively in clinical studies such as tribute, jbr.10, and ifct-0002, they were shown not to be significantly associated with chemoresistance, nor to be predictive of a differential benefit from adjuvant chemotherapy 57,79,80. A recent prospective biomarker-driven study conducted in 889 patients included in a phase iii trial comparing placebo with sequential erlotinib maintenance in unresectable nsclc (saturn, BO18192) showed that the presence of KRAS mutations was not predictive for erlotinib efficacy and was significantly associated with reduced progression-free survival 81.
2.5. BRAF
BRAF encodes a nonreceptor serine/threonine kinase that is a member of the Ras/mapk signaling pathway downstream of Ras protein. Upon activation, BRAF directly phosphorylates MEK, which in turns phosphorylates ERK, thereby regulating cellular responses to growth signals 82,83.
Davies et al. were the first to identify somatic activating mutations in the BRAF gene by screening 923 cancer samples. Missense mutations were found in approximately 60% of melanomas and 15% of colorectal cancers. Furthermore, mutations of BRAF were identified in other human cancers at smaller percentages, including in nsclcs at 3%. The same study also showed that most BRAF mutations (89%) substitute a Glu for Val at residue 600 (V600E) in the activation segment of the kinase domain. Interestingly, all BRAF mutations found in nsclcs during the study were non-V600E; by contrast, in melanoma, 91% of mutations involved V600E 83. Accordingly, Brose et al. identified BRAF mutations in 5 of 179 nsclc cases (3%) and found that all mutated samples but 1 had non-V600E mutations 64.
Recently, Caucasian patients with nsclc were investigated to determine the prevalence, distribution, and prognostic role of BRAF mutations. In this large cohort, mutations of BRAF were found predominantly in lung adcs (97.3%), with approximately 57% being V600E and 43% being non-V600E. The V600E mutations were demonstrated to be significantly more prevalent in women than in men, and in never-smokers than in smokers or former smokers, although all non-V600E mutations were found in tobacco users. Furthermore, V600E-mutated nsclc showed a more aggressive tumour histotype characterized by micropapillary features and associated with poor prognosis 84. Similarly, Paik et al. found that the frequency of non-V600E mutations was higher in nsclc than in melanoma, which could reflect a carcinogenic effect caused by tobacco 85.
BRAF mutations were shown to be mutually exclusive with EGFR mutations within exons 18–21, KRAS codon 12 mutations, ERBB2 codon 20 mutations, and translocations in ALK 85,86.
3. FUTURE OF NSCLC
In analyzing the future of molecular testing and adoption of personalized treatments in nsclc, four aspects need consideration:
Implementing strategies for routine testing of known biomarkers associated with specific treatments (currently EGFR and ALK)
Identifying new biomarkers and their potential inhibitors
Understanding the development of resistance by tumours
Funding research in lung oncology
Identification of defects in lung cancer tyrosine kinase signalling has been accomplished through the phosphoproteomic approach, revealing (in addition to the already known oncogenic kinases egfr and c-Met, and Alk and Ros fusion proteins) other activated tyrosine kinases, including PdgfrA and Ddr1, not previously implicated in the genesis of nsclc 87.
Several other molecules, such as the receptor tyrosine kinase c-Met and its ligands 15,87, hepatocyte growth factor, vascular endothelial growth factor and its receptor, her2, pik3ca, braf, and insulin-like growth factor 1 receptor (igf-1r) that are involved in cell signalling, are currently being investigated.
Abnormalities in the receptor tyrosine kinase c-Met and pik3ca have been shown to play a role in predicting resistance to tki therapy 15,88.
Alterations in her2, with different prevalences depending on the technical assay used, have been demonstrated in nsclc 89. Those observations led the authors to the conclusion that inhibitory monoclonal antibodies might play a role in the treatment of a subgroup of nsclc patients.
Evidence has also implicated igf-1r, its ligands igf-1 and -2, and their related downstream signalling in the development of cancer and also in a mechanism of resistance development in egfr-targeted therapy 90,91. The development of a potential therapeutic strategy targeting igf-1r was initiated by the development of a human antibody that binds the receptor with high affinity, consequently inhibiting ligand attachment, and preventing downstream signalling of the mapk/pi3k/Akt pathways. However, the association of a monoclonal antibody to igf-1r and erlotinib in unselected nsclc patients did not appear beneficial, likely because of high drug toxicity, which prevented optimal dosage 92–94.
Several others potential biomarkers of interest are thought to have a predominant role as indicators of chemotherapy sensitivity. That group includes Ki-67, p27, p16, the cyclin-dependent kinases, ercc1, brca, tubulin iii, Rrm1, and Tp53. Recently, a consensus meeting of Canadian lung cancer oncologists and pathologists found that the evidence is currently insufficient to support routine testing for those markers 95.
Other approaches are also been studied, and among those, mage-a3 (melanoma antigen-A3) deserves to be mentioned. The phase iii study magrit (mage-a3 as Adjuvant Non-Small Cell Lung Cancer Immunotherapy) is investigating the efficacy of mage-a3 antigen-specific cancer immunotherapy in preventing cancer relapse, when treatment is given after tumour resection in patients with mage-a3–positive stages ib, ii, and iiia nsclc. Approximately 30% of nsclc tumours are mage-a3–positive. A tumour-specific antigen, mage-a3 is expressed by a variety of cancer cells, but not by normal ones. The strict tumour-specific expression of mage-a3 has prompted immunotherapeutic trials 96. Non-small-cell lung carcinoma is among the tumours that express mage-a3.
Today, the general consensus among oncologists appears to be that, whenever possible, tissue rather than cytology specimens should be obtained in nsclc. However, it is possible to evaluate EGFR mutation status by drawing blood from patients with nsclc, particularly those at advanced stages. The test is accomplished with the use of a microfluidic device containing microposts coated with antibodies against epithelial cells. The EGFR mutational analysis is performed on dna recovered from circulating tumour cells 97.
Research funding is crucial for the development of any science. Unfortunately, the reality of funding in lung research is far from optimal. The excerpt that follows is reproduced from a recent article published by The New York Times that emphasizes the perennial underfunding of cancer research 98:
The big loser in the cancer funding race is lung cancer. It is the biggest cancer killer in the country, yet on a per-death basis receives the least [U.S. National Cancer Institute] funding among major cancers. In 2006, the NCI spent $1,518 for each new case of lung cancer and $1,630 for each lung cancer death, according to data from the institute and the American Cancer Society.
Among the big cancers, breast cancer receives the most funding per new case, $2,596 — and by far the most money relative to each death, $13,452. Notably, prostate cancer, the most common cancer, receives the least funding per new case at just $1,318. But on a per-death basis it ranks second, with $11,298 in NCI funds.
The situation described likely derives from a lack of public mobilization; lung cancer nongovernmental organizations are scarce. Survivors represent a small fraction of the patients affected by the disease, and they are frequently dogged by the “guilt effect” for having smoked and therefore having voluntarily exposed themselves to the potential risks. However, there is hope that the development of new personalized strategies in nsclc, based on targeting specific molecular alterations, will eventually lead to prolonged survival. Those survival improvements in turn will influence public awareness. Consequently, those two factors could synergistically lead to heightened interest in pulmonary research, bringing about changes that will perhaps forever alter the history of human lung cancers.
4. ACKNOWLEDGMENTS
The authors thank Anke van Rijk phd for providing the ALK fish picture, George Chong phd for contributions related to the molecular techniques used for the detection of mutations in nsclc, and Katerina Ntapolias for technical support.
Footnotes
Vysis ALK Break Apart FISH Probe Kit, with the Vysis Paraffin Pretreatment IV and Post Hybridization Wash Buffer Kit, ProbeChek ALK Negative Control Slides, and ProbeChek ALK Positive Control Slides.
5. CONFLICT OF INTEREST DISCLOSURES
GDAB has consulted for Pfizer and Boehringer Ingelheim. AS has consulted for AstraZeneca and Amgen. EFB has no relevant financial conflicts of interest to declare.
6. REFERENCES
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Reungwetwattana T, Weroha SJ, Molina JR. Oncogenic pathways, molecularly targeted therapies, and highlighted clinical trials in non-small-cell lung cancer (nsclc) Clin Lung Cancer. 2011. [Epub ahead of print] [DOI] [PubMed]
- 3.Johnson DH, Fehrenbacher L, Novotny WF, et al. Randomized phase ii trial comparing bevacizumab plus carboplatin and paclitaxel with carboplatin and paclitaxel alone in previously untreated locally advanced or metastatic non-small-cell lung cancer. J Clin Oncol. 2004;22:2184–91. doi: 10.1200/JCO.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 4.Scagliotti GV, Parikh P, von Pawel J, et al. Phase iii study comparing cisplatin plus gemcitabine with cisplatin plus pemetrexed in chemotherapy-naive patients with advanced-stage non-small-cell lung cancer. J Clin Oncol. 2008;26:3543–51. doi: 10.1200/JCO.2007.15.0375. [DOI] [PubMed] [Google Scholar]
- 5.Travis WD, Brambilla E, Van Schil P, et al. Paradigm shifts in lung cancer as defined in the new iaslc/ats/ers lung adenocarcinoma classification. Eur Respir J. 2011;38:239–43. doi: 10.1183/09031936.00026711. [DOI] [PubMed] [Google Scholar]
- 6.Blons H, Côté JF, Le Corre D, et al. Epidermal growth factor receptor mutation in lung cancer are linked to bronchioloalveolar differentiation. Am J Surg Pathol. 2006;30:1309–15. doi: 10.1097/01.pas.0000213285.65907.31. [DOI] [PubMed] [Google Scholar]
- 7.Shaw AT, Yeap BY, Mino–Kenudson M, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4–ALK. J Clin Oncol. 2009;27:4247–53. doi: 10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finberg KE, Sequist LV, Joshi VA, et al. Mucinous differentiation correlates with absence of EGFR mutation and presence of KRAS mutation in lung adenocarcinomas with bronchioloalveolar features. J Mol Diagn. 9:320–6. doi: 10.2353/jmoldx.2007.060182. 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salomon DS, Brandt R, Ciardiello F, Normanno N. Epidermal growth factor–related peptides and their receptors in human malignancies. Crit Rev Oncol Hematol. 1995;19:183–232. doi: 10.1016/1040-8428(94)00144-I. [DOI] [PubMed] [Google Scholar]
- 10.Ladanyi M, Pao W. Lung adenocarcinoma: guiding egfr-targeted therapy and beyond. Mod Pathol. 2008;21(suppl 2):S16–22. doi: 10.1038/modpathol.3801018. [DOI] [PubMed] [Google Scholar]
- 11.Bell DW, Brannigan BW, Matsuo K, et al. Increased prevalence of EGFR-mutant lung cancer in women and in East Asian populations: analysis of estrogen-related polymorphisms. Clin Cancer Res. 2008;14:4079–84. doi: 10.1158/1078-0432.CCR-07-5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keedy VL, Temin S, Somerfield MR, et al. American Society of Clinical Oncology provisional clinical opinion: epidermal growth factor receptor (EGFR) mutation testing for patients with advanced non-small-cell lung cancer considering first-line egfr tyrosine kinase inhibitor therapy. J Clin Oncol. 2011;29:2121–7. doi: 10.1200/JCO.2010.31.8923. [DOI] [PubMed] [Google Scholar]
- 13.Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–8. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 14.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the egfr kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ErbB3 signaling. Science. 2007;316:1039–43. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 16.Liu Y, Liu B, Li XY, et al. A comparison of arms and direct sequencing for EGFR mutation analysis and tyrosine kinase inhibitors treatment prediction in body fluid samples of non-small-cell lung cancer patients. J Exp Clin Cancer Res. 2011;30:111. doi: 10.1186/1756-9966-30-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Billah S, Stewart J, Staerkel G, Chen S, Gong Y, Guo M. EGFR and KRAS mutations in lung carcinoma: molecular testing by using cytology specimens. Cancer Cytopathol. 2011;119:111–17. doi: 10.1002/cncy.20151. [DOI] [PubMed] [Google Scholar]
- 18.Chowdhuri SR, Xi L, Pham TH, et al. EGFR and KRAS mutation analysis in cytologic samples of lung adenocarcinoma enabled by laser capture microdissection. Mod Pathol. 2011. [Epub ahead of print] [DOI] [PMC free article] [PubMed]
- 19.Kanaji N, Bandoh S, Ishii T, et al. Detection of epidermal growth factor receptor mutations in a few cancer cells from transbronchial cytologic specimens by reverse transcriptasepolymerase chain reaction. Mol Diagn Ther. 2011;15:353–9. doi: 10.1007/BF03256471. [DOI] [PubMed] [Google Scholar]
- 20.Rekhtman N, Brandt SM, Sigel CS, et al. Suitability of thoracic cytology for new therapeutic paradigms in non-small cell lung carcinoma: high accuracy of tumour subtyping and feasibility of egfr and kras molecular testing. J Thorac Oncol. 2011;6:451–8. doi: 10.1097/JTO.0b013e31820517a3. [DOI] [PubMed] [Google Scholar]
- 21.Pao W, Ladanyi M. Epidermal growth factor receptor mutation testing in lung cancer: searching for the ideal method. Clin Cancer Res. 2007;13:4974–83. doi: 10.1158/1078-0432.CCR-07-1387. [DOI] [PubMed] [Google Scholar]
- 22.Marchetti A, Martella C, Felicioni L, et al. EGFR mutations in non-small-cell lung cancer: analysis of a large series of cases and development of a rapid and sensitive method for diagnostic screening with potential implications on pharmacologic treatment. J Clin Oncol. 2005;23:857–65. doi: 10.1200/JCO.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 23.Zhou C, Ni J, Zhao Y, Su B. Rapid detection of epidermal growth factor receptor mutations in non-small cell lung cancer using real-time polymerase chain reaction with TaqMan-mgb probes. Cancer J. 2006;12:33–9. doi: 10.1097/00130404-200601000-00007. [DOI] [PubMed] [Google Scholar]
- 24.Matsukuma S, Yoshihara M, Kasai F, et al. Rapid and simple detection of hot spot point mutations of epidermal growth factor receptor, BRAF, and NRAS in cancers using the loop-hybrid mobility shift assay. J Mol Diagn. 2006;8:504–12. doi: 10.2353/jmoldx.2006.060030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yatabe Y, Hida T, Horio Y, Kosaka T, Takahashi T, Mitsudomi T. A rapid, sensitive assay to detect EGFR mutation in small biopsy specimens from lung cancer. J Mol Diagn. 2006;8:335–41. doi: 10.2353/jmoldx.2006.050104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan Q, Pao W, Ladanyi M. Rapid polymerase chain reaction–based detection of epidermal growth factor receptor gene mutations in lung adenocarcinomas. J Mol Diagn. 2005;7:396–403. doi: 10.1016/S1525-1578(10)60569-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomas RK, Baker AC, Debiasi RM, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39:347–51. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 28.Tanaka T, Nagai Y, Miyazawa H, et al. Reliability of the peptide nucleic acid–locked nucleic acid polymerase chain reaction clamp-based test for epidermal growth factor receptor mutations integrated into the clinical practice for non-small cell lung cancers. Cancer Sci. 2007;98:246–52. doi: 10.1111/j.1349-7006.2006.00377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimura H, Kasahara K, Kawaishi M, et al. Detection of epidermal growth factor receptor mutations in serum as a predictor of the response to gefitinib in patients with non-small-cell lung cancer. Clin Cancer Res. 2006;12:3915–21. doi: 10.1158/1078-0432.CCR-05-2324. [DOI] [PubMed] [Google Scholar]
- 30.Cohen V, Agulnik JS, Jarry J, et al. Evaluation of denaturing high-performance liquid chromatography as a rapid detection method for identification of epidermal growth factor receptor mutations in non-small-cell lung cancer. Cancer. 2006;107:2858–65. doi: 10.1002/cncr.22331. [DOI] [PubMed] [Google Scholar]
- 31.Thomas RK, Nickerson E, Simons JF, et al. Sensitive mutation detection in heterogeneous cancer specimens by massively parallel picoliter reactor sequencing. Nat Med. 2006;12:852–5. doi: 10.1038/nm1437. [DOI] [PubMed] [Google Scholar]
- 32.Asano H, Toyooka S, Tokumo M, et al. Detection of EGFR gene mutation in lung cancer by mutant-enriched polymerase chain reaction assay. Clin Cancer Res. 2006;12:43–8. doi: 10.1158/1078-0432.CCR-05-0934. [DOI] [PubMed] [Google Scholar]
- 33.Hoshi K, Takakura H, Mitani Y, et al. Rapid detection of epidermal growth factor receptor mutations in lung cancer by the SMart-Amplification Process. Clin Cancer Res. 2007;13:4974–83. doi: 10.1158/1078-0432.CCR-07-0509. [DOI] [PubMed] [Google Scholar]
- 34.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4–ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–6. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 35.Morris SW, Kirstein MN, Valentine MB, et al. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin’s lymphoma. Science. 1994;263:1281–4. doi: 10.1126/science.8122112. [Erratum in: Science 1995;267:316–17] [DOI] [PubMed] [Google Scholar]
- 36.Shiota M, Fujimoto J, Semba T, et al. Hyperphosphorylation of a novel 80 kDa protein–tyrosine kinase similar to Ltk in a human Ki-1 lymphoma cell line, AMS3. Oncogene. 1994;9:1567–74. [PubMed] [Google Scholar]
- 37.Pulford K, Morris SW, Turturro F. Anaplastic lymphoma kinase proteins in growth control and cancer. J Cell Physiol. 2004;199:330–58. doi: 10.1002/jcp.10472. [DOI] [PubMed] [Google Scholar]
- 38.Trinei M, Lanfrancone L, Campo E, et al. A new variant anaplastic lymphoma kinase (Alk)–fusion protein (atic-alk) in a case of ALK-positive anaplastic large cell lymphoma. Cancer Res. 2000;60:793–8. [PubMed] [Google Scholar]
- 39.Touriol C, Greenland C, Lamant L, et al. Further demonstration of the diversity of chromosomal changes involving 2p23 in ALK-positive lymphoma: 2 cases expressing ALK kinase fused to CLTCL (clathrin chain polypeptide-like) Blood. 2000;95:3204–7. [PubMed] [Google Scholar]
- 40.Lamant L, Dastugue N, Pulford K, Delsol G, Mariamé B. A new fusion gene TPM3–ALK in anaplastic large cell lymphoma created by a (1;2)(q25;p23) translocation. Blood. 1999;93:3088–95. [PubMed] [Google Scholar]
- 41.Hernández L, Pinyol M, Hernández S, et al. TRK-fused gene (TFG) is a new partner of ALK in anaplastic large cell lymphoma producing two structurally different TFG–ALK translocations. Blood. 1999;94:3265–8. [PubMed] [Google Scholar]
- 42.Lawrence B, Perez–Atayde A, Hibbard MK, et al. TPM3–ALK and TPM4–ALK oncogenes in inflammatory myofibroblastic tumors. Am J Pathol. 2000;157:377–84. doi: 10.1016/S0002-9440(10)64550-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi YL, Takeuchi K, Soda M, et al. Identification of novel isoforms of the EML4–ALK transforming gene in non-small cell lung cancer. Cancer Res. 2008;68:4971–6. doi: 10.1158/0008-5472.CAN-07-6158. [DOI] [PubMed] [Google Scholar]
- 44.Takeuchi K, Choi YL, Togashi Y, et al. KIF5B–ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK- positive lung cancer. Clin Cancer Res. 2009;15:3143–9. doi: 10.1158/1078-0432.CCR-08-3248. [DOI] [PubMed] [Google Scholar]
- 45.Lin E, Li L, Guan Y, et al. Exon array profiling detects EML4–ALK fusion in breast, colorectal, and non-small cell lung cancers. Mol Cancer Res. 2009;7:1466–76. doi: 10.1158/1541-7786.MCR-08-0522. [DOI] [PubMed] [Google Scholar]
- 46.Camidge DR, Kono SA, Flacco A, et al. Optimizing the detection of lung cancer patients harboring anaplastic lymphoma kinase (ALK) gene rearrangements potentially suitable for Alk inhibitor treatment. Clin Cancer Res. 2010;16:5581–90. doi: 10.1158/1078-0432.CCR-10-0851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Inamura K, Takeuchi K, Togashi Y, et al. EML4–ALK lung cancers are characterized by rare other mutations, a TTF-1 cell lineage, an acinar histology, and young onset. Mod Pathol. 2009;22:508–15. doi: 10.1038/modpathol.2009.2. [DOI] [PubMed] [Google Scholar]
- 48.Rodig SJ, Mino–Kenudson M, Dacic S, et al. Unique clinicopathologic features characterize ALK-rearranged lung adenocarcinoma in the Western population. Clin Cancer Res. 2009;15:5216–23. doi: 10.1158/1078-0432.CCR-09-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koivunen JP, Mermel C, Zejnullahu K, et al. EML4–ALK fusion gene and efficacy of an Alk kinase inhibitor in lung cancer. Clin Cancer Res. 2008;14:4275–83. doi: 10.1158/1078-0432.CCR-08-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.United States Department of Health and Human Services, Food and Drug Administration (fda) Crizotinib [Web page] Silver Spring, MD: FDA; 2011. [Available at: http://www.fda.gov/AboutFDA/CentersOffices/OfficeofMedicalProductsandTobacco/CDER/ucm270058.htm; cited March 29, 2012] [Google Scholar]
- 51.Kim H, Yoo SB, Choe JY, et al. Detection of ALK gene rearrangement in non-small cell lung cancer: a comparison of fluorescence in situ hybridization and chromogenic in situ hybridization with correlation of Alk protein expression. J Thorac Oncol. 2011;6:1359–66. doi: 10.1097/JTO.0b013e31821cfc73. [DOI] [PubMed] [Google Scholar]
- 52.Boland JM, Erdogan S, Vasmatzis G, et al. Anaplastic lymphoma kinase immunoreactivity correlates with ALK gene rearrangement and transcriptional up-regulation in non-small cell lung carcinomas. Hum Pathol. 2009;40:1152–8. doi: 10.1016/j.humpath.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 53.Mino–Kenudson M, Chirieac LR, Law K, et al. A novel, highly sensitive antibody allows for the routine detection of ALK-rearranged lung adenocarcinomas by standard immunohistochemistry. Clin Cancer Res. 2010;16:1561–71. doi: 10.1158/1078-0432.CCR-09-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martelli MP, Sozzi G, Hernandez L, et al. EML4–ALK rearrangement in non-small cell lung cancer and non-tumor lung tissues. Am J Pathol. 2009;174:661–70. doi: 10.2353/ajpath.2009.080755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kranenburg O. The KRAS oncogene: past, present, and future. Biochim Biophys Acta. 2005;1756:81–2. doi: 10.1016/j.bbcan.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 57.Loriot Y, Mordant P, Deutsch E, Olaussen KA, Soria JC. Are ras mutations predictive markers of resistance to standard chemotherapy? Nat Rev Clin Oncol. 2009;6:528–34. doi: 10.1038/nrclinonc.2009.106. [DOI] [PubMed] [Google Scholar]
- 58.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–80. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Capella G, Cronauer–Mitra S, Pienado MA, Perucho M. Frequency and spectrum of mutations at codons 12 and 13 of the c-K-ras gene in human tumors. Environ Health Perspect. 1991;93:125–31. doi: 10.1289/ehp.9193125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [Erratum in: Nat Rev Cancer 2007;7:563] [DOI] [PubMed] [Google Scholar]
- 61.Roberts PJ, Der CJ. Targeting the Raf–Mek–Erk mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007;26:3291–310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- 62.Ihle NT, Byers LA, Kim ES, et al. Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. J Natl Cancer Inst. 2012;104:228–39. doi: 10.1093/jnci/djr523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Siegfried JM, Gillespie AT, Mera R, et al. Prognostic value of specific KRAS mutations in lung adenocarcinomas. Cancer Epidemiol Biomarkers Prev. 1997;6:841–7. [PubMed] [Google Scholar]
- 64.Brose MS, Volpe P, Feldman M, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 2002;62:6997–7000. [PubMed] [Google Scholar]
- 65.Roberts PJ, Stinchcombe TE, Der CJ, Socinski MA. Personalized medicine in non-small-cell lung cancer: is KRAS a useful marker in selecting patients for epidermal growth factor receptor–targeted therapy? J Clin Oncol. 2010;28:4769–77. doi: 10.1200/JCO.2009.27.4365. [DOI] [PubMed] [Google Scholar]
- 66.Aviel–Ronen S, Blackhall FH, Shepherd FA, Tsao MS. K-ras mutations in non-small-cell lung carcinoma: a review. Clin Lung Cancer. 2006;8:30–8. doi: 10.3816/CLC.2006.n.030. [DOI] [PubMed] [Google Scholar]
- 67.Herbst RS, Kelly K, Chansky K, et al. Phase ii selection design trial of concurrent chemotherapy and cetuximab versus chemotherapy followed by cetuximab in advanced-stage non-small-cell lung cancer: Southwest Oncology Group study S0342. J Clin Oncol. 2010;28:4747–54. doi: 10.1200/JCO.2009.27.9356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Graziano SL, Gamble GP, Newman NB, et al. Prognostic significance of K-ras codon 12 mutations in patients with resected stage i and ii non-small-cell lung cancer. J Clin Oncol. 1999;17:668–75. doi: 10.1200/JCO.1999.17.2.668. [DOI] [PubMed] [Google Scholar]
- 69.Li M, Liu L, Liu Z, et al. The status of KRAS mutations in patients with non-small cell lung cancers from mainland China. Oncol Rep. 2009;22:1013–20. doi: 10.3892/or_00000529. [DOI] [PubMed] [Google Scholar]
- 70.Riely GJ, Kris MG, Rosenbaum D, et al. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin Cancer Res. 2008;14:5731–4. doi: 10.1158/1078-0432.CCR-08-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mao C, Qiu LX, Liao RY, et al. KRAS mutations and resistance to egfr-tkis treatment in patients with non-small cell lung cancer: a meta-analysis of 22 studies. Lung Cancer. 2010;69:272–8. doi: 10.1016/j.lungcan.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 72.Marks JL, Broderick S, Zhou Q, et al. Prognostic and therapeutic implications of EGFR and KRAS mutations in resected lung adenocarcinoma. J Thorac Oncol. 2008;3:111–16. doi: 10.1097/JTO.0b013e318160c607. [DOI] [PubMed] [Google Scholar]
- 73.Raponi M, Winkler H, Dracopoli NC. KRAS mutations predict response to EGFR inhibitors. Curr Opin Pharmacol. 2008;8:413–18. doi: 10.1016/j.coph.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 74.Lièvre A, Bachet JB, Le Corre D, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–5. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 75.Karapetis CS, Khambata–Ford S, Jonker DJ, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–65. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 76.Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–34. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 77.Slebos RJ, Kibbelaar RE, Dalesio O, et al. K-ras oncogene activation as a prognostic marker in adenocarcinoma of the lung. N Engl J Med. 1990;323:561–5. doi: 10.1056/NEJM199008303230902. [DOI] [PubMed] [Google Scholar]
- 78.Mascaux C, Iannino N, Martin B, et al. The role of ras oncogene in survival of patients with lung cancer: a systematic review of the literature with meta-analysis. Br J Cancer. 2005;92:131–9. doi: 10.1038/sj.bjc.6602258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Eberhard DA, Johnson BE, Amler LC, et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol. 2005;23:5900–9. doi: 10.1200/JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
- 80.Tsao MS, Aviel–Ronen S, Ding K, et al. Prognostic and predictive importance of p53 and ras for adjuvant chemotherapy in non small-cell lung cancer. J Clin Oncol. 2007;25:5240–7. doi: 10.1200/JCO.2007.12.6953. [DOI] [PubMed] [Google Scholar]
- 81.Brugger W, Triller N, Blasinska–Morawiec M, et al. Prospective molecular marker analyses of egfr and kras from a randomized, placebo-controlled study of erlotinib maintenance therapy in advanced non-small-cell lung cancer. J Clin Oncol. 2011;29:4113–20. doi: 10.1200/JCO.2010.31.8162. [DOI] [PubMed] [Google Scholar]
- 82.Wan PT, Garnett MJ, Roe SM, et al. Mechanism of activation of the Raf–Erk signaling pathway by oncogenic mutations of B-raf. Cell. 2004;116:855–67. doi: 10.1016/S0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 83.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 84.Marchetti A, Felicioni L, Malatesta S, et al. Clinical features and outcome of patients with non-small-cell lung cancer harboring BRAF mutations. J Clin Oncol. 2011;29:3574–9. doi: 10.1200/JCO.2011.35.9638. [DOI] [PubMed] [Google Scholar]
- 85.Paik PK, Arcila ME, Fara M, et al. Clinical characteristics of patients with lung adenocarcinomas harboring BRAF mutations. J Clin Oncol. 2011;29:2046–51. doi: 10.1200/JCO.2010.33.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kobayashi M, Sonobe M, Takahashi T, et al. Clinical significance of BRAF gene mutations in patients with non-small cell lung cancer. Anticancer Res. 2011;31:4619–23. [PubMed] [Google Scholar]
- 87.Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 88.Ludovini V, Bianconi F, Pistola L, et al. Optimization of patient selection for egfr-tkis in advanced non-small cell lung cancer by combined analysis of KRAS, PIK3CA, MET, and non-sensitizing EGFR mutations. Cancer Chemother Pharmacol. 2012 doi: 10.1007/s00280-012-1829-7. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 89.Pellegrini C, Falleni M, Marchetti A, et al. her-2/neu alterations in non-small cell lung cancer: a comprehensive evaluation by real time reverse transcription–pcr, fluorescence in situ hybridization, and immunohistochemistry. Clin Cancer Res. 2003;9:3645–52. [PubMed] [Google Scholar]
- 90.Jacobs CI. A review of the role of insulin-like growth factor 2 in malignancy and its potential as a modifier of radiation sensitivity. Clin Oncol (R Coll Radiol) 2008;20:345–52. doi: 10.1016/j.clon.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 91.Guix M, Faber AC, Wang SE, et al. Acquired resistance to egfr tyrosine kinase inhibitors in cancer cells is mediated by loss of igf-binding proteins. J Clin Invest. 2008;118:2609–19. doi: 10.1172/JCI34588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ramalingam SS, Spigel DR, Chen D, et al. Randomized phase ii study of erlotinib in combination with placebo or R1507, a monoclonal antibody to insulin-like growth factor-1 receptor, for advanced-stage non-small-cell lung cancer. J Clin Oncol. 2011;29:4574–80. doi: 10.1200/JCO.2011.36.6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Weickhardt A, Doebele R, Oton A, et al. A phase i/ii study of erlotinib in combination with the anti-insulin-like growth factor-1 receptor monoclonal antibody IMC-A12 (cixutumumab) in patients with advanced non-small cell lung cancer. J Thorac Oncol. 2012;7:419–26. doi: 10.1097/JTO.0b013e31823c5b11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cohen RB, Langer CJ, Simon GR, et al. A phase i/randomized phase ii, non-comparative, multicenter, open label trial of CP-547,632 in combination with paclitaxel and carboplatin or paclitaxel and carboplatin alone as first-line treatment for advanced non-small cell lung cancer (nsclc) Cancer Chemother Pharmacol. 2007;60:81–9. doi: 10.1007/s00280-006-0352-0. [DOI] [PubMed] [Google Scholar]
- 95.Ellis PM, Blais N, Soulieres D, et al. A systematic review and Canadian consensus recommendations on the use of biomarkers in the treatment of non-small cell lung cancer. J Thorac Oncol. 2011;6:1379–91. doi: 10.1097/JTO.0b013e318220cb8e. [DOI] [PubMed] [Google Scholar]
- 96.Sang M, Lian Y, Zhou X, Shan B. mage-a family: attractive targets for cancer immunotherapy. Vaccine. 2011;29:8496–500. doi: 10.1016/j.vaccine.2011.09.014. [DOI] [PubMed] [Google Scholar]
- 97.Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in egfr in circulating lung-cancer cells. N Engl J Med. 2008;359:366–77. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Parker–Pope T. Cancer funding: does it add up? [“Well” blog post] The New York Times. Mar 6, 2008. 12:21 pm. [Available at: http://well.blogs.nytimes.com/2008/03/06/cancer-fundingdoes-it-add-up/; cited March 29,2012]