Abstract
New drugs such as pemetrexed, the epidermal growth factor receptor (egfr) tyrosine kinase inhibitors, and the Alk inhibitor crizotinib have recently enabled progress in the management of advanced non-small-cell lung cancer (nsclc). More drugs, especially Met inhibitors, will follow. However, the benefits of these agents are not uniform across the spectrum of nsclc, and optimizing their utility requires some degree of subgrouping of nsclc by the presence or absence of certain biomarkers.
The biomarkers of current or imminent value are EGFR and KRAS mutational status, ALK rearrangements, and MET immunohistochemistry. As a predictor of benefit for anti-egfr monoclonal antibodies, EGFR immunohistochemistry is also of potential interest.
Some of the foregoing biomarkers (EGFR, ALK, MET) are direct drivers of the malignant phenotype. As such, they are, quite rationally, the direct targets of inhibitory drugs. However, KRAS, while definitely a driver, has resisted attempts at direct pharmacologic manipulation, and its main value might lie in its role as part of an efficient testing algorithm, because KRAS mutations appear to exclude EGFR and ALK mutations. The indirect value of KRAS in determining sensitivity to other targeted agents or to pemetrexed remains controversial. The other biomarkers (EGFR, ALK, MET) may also have indirect value as predictors of sensitivity to chemotherapy in general, to pemetrexed specifically, and to radiotherapy and molecularly targeted agents.
These biomarkers have all enabled the co-development of new drugs with companion diagnostics, and they illustrate the paradigm that will govern progress in oncology in the immediate future. However, in nsclc, the acquisition of sufficient biopsy material remains a stubborn obstacle to the evolution of novel targeted therapies.
Keywords: nsclc, lung cancer, EGFR, ALK, KRAS, prognosis, prediction
1. INTRODUCTION
Metastatic non-small-cell lung cancer (nsclc) remains, with rare surgical exceptions, incurable. Pending radical new solutions, scientific progress is currently channelled into the conversion of this rapidly lethal disease into a chronic illness. How to make that conversion is conventionally believed to involve “personalized medicine”: Tumour biopsies are tested for certain causative and characteristic molecular lesions (“targets”), guiding the selection of customized drugs designed to directly interact with and inhibit those targets.
This paradigm, based on the concept of causality, is deeply seductive, given that it appears to offer the prospect of both efficacy and lack of toxicity. It hews to a fundamentally rational worldview as suggested by its common appellation, “targeted therapy.” The molecular lesion is meant to be causally responsible for maintenance of the malignant phenotype and also distinctive, even uniquely characteristic, of the cancer cells. Hence the prospects for both tumour control and selectivity.
The foregoing perspective, while undoubtedly simplistic, nonetheless provides a framework for how four key genes—EGFR, ALK, MET, and KRAS—will increasingly influence the management of metastatic nsclc. Those genes, when altered in measurable ways, unquestionably contribute to the pathogenesis of nsclc, and as such, are widely agreed to be “drivers” 1. It is rational to seek to inhibit them, even to hope that drugs can be designed that will selectively block the oncogenic varieties while sparing their normal counterparts. But there is no guarantee that oncogenic variants are necessarily druggable, or if they are, that the cancer cell will not eventually find a way around the inhibition. Furthermore, those genes, although not necessarily mutated sensu stricto, may, through altered expression, nonetheless still contribute to the cancer, thus reducing the prospects for selectivity.
Additionally, genetic aberrations such as these may convey useful information beyond the notion of the direct target. Broadly, that information can be classified as prognostic (foreknowledge of probable events in the absence of therapy, which may continue to influence outcomes regardless of therapy) and predictive (indicating the prospects for success of particular therapies), and might be mechanistically related to the aberrant gene, but might also be purely empirical—that is, exhibiting no obvious causal relationship. These genetic alterations, then, are “biomarkers” sensu lato, and their utility extends into predicting the future clinical course (even absent therapy) and the selection of drugs (whether those drugs target those particular genes directly or not).
It is better, therefore, to approach EGFR, ALK, MET, and KRAS and the entire expanding suite of molecular drivers 2 as biomarkers in the broad sense, and not just as direct targets, although the latter status is clearly of major importance.
Although the present review focuses more on the biomarker utility of these genes and less on the technicalities of their measurement, we must emphasize that the acquisition of adequate biopsy material remains problematic in the management of metastatic nsclc. That problem can partly be addressed by educating respirologists, interventional radiologists, and thoracic surgeons, but sometimes there is no possibility of obtaining other than scant tissue. The reasons include hazard, technical factors, access, patient refusal, and avoidance of delay.
In the event that the clinician’s hand is forced, we therefore provide information correlating the foregoing biomarkers with (usually available) clinical, pathologic, and demographic characteristics. Emphatically, however, it is better to make therapeutic decisions on the basis of a direct test. However, as a definitive solution to this problem, reliable testing based on blood work (that is, analysis of circulating tumour cells or plasma dna) should soon become available 3,4.
2. EGFR
In the early 1960s, Stanley Cohen isolated the mitogen “epidermal growth factor” (egf) from murine salivary gland 5. In 1973, the egf receptor (egfr) was described 6; this receptor was later appreciated as the first of a family of 4 human epidermal tyrosine kinase receptors (her1–4) 7, attended by a broad spectrum of ligands besides egf, participating in a multifaceted and adaptive signalling network 8 subserving growth and survival. EGFR, cloned and isolated in 1984 9, encodes a 1210-amino-acid transmembrane protein, including an extracellular ligand-binding ectodomain, an anchoring transmembrane domain, and a submembrane tyrosine kinase domain. Ligand activation involves homo-dimerization (or hetero-dimerization with other her family members), and then activation of the tyrosine kinase domain, resulting in tyrosine autophosphorylation, which enables engagement with 6 or more signalling pathways subserving “cell fate decisions” 8, including the pi3k/Akt and Erk pathways of particular interest in oncology.
Dysregulation of egfr contributes to a range of cancers and occurs in various ways 10. In nsclc, the most important are activating EGFR mutations and increased protein expression. Either dysregulation may possibly be associated with increased gene copy number. The uncommon EGFRvIII mutation has also been detected in a few squamous cell lung cancers 1. However it arises, dysregulated EGFR activation promotes the malignant phenotype by mediating cell proliferation, raising the apoptotic threshold, increasing cellular motility (and hence metastasis), enhancing neoangiogenesis, and conferring resistance to chemotherapy and radiation.
Although earlier efforts at predicting anti-egfr therapeutic sensitivity focused on egfr protein overexpression and EGFR gene copy number increment, the most important parameter is whether an activating EGFR mutation is present. The mutations are almost exclusively found in lung adenocarcinomas; they are more common in never-smokers or light exsmokers, women, and patients of East Asian origin. In this demographic, 60%–70% of patients will have a detectable mutation in EGFR. Caucasian smokers or ex-smokers with adenocarcinomas have an 8% incidence—enough to mandate testing. All patients with adenocarcinomas should be tested for EGFR mutation (Table i) 11–13, although that dictum may need to be softened depending on immunophenotyping. Mutations are associated mainly with papillary and micropapillary adenocarcinomas or non-mucinous bronchioloalveolar adenocarcinomas (rarely with solid adenocarcinomas) and seem mostly to require an immunophenotype positive for thyroid transcription factor 1 (ttf-1).
TABLE I.
| Variable | Value by locale | |
|---|---|---|
| East Asiaa | Western worldb | |
| Studies (n) | 6 | 2 |
| Patients (n) | 814 | 116 |
| Never-smokers, EGFR M+ (%) | 70 | 37 |
| Ever-smokers, EGFR M+ (%) | 29 | 8 |
Japan, Korea, Taiwan, Hong Kong.
United States, Australia.
Nearly all activating EGFR mutations occur in exons 18–21. The most important are deletions within exon 19 (more than 20 variants) and point (missense) mutations in exon 21 (usually L858R, occasionally L861Q or L861R). Very occasionally, point mutations involve exon 18 (for example, G719C and others at G719). Generally the tyrosine kinase domain is affected, probably leading to increased atp binding, with enhanced (and ligand-independent) downstream signalling, especially via the Akt and stat pathways, affecting cell survival 14. The resulting condition (“oncogene addiction”) is characterized by a dependency of the cancer cell on the mutation. Also implicated is the Erk1/2 pathway, essential to cellular proliferation 15. The benefits of egfr blockade may ultimately be mediated by a shift toward apoptosis in the balance of the pro- and anti-apoptotic members of the Bcl-2 family of proteins.
The centrality of egfr signalling has led to intensive efforts to design therapies aimed at blockade. Two approaches have proved successful: antiegfr monoclonal antibodies against the extracellular ligand-binding domain, and small-molecule tyrosine kinase inhibitors (tkis) to block binding of atp (upon which signalling depends). The latter have proved much more valuable in nsclc, although egfr antibodies have also demonstrated activity.
Curiously, small-molecule tkis (gefitinib and erlotinib) were designed before the elucidation, in 2004 by three American groups, of the EGFR mutation 2,16,17. The small subset of metastatic nsclc patients who had responded dramatically to singleagent tki therapy prompted the search for an explanation, culminating in discovery of the mutations. These mutations not only confer oncogene addiction, but also fortuitously show markedly increased affinity for gefitinib or erlotinib because of residue repositioning around the binding cleft 18. It soon became apparent that almost all the dramatic responses had occurred in patients whose cancers harboured one of these activating (and sensitizing) EGFR mutations; however, egfr-tki can also, to a lesser extent, benefit patients without those mutations: that is, the EGFR “wild-type” (EGFR WT) patients, whose cancers are presumably driven by upregulated signalling (from overexpression of the normal protein, for instance).
Small (mainly East Asian) studies of egfr-tki monotherapy with gefitinib rapidly confirmed high objective response rates (55%–91%) in patients with cancers harbouring a mutation 19–26. A large nonrandomized 217-patient Spanish-led experience 27 with erlotinib was published in 2009. The objective response rate (orr) of 70.6%, the progressive disease rate of just 10.2%, the prolonged progression-free survival (pfs) of 14 months, and the overall survival (os) of 27 months suggested that responsiveness in mutationpositive patients was not a function of ethnicity and that erlotinib might be superior to gefitinib. Furthermore, Caucasian patients demonstrated a spectrum of EGFR mutational subtypes similar to those seen in East Asian patients. Those phase ii trials led to six large randomized trials comparing first-line egfr-tki with then-standard platinum-doublet third-generation chemotherapy in proven EGFR mutation–positive patients (EGFR M+) or in populations enriched for mutation positivity (Table ii).
TABLE II.
Randomized trials of chemotherapy compared with epidermal growth factor receptor (egfr) tyrosine kinase inhibitor (tki) in mutation-positive patients
| Reference (study name) | Regimen | orr (%) | Statistic | p Value | pfs (months) | hr | p Value | os (months) | hr | p Value |
|---|---|---|---|---|---|---|---|---|---|---|
| Lee et al., 200928 ; Ku et al., 2011 29 (First-signal) | Cisplatin–gemcitabine vs. gefitinib | 38 | 0.002 | 6.7 | 0.0084 | 26.5 | hr? | 0.648 | ||
| 85 | 8.4 | 30.6 | ||||||||
| Maemondo et al., 201030 (nej 002) | Carboplatin–paclitaxel vs. gefitinib | 31 | <0.001 | 5.4 | 0.30 | <0.001 | 23.6 | Not available | 0.31 | |
| 74 | 10.8 | 30.5 | ||||||||
| Mitsudomi et al., 201031 (wjtog 3405) | Cisplatin–docetaxel vs. gefitinib | 32 | <0.001 | 6.3 | 0.49 | <0.0001 | Not reached | 1.64 | 0.211 | |
| 62 | 9.2 | 30.9 | ||||||||
| Fukuoka et al., 201132 ; Mok, 2011a (ipass) | Carboplatin–paclitaxel vs. gefitinib | 47 | <0.001 | 6.3 | 0.48 | <0.001 | 21.9 | 1.0 | 0.99 | |
| 71 | 9.5 | 21.6 | ||||||||
| Zhou et al., 201133 (optimal) | Carboplatin–gemcitabine vs. erlotinib | 36 | <0.0001 | 4.6 | 0.16 | <0.0001 | Not available | Not available | ||
| 83 | 13.1 | |||||||||
| Rossell et al., 201234 (eurtac) | Platinum–gemcitabine or platinum–docetaxel vs. erlotinib | 15 | or: 7.5 | <0.0001 | 5.2 | 0.37 | <0.0001 | 19.5 | 1.047 | 0.87 |
| 58 | 9.7 | 19.3 |
Mok T. Novel therapies [part of mini-symposium M12]. Presented at the 14th World Conference on Lung Cancer; Amsterdam, Netherlands; July 3–7, 2011.
orr = objective response rate; pfs = progression-free survival; hr = hazard ratio; os = overall survival; or = odds ratio.
The randomized studies (ipass, wjtog 3405, nej002, First-signal, optimal, and eurtac) uniformly revealed that, compared with chemotherapy, first-line tki consistently resulted in a higher orr and longer pfs; however, os was not prolonged because of extensive crossover from chemotherapy to tki upon progression. Because tki and chemotherapy appear non-cross-resistant, those who receive a second-line tki benefit as much as those who receive it in the first line 27,35. However, because of unavoidable attrition (35% in ipass), it is desirable to treat with a tki up front if possible in EGFR M+ patients, notwithstanding a modest delay to secure a test result. However, as revealed by ipass, the one trial to accrue and analyze both EGFR M+ and WT patients, the opposite is even more true. Clearly, in EGFR WT disease (approximately 40% of the East Asian ipass population of never-smokers or light ex-smokers), gefitinib appears virtually devoid of useful activity (orr: 1.1%) and may be associated with passive harm because of the opportunity cost of delaying active chemotherapy.
In the trials, patients with exon 19 deletions and exon 21 point mutations did not have markedly different outcomes on tki (the former perhaps conferring a modestly better outcome). Also, erlotinib is probably not markedly different from gefitinib in outcome, and (from ipass) EGFR mutation positivity is prognostic for inherently longer survival. There is a suggestion (again from ipass) that, compared with EGFR WT patients, those who are EGFR M+ respond better to chemotherapy, although the orr in ipass for M+ patients (47%) was outside the range for chemotherapy in the other five randomized trials (15%–37%). However, ipass did establish that first-line chemotherapy in EGFR M+ patients was much more active than tki in EGFR WT patients, implying that EGFR-unknown patients should receive first-line chemotherapy rather than “empirical tki.”
A subsequent randomized trial of post-chemotherapy maintenance erlotinib compared with placebo (saturn) exhibited a dramatic benefit for the EGFR M+ subset [hazard ratio (hr): 0.10] in pfs, but not in os, again because of crossover. Interestingly, the EGFR WT patients did experience an os advantage—but only if the best response on prior first-line chemotherapy was stable disease, not complete or partial response 36.
Mutations in EGFR also occur in exon 20, especially T790M, which inserts a bulky methionine over the atp binding cleft, blocking access to first-generation egfr-tki (but not to atp) 37. This “gatekeeper” T790M mutation occurs only within a pre-existing sensitizing mutation, either del 19 or exon 21, and seemingly causes up to 50% of the resistance inevitably occurring in all EGFR M+ patients on first-generation tki (gefitinib or erlotinib). Novel egfr-tki (for example, afatinib, dacomitinib) bind despite the T790M mutation, but can also bind to other her receptors. These drugs can undoubtedly benefit patients failed by first-generation tki, but whether T790M binding is responsible remains uncertain. The lux-Lung series of trials with afatinib are illustrative; lux-Lung 1 randomized patients who had received prior platinum chemotherapy and who had progressed after 12 or more weeks on erlotinib or gefitinib to either afatinib or placebo. The pfs, orr, and symptom control outcomes strongly favoured afatinib, but os was not significantly different (79% of patients on the placebo arm received further lines of treatment). The large single-arm phase ii lux-Lung 2 trial included EGFR M+ patients at either first or second line. The orr was 60%, but the pfs was an impressive 14 months. Results of lux-Lung 3, which is now accrued and which randomized EGFR M+ patients in the first line to afatinib or cisplatin–pemetrexed, are imminent and could lead to regulatory application.
Other resistance mechanisms to tki in M+ patients include MET amplification (5%–20%) and, occasionally, epithelial–mesenchymal transition and even transformation to a small-cell phenotype 38. However, progressive disease on a first-generation tki according to the formal Response Evaluation Criteria in Solid Tumors does not necessarily mean exhausted utility, because abrupt tki cessation can, in about 20% of patients, induce a significant “flare” phenomenon that responds to immediate re-introduction of the same tki 39. Also, rechallenge with the same tki after a “holiday” (during which chemotherapy may be given) is increasingly recognized as valuable 40,41. There is an unmet need for biomarkers to guide the management of patients who experience technical progressive disease in front-line tki, and there is evidence that resistance may differentially affect some metastases and not others—that is, clonal metastasis 42,a.
Updated ipass biomarker analysis 32 clearly showed that measurement of EGFR gene copy number by fluorescence in-situ hybridization (fish) or of egfr expression by immunohistochemistry (ihc) does not substitute for a mutation test. However, high copy number or ihc expression seems to be a weak surrogate for EGFR mutation positivity.
The ncic br.21 trial enrolled second- or third-line metastatic nsclc patients who had exhausted their chemotherapy options. It showed an os benefit for erlotinib compared with placebo. A limited biomarker analysis suggested that high EGFR copy number by fish (because of either gene amplification or high polysomy) 43 predicted a higher orr (21% vs. 5%) and an improved os benefit from erlotinib (hr: 0.43 vs. 0.80 in fish-negative patients). The fish-positive control subjects had the worst os, but the most benefit from erlotinib, and compared with mutational status, fish seemed to influence os more 44.
Erlotinib was administered to more than 7000 patients in the large, open-label trust study (0–2 prior chemotherapies), with the German centres reporting their biomarker data. EGFR mutations and fish positivity predicted response. Positivity by fish also predicted pfs and os. The egfr ihc positivity weakly correlated with pfs and os. Interestingly, 22% of patients were both ihc-positive and fish-positive; about half to two thirds were ihc-positive, but fish-negative; and 11%–21% were ihc-negative and fish-negative, independent of histology 45.
In nonrandomized studies such as trust, and even in randomized studies not using a placebo control, it is impossible to disentangle prognostic and predictive factors for pfs and os; in this respect, br.21 is highly valuable—as is isel46,47, a similar study that compared gefitinib with placebo, but in a more refractory population. In isel, which showed a nonsignificant benefit for gefitinib compared with placebo, high EGFR copy number predicted an os treatment effect (hr: 0.61 compared with placebo). An interaction test was significant (p = 0.045), indicating a genuinely different effect by copy number. The same applied to ihc status. EGFR mutations substantially predicted response (37.5% vs. 2.6%), but the data were too few to adjudicate survival effects. Results in the isel placebo group also suggested that fish positivity was an adverse prognostic indicator (median survival time: 4.5 months vs. 6.4 months; hr: 1.41).
The br.21, trust, and isel trials seem to imply utility for fish and ihc as well as for EGFR mutational status, especially in Caucasian patients, in whom fish positivity is more common than is mutation in unselected patients. In isel, 30.8% were fish-positive and 12.1% were M+. Of the entire population, 20.2% were East Asian. In br.21 (only 12% East Asian), 38% were fish-positive and 18% were EGFR M+.
Ellis et al. performed a meta-analysis on the br.21 and saturn trials, two post-first-line trials, each with a placebo arm. Those authors concluded that egfr ihc positivity is prognostic (weakly) for longer pfs and os, that EGFR fish status was not prognostic, and that EGFR mutations may be prognostic for os (perhaps confounded by crossover). Neither ihc nor fish were recommended for “routine” prediction of erlotinib sensitivity; mutation positivity implied a better pfs on erlotinib, but mutation negativity did not preclude a benefit, and therefore EGFR mutation testing was not valuable after the first line 48. In that analysis, some results for ihc, fish (especially), and mutation status appeared to be discrepant between br.21 and saturn. In particular, fish positivity was both predictive and negatively prognostic in br.21, but not in saturn. Notably, the br.21 and saturn patient populations were dissimilar.
The utility of fish in the context of egfr-tki, especially in EGFR WT patients of any histology, should not be discounted for both prognosis and prediction.
Anti-egfr monoclonal antibodies, especially cetuximab, added to chemotherapy in metastatic nsclc generally produce modestly positive results. The flex study considered the addition of cetuximab to cisplatin–vinorelbine in egfr ihc-positive metastatic nsclc. Median os was increased by 1.2 months (hr: 0.871; p = 0.044) 49. However, application of a scoring system (“H-score,” continuous scale 0–300) revealed that 31% scored high (>200) and that the high-scoring patients (either histology) monopolized the os benefit (9.6 months vs. 12.0 months; hr: 0.73; p = 0.01). The low-score hr was 0.99. The interaction test was significant (p = 0.044) 50. The Southwest Oncology Group 0819 study is attempting to prospectively confirm that result with cetuximab and carboplatin–paclitaxel–bevacizumab.
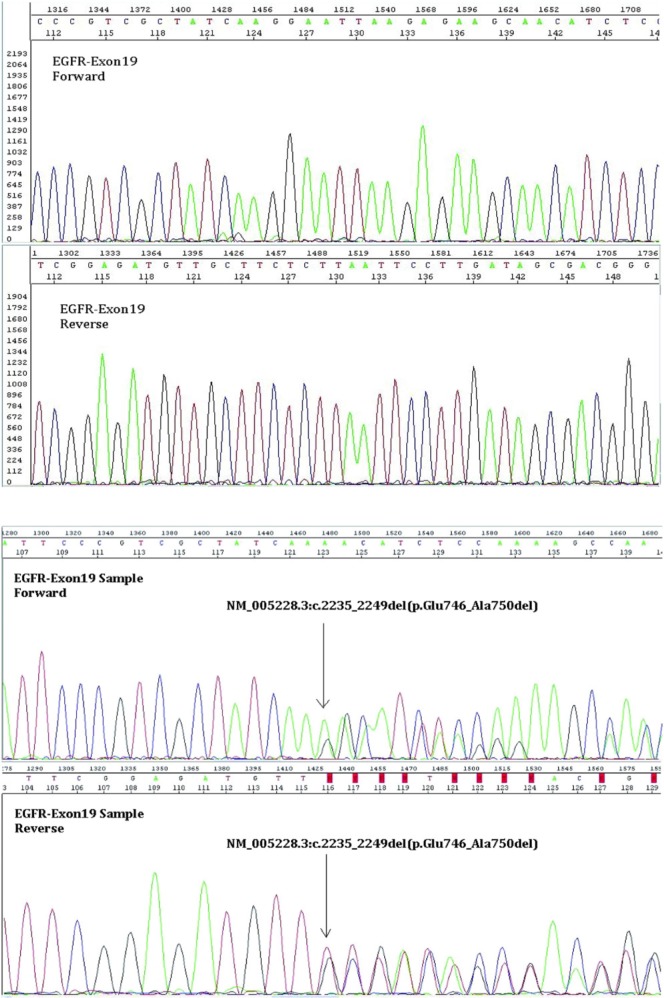
Technical aspects of EGFR testing are beyond our scope 51; however, microdissection and sequencing (Figure 1) may represent the current clinical standard. Allele-specific amplification—for example, Scorpions ARMS (DxS Limited, Manchester, U.K.)—is an alternative. Experimental mutation-specific antibodies are highly specific (97%–100%) and moderately sensitive (74.2%–100%) 52–55. Detection of mutations in circulating tumour cells 56,57 or even circulating dna 58,59 is rapidly being perfected.
FIGURE 1.
The dna extracted from the macrodissected tissue specimen was amplified using primers specific for exon 19 of the EGFR gene. The polymerase chain reaction product was then purified and sequenced in both directions using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, U.S.A.). The sequences obtained were then compared with the U.S. National Center for Biotechnology Information reference sequence (EGFR: NM_005228.3). In this case, a deletion is observed to span nucleotides 2235–2249, which results in the deletion of amino acids at positions 746–750 (inclusive) in exon 19 of the EGFR gene.
In Canada (Table iii), EGFR testing has been centralized in 5 laboratories, which might use different methodologies (for example, restriction fragment length polymorphism analysis, sequencing; Figure 1) and for which a minimum of five 5-μm sections are required, each with more than 100 tumour cells per section, and with the tumour cells representing more than 25% of the nucleated cells. Specimens are preferably microdissected and are better derived from core biopsies, although cell blocks and generous fine-needle aspirates may be adequate. Neither ihc nor fish are routinely obtained.
TABLE III.
Testing for EGFR mutation and selected uses of epidermal growth factor receptor inhibitors by province in Canada
| Province | EGFR testing | First-line gefitinib (EGFR M+) | Maintenance erlotiniba |
|---|---|---|---|
| BC | √ | √ | √ |
| AB | √ | √ | X |
| SK | AZb | Case by case | X |
| MB | AZ | Case by case | X |
| ON | AZ | √ | X |
| QC | √ | √ | X |
| NS | AZ | X | X |
| NL | AZ | X | X |
| PE | AZ | X | X |
Not restricted by EGFR status.
Paid for by pharmaceutical company.
AZ = AstraZeneca Canada.
3. ALK
As a driver oncogene, ALK (the anaplastic lymphoma kinase gene) was initially discovered in a chromosomal rearrangement in anaplastic large-cell lymphoma 60. In 2007, Soda et al. described ALK activation in a subset of nsclc that exhibited a “small inversion” in chromosome 2, fusing the normally separated EML4 (echinoderm microtubule-associated protein-like 4 gene) with ALK 61. This EML4–ALK fusion transcript was detected in 5 of 75 Japanese nsclc patients and in none of 261 patients with “other” cancers. Interestingly, although some EGFR (and KRAS) mutations were also found in the nsclc cohort, none overlapped with the patients positive for EML4–ALK. The oncogenicity of the transcript was confirmed by transfection of expression plasmids into 3T3 cells, transforming them and subsequently showing tumorigenicity in nude mice. Although variants of the fusion transcript have been identified, in each case oncogenicity requires intact kinase function of ALK.
It was soon revealed that EML4–ALK lung carcinogenesis extended beyond Asia, characteristically occurring in middle-aged patients, usually neversmokers of either sex, and presenting as adenocarcinoma, especially the acinar histology in East Asia or the signet-ring or cribriform morphology in the West. This variant is always positive for ttf-1 62,63. Furthermore, mutual exclusivity between EML4–ALK and EGFR and KRAS mutations has been confirmed 64.
The interest in EML4–ALK that has elevated its importance above its 2.5% incidence in nsclc is its relatively specific and well-tolerated inhibitor, crizotinib. Crizotinib, originally in development as a Met inhibitor 65, is also a potent Alk inhibitor. Entering human studies in 2006, the maximum tolerated dose was established as 250 mg twice daily. While that trial was open, the Morris et al. study was published, and the first EML4–ALK patient enrolled (receiving 300 mg orally, twice daily) enjoyed a rapid and dramatic response. Subsequently, intensive efforts were made to recruit nsclc patients based on ALK rearrangements, and a high orr was confirmed (10 responders in the first 19 patients), as reported in 2009 66. A further trial in 82 ALK-rearranged nsclc patients appeared in 2010, showing a 57% orr and 33% of patients with stable disease 67. Further trials reported in 2011 that involved 119 (A8081001) and 136 (profile 1005) patients led to conditional approval of crizotinib (Xalkori: Pfizer, Mission, KS, U.S.A.) by the U.S. Food and Drug Administration (fda) 67,68 and by Health Canada more recently.
In those 255 patients (median age: 51 years; 48% men; 63% Caucasian, 30% Asian; 70% never-smokers, 28% former smokers; 96.5% with adenocarcinoma), the orrs were 61% (A8081001) and 50% (profile 1005b). The pfs is expected to be ± 10 months (A8081001), with the os still uncertain. In patients progressing on the chemotherapy arm of the randomized second-line trial of pemetrexed or docetaxel compared with crizotinib (profile 1007), profile 1005 confirmed a very high orrb. An ongoing phase iii trial, profile 1014, is investigating first-line crizotinib compared with platin–pemetrexed.
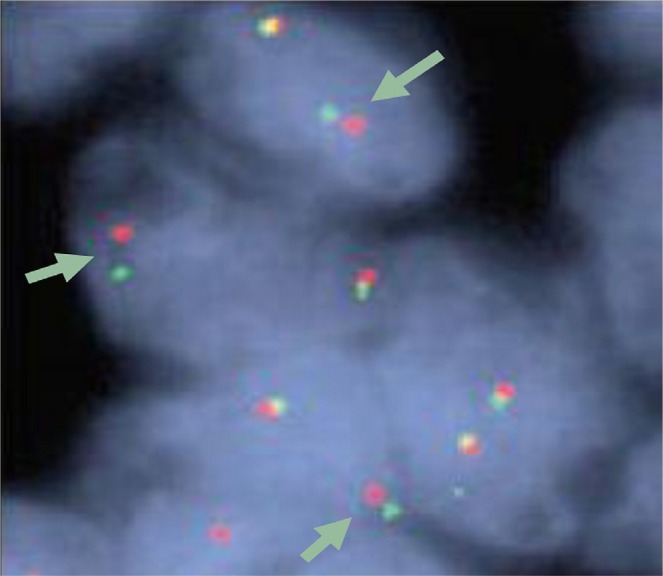
The best detection method for ALK rearrangements in nsclc is debatable. The current clinical standard—the Break Apart fish Probe kit (Abbott Molecular, Abbott Park, IL, U.S.A.), Figure 2—uses fluorescent green (5′) and red (3′) signals on loci in chromosome 2, normally so close together that they may fuse visually. Positivity consists of separation of these two markers by more than 2 signal diameters, or a red signal alone, in more than 15% tumour cells, counting more than 50 tumour cells. This is the companion assay approved by the fda with crizotinib. Suitable for formalin-fixed, paraffin-embedded specimens, it is technically demanding and expensive, encouraging development of alternative methodologies, for example, reverse-transcriptase polymerase chain reaction (requiring knowledge of known fusion variants), dna sequencing, or ihc. Immunohistochemistry, potentially with augmentation, may become a standard-of-care, high concordance with fish having been established for ihc 3+ or ihc 0 69. Intermediate ihc scores may, however, still require fish. Several different antibodies are in development 64.
FIGURE 2.
An example of a positive test with the Break Apart fish Probe kit (Abbott Molecular, Abbott Park, IL, U.S.A.). A red and a green probe are hybridized to regions flanking the ALK translocation breakpoint; these probes will be separated by an intervening fusion of a translocated fragment (for example, EML4). That intervening fusion can be clearly seen here in several cells (arrows). In the other (normal) allele, the red and green probes are not separated, even appearing yellow (an artefact of visual overlap). The assay is designated positive if 15% or more of 50 or more cell nuclei demonstrate the split signal or an isolated red signal.
Already, a crizotinib resistance mechanism has been identified, a “gatekeeper” mutation L1196M 70–72. The gene ROS is also a target of crizotinib. Activation of ROS can be found in about 1.7% of nsclc and can be assayed for; crizotinib appears to have marked activity in these cases 73.
Pemetrexed may have exceptional activity in ALK-rearranged nsclc 74, with a response (in monotherapy or in combination with a platin) of 42% and a pfs of 9 months. Other publications have appeared in support 75,76, but more recently, those findings have been questioned. The ongoing profile studies should be informative.
4. KRAS
The ras oncogenes were identified as cellular homologues of the Harvey and Kirsten strains of a mouse sarcoma virus 77. Normally, Ras functions in signal transduction downstream of transmembrane receptor tyrosine kinases, especially egfr, to which it is recruited by adaptor molecules, after binding of growth factors such as egf and transforming growth factor α to the receptor tyrosine kinase. Activation of Ras occurs through gtp binding, and as an intrinsic gtpase, it catalyzes gtp breakdown, enabling (then switching off) downstream signalling predominantly via the Raf/Mek/Erk downstream signal transduction pathway (the classical mapk pathway). Erk activates transcription of genes mediating mitosis (and cell survival). At least 9 other pathways may be stimulated by Ras, including pi3k/Akt, a survival pathway 78,79.
Of the KRAS mutations in nsclc, 97% occur in exon 2, codon 12 or 13 80. These missense mutations impair the functionality of ras gtpase, locking the Ras signalling in active mode. Paradoxically, although the mutations inactivate Ras, the result is persistent signal activation. That persistence is one reason that Ras has been difficult to “drug”; it requires reactivation, not inactivation, to switch the signalling off.
Mutations of ras in nsclc occur predominantly in “smoking adenocarcinoma” patients (30%–40%, Table i). In those patients, the mutations are G-to-T or G-to-C transversions (that is, pyrimidine swapped for purine); recently however, in never-smokers with adenocarcinoma, “transition” mutations [G to A (purine for purine)] have occasionally been found (approximately 15%), also probably oncogenic 81. The mutation subtype may alter downstream signal activation, with potential implications 82 for prognosis. This heterogeneity may explain some of the conflicting data that characterize KRAS clinical research.
Currently, KRAS itself remains undruggable despite decades of effort. Attention has recently focused on inhibition of the Ras-contingent downstream signalling (especially Raf and Erk) or exploitation of synthetic lethality 83.
Whether KRAS mutations influence egfr-tki responsiveness is contentious, and current Canadian recommendations discourage ras testing 48. Studies indicating no benefit 44,84 have to be balanced by studies indicating that KRAS mutations are compatible with some benefit in, for example, maintenance—as with saturn 85. However, KRAS mutations may indicate a short pfs in the control arm and may therefore be adversely prognostic regardless of treatment.
The negative predictive effect of KRAS for treatment with anti-egfr antibodies in colorectal cancer does not carry over to nsclc treated with cetuximab; consider flex, for example, in which KRAS mutations were neither predictive nor prognostic 86. However, KRAS mutations may sensitize tumours to antifolates such as pemetrexed 74,87, possibly by upregulation of mir-181c, a micro rna that can downregulate KRAS. Those observations require confirmation, given the high frequency of KRAS mutations in adenocarcinoma associated with smoking.
Currently, the chief value of KRAS lies in providing information about the other biomarkers that are directly druggable—that is, EGFR and ALK. The presence of mutated KRAS rules out ALK and EGFR, and KRAS may therefore form part of an efficient pathway in a testing algorithm.
5. MET
Met is a receptor tyrosine kinase often expressed in epithelium. Its paracrine ligand, hepatocyte growth factor (“scatter factor”), is produced by stromal cells. Met signals via Ras, pi3k/Akt, and stat, affecting mitosis, survival, angiogenesis, migration, invasion, and as implied, mesenchymal–epithelial transversion. Upregulation in cancer cells results in “invasive growth” 88. Amplification of MET is documented in 4.1% of North American lung adenocarcinomas, but MET overexpression maybe more commonc. Mutations in MET occur rarely.
Upregulation of MET may depend on prior exposure to therapy and may mediate resistance to it. Several studies indicate that MET amplification is responsible for ±20% of resistance to egfr-tki 89–92, prompting the development of Met-inhibitory strategies. Tivantinib (ARQ 197) is currently in phase iii trial (marquee) based on a successful randomized phase ii study (erlotinib ± tivantinib). Non-squamous and KRAS M+ patients benefited most. MetMAb (Hoffmann–La Roche, Mississauga, ON), an anti-Met monoclonal antibody, achieved significant pfs and os benefit in a randomized phase ii trial (oam 4558g) with a similar “erlotinib ± experimental drug” design, but only in high expressors of MET (Met ihc 2+ or 3+). Detection by ihc (that is, expression) may be more reliable than detection by fish (that is, amplification) in predicting MetMAb benefit. The effect in low expressors of Met appeared actually harmful, highlighting the importance of a companion diagnostic as MetMAb proceeds into phase iii.
Crizotinib, although approved for ALK-rearranged metastatic nsclc, is also a good Met inhibitor. An anecdotal report 93 of a rapid, durable response to crizotinib in a MET-amplified nsclc patient with normal ALK, suggests that crizotinib may be suitable for that situation as well as for ALK rearrangements, as already shown for other types of cancer with MET amplification 94,95.
MET will likely be the next major biomarker in metastatic nsclc, given the speed with which the foregoing drugs (and others) 88 are approaching the clinic. How best to integrate them into the increasingly complex metastatic nsclc algorithm will require substantial investment, but will likely pay major dividends.
6. SUMMARY
EGFR and ALK are biomarkers of current relevance in the management of non-squamous metastatic nsclc and definitely predict a higher likelihood of benefit from egfr-tki and crizotinib respectively. Across Canada, efforts to promote access to testing require intensification. KRAS testing remains controversial—but interesting in the research setting and in testing algorithms as an efficiency tactic, because KRAS mutations are common and almost entirely rule out EGFR mutations and ALK rearrangements. MET amplification—or more likely, Met ihc—is required to optimize the development and clinical deployment of Met-directed therapies. Subject to confirmation, egfr ihc (“H-score”) might allow for the selection of patients benefiting from anti-egfr monoclonal antibodies such as cetuximab.
The problem of inconsistent access to adequate tissue remains an important obstacle to the evolution of personalized medicine in metastatic nsclc. The solution lies partly in the ongoing development of serum-based molecular assays, but for now, it lies in the education of interventional radiologists, thoracic surgeons, and respirologists, because optimal treatment of metastatic nsclc is highly contingent on an adequate biopsy.
Footnotes
Zhong WZ. Genomic heterogeneity between primary tumor and its metastases. Presented at the 3rd International Thoracic Oncology Congress Dresden; Dresden, Germany; September 13–15, 2012.
Riely GJ, Kim DW, Crino L, et al. Phase 2 data for crizotinib (PF-02341066) in ALK-positive advanced non-small cell lung cancer (nsclc): profile 1005 [abstract 1618]. Presented at the 14th World Conference on Lung Cancer; Amsterdam, Netherlands; July 3–7, 2011.
Varella–Garcia M, Iafrate J, Pao W, et al. ALK fusion and MET amplification as molecular biomarkers and therapeutic targets in advanced lung adenocarcinomas in the Lung Cancer Mutation Consortium [abstract 1348]. Presented at the 14th World Conference on Lung Cancer; Amsterdam, Netherlands; July 3–7, 2011.
7. CONFLICT OF INTEREST DISCLOSURES
The authors declare consultancy work for Lilly and Roche; work on advisory boards for Pfizer, AstraZeneca, and Roche; membership in a speakers’ bureau for Lilly.
8. REFERENCES
- 1.Cheng L, Alexander RE, Maclennan GT, et al. Molecular pathology of lung cancer: key to personalized medicine. Mod Pathol. 2012;25:347–69. doi: 10.1038/modpathol.2011.215. [DOI] [PubMed] [Google Scholar]
- 2.Pao W, Miller V, Zakowski M, et al. egf receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumours to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–11. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aung KL, Board RE, Ellison G, et al. Current status and future potential of somatic mutation testing from circulating free dna in patients with solid tumours. HUGO J. 2010;4:11–21. doi: 10.1007/s11568-011-9149-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goto K, Ichinose Y, Ohe Y, et al. Epidermal growth factor receptor mutation status in circulating free dna in serum: from ipass, a phase iii study of gefitinib or carboplatin/paclitaxel in non-small cell lung cancer. J Thorac Oncol. 2012;7:115–21. doi: 10.1097/JTO.0b013e3182307f98. [DOI] [PubMed] [Google Scholar]
- 5.Cohen S. Isolation of a mouse submaxillary gland protein accelerating incisor eruption and eyelid opening in the newborn animal. J Biol Chem. 1962;237:1555–62. [PubMed] [Google Scholar]
- 6.Hollenberg MD, Cuatrecasas P. Epidermal growth factor: receptors in human fibroblasts and modulation of action by cholera toxin. Proc Natl Acad Sci U S A. 1973;70:2964–8. doi: 10.1073/pnas.70.10.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burgess AW. EGFR family: structure physiology signalling and therapeutic targets. Growth Factors. 2008;26:263–74. doi: 10.1080/08977190802312844. [DOI] [PubMed] [Google Scholar]
- 8.Avraham R, Yarden Y. Feedback regulation of EGFR signalling: decision making by early and delayed loops. Nat Rev Mol Cell Biol. 2011;12:104–17. doi: 10.1038/nrm3048. [DOI] [PubMed] [Google Scholar]
- 9.Ullrich A, Coussens L, Hayflick JS, et al. Human epidermal growth factor receptor cdna sequence and aberrant expression of the amplified gene in A431 epidermoid carcinoma cells. Nature. 1984;309:418–25. doi: 10.1038/309418a0. [DOI] [PubMed] [Google Scholar]
- 10.Mendelsohn J, Baselga J. Status of epidermal growth factor receptor antagonists in the biology and treatment of cancer. J Clin Oncol. 2003;21:2787–99. doi: 10.1200/JCO.2003.01.504. [DOI] [PubMed] [Google Scholar]
- 11.Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97:339–46. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 12.Tam IY, Chung LP, Suen WS, et al. Distinct epidermal growth factor receptor and KRAS mutation patterns in non-small cell lung cancer patients with different tobacco exposure and clinicopathologic features. Clin Cancer Res. 2006;12:1647–53. doi: 10.1158/1078-0432.CCR-05-1981. [DOI] [PubMed] [Google Scholar]
- 13.Bae NC, Chae MH, Lee MH, et al. EGFR, ERBB2, and KRAS mutations in Korean non-small cell lung cancer patients. Cancer Genet Cytogenet. 2007;173:107–13. doi: 10.1016/j.cancergencyto.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 14.Sordella R, Bell DW, Haber DA, Settleman J. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305:1163–7. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- 15.Takeuchi K, Ito F. egf receptor in relation to tumor development: molecular basis of responsiveness of cancer cells to egfr-targeting tyrosine kinase inhibitors. FEBS J. 2010;277:316–26. doi: 10.1111/j.1742-4658.2009.07450.x. [DOI] [PubMed] [Google Scholar]
- 16.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 17.Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 18.Okamoto I. Epidermal growth factor receptor in relation to tumor development: egfr-targeted anticancer therapy. FEBS J. 2010;277:309–15. doi: 10.1111/j.1742-4658.2009.07449.x. [DOI] [PubMed] [Google Scholar]
- 19.Inoue A, Suzuki T, Fukuhara T, et al. Prospective phase ii study of gefitinib for chemotherapy-naive patients with advanced non-small-cell lung cancer with epidermal growth factor receptor gene mutations. J Clin Oncol. 2006;24:3340–6. doi: 10.1200/JCO.2005.05.4692. [DOI] [PubMed] [Google Scholar]
- 20.Asahina H, Yamazaki K, Kinoshita I, et al. A phase ii trial of gefitinib as first-line therapy for advanced non-small cell lung cancer with epidermal growth factor receptor mutations. Br J Cancer. 2006;95:998–1004. doi: 10.1038/sj.bjc.6603393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sutani A, Nagai Y, Udagawa K, et al. Gefitinib for non-small-cell lung cancer patients with epidermal growth factor receptor gene mutations screened by peptide nucleic acid-locked nucleic acid pcr clamp. Br J Cancer. 2006;95:1483–9. doi: 10.1038/sj.bjc.6603466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshida K, Yatabe Y, Park JY, et al. Prospective validation for prediction of gefitinib sensitivity by epidermal growth factor receptor gene mutation in patients with non-small cell lung cancer. J Thorac Oncol. 2007;2:22–8. doi: 10.1097/01243894-200701000-00006. [DOI] [PubMed] [Google Scholar]
- 23.Sunaga N, Tomizawa Y, Yanagitani N, et al. Phase ii prospective study of the efficacy of gefitinib for the treatment of stage iii/iv non-small cell lung cancer with EGFR mutations, irrespective of previous chemotherapy. Lung Cancer. 2007;56:383–9. doi: 10.1016/j.lungcan.2007.01.025. [DOI] [PubMed] [Google Scholar]
- 24.Tamura K, Okamoto I, Kashii T, et al. Multicentre prospective phase ii trial of gefitinib for advanced non-small cell lung cancer with epidermal growth factor receptor mutations: results of the West Japan Thoracic Oncology Group trial (wjtog 0403) Br J Cancer. 2008;98:907–14. doi: 10.1038/sj.bjc.6604249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sequist LV, Martins RG, Spigel D, et al. First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. J Clin Oncol. 2008;26:2442–9. doi: 10.1200/JCO.2007.14.8494. [DOI] [PubMed] [Google Scholar]
- 26.Sugio K, Uramoto H, Onitsuka T, et al. Prospective phase ii study of gefitinib in non-small cell lung cancer with epidermal growth factor receptor gene mutations. Lung Cancer. 2009;64:314–18. doi: 10.1016/j.lungcan.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 27.Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361:958–67. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 28.Lee JS, Park K, Kim SW, et al. A randomized phase iii study of gefitinib versus standard chemotherapy (gemcitabine plus cisplatin) as a first-line treatment for never-smokers with advanced or metastatic adenocarcinoma of the lung [abstract PRS.4] J Thorac Oncol. 2009;4(suppl 1):S283. [Google Scholar]
- 29.Ku GY, Haaland BA, de Lima Lopes G., Jr Gefitinib vs chemotherapy as first-line therapy in advanced non-small cell lung cancer: meta-analysis of phase iii trials. Lung Cancer. 2011;74:469–73. doi: 10.1016/j.lungcan.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 30.Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–8. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 31.Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (wjtog 3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121–8. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 32.Fukuoka M, Wu YL, Thongprasert S, et al. Biomarker analyses and final overall survival results from a phase iii, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non-small-cell lung cancer in Asia (ipass) J Clin Oncol. 2011;29:2866–74. doi: 10.1200/JCO.2010.33.4235. [DOI] [PubMed] [Google Scholar]
- 33.Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation–positive non-small-cell lung cancer (optimal, ctong-0802): a multicenter, open-label, randomized, phase 3 study. Lancet Oncol. 2011;12:735–42. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 34.Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation–positive non-small-cell lung cancer (erutac): a multicenter, open-label, randomized phase 3 trial. Lancet Oncol. 2012;13:239–46. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 35.Wu JY, Yu CJ, Shih JY, Yang CH, Yang PC. Influence of first-line chemotherapy and EGFR mutations on second-line gefitinib in advanced non-small cell lung cancer. Lung Cancer. 2010;67:348–54. doi: 10.1016/j.lungcan.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 36.Cappuzzo F, Ciuleanu T, Stelmakh L, et al. Erlotinib as maintenance treatment in advanced non-small-cell lung cancer: a multicentre, randomized, placebo-controlled phase 3 study. Lancet Oncol. 2010;11:521–9. doi: 10.1016/S1470-2045(10)70112-1. [DOI] [PubMed] [Google Scholar]
- 37.Yun CH, Mengwasser KE, Toms AV, et al. The T790M mutation in egfr kinase causes drug resistance by increasing the affinity for atp. Proc Natl Acad Sci U S A. 2008;105:2070–5. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sequist LV, Waltman BA, Dias–Santagata D, et al. Genotypic and histological evolution of lung cancer acquiring resistance to egfr inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chaft JE, Oxnard GR, Sima CS, Kris MG, Miller VA, Riely GJ. Disease flare after tyrosine kinase inhibitor discontinuation in patients with EGFR-mutant lung cancer and acquired resistance to erlotinib or gefitinib: implications for clinical trial design. Clin Cancer Res. 2011;17:6298–303. doi: 10.1158/1078-0432.CCR-11-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Becker A, Crombag L, Heideman DA, et al. Retreatment with erlotinib: regain of tki sensitivity following a drug holiday for patients with nsclc who initially responded to egfr-tki treatment. Eur J Cancer. 2011;47:2603–6. doi: 10.1016/j.ejca.2011.06.046. [DOI] [PubMed] [Google Scholar]
- 41.Oxnard GR, Janjigian YY, Arcila ME, et al. Maintained sensitivity to egfr tyrosine kinase inhibitors in EGFR-mutant lung cancer recurring after adjuvant erlotinib or gefitinib. Clin Cancer Res. 2011;17:6322–8. doi: 10.1158/1078-0432.CCR-11-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park S, Holmes–Tisch AJ, Cho EY, et al. Discordance of molecular biomarkers associated with epidermal growth factor receptor pathway between primary tumors and lymph node metastasis in non-small cell lung cancer. J Thorac Oncol. 2009;4:809–15. doi: 10.1097/JTO.0b013e3181a94af4. [DOI] [PubMed] [Google Scholar]
- 43.Hirsch FR, Varella–Garcia M, Bunn PA, Jr, et al. Molecular predictors of outcome with gefitinib in a phase iii placebo-controlled study in advanced non-small-cell lung cancer. J Clin Oncol. 2006;24:5034–42. doi: 10.1200/JCO.2006.06.3958. [DOI] [PubMed] [Google Scholar]
- 44.Zhu CQ, da Cunha Santos G, Ding K, et al. Role of KRAS and EGFR as biomarkers of response to erlotinib in National Cancer Institute of Canada Clinical Trials Group Study br.21. J Clin Oncol. 2008;26:4268–75. doi: 10.1200/JCO.2007.14.8924. [DOI] [PubMed] [Google Scholar]
- 45.Schneider CP, Heigener D, Schott-von-Römer K, et al. Epidermal growth factor receptor–related tumor markers and clinical outcomes with erlotinib in non-small cell lung cancer: an analysis of patients from German centers in the trust study. J Thorac Oncol. 2008;3:1446–53. doi: 10.1097/JTO.0b013e31818ddcaa. [DOI] [PubMed] [Google Scholar]
- 46.Thatcher N, Chang A, Parikh P, Rodrigues Pereira J, et al. Gefitinib plus best supportive care in previously treated patients with refractory advanced non-small-cell lung cancer: results from a randomised, placebo-controlled, multicentre study (Iressa Survival Evaluation in Lung Cancer) Lancet. 2005;366:1527–37. doi: 10.1016/S0140-6736(05)67625-8. [DOI] [PubMed] [Google Scholar]
- 47.Hirsch FR, Varella–Garcia M, Bunn PA, Jr, et al. Molecular predictors of outcome with gefitinib in a phase iii placebo-controlled study in advanced non-small-cell lung cancer. J Clin Oncol. 2006;24:5034–42. doi: 10.1200/JCO.2006.06.3958. [DOI] [PubMed] [Google Scholar]
- 48.Ellis PM, Blais N, Soulieres D, et al. A systematic review and Canadian consensus recommendations on the use of biomarkers in the treatment of non-small cell lung cancer. J Thorac Oncol. 2011;6:1379–91. doi: 10.1097/JTO.0b013e318220cb8e. [DOI] [PubMed] [Google Scholar]
- 49.Pirker R, Pereira JR, Szczesna A, et al. Cetuximab plus chemotherapy in patients with advanced non-small-cell lung cancer (flex): an open-label randomised phase iii trial. Lancet. 2009;373:1525–31. doi: 10.1016/S0140-6736(09)60569-9. [DOI] [PubMed] [Google Scholar]
- 50.Pirker R, Pereira JR, von Pawel J, et al. Epidermal growth factor receptor (EGFR) expression as a predictor for survival of first line chemotherapy plus cetuximab in flex study patients with advanced non-small cell lung cancer (nsclc) Lancet Oncol. 2012;13:33–42. doi: 10.1016/S1470-2045(11)70318-7. [DOI] [PubMed] [Google Scholar]
- 51.Pirker R, Herth FJ, Kerr KM, et al. Consensus for EGFR mutation testing in non-small cell lung cancer: results from a European workshop. J Thorac Oncol. 2010;5:1706–13. doi: 10.1097/JTO.0b013e3181f1c8de. [DOI] [PubMed] [Google Scholar]
- 52.Yu J, Kane S, Wu J, et al. Mutation-specific antibodies for the detection of EGFR mutations in non-small-cell lung cancer. Clin Cancer Res. 2009;15:3023–8. doi: 10.1158/1078-0432.CCR-08-2739. [DOI] [PubMed] [Google Scholar]
- 53.Brevet M, Arcila M, Ladanyi M. Assessment of EGFR mutation status in lung adenocarcinoma by immunohistochemistry using antibodies specific to the two major forms of mutant EGFR. J Mol Diagn. 2010;12:169–76. doi: 10.2353/jmoldx.2010.090140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kawahara A, Yamamoto C, Nakashima K, et al. Molecular diagnosis of activating EGFR mutations in non-small cell lung cancer using mutation-specific antibodies for immunohistochemical analysis. Clin Cancer Res. 2010;16:3163–70. doi: 10.1158/1078-0432.CCR-09-3239. [DOI] [PubMed] [Google Scholar]
- 55.Kato Y, Peled N, Wynes MW, et al. Novel epidermal growth factor receptor mutation-specific antibodies for non-small cell lung cancer: immunohistochemistry as a possible screening method for epidermal growth factor receptor mutations. J Thorac Oncol. 2010;5:1551–8. doi: 10.1097/JTO.0b013e3181e9da60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–77. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sequist LV, Nagrath S, Toner M, Haber DA, Lynch TJ. The ctc-chip: an exciting new tool to detect circulating tumor cells in lung cancer patients. J Thorac Oncol. 2009;4:281–3. doi: 10.1097/JTO.0b013e3181989565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yung TK, Chan KC, Mok TS, Tong J, To KF, Lo YM. Single-molecule detection of epidermal growth factor receptor mutations in plasma by microfluidics digital pcr in non-small cell lung cancer patients. Clin Cancer Res. 2009;15:2076–84. doi: 10.1158/1078-0432.CCR-08-2622. [DOI] [PubMed] [Google Scholar]
- 59.Bai H, Mao L, Wang HS, et al. Epidermal growth factor receptor mutations in plasma dna samples predict tumor response in Chinese patients with stages iiib to iv non-small-cell lung cancer. J Clin Oncol. 2009;27:2653–9. doi: 10.1200/JCO.2008.17.3930. [DOI] [PubMed] [Google Scholar]
- 60.Morris SW, Kirstein MN, Valentine MB, et al. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin’s lymphoma. Science. 1994;263:1281–4. doi: 10.1126/science.8122112. [DOI] [PubMed] [Google Scholar]
- 61.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4–ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–6. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 62.Inamura K, Takeuchi K, Togashi Y, et al. EML4–ALK lung cancers are characterized by rare other mutations, a ttf-1 cell lineage, an acinar histology, and young onset. Mod Pathol. 2009;22:508–15. doi: 10.1038/modpathol.2009.2. [DOI] [PubMed] [Google Scholar]
- 63.Shaw AT, Yeap BY, Mino–Kenudson M, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbour EML4–ALK. J Clin Oncol. 2009;27:4247–53. doi: 10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Subramanian J, Corrales L, Soulieres D, Morgensztern D, Govindan R. Summary of presentations from the 46th Annual Meeting of the American Society of Clinical Oncology (2010) focus on tumor biology and biomarkers related to lung cancer. J Thorac Oncol. 2011;6:399–403. doi: 10.1097/JTO.0b013e318200f972. [DOI] [PubMed] [Google Scholar]
- 65.Ou SH. Crizotinib: a novel and first-in-class multitargeted tyrosine kinase inhibitor for the treatment of anaplastic lymphoma kinase rearranged non-small cell lung cancer and beyond. Drug Des Devel Ther. 2011;5:471–85. doi: 10.2147/DDDT.S19045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kwak EL, Camidge DR, Clark J, et al. Clinical activity observed in a phase i dose escalation trial of an oral c-Met and Alk inhibitor, PF-02341066 [abstract 3509] J Clin Oncol. 2009;27 [Available online at: http://www.asco.org/ASCOv2/Meetings/Abstracts?&vmview=abst_detail_view&confID=65&abstractID=30947; cited May 23, 2012] [Google Scholar]
- 67.Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Crino L, Kim DW, Riely GJ, et al. Initial phase ii results with crizotinib in advanced ALK-positive non-small cell lung cancer (nsclc): profile 1005 [abstract 7514] J Clin Oncol. 2011;29 [Available online at: http://www.asco.org/ASCOv2/Meetings/Abstracts?&vmview=abst_detail_view&confID=102&abstractID=81844; cited May 23, 2012] [Google Scholar]
- 69.Yang P, Kulig K, Boland JM, et al. Worse disease-free survival in never-smokers with ALK+ lung adenocarcinoma. J Thorac Oncol. 2012;7:90–7. doi: 10.1097/JTO.0b013e31823c5c32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Choi YL, Soda M, Yamashita Y, et al. EML4–ALK mutations in lung cancer that confer resistance to Alk inhibitors. N Engl J Med. 2010;363:1734–9. doi: 10.1056/NEJMoa1007478. [DOI] [PubMed] [Google Scholar]
- 71.Katayama R, Khan TM, Benes C, et al. Therapeutic strategies to overcome crizotinib resistance in non-small cell lung cancers harboring the fusion oncogene EML4–ALK. Proc Natl Acad Sci U S A. 2011;108:7535–40. doi: 10.1073/pnas.1019559108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sakamoto H, Tsukaguchi T, Hiroshima S, et al. CH5424802, a selective Alk inhibitor capable of blocking the resistant gatekeeper mutant. Cancer Cell. 2011;19:679–90. doi: 10.1016/j.ccr.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 73.Bergethon K, Shaw AT, Ou SH, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol. 2012;30:863–70. doi: 10.1200/JCO.2011.35.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Camidge DR, Kono SA, Lu X, et al. Anaplastic lymphoma kinase gene rearrangements in non-small cell lung cancer are associated with prolonged progression-free survival on pemetrexed. J Thorac Oncol. 2011;6:774–80. doi: 10.1097/JTO.0b013e31820cf053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Takeda M, Okamoto I, Sakai K, et al. Successful long-term treatment with pemetrexed of nsclc associated with EML4–ALK and low thymidylate synthase expression. Clin Lung Cancer. 2012;13:157–9. doi: 10.1016/j.cllc.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 76.Lee JO, Kim TM, Lee SH, et al. Anaplastic lymphoma kinase translocation: a predictive biomarker of pemetrexed in patients with non-small cell lung cancer. J Thorac Oncol. 2011;6:1474–80. doi: 10.1097/JTO.0b013e3182208fc2. [DOI] [PubMed] [Google Scholar]
- 77.Chang EH, Gonda MA, Ellis RW, Scolnick EM, Lowy DR. Human genome contains four genes homologous to transforming genes of Harvey and Kirsten murine sarcoma viruses. Proc Natl Acad Sci U S A. 1982;79:4848–52. doi: 10.1073/pnas.79.16.4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vakiani E, Solit DB. KRAS and BRAF: drug targets and predictive biomarkers. J Pathol. 2011;223:219–29. doi: 10.1002/path.2796. [DOI] [PubMed] [Google Scholar]
- 79.Xu N, Lao Y, Zhang Y, Gillespie DA. Akt: a double-edged sword in cell proliferation and genome stability. J Oncol. 2012;2012:951724. doi: 10.1155/2012/951724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Forbes S, Clements J, Dawson E, et al. cosmic 2005. Br J Cancer. 2006;94:318–22. doi: 10.1038/sj.bjc.6602928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Riely GJ, Kris MG, Rosenbaum D, et al. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin Cancer Res. 2008;14:5731–4. doi: 10.1158/1078-0432.CCR-08-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ihle NT, Byers LA, Kim ES, et al. Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. J Natl Cancer Inst. 2012;104:228–39. doi: 10.1093/jnci/djr523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Luo J, Emanuele MJ, Li D, et al. A genome-wide rnai screen identifies multiple synthetic lethal interactions with the ras oncogene. Cell. 2009;137:835–48. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Eberhard DA, Johnson BE, Amler LC, et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol. 2005;23:5900–9. doi: 10.1200/JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
- 85.Brugger W, Triller N, Blasinska–Morawiec M, et al. Prospective molecular marker analyses of EGFR and KRAS from a randomized, placebo-controlled study of erlotinib maintenance therapy in advanced non-small-cell lung cancer. J Clin Oncol. 2011;29:4113–20. doi: 10.1200/JCO.2010.31.8162. [DOI] [PubMed] [Google Scholar]
- 86.O’Byrne KJ, Gatzemeier U, Bondarenko I, et al. Molecular biomarkers in non-small-cell lung cancer: a retrospective analysis of data from the phase 3 flex study. Lancet Oncol. 2011;12:795–805. doi: 10.1016/S1470-2045(11)70189-9. [DOI] [PubMed] [Google Scholar]
- 87.Bacus S. KRAS mutation and amplification status predicts sensitivity to antifolate therapies in non-small cell lung cancer [abstract PR-2] Mol Cancer Ther. 2011;10(suppl 1) [Google Scholar]
- 88.Feng Y, Thiagarajan PS, Ma PC. Met signaling: novel targeted inhibition and its clinical development in lung cancer. J Thorac Oncol. 2012;7:459–67. doi: 10.1097/JTO.0b013e3182417e44. [DOI] [PubMed] [Google Scholar]
- 89.Ma PC, Tretiakova MS, MacKinnon AC, et al. Expression and mutational analysis of MET in human solid cancers. Genes Chromosomes Cancer. 2008;47:1025–37. doi: 10.1002/gcc.20604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ErbB3 signaling. Science. 2007;316:1039–43. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 91.Cappuzzo F, Jänne PA, Skokan M, et al. MET increased gene copy number and primary resistance to gefitinib therapy in non-small-cell lung cancer patients. Ann Oncol. 2009;20:298–304. doi: 10.1093/annonc/mdn635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A. 2007;104:20932–7. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ou SH, Kwak EL, Siwak–Tapp C, et al. Activity of crizotinib (PF02341066), a dual mesenchymal–epithelial transition (MET) and anaplastic lymphoma kinase (ALK) inhibitor, in a non-small cell lung cancer patient with de novo MET amplification. J Thorac Oncol. 2011;6:942–6. doi: 10.1097/JTO.0b013e31821528d3. [DOI] [PubMed] [Google Scholar]
- 94.Lennerz JK, Kwak EL, Ackerman A, et al. MET amplification identifies a small and aggressive subgroup of esophagogastric adenocarcinoma with evidence of responsiveness to crizotinib. J Clin Oncol. 2011;29:4803–10. doi: 10.1200/JCO.2011.35.4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chi A, Kwak EL, Clark JW, et al. Clinical improvement and rapid radiographic regression induced by a Met inhibitor in a patient with MET-amplified glioblastoma [abstract 2072] J Clin Oncol. 2011;29 doi: 10.1200/JCO.2011.38.4586. [Available online at: http://www.asco.org/ASCOv2/Meetings/Abstracts?&vmview=abst_detail_view&confID=102&abstractID=82407; cited May 23, 2012] [DOI] [PubMed] [Google Scholar]