Abstract
The HLA-C*07:01:01G allele group consists of three nonsynonymous alleles, C*07:01:01, C*07:06 and C*07:18, plus C*07:01:02, which is synonymous to C*07:01:01. All of these alleles have identical exons 2, 3 and 4, but differ in exons 5 or 6. Therefore routine sequence-based typing (SBT) of exons 2 and 3 is unable to resolve these subtypes, resulting in ambiguous typing results in population and disease cohort studies. In the present study, we fully characterized C*07:01:01G subtypes in European and African Americans and examined their relative frequency distributions. In European Americans C*07:01:01G is predominantly represented by C*07:01:01 (94.4%), whereas C*07:01:02 (1.1%) and C*07:18 (4.5%) were detected relatively infrequently. In African Americans C*07:18 (42.4%) showed a high frequency similar to that of C*07:01:01 (44.7%) whereas C*07:06 was detected at a low frequency (4.7%). C*07:06 was found exclusively on B*44:03 carrying haplotypes in both ethnic groups, but C*07:18 showed multiple linkage relationships with HLA-B. These results demonstrate that C*07:01:01G as defined by routine SBT is a heterogeneous group of alleles, especially among individuals of African origin. If C*07:01:01G subtypes prove to bear divergent functional significance, it would be necessary to include these subtypes in routine HLA-C typing for clinical transplantation and disease association studies.
Keywords: HLA-C locus, allele diversity, C*07:01:01G, allele combination, sequence-based typing (SBT)
1. Introduction
HLA-C*07 is one of the most common, divergent, and polymorphic HLA-C lineages with 281 synonymous, non-synonymous and null alleles recognized so far (http://www.ebi.ac.uk/imgt/hla/, Release 3.7.0, 12-January-2012). C*07:01:01G is the name for the allele group comprising eight subtypes sharing identical exons 2 and 3. Four of them, C*07:01:01, C*07:01:02, C*07:06 and C*07:18 share an identical nucleotide sequence from exon 1 through exon 4 but differ in exons 5 or 6. The other four, C*07:01:09, C*07:52, C*07:153 and C*07:166, differ from C*07:01:01 only in exon 4. According to the National Marrow Donor Program (NMDP, http://bioinformatics.nmdp.org/) database, C*07:01:01G was commonly detected in all major populations in the US: 17% in European Americans, 12% in African Americans, 10% in Hispanics, and 4% in Asian and Pacific Islanders. Three of the C*07:01:01G subtypes, C*07:01:01, C*07:06 and C*07:18 are on the ASHI CWD allele list.
The genes encoding C*07:01:01, C*07:01:02, C*07:06 and C*07:18 share an identical sequence from exon 1 through exon 4 but differ in exons 5 and 6 (Figure 1), which encode the transmembrane segment and cytoplasmic tail of the HLA-C molecule, respectively [1]. Though little is known about the functional significance of the polymorphisms outside of the peptide binding regions (PBR), the potential effect of amino acid variations in the transmembrane segment and cytoplastic tail on HLA expression and intracellular signalling, as well as their potential influence on transplantation and disease association, have drawn interest [2-4].
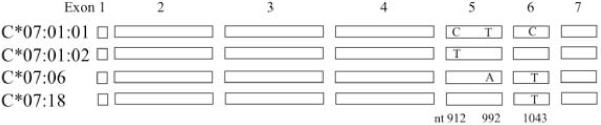
Fig. 1.
Nucleotide variations of C*07:01:01G subtypes detected in the present study. The four subtypes in the C*07:01:01G allele group share identical exons 1 through 4 and 7 but differ in three positions in exons 5 and 6.
C*07:06 was first identified in 1996 by cDNA cloning, where only the coding sequence was published [5]. The full-length sequence of C*07:06 has been characterized more recently in greater detail, and the allele was found to have two non-synonymous substitutions relative to C*07:01:01 at nt992 T>A in exon 5 and nt1043 C>T in exon 6, plus four additional substitutions in intron 3, intron 4, and the 3′-UTR [6]. The coding sequence of C*07:18, which differs from C*07:01:01 by a single nucleotide at nt1043 C>T in exon 6, was first detected in 2003 [4]. In a recent study C*07:18 was indicated as one of the risk factors for severe cutaneous adverse reactions (SCAR), a condition of severe hypersensitivity to a variety of medicines [7]. C*07:01:02 differs from C*07:01:01 by a single synonymous substitution at nt912 C>T in exon 5.
Routine HLA class I tying has focused on the 4-digit PBR polymorphism encoded by exons 2 and 3, whereas other coding and non-coding polymorphisms of these genes have largely been neglected. Therefore, little information has been generated about the frequency distribution of C*07:01:01G subtypes from population and disease-association studies [8-10], though C*07:01:01, C*07:06 and C*07:18 are all on the ASHI CWD allele list [11]. Previous studies did show strong linkage disequilibrium relationships of C*07:01:01G subtypes with HLA-B alleles. In particular, C*07:06 and C*07:18 were found to be associated with B*44 and B*58:01, respectively [6, 12, 13], the latter of which associates with protection against HIV [14].
We have recently characterized C*07:01:01G subtypes in Northern and Southern Chinese populations and found that C*07:01:01 and C*07:06 were both represented with C*07:06 having a slightly higher frequency. C*07:18 was completely absent from the 1795 individuals tested [6]. Interestingly, C*07:18 was detected as the second most common C*07:01:01G allele after C*07:01:01 in Ugandans from East Africa, whereas C*07:06 was only found in a single individual of the 175 subjects examined [12]. These limited studies suggest that the distribution of C*07:01:01G subtypes is highly divergent across ethnic groups. Greater knowledge of the subtype distribution may carry significance in transplantation, population, and disease association studies.
In the present study we fully characterize C*07:01:01G subtypes and investigate their relative frequencies in African American and European American subjects, providing an estimate of how much of the diversity in this grouping was actually missed in previous HLA typing across population and disease studies.
2. Materials and Methods
2.1. Study subjects
A total of 226 C*07:01:01G-positive individuals from cohorts we genotyped previously for exons 2 and 3 were selected for full characterization of C*07:01:01G subtypes. The details of the cohorts have been previously described [15]. The study subjects were grouped into two panels for separate analyses. The first panel consisted of randomly selected C*07:01:01G-positive individuals of 80 European Americans and 81 African Americans for estimating the relative frequencies of C*07:01:01G subtypes in the two cohorts. The second panel were individuals with both C*07:01:01G and B*44, and was used to evaluate the previously reported linkage relationship of C*07:06 with B*44. The second panel included 11 B*44 positive individuals from the first panel of randomly selected C*07:01:01G-positive European or African Americans plus an additional 65 individuals of mixed ethnicities that were selected specifically for having both C*07:01:01G and B*44. A total of 76 individuals (59 European Americans, 8 African Americans, 5 Hispanics, 2 Asians and 2 of unknown ethnicity) in the second panel were available for examining the linkage relationship between C*07:06 and B*44.
The study subjects have been previously typed for HLA-C by routine SBT of exons 2 and 3. These subjects were retyped for HLA-C by sequencing exons 2 through 6 in the present study.
2.2. Sequencing of exons 2, 3 and 4
Genomic DNA was used to examine HLA-C variation by SBT in this study. PCR amplification of exons 2, 3 and 4 in a single amplicon was achieved using HLA-C specific primers that match the sequences in the 5′UTR and intron 4, generating a PCR fragment of about 2000 bp in size. The PCR primers and conditions have been described previously [6]. PCR amplicons were purified using the Omega Mag-Bind® EZPure commercial kit. Sequencing reactions of exons 2, 3 and 4 were performed separately in both orientations using published primers (exon 2: Forward 5′-GGGTCTCAGCCMCTCCTC-3′, Reverse 5′-GCC GTC CGT GGG GGA TG-3′; exon 3: Forward 5′-GCCCCAGTCRCCTTTAC-3′, Reverse 5′-TTCCTCCCCTCCTCGTG-3′; exon 4: Forward 5′-TTC TCA GGA TRG TCA CAT G-3′, Reverse 5′-CCYCATYCCCCTCCTTAC-3′) [16]. Sequences were analyzed on an ABI 3730XL DNA Sequencer (Applied Biosystem, Foster City, CA).
2.3. Sequencing of exons 5 and 6
Sequencing of exons 5 and 6 followed our in-house SBT protocol [6] with a modification to the forward PCR primer. An amplicon of 1050 bp was obtained using primers spanning exons 5 through 8 (forward 5′-GTA AGG AGG GGR ATG RGG GGT-3′; reverse 5′-AAT CCT GCA TCT CAG TCC CAC-3′). PCR was carried out in a 25 μl volume containing 12.5 μl of 2×GC Buffer, 0.2 μl of each dNTP (25 mM), 1 μl of each PCR primer (10 μM), 100 ng of genomic DNA, and 2.5 U of Genomic LA Taq polymerase (Clontech, 1290 Terra Bella Avenue Mountain View, CA 94043,USA). PCR conditions were: 95°C for 2 min followed by 35 cycles of 30 sec. at 95°C, 30 sec. at 62°C, and 1.5 min. at 72°C, and a final extension at 72°C for 15 minutes. Sequencing was performed using the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit and our in-house sequencing primers (exon 5: forward 5′-GTCAGGGCTGAGGCTTG-3′, reverse 5′-GATGGTGCTTCCAGTAAC-3′; exon 6: forward 5′-TCCAAGACTAGGAGGTTC-3′, Reverse 5′-AAAAAGACCTGGTCAGAG-3′).
2.4. Assignment of HLA-C alleles and HLA-B/C haplotypes
HLA-C alleles were assigned with the help of the ASSIGN 3.5 HLA typing software (Conexio Genomics, Fremantle, Western Australia). Since family members were not available for segregation analysis, all C*07:01:01G-related HLA-B/C haplotypes described in this study were elucidated by maximum likelihood estimation based on our HLA-typed cohorts, including 2620 European American and 1123 African American individuals (unpublished data).
3. Results
3.1. Relative distribution of C*07:01:01G subtypes in European and African Americans
Based on the sequencing data of exons 2 through 6, four C*07:01:01G subtypes were detected in our sample of randomly selected C*07:01:01G-positive subjects (n = 161). These subtypes showed highly divergent frequency distributions between European and African American groups as shown in Table 1. C*07:01:01 dominated the C*07:01:01G grouping in European Americans, whereas C*07:01:02 was detected only in one of these 81 subjects. C*07:18 was detected in four individuals and C*07:06 was absent. African Americans, however, showed a greater level of diversity in the C*07:01:01G grouping, in that C*07:01:01 and C*07:18 were both commonly detected with similar frequencies (44.7% and 42.4%, respectively). C*07:01:02 and C*07:06 were also present, though at lower frequencies (8.2% and 4.7%, respectively). The other four non-CWD subtypes, i.e. C*07:01:09, C*07:52, C*07:153 and C*07:166, were not detected in this study.
Table 1. Relative frequencies of C*07:01:01G subtypes in C*07:01:01G-positive European and African Americans.
| European Americans (N = 89*) | African Americans (N = 85*) | |||
|---|---|---|---|---|
| Allele | n# | % | n# | % |
| C*07:01:01 | 84 | 94.4 | 38 | 44.7 |
| C*07:01:02 | 1 | 1.1 | 7 | 8.2 |
| C*07:06 | 0 | 0 | 4 | 4.7 |
| C*07:18 | 4 | 4.5 | 36 | 42.4 |
Total copy number of C*07:01:01G detected in 81 European Americans and 80 African Americans.
3.2. C*07:01:02 association with HLA-B*57:03 and B*49:01
A total of nine copies of C*07:01:02 (1 European American, 7 African Americans and 1 of unknown ethnicity) were observed in the all samples combined (i.e. both the randomly selected C*07:01:01G plus the selected B*44/C*07:01:01G group, n = 226). Six of them were found on B*57:03 haplotypes and the other two on B*49:01 haplotypes. The remaining 35 B*49:01-positive individuals all associated with C*07:01:01.
3.3. C*07:06 association with B*44:03
A panel of 76 individuals with both B*44 and C*07:01:01G (59 European Americans, 8 African Americans, 5 Hispanics, 2 Asians and 2 of unknown ethnicity) were examined to confirm the previously reported linkage relationship of C*07:06 with B*44 (Table 2). Three B*44 subtypes, B*44:02 (n = 27, including one homozygote), B*44:03 (n = 48), and B*44:10 (n = 1), along with all four C*07:01:01G subtypes were represented in the panel of 76 B*44 positive individuals. Among the 28 samples with B*44:02 or B*44:10, C*07:01:01 was the only C*07:01:01G subtype detected. However, only two of these 27 individuals carried the B*44:02-C*07:01:01G haplotype, whereas all the others carried B*44:02 and C*07:01:01G alleles on separate haplotypes. Among the 48 B*44:03-positive samples (36 European Americans, 6 African Americans, 2 Hispanics, 2 Asians and 2 of unknown ethnicity. most carried C*07:01:01 (n = 33; 69%), but C*07:01:02, C*07:06 and C*07:18 were also represented. Thirteen of the 48 samples actually carried the B*44:03-C*07:01:01G haplotype, 11 of which were C*07:06 and two of which were C*07:01:01. The remaining 35 individuals carried B*44:03 and C*07:01:01G on separate haplotypes. Thus, 100% of C*07:06 alleles were found on the B*44:03 positive haplotypes and 85% of C*07:01:01G alleles on the B*44:03 positive haplotypes were actually C*07:06. This linkage relationship between B*44:03 and C*07:06 was true in all ethnic groups examined.
Table 2. C*07:01:01G subtypes in B*44-positive individuals.
| C*07:01:01G subtype | B*44 | B*44:02 | B*44:03 | B*44:03-C*07:01:01G haplotype |
|---|---|---|---|---|
| N = 76 | N = 27 | N = 48 | N = 13 | |
| C*07:01:01 | 61 (80.2%) | 27 (100%) | 33 (68.8%) | 2 (15.4) |
| C*07:01:02 | 1 (1.3%) | 0 | 1 (2.2%) | 0 |
| C*07:06 | 11 (14.5%) | 0 | 11 (22.9%) | 11 (84.6%) |
| C*07:18 | 3 (4%) | 0 | 3 (6.3%) | 0 |
Total copy number of C*07:01:01G detected in 81 European Americans and 80 African Americans.
3.4. C*07:18 association with B*58:01
C*07:18 showed a dominant linkage relationship with B*58:01 in both European and African American groups. All four copies of C*07:18 detected in European Americans were associated with B*58:01 and they were the only four samples having both C*07:01:01G and B*58:01. Eighteen of the 33 copies (including one homozygote) of C*07:18 detected in African Americans were associated with B*58:01, which accounted for all but one of the B*58:01-C*07:01:01G haplotypes. The remaining 15 copies of C*07:18 were associated with B*57:03 (n = 8), B*07:02 (n = 3), B*08:01 (n = 2), B*35:01 (n = 1) and B*42:01 (n = 1).
4. Discussion
High-resolution HLA-C typing has important implications for bone marrow transplantation [17-19] and population studies [20, 21]. Routine HLA-C SBT has been limited to polymorphisms within exons 2 and 3 that encode the antigen-binding domains. Alleles indistinguishable by routine SBT due to synonymous or nonsynonymous substitutions outside of exons 2-3 have collectively been assigned the name of the lowest numbered allele of the group and tagged with a “G” (e.g. “C*07:01:01G”, (http://www.ebi.ac.uk/imgt/hla/pdf/ambiguity_v370.pdf). C*07:01:01, C*07:06 and C*07:18 are present on the NMDP list of CWD alleles, but their frequencies across different populations are largely unknown. There is growing interest in the potential significance of polymorphisms in the transmembrane segment, cytoplastic tail, 3′UTR, and 5′UTR of HLA class I, and full characterization of HLA-C alleles may provide new insights into HLA associations with certain diseases, as exemplified with 3′UTR variation in HLA-C with HIV disease [22]. Here, we have characterized the C*07:01:01G alleles based on sequence polymorphisms within exons 2 through 6 in order to resolve the relative frequency distribution of C*07:01:01G subtypes in European and African American subjects, along with their linkage relationships with HLA-B.
C*07:01:01G is the most common HLA-C allele in the NMDP database of mixed ethnicities [20]. The present study demonstrated that in European and African Americans combined, C*07:01:01G has considerable heterogeneity with four subtypes represented. C*07:01:01G is overwhelmingly dominated by C*07:01:01 in European Americans, accounting for 94% of the C*07:01:01G frequency. C*0706 was not detected in the group of 81 randomly selected C*07:01 positive European American individuals, indicating that it is not common in this population. However, it is not completely absent since it was detected in the panel of European Americans selected for both B*44 and C*07:01:01G, in which C*07:06 was enriched due to its linkage disequilibrium with B*44:03. A previous study involving 42 Dutch individuals with C*07:01:01G also found C*07:01:01 to be the dominant subtype with only one C*07:06-positive individual [23].
African Americans showed a much higher level of diversity in C*07:01:01G, with C*07:01:01 accounting for only about half of the grouping, the other half being shared between C*07:18, C*07:06 and C*07:01:02. In particular, C*07:18 was nearly as frequent as C*07:01:01, which is in accordance with a previous study in a Ugandan population, where C*07:18 was found to be the second most common C*07:01:01G allele (about half of the frequency of C*07:01:01) [12]. C*07:06 is a rare allele in both European and African Americans, but in Chinese this allele is even more common than C*07:01:01, although the overall frequency of C*07:01:01G in Chinese is much lower than in Europeans and Africans [6]. Both the present results and previous studies strongly argue that C*07:01:01G detected by routine SBT cannot be assumed to be C*07:01:01. Overall, the data indicate that high-resolution typing protocols should include C*07:01:01G subtyping for C*07:01:01G-positive samples, particularly for individuals of African and Asian origins.
Because of the short physical distance separating HLA-B and -C in the MHC, the alleles of the two loci generally exhibit strong linkage disequilibrium. C*07, as one of the most common HLA-C allelic groups, is known to have multiple linkage relationships with B alleles and the extended haplotypes. C*07:01:01G subtypes detected both in the present and previous studies show clear and distinct linkage relationships with HLA-B alleles, though the HLA-B/C haplotypes examined in this study were elucidated from population data instead of family analysis. Several of the previously described haplotypes of HLA-B and C*07:01:01G can now be attributed to different C*07:01:01G subtypes. For example, most of the B*44:03 association with C*07:01:01G can now be attributed to C*07:06. Similarly, almost all of the B*58:01 association with C*07:01:01G in both European and African Americans can be attributed to C*07:18. The linkage relationship reflects the evolutionary history of C*07:01:01G subtypes. C*07:06 was apparently generated on a B*44:03 haplotype with a relatively short history since it still maintains the single linkage relationship with HLA-B*44:03. C*07:18 was probably generated on a B*58:01 haplotype. Given its relatively high frequency in African populations and multiple linkage relationships with HLA-B, C*07:18 is likely to have had a fairly long existence. Knowledge of the linkage relationship may help to identify the C*07:01:01G-positive individuals who are likely to have a non-C*07:01:01 subtype in retrospective inspection of old data. For instance, on the basis of present results, 85% of B*44:03 linked C*07:01:01G will be C*07:06 and the chance of detecting C*07:06 on haplotypes linked with other HLA-B alleles (including B*44:02) is small. Similarly almost all (95%) B*58:01-linked C*07:01:01G will be C*07:18, though about 45% of C*07:18 detected in African Americans could be linked with other HLA-B alleles.
Traditionally the focus of HLA typing is on the PBR, but in recent years the functional significance of genetic polymorphisms outside of the PBR has drawn much interest. Examples include the effect of the sequence variation in regions coding for the transmembrane segment, cytoplasmic tail and the 3′-UTR on HLA expression and cytoplasmic signal transduction. Such variation that has an influence on function of the class I molecule may directly impact disease pathogenesis [2-4, 22]. C*07:01:01G is a common allele grouping with indistinguishable subtypes by routine HLA typing. The four C*07:01:01G subtypes, in particular the three non-synonymous alleles defined by variations outside of exons 2 and 3, were each detected as a dominant C*07:01:01G subtype in certain populations (i.e. C*07:01:01 in Europeans, C*07:06 in Asians and C*07:18 along with C*07:01:01 in Africans). If indeed these subtypes confer functional differences, their characterization may provide further insights into the effect of HLA matching for clinical transplantation and HLA association with human disease.
Acknowledgements
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This Research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. This study was supported by the research fund of Guangdong Science & Technology Department (Research Project number: 2008B030301277).
ABBREVIATIONS
- HLA
human leukocyte antigen
- ASHI
American Society for Histocompatibility and Immunogenetics
- CWD
common and well-documented
- NMDP
National Marrow Donor Program
- SBT
Sequence based typing
- PBR
peptide binding region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Robinson J, et al. IMGT/HLA database--a sequence database for the human major histocompatibility complex. Tissue Antigens. 2000;55(3):280–7. doi: 10.1034/j.1399-0039.2000.550314.x. [DOI] [PubMed] [Google Scholar]
- 2.Skov S, et al. Activation of Stat-3 is involved in the induction of apoptosis after ligation of major histocompatibility complex class I molecules on human Jurkat T cells. Blood. 1998;91(10):3566–73. [PubMed] [Google Scholar]
- 3.Skov S. Intracellular signal transduction mediated by ligation of MHC class I molecules. Tissue Antigens. 1998;51(3):215–23. [PubMed] [Google Scholar]
- 4.Delfino L, Morabito A, Ferrara GB. HLA-C sequence based typing: nucleotide analysis from exon 1 through exon 8. Identification of a new allele: Cw*0718. Tissue Antigens. 2003;62(5):418–25. doi: 10.1034/j.1399-0039.2003.00110.x. [DOI] [PubMed] [Google Scholar]
- 5.Vilches C, et al. Molecular cloning of two new HLA-C alleles: Cw*1801 and Cw*0706. Tissue Antigens. 1996;48(6):698–702. doi: 10.1111/j.1399-0039.1996.tb02694.x. [DOI] [PubMed] [Google Scholar]
- 6.Deng Z, et al. HLA-C polymorphisms and PCR dropout in exons 2 and 3 of the Cw*0706 allele in sequence-based typing for unrelated Chinese marrow donors. Hum Immunol. 2010;71(6):577–81. doi: 10.1016/j.humimm.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 7.Kazeem GR, et al. High-resolution HLA genotyping and severe cutaneous adverse reactions in lamotrigine-treated patients. Pharmacogenet Genomics. 2009;19(9):661–5. doi: 10.1097/FPC.0b013e32832c347d. [DOI] [PubMed] [Google Scholar]
- 8.Maiers M, Gragert L, Klitz W. High-resolution HLA alleles and haplotypes in the United States population. Hum Immunol. 2007;68(9):779–88. doi: 10.1016/j.humimm.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 9.Hong W, et al. Distributions of HLA class I alleles and haplotypes in Northern Han Chinese. Tissue Antigens. 2005;66(4):297–304. doi: 10.1111/j.1399-0039.2005.00474.x. [DOI] [PubMed] [Google Scholar]
- 10.Cao K, et al. Analysis of the frequencies of HLA-A, B, and C alleles and haplotypes in the five major ethnic groups of the United States reveals high levels of diversity in these loci and contrasting distribution patterns in these populations. Hum Immunol. 2001;62(9):1009–30. doi: 10.1016/s0198-8859(01)00298-1. [DOI] [PubMed] [Google Scholar]
- 11.Cano P, et al. Common and well-documented HLA alleles: report of the Ad-Hoc committee of the american society for histocompatiblity and immunogenetics. Hum Immunol. 2007;68(5):392–417. doi: 10.1016/j.humimm.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 12.Kijak GH, et al. HLA class I allele and haplotype diversity in Ugandans supports the presence of a major east African genetic cluster. Tissue Antigens. 2009;73(3):262–9. doi: 10.1111/j.1399-0039.2008.01192.x. [DOI] [PubMed] [Google Scholar]
- 13.van der Vlies SA, et al. Strong association between HLA-Cw*0706 and HLA-B*44032 in the Bubi population from Equatorial Guinea. Tissue Antigens. 2000;55(1):57–60. doi: 10.1034/j.1399-0039.2000.550110.x. [DOI] [PubMed] [Google Scholar]
- 14.Kiepiela P, et al. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432(7018):769–75. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- 15.Gao X, et al. AIDS restriction HLA allotypes target distinct intervals of HIV-1 pathogenesis. Nat Med. 2005;11(12):1290–2. doi: 10.1038/nm1333. [DOI] [PubMed] [Google Scholar]
- 16.Zeng JQ, et al. An analysis of the reason for HLA-C allele dropout in five samples by sequence-based typing. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2009;26(5):562–6. doi: 10.3760/cma.j.issn.1003-9406.2009.05.019. [DOI] [PubMed] [Google Scholar]
- 17.Flomenberg N, et al. Impact of HLA class I and class II high-resolution matching on outcomes of unrelated donor bone marrow transplantation: HLA-C mismatching is associated with a strong adverse effect on transplantation outcome. Blood. 2004;104(7):1923–30. doi: 10.1182/blood-2004-03-0803. [DOI] [PubMed] [Google Scholar]
- 18.Petersdorf EW, et al. Limits of HLA mismatching in unrelated hematopoietic cell transplantation. Blood. 2004;104(9):2976–80. doi: 10.1182/blood-2004-04-1674. [DOI] [PubMed] [Google Scholar]
- 19.Lee SJ, et al. High-resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood. 2007;110(13):4576–83. doi: 10.1182/blood-2007-06-097386. [DOI] [PubMed] [Google Scholar]
- 20.Turner S, et al. Sequence-based typing provides a new look at HLA-C diversity. J Immunol. 1998;161(3):1406–13. [PubMed] [Google Scholar]
- 21.Lebedeva TV, et al. Emerging new alleles suggest high diversity of HLA-C locus. Tissue Antigens. 2005;65(1):101–6. doi: 10.1111/j.1399-0039.2005.00333.x. [DOI] [PubMed] [Google Scholar]
- 22.Kulkarni S, et al. Differential microRNA regulation of HLA-C expression and its association with HIV control. Nature. 2011;472(7344):495–8. doi: 10.1038/nature09914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Vlies SA, Voorter CE, van den Berg-Loonen EM. A reliable and efficient high resolution typing method for HLA-C using sequence-based typing. Tissue Antigens. 1998;52(6):558–68. doi: 10.1111/j.1399-0039.1998.tb03087.x. [DOI] [PubMed] [Google Scholar]