Abstract
Multiple human breast and rat mammary carcinoma susceptibility (Mcs) alleles have been identified. Wistar Kyoto (WKY) rats are resistant to developing mammary carcinomas, while Wistar Furth (WF) females are susceptible. Gene transcripts at Mcs5a1, Mcs5a2, and Mcs5c are differentially expressed between resistant WKY and susceptible WF alleles in immune-system tissues. We hypothesized that immune-related gene transcript profiles are genetically determined in mammary carcinoma resistant and susceptible mammary glands. Low-density QPCR arrays were used to compare inflammation related genes between mammary carcinoma resistant WKY and susceptible WF females. Mammary gland gene transcript levels predicted to be different based on arrays were tested in independent samples. In total, twenty females per strain were exposed to 7,12-dimethylbenz(a)anthracene (DMBA) to induce mammary carcinogenesis. Twelve age-matched controls per strain without DMBA were included to determine main effects of DMBA-exposure. Significant (ANOVA P ≤ 0.01) effects of strain on mammary gland transcript level were observed for Cx3cl1, Il11ra, Il4, C3, Ccl20, Ccl11, Itgb2, Cxcl12, and Cxcr7. Significant effects of DMBA-exposure were observed for Cx3cl1, Il11ra, Cxcr4, Il4ra, and Il4. Strain and DMBA-exposure interaction effects were significant for Cx3cl1. Transcript levels of Cxcr7 relative to Cxcr4 were modified differently by DMBA in mammary carcinoma resistant and susceptible strains. In conclusion, several genetically-determined differences in cytokine, chemokine, and receptor gene transcript levels were identified between mammary carcinoma susceptible and resistant mammary glands, which may be indicative of cell populations and activities that suppress mammary carcinogenesis in resistant genotypes.
Keywords: breast cancer susceptibility, rat mammary carcinogenesis, inflammation, Ccl20, Cxcr7
Human breast and rat mammary cancer susceptibility are complex phenotypes. A majority of breast cancer risk associated low penetrance polymorphisms are located in non-coding DNA [1], suggesting genetic variation in gene-regulatory mechanisms is important for determining susceptibility. Mammary carcinoma resistant Wistar Kyoto (WKY) and susceptible Wistar Furth (WF) rat strains have been used to identify rat mammary carcinoma susceptibility (Mcs) loci [2]. Some rat Mcs loci are known to be concordant orthologs of human breast cancer risk associated or potentially associated alleles [3, 4]. F-box protein 10 (Fbxo10), FERM and PDZ domain containing 1 (Frmpd1), and tenascin c (Tnc), which are respectively located at rat Mcs5a1, Mcs5a2, and Mcs5c, are differentially expressed between susceptible and resistant rat alleles in immune cells and tissues [3, 5, 6].
Biological differences between breast cancer susceptibility genotypes that exist prior to the formation of palpable tumors are important to identify. Additionally, mechanisms identified in cancer resistant rats may be good targets for cancer prevention in humans. Inflammatory responses play decisive roles at different stages of tumor development, including initiation, promotion, malignant conversion, invasion, and metastasis [7]. Immune-mediated inflammatory cells, cytokines, and chemokines are associated with breast cancer risk, prognosis, and metastatsis [8]. We hypothesized that additional immune system genes may be differentially expressed between mammary cancer resistant WKY and susceptible WF rats. We report and compare genetically determined mammary gland transcript-level differences in inflammatory cytokines, chemokines, and receptors between these genotypes.
We used low-density QPCR arrays that contained eighty-four rat inflammatory cytokine and chemokine genes followed by validation in independent samples. One QPCR array per rat sample (N=16 arrays) was used to compare transcript levels between cancer susceptible (n = 8 WF) and resistant (n = 8 WKY) females that were twelve weeks of age and had received a single oral dose of DMBA to induce mammary carcinogenesis. This age was chosen because it is after acute DMBA toxicity and before frank carcinomas are detectable by palpation in cancer susceptible WF mammary glands. Inflammatory cytokines, chemokines, and receptors that were differentially expressed (P < 0.05) between these strains in this experiment were: Cx3cl1, Il4, C3, Cxcl12/Sdf1, and Cxcl4/Pf4. Those approaching significant differential expression (P > 0.05 and < 0.25) between resistant and susceptible mammary glands were: Ccl20, Ccl11/eotaxin, Itgb2/Cd18, Il11, Il8rb, and Cxcl1. Complete gene names are listed in Table S1.
Further testing was completed to determine which predictions between strains were valid, and to compare gene transcript levels between mammary carcinogen induced (DMBA) and age-matched unexposed mammary glands. Receptors for chemokine Cxcl12 (Cxcr4, Cxcr7) and cytokines Il11 (Il11ra) and Il4 (Il4ra) were not present on QPCR arrays used; therefore, transcript levels of these receptors were tested in this experiment. Receptors of other cytokines and chemokines that data suggested to be different were present on QPCR arrays, and were not significantly different between DMBA-exposed susceptible and resistant rat mammary glands.
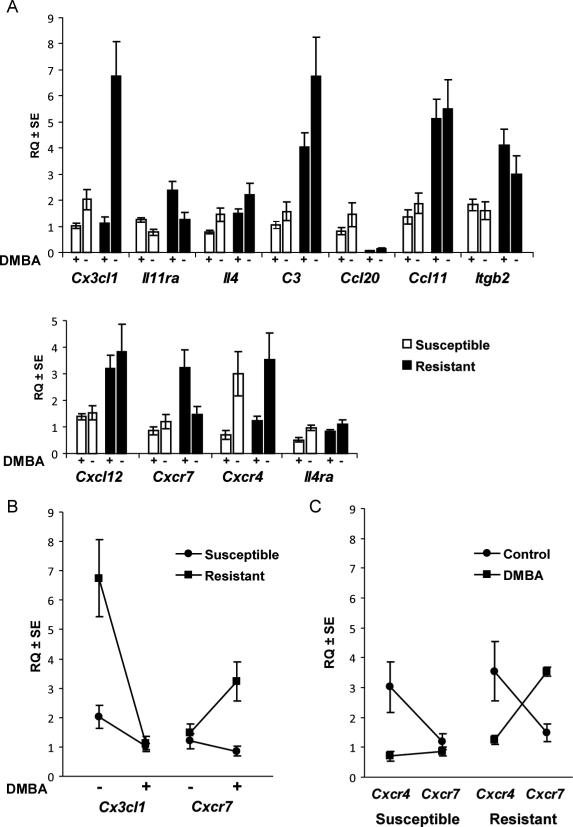
We used mammary gland total RNA collected from a second group of twelve-week old female rats that were administered DMBA independently of females used in our QPCR array experiment. Mammary carcinoma susceptible (n = 24 WF) and resistant (n = 24 WKY) females were randomly assigned to two exposure groups, with DMBA (n = 24 females) and without (n = 24 females). Samples from DMBA-exposed females used on low-density QPCR arrays were also tested with primers designed for this experiment and statistically analyzed with samples from this second group of females. Quantitative PCR results shown in Figure 1A were analyzed by two-way ANOVA for independent effects of strain and DMBA-exposure, and an interaction effect (Table 1). Cytokines, chemokines, and receptors with a statistically significant effect of strain were Cx3cl1 (P < 0.0001), Il11ra (P = 0.0023), Il4 (P = 0.0025), C3 (P < 0.0001), Ccl20 (P < 0.0001), Ccl11/eotaxin (P < 0.0001), Itgb2/Cd18 (P = 0.0018), Cxcl12/Sdf1 (P = 0.0004), and Cxcr7 (P = 0.0112). With exception to Ccl20, transcripts that were significantly different were 1.7 to 4-fold higher in cancer resistant compared to susceptible mammary glands. Chemokine Ccl20 transcript levels were 11.5-fold higher in mammary glands from susceptible compared to resistant females.
Figure 1. Inflammatory cytokine, chemokine, and receptor transcript levels in mammary carcinoma susceptible WF and resistant WKY mammary glands with DMBA and without.
Graphed are means ± SEs of relative quantification (RQ) of genes indicated on x-axes. Rplp2 levels were used to standardize targets. Twenty females per strain were given DMBA, and twelve aged matched controls per strain were included. A, inflammatory cytokine, chemokine, and receptor mammary gland transcript levels in twelve week old mammary carcinoma susceptible WF (unfilled bars) and resistant WKY (filled bars) females with DMBA [+] or without [-]. B, strain by DMBA-exposure interactions were statistically significant for Cx3cl1 (P < 0.0001) and potentially significant for Cxcr7 (P = 0.0435). Compared to mammary cancer susceptible WF mammary glands (circles), Cx3cl1 expression was significantly higher in mammary glands from resistant WKY rats (squares) not exposed to DMBA (P = 0.0034). There was no difference in Cx3cl1 transcript level between strains in mammary carcinogen induced (DMBA +) females (P = 0.7483). Cxcr7 levels were significantly higher in mammary cancer resistant WKY rats (squares) compared susceptible WF females (circles) with DMBA (P = 0.0027), but not without (P = 0.4966). C, mammary gland Cxcr4 and Cxcr7 expression in DMBA-exposed (squares) and un-exposed (circles) females compared between mammary carcinoma resistant WKY and susceptible WF rat strains. Cxcr4 expression was higher than Cxcr7 in both strains (P = 0.0367 [WF] and 0.0606 [WKy]) when control females not exposed to mammary carcinogen were evaluated. Cxcr7 levels were significantly higher than Cxcr4 levels in the mammary cancer resistant strain (WKY) following DMBA exposure (P = 0.0059). Cxcr7 and Cxcr4 transcript levels were similar in mammary cancer susceptible WF females following DMBA exposure (P = 0.4673).
Table 1.
ANOVA Summary and Fold Differences in Mean Cytokine, Chemokine, and Receptor Transcript Levels Between Mammary Carcinoma Resistant WKY and Susceptible WF Rat Strains With DMBA and Age-Matched Controls Without
| Gene Transcript | ANOVA P-Value* | Transcript Level Fold-Difference (Resistant/Susceptible) | Transcript Level Fold-Difference (DMBA/Control) | ||||
|---|---|---|---|---|---|---|---|
| Strain (S) | Exposure (E) | G × E | DMBA | Control | Combined | ||
| Cx3cl1 | <0.0001 | <0.0001 | <0.0001 | 1.09 | 3.33 | 2.28 | 0.24 |
| Il11ra | 0.0023 | 0.0024 | 0.2053 | 1.90 | 1.62 | 1.84 | 1.82 |
| Il4 | 0.0025 | 0.0036 | 0.9258 | 1.91 | 1.52 | 1.73 | 0.61 |
| C3 | <0.0001 | 0.0238 | 0.1204 | 3.84 | 4.30 | 4.06 | 0.60 |
| Ccl20 | <0.0001 | 0.0557 | 0.1182 | 0.08 | 0.09 | 0.09 | 0.57 |
| Ccl11/eotaxin | <0.0001 | 0.5322 | 0.9123 | 3.77 | 2.92 | 3.41 | 0.87 |
| Itgb2/Cd18 | 0.0018 | 0.2305 | 0.4324 | 2.24 | 1.86 | 2.11 | 1.29 |
| Cxcl12/sdf1 | 0.0004 | 0.4826 | 0.6529 | 2.29 | 2.49 | 2.37 | 0.85 |
| Cxcr7 | 0.0112 | 0.1688 | 0.0435 | 3.76 | 1.23 | 2.62 | 1.59 |
| Cxcr4 | 0.2897 | <0.0001 | 0.9889 | 1.78 | 1.18 | 1.46 | 0.29 |
| Il4ra | 0.0573 | 0.0026 | 0.4653 | 1.61 | 1.15 | 1.42 | 0.64 |
| Cxcl4/Pf4 | 0.0552 | 0.3317 | 0.2880 | 1.74 | 1.21 | 1.55 | 1.21 |
| Il11 | 0.0617 | 0.7137 | 0.7956 | 1.29 | 1.22 | 1.26 | 1.04 |
| Il8rb | 0.7704 | 0.0137 | 0.5604 | 1.04 | 0.91 | 0.99 | 0.73 |
| Cxcl1 | 0.1177 | 0.0354 | 0.1226 | 0.99 | 0.41 | 0.62 | 0.43 |
DMBA: 7,12-Dimethylbenz(a)anthracene given at 50-55 days of age; QPCR was used to compare transcript levels between rat mammary glands from 12 week old mammary carcinoma resistant (WKY) and susceptible (WF) virgin females with DMBA and controls without DMBA
P-Values from two-way ANOVA of gene transcript level by DMBA exposure (with or without), mammary carcinoma susceptibility genotype (resistant or susceptible), and interaction (S × E).
Cytokines, chemokines, and receptors with a significant effect of DMBA-exposure (with or without) on mammary gland transcript level were Cx3cl1 (P < 0.0001), Il11ra (P = 0.0024), Il4 (P = 0.0036), Cxcr4 (P < 0.0001), Il4ra (P = 0.0026). Transcript level of Il11ra was 1.8 fold higher with DMBA compared to without. All other transcripts with a significant exposure effect were decreased 24-64% with DMBA compared to without.
A strain by DMBA-exposure interaction was statistically significant for Cx3cl1 (P < 0.0001; Table 1). Mammary gland Cx3cl1 levels between resistant and susceptible females shown in Figure 1B were 3.3-fold higher in mammary carcinoma resistant females without DMBA (P = 0.0034), but were not different between strains with DMBA (P = 0.7483). The source of a potential Cxcr7 strain by exposure interaction (P = 0.0435) was a Cxcr7 transcript level increase (Figure 1B) within resistant mammary glands exposed to DMBA (P = 0.0328, one-tailed t-test), and an observed decrease in susceptible mammary glands (P = 0.1238, one-tailed t-test).
CXCR4 and CXCR7 are both G-protein coupled receptors of CXCL12. It seemed from our data that potential complexity existed between mammary cancer susceptible and resistant genotypes for these two receptors. We completed a posteriori tests to determine if interactions existed between these two genes. Mammary gland Cxcr4 and Cxcr7 transcript level was used as the dependent variable in two-way ANOVA. Gene (P = 0.0137), DMBA-exposure (P = 0.0001), and the interaction (P = 0.0036) were statistically significant when mammary cancer susceptible WF females were analyzed. The gene by exposure interaction only was significant (P = 0.0013) when mammary cancer resistant Cxcr4 and Cxcr7 transcript levels were used as the dependent variable. As displayed in Figure 1C, levels of Cxcr7 relative to Cxcr4 were lower in both susceptible (P = 0.0367) and resistant (P = 0.0606) mammary glands that had not been exposed to DMBA; however, four weeks after DMBA exposure Cxcr7 levels were higher than Cxcr4 levels in resistant mammary glands (P = 0.0059). This was due to a drop in Cxcr4 and an increase in Cxcr7 in resistant mammary glands following carcinogen exposure. As in the resistant strain, levels of Cxcr4 dropped in susceptible mammary glands following DMBA exposure, but Cxcr7 levels did not increase relative to Cxcr4 levels (P = 0.4673).
In summary, transcript levels of Cx3cl1, Il11ra, Il4, C3, Ccl11, Itgb2, Cxcl12, and, Cxcr7 were higher, and Ccl20 levels were lower in mammary carcinoma resistant compared to susceptible mammary glands. These genetically determined differences in cytokine, chemokine, and receptor profiles between susceptible and resistant genotypes are anticipated to lead to differences in mammary gland stromal constitutes and activities. Further, these differences may be important for a cancer resistant mammary gland to launch an immediate response to carcinogenesis. Cytokine and chemokine transcript profiles between mammary carcinoma resistant and susceptible mammary glands suggest that lymphocytes, eosinophils, neutrophils, denditric, and NK cells may be contributing to susceptibility differences between these strains. In support of our results, Smits et al. reported that genetically determined mammary gland γδT cell population differences exist between mammary carcinoma resistant WF.WKY-Mcs5a congenic and susceptible females [9]. Chemokine CX3CL1/fractalkine is produced by a variety of cells, recruits Th effector cells, and promotes the adhesion of monocytes to endothelial cells [10]. Chemokine CCL20 is expressed in cytokeratin positive cells and potentially recruits dendritic cell precursors [11]. Depending on subtype, dendritic cells may inhibit or promote tumorigenesis [12].
Chemokine CCL11/eotaxin attracts eosinophils, which are important for proper mammary gland development as Ccl11-/- mice have deficiencies in mammary gland branching and terminal end bud formation [13]. Blomqvist and colleagues reported that eosinophils were not detected in any of over 200 hematoxylin-eosin stained invasive breast cancer tissues [14]. Another link to a potential for eosinophil mediated cancer resistance is Il4, a pleiotropic, anti-inflammatory, and anti-tumorigenic cytokine produced by Th2 CD4+ T cells, eosinophils, basophils, mast cells, and NKT cells. Wu et al. discovered that eosinophils are the major Il4 producing cells in mouse white adipose tissue [15]. Another potential source of increased Il4 transcript levels in WKY cancer resistant mammary glands may be Il4 producing γδT cells, which are at higher levels in WF.WKY-Mcs5a compared to cancer susceptible mammary glands [9]. Importantly, IL4 may also promote tumorigenesis depending on the source, dose, and window of expression of IL4 [16].
To date, none of the genes identified in this study have been determined to be rat mammary carcinoma susceptibility genes. The WKY rat strain used in this study is completely resistant to developing mammary carcinomas; however, it does harbor increased susceptibility alleles that include Mcs5b and Mcs8 [2, 17]. Therefore, additional work will be required to determine which transcripts represent a favorable condition when at a higher level and which might be unfavorable. Studies will also be required to determine if these genes segregate with mammary carcinoma susceptibility. Interestingly, none of the genetically determined transcript profiles identified in this study are located within known WKY resistant-rat Mcs QTLs (Figure S1). This suggests that if these gene transcripts do contribute to mammary cancer susceptibility, either they are controlled in trans by known Mcs QTLs or unknown WKY Mcs QTLs exist. It is noteworthy that even though Cxcl1 and Cxcl4 did not meet our criterion for statistical significance, these transcripts are located within Mcs8. Further investigation of Cxcl1 and Cxcl4 is warranted as subtle transcript level differences have been associated with Mcs5a1, Mcs5a2, and Mcs5c alleles [3, 5]. Interleukin 11ra is located within rat Estrogen induced mammary cancer-8, a QTL identified using the ACI rat strain, which is susceptible to 17β-estradiol induced mammary cancer [18].
Human orthologous loci containing these genes are good candidates to test in genome-targeted breast cancer risk association studies. Epidemiological evidence indicates that C3, an estrogen responsive gene, is associated with increased breast cancer risk in nulliparous compared to parous women [19]. Genetic variation at ITGβ2/CD18 and CXCL12 loci has shown association or potential-association with breast cancer susceptibility [20, 21]. Breast cancer cells that express ITGβ2 are recognized and killed my natural killer cells [22]. Chemokine gene CXCL12 is estrogen regulated and the gene product attracts naïve T cells and monocytes [23].
In our study transcript levels of Cx3cl1, Il11ra, Il4, Cxcr4, and Il4ra were significantly different between DMBA-exposed and unexposed mammary glands. These differences are suggestive of responses to early stages of mammary carcinogenesis, as mammary gland transcript levels were evaluated before frank carcinomas are detectable in mammary carcinoma susceptible rats.
In conclusion, we identified multiple inflammation-related cytokines, chemokines, and receptors with different transcript levels between mammary carcinoma susceptible and resistant mammary glands. We also uncovered an interaction between Cxcr4 and Cxcr7 transcript levels in a carcinoma resistant mammary gland. A majority of differentially expressed gene transcripts identified (all except Ccl20) were higher in resistant compared to susceptible mammary glands, suggesting that mammary cancer resistance is an active process. Although some gene transcript levels decreased in response to DMBA they remained higher in resistant compared to susceptible genotypes. Our results support a hypothesis that genetically determined differences in immune-cell populations or activities exist within mammary glands in response to events related to carcinogenesis.
Materials and Methods
Mammary carcinoma susceptible Wistar Furth (WF/NHsd) and resistant Wistar Kyoto (WKY/NHsd) virgin female rats were purchased from Harlan (Indianapolis, IN). Procedures involving rodents were carried out in accordance to protocols approved by the University of Louisville Animal Care and Use Committee. Rats were housed in an AAALAC approved facility with a 12 hour light/dark cycle and ad libitum food/water. To induce mammary carcinogenesis, 7,12-Dimethylbenz(a)anthracene (DMBA) was dissolved in sesame oil at 20 mg/mL and 65 mg DMBA/KG body mass was administered orally by gavage between 50-55 days of age.
Right cranial inguinal mammary glands with lymph nodes removed were excised four weeks following DMBA administration, flash frozen in liquid nitrogen, and stored at -80° C. Total RNA was isolated from mammary gland tissue homogenates using TRI reagent (Molecular Research Center), potential genomic DNA contamination was reduced using TURBO DNase (Life Technologies), and one g RNA was used to generate cDNA using SuperScript III reverse transcriptase (Life Technologies). One rat inflammatory cytokine QPCR array (SABiosciences PN PARN011, Qiagen) per sample was used to screen 84 genes involved in mediating an inflammatory response. Validation by SYBR-green (Life Technologies) QPCR was completed using primers designed with Primer-3 [24] and independent groups of WF and WKY rats with DMBA and without. Primers used for QPCR are provided in Table S2. Relative quantification (RQ) values were used to compare transcript levels. RQ is the expression level of a nucleic acid sequence (target) in an experimental sample relative to a calibrator sample. Rat Rplp2 was used as an endogenous control to standardize target levels.
Statistical significance level of each low-density array comparison was determined using a Student's t-test and SA Biosciences software (Qiagen). Statistical analysis of data to validate predictions based on low-density arrays was performed by two-way ANOVA. Here RQ of gene transcript level was the dependent variable. Strain (WKY, WF) and DMBA-exposure (with, without) were independent variables. Two-way ANOVA was also used to test for an interaction between Cxcr4 and Cxcr7 gene transcript levels. RQ of gene transcript level was the dependent variable. Gene (Cxcr4, Cxcr7) and DMBA-exposure (with, without) were independent variables. To reduce potential for false-positives a p value ≤0.01 was considered statistically significant. Student's t-tests were used for a posteriori comparisons of transcript levels between two groups. Unless denoted otherwise, significance level probabilities are two-tailed.
Supplementary Material
Highlights.
We compared inflammatory genes in a rat genetic model of human breast cancer.
Most differentially expressed transcripts were higher in resistant mammary glands.
Transcripts different in carcinogen exposed/un-exposed females were identified.
Mammary Cxcr7 was higher relative to Cxcr4 in carcinogen exposed resistant females.
Acknowledgments
The authors thank Aaron Puckett for his technical assistance with rodent procedures. This work was supported by the National Institutes of Health grants CA137052, ES014443, and RR024489. J.S. was supported by the University of Louisville Integrated Program in Biomedical Sciences. Sponsors had no role in this research beyond funding.
Abbreviations
- DMBA
7,12-dimethylbenz(a)anthracene
- Mcs
mammary carcinoma susceptibility
- RQ
relative quantification
- WKY
Wistar Kyoto
- WF
Wistar Furth
- QPCR
quantitative polymerase chain reaction
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hindorff LA, Gillanders EM, Manolio TA. Genetic architecture of cancer and other complex diseases: lessons learned and future directions. Carcinogenesis. 2011;32:945–954. doi: 10.1093/carcin/bgr056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lan H, Kendziorski CM, Haag JD, Shepel LA, Newton MA, Gould MN. Genetic loci controlling breast cancer susceptibility in the Wistar-Kyoto rat. Genetics. 2001;157:331–9. doi: 10.1093/genetics/157.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Samuelson DJ, Hesselson SE, Aperavich BA, Zan Y, Haag JD, Trentham-Dietz A, et al. Rat Mcs5a is a compound quantitative trait locus with orthologous human loci that associate with breast cancer risk. Proc Natl Acad Sci U S A. 2007;104:6299–304. doi: 10.1073/pnas.0701687104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanders J, Haag JD, Samuelson DJ. Physical Confirmation and Mapping of Overlapping Rat Mammary Carcinoma Susceptibility QTLs, Mcs2 and Mcs6. PLoS ONE. 2011;6:e19891. doi: 10.1371/journal.pone.0019891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Veillet AL, Haag JD, Remfert JL, Meilahn AL, Samuelson DJ, Gould MN. Mcs5c: A Mammary Carcinoma Susceptibility Locus Located in a Gene Desert that Associates with Tenascin C Expression. Cancer Prevention Research. 2011;4:97–106. doi: 10.1158/1940-6207.CAPR-10-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smits BMG, Traun BD, Devries TL, Tran A, Samuelson D, Haag JD, et al. An insulator loop resides between the synthetically interacting elements of the human/rat conserved breast cancer susceptibility locus MCS5A/Mcs5a. Nucleic Acids Research. 2012;40:132–47. doi: 10.1093/nar/gkr610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–99. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Standish LJ, Sweet ES, Novack J, Wenner CA, Bridge C, Nelson A, et al. Breast cancer and the immune system. J Soc Integr Oncol. 2008;6:158–68. [PMC free article] [PubMed] [Google Scholar]
- 9.Smits BM, Sharma D, Samuelson DJ, et al. The non-protein coding breast cancer susceptibility locus Mcs5a acts in a non-mammary cell-autonomous fashion through the immune system and modulates T-cell homeostasis and functions. Breast Cancer Res. 2011;13:R81. doi: 10.1186/bcr2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bazan JF, Bacon KB, Hardiman G, et al. A new class of membrane-bound chemokine with a CX3C motif. Nature. 1997;385:640–44. doi: 10.1038/385640a0. [DOI] [PubMed] [Google Scholar]
- 11.Thomachot MC, Bendriss-Vermare N, Massacrier C, et al. Breast carcinoma cells promote the differentiation of CD34+ progenitors towards 2 different subpopulations of dendritic cells with CD1a(high)CD86(-)Langerin- and CD1a(+)CD86(+)Langerin+ phenotypes. Int J Cancer. 2004;110:710–20. doi: 10.1002/ijc.20146. [DOI] [PubMed] [Google Scholar]
- 12.Kees T, Egeblad M. Innate immune cells in breast cancer. J Mammary Gland Biology and Neoplasia. 2011;16:189–203. doi: 10.1007/s10911-011-9224-2. [DOI] [PubMed] [Google Scholar]
- 13.Gouon-Evans V, Rothenberg ME, Pollard JW. Postnatal mammary gland development requires macrophages and eosinophils. Development. 2000;127:2269–82. doi: 10.1242/dev.127.11.2269. [DOI] [PubMed] [Google Scholar]
- 14.Amini RM, Aaltonen K, Nevanlinna H, et al. Mast cells and eosinophils in invasive breast carcinoma. BMC Cancer. 2007;7:165. doi: 10.1186/1471-2407-7-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu D, Molofsky AB, Liang HE, et al. Eosinophils sustain adipose alternatively activated macrophages associated with glucose homeostasis. Science. 2011;332:243–7. doi: 10.1126/science.1201475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z, Chen L, Qin Z. Paradoxical Roles of IL-4 in Tumor Immunity. Cellular & Molecular Immunology. 2009;6:415–22. doi: 10.1038/cmi.2009.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samuelson DJ, Aperavich BA, Haag JD, Gould MN. Fine mapping reveals multiple loci and a possible epistatic interaction within the mammary carcinoma susceptibility quantitative trait locus, Mcs5. Cancer Res. 2005;65:9637–42. doi: 10.1158/0008-5472.CAN-05-1498. [DOI] [PubMed] [Google Scholar]
- 18.Schaffer BS, Lachel CM, Pennington KL, Murrin CR, Strecker TE, Tochacek M, et al. Genetic bases of estrogen-induced tumorigenesis in the rat: mapping of loci controlling susceptibility to mammary cancer in a Brown Norway x ACI intercross. Cancer Res. 2006;66:7793–800. doi: 10.1158/0008-5472.CAN-06-0143. [DOI] [PubMed] [Google Scholar]
- 19.Balogh GA, Russo IH, Spittle C, Heulings R, Russo J. Immune-surveillance and programmed cell death-related genes are significantly overexpressed in the normal breast epithelium of postmenopausal parous women. Int J Oncol. 2007;31:303–12. doi: 10.3892/ijo.31.2.303. [DOI] [PubMed] [Google Scholar]
- 20.Lee JY, Park AK, Lee KM, Park SK, Han S, Han W, et al. Candidate gene approach evaluates association between innate immunity genes and breast cancer risk in Korean women. Carcinogenesis. 2009;30:1528–31. doi: 10.1093/carcin/bgp084. [DOI] [PubMed] [Google Scholar]
- 21.Shen W, Cao X, Xi L, Deng L. CXCL12 G801A polymorphism and breast cancer risk: a meta-analysis. Mol Biol Rep. 2012;39:2039–44. doi: 10.1007/s11033-011-0951-7. [DOI] [PubMed] [Google Scholar]
- 22.Cooley S, Burns LJ, Repka T, Miller JS. Natural killer cell cytotoxicity of breast cancer targets is enhanced by two distinct mechanisms of antibody-dependent cellular cytotoxicity against LFA-3 and HER2/neu. Exp Hematol. 1999;27:1533–41. doi: 10.1016/s0301-472x(99)00089-2. [DOI] [PubMed] [Google Scholar]
- 23.Tsutsumi A, Okada H, Nakamoto T, Okamoto R, Yasuda K, et al. Estrogen induces stromal cell-derived factor 1 (SDF-1/CXCL12) production in human endometrial stromal cells: a possible role of endometrial epithelial cell growth. Fertility and Sterility. 2011;95:444–7. doi: 10.1016/j.fertnstert.2010.08.037. [DOI] [PubMed] [Google Scholar]
- 24.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–86. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.