Abstract
The ability to sense mechanical forces is vital to cell physiology. Yet, the molecular basis of mechano-signaling remains unclear. Previous studies have shown that zyxin, a focal adhesion protein, is recruited at force-bearing sites on the actin cytoskeleton and, therefore, identifies zyxin as a mechano-sensing protein candidate. Furthermore, zyxin accumulation at force-bearing sites requires the LIM domain located at the C-terminus of zyxin. The zyxin LIM domain consists of three LIM motifs, each containing two zinc-binding sites. Since individual LIM motifs do not accumulate at focal adhesions or force-bearing sites, we hypothesize that multiple zyxin LIM domains increase force sensitivity. Using a miniature force sensor and GFP-tagged LIM variants, we quantified the relationship between single, tandem dimer and trimer LIM protein localization and traction forces. While the presence of extra LIM domains affected VASP recruitment to focal adhesions, force sensitivity was not enhanced over the single LIM domain. Therefore, zyxin force sensitivity is optimal with a single LIM domain, while additional LIM domains fail to enhance force sensitivity.
Keywords: cell migration, focal adhesion, zyxin, LIM domain, traction force, force sensor
INTRODUCTION
Physical force is an integral component of the cellular microenvironment, and serves as a signaling mechanism to complement chemically-mediated signaling. Such mechanical perturbations originate from an elastic extracellular matrix, neighboring cells, or shear stress exerted by fluid flow. Numerous types of cells have been shown to sense physical forces, e.g. stiffness of extracellular matrix, which in turn alter cell migration, stem cell differentiation and cancer cell invasion [1]. Yet, the molecular mechanisms by which cells convert mechanical signal to chemical signal remains ambiguous.
Zyxin is a focal adhesion protein with unique binding sites for Ena/VASP and α-actinin at the N-terminus and the LIM domain located at the C-terminus of the protein. Zyxin localizes at focal adhesions, actin stress fibers and cell-cell contacts. The unique feature of zyxin is that zyxin preferentially binds to force-bearing actin stress fibers. For example, when the actin stress fibers are stretched by a cantilever of an atomic force microscope [2] or prodded by a micropipette [3], zyxin rapidly accumulates at the sites of mechanical perturbation. Zyxin is thought to recognize the actin filaments under tension, possibly through severed filaments with exposed actin tips, and facilitate the repair of actin stress fibers [3]. In addition, zyxin accumulation also correlates with traction force exerted by migrating cells [4]. These results suggest that zyxin responds to both externally applied and internally generated forces, and zyxin may serve as a key component of mechano-sensing module for various cellular processes.
The ability of zyxin to accumulate at focal adhesions or force-bearing sites is solely due to the C-terminus LIM domain. This LIM domain consists of three motifs containing two zinc binding loops or zinc fingers where zinc binding is essential for the structure of the LIM domain [5]. Previous studies have demonstrated that all three zyxin LIM motifs are required for the efficient localization of zyxin at focal adhesions and force-bearing sites [4,6], suggesting that the force sensing function of zyxin requires the complete zyxin LIM domain. Interestingly, unlike zyxin, other LIM containing proteins have more than three LIM motifs [5], though how varying numbers of LIM motifs may alter protein function or force sensitivity is not known.
To further analyze the force-sensitivity of the zyxin LIM domain, we generated artificial LIM constructs that consist of multiple zyxin LIM domains. We hypothesize that increasing the number of LIM domains will increase force-sensitivity. Using a miniature force sensor, we quantified the accumulation of LIM variants at force-bearing sites and found that extra LIM domains do not enhance force sensitivity. Our results suggest that the single zyxin LIM domain has the optimal force-sensitivity, and that the binding partner of LIM domain most likely recognizes all three LIM motifs.
MATERIALS & METHODS
Cell lines and reagents
MDCK GII cells were cultured in DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% FBS (Atlanta Biologicals, Lawrenceville, GA). For western blot or immune-fluorescence analysis, the antibodies used were GFP (Invitrogen, Carlsbad, CA), VASP (BD Biosciences, San Jose, CA) and tubulin (Sigma-Aldrich, St. Louis, MO). For western blot, the signals on the nitrocellulose membrane were detected by chemiluminescence with an enhanced ECL reagent (Pierce Biotechnology).
LIM constructs
The LIM sequences were PCR amplified from the single LIM (338-572AA) plasmid using the following primers: ACTCAGATCTCGAGCGCCACCATGGAGAACCAAAACCAG (sense) and GGCGAATTCGAAGCTTCGCGTCTGGGCTCTAGC (anti-sense) for the tandem dimer LIM sequence, and ATCCGCTAGCGCTCGCCACCATGGAGAACCAAAACCAG (sense) and TGAGCTCGAGATCTGGCGTCTGGGCTCTAGC (anti-sense) for the tandem trimer LIM sequence. The PCR products were digested with XhoI/EcoRI (tandem dimer LIM) or NheI/BglII (tandem trimer LIM), then ligated into the GFP-tagged single LIM plasmid (a gift from Dr. Hiroaki Hirata, Nagoya University). The LIM inserts were sequence verified. Using Lipofectamine 2000 and G418 (Invitrogen, Carlsbad,CA), MDCK cells were stably transfected with the GFP-tagged LIM plasmids.
VASP co-localization analysis
The accumulation of VASP at focal adhesions was quantified using ImageJ. First, focal adhesions were identified by a local maximum search algorithm using the GFP channel, then assigned individual ROIs. Second, the intensities of zyxin, LIM variants and VASP in focal adhesions were quantified. The numbers of focal adhesions analyzed are 614, 440, 490, and 326 for GFP-tagged zyxin, single LIM, tandem dimer LIM, and tandem trimer LIM expressing cells, respectively. All images were taken at the same exposure using identical optics.
Live-cell confocal microscopy
All samples were imaged on a Zeiss Axio Observer equipped with a Yokogawa spinning disk confocal system, a 40× objective, 488 and 561nm solid-state lasers, and a CoolSNAP HQ camera. The microscope system was controlled and automated by Slidebook software (Intelligent Imaging Innovations, Denver, CO, USA). Live cells were imaged on glass bottom dishes (MatTek, Ashland, MA, USA) in a temperature-controlled chamber at 37°C.
Micro-fabrication of miniature force sensor
Fabrication of micro-pillar arrays was described previously [4]. Briefly, the micro-pillar master was etched with deep reactive ion etcher and consisted of dimensions of 2 µm in pillar diameter, 6 µm in height, and 4 µm in pitch (AdvancedMEMS, Berkeley, CA, USA). Using Polydimethylsiloxane (PDMS), the negative mold was casted, then PDMS pillars were fabricated. A droplet of 100 µg/mL fibronectin (BD Biosciences, San Jose, CA, USA) solution spiked with rhodamine fibronectin (Cytoskeleton, Denver, CO, USA) in 1:5 ratio was deposited on the pillar tips for 10 minutes, then the pillars were wetted in PBS with 1% bovine serum albumin and 0.05% Triton X-100. The pillars were then washed with PBS and the growth media. This approach created a fibronectin mesh on the pillar array, making it easier for cells to adhere and spread. Due to re-organization of fibronectin by migrating cells, some cells were able to deflect distant pillars using fibronectin bundles. We avoided this phantom deflection in our analysis. The traction force was calculated based on the displacement of pillar tips from the original positions and pillar stiffness of 2.5 MPa. To quantify the GFP intensity, we defined a ROI for each pillar and thresholded to identify the GFP positive puncta, which was normalized to corresponding background intensity using ImageJ. The numbers of cells analyzed were 20, 12, and 3 for single, tandem dimer, and trimer LIM expressing cells. See detailed protocol in reference [4].
RESULTS AND DISCUSSION
Expression and localization of tandem LIM proteins
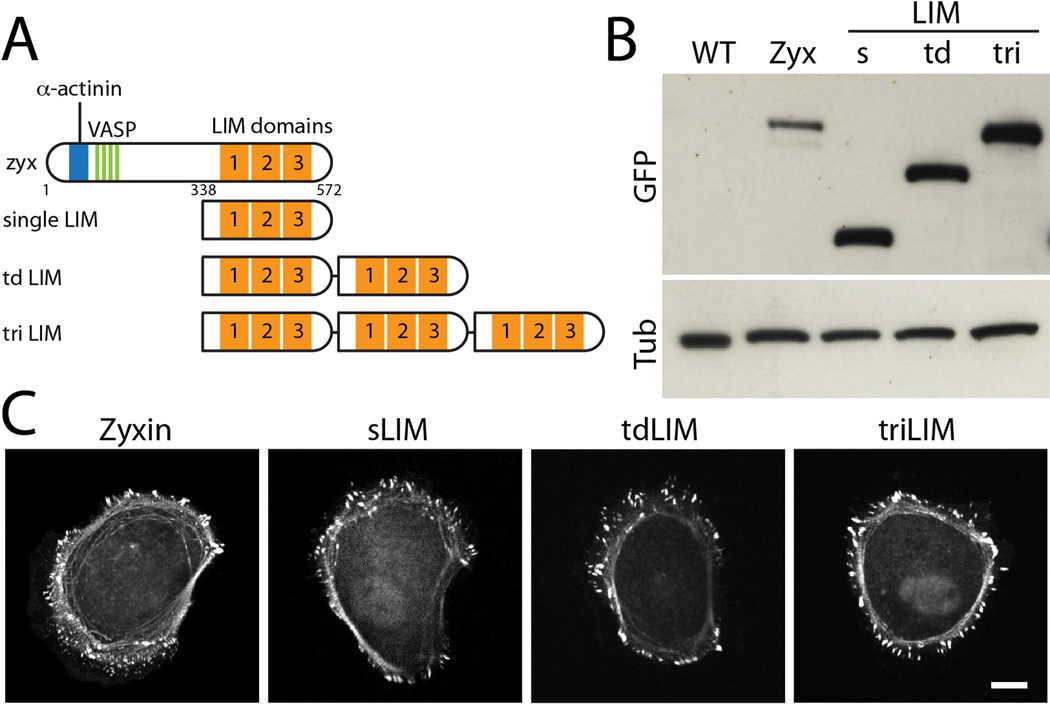
Since the LIM domain is solely responsible for force-sensitive accumulation of zyxin [4], we hypothesized that the force-sensitive accumulation of LIM domain proteins would be enhanced by the presence of multiple LIM sequences. Based on the GFP-tagged human zyxin LIM domain (338-572AA), we generated tandem dimer and trimer LIM constructs (Figure 1A), then transfected these plasmids, and isolated MDCK cell lines stably expressing these genes (Figure 1B). Using a GFP antibody, we compared the levels of zyxin LIM variants in stable cell lines and selected clones expressing similar levels of LIM proteins (Figure 1B). This approach allowed us to analyze how multiple LIM domains at a given quantity of LIM proteins affect force-sensitive localization.
Figure 1. MDCK cells expressing tandem zyxin LIM plasmids.
(A) Schematic of zyxin and tandem LIM sequences. (B) Western blot of stable MDCK cell lines expressing no exogenous zyxin (WT), the full-length zyxin (Zyx), and single (s), tandem dimer (td), and tandem trimer (tri) LIM sequence. All constructs are tagged with GFP. The membranes were blotted with GFP (top) and tubulin (bottom). (C) Live-cell images of MDCK cells expressing GFP-tagged full-length zyxin, single LIM, tandem dimer LIM, and tandem trimer LIM constructs. Scale bar 10 µm.
Exogenous GFP-tagged zyxin proteins localized to focal adhesions and along stress fibers of single MDCK cells (Figure 1C). This localization is similar to endogenous zyxin proteins observed through zyxin antibody staining [7], suggesting that the GFP tag does not compromise its localization. When the N-terminus of zyxin was truncated and only the LIM domain was expressed, the single LIM domain localized to both focal adhesions and along actin stress fibers (Figure 1C). Both tandem dimer and trimer LIM proteins localized at focal adhesions and along stress fibers similarly to GFP-tagged zyxin and single LIM proteins (Figure 1C). These results suggest that the presence of multiple LIM domains does not prevent focal adhesion and actin stress fiber localization.
The LIM proteins displace VASP from focal adhesions
The exogenous expression of zyxin LIM domain was shown to displace endogenous zyxin from focal adhesions. Due to the lack of a VASP binding site in the LIM domain, the zyxin LIM domain is unable to recruit VASP to focal adhesions [7,8,9,10]. VASP is an actin anti-capping protein that directly binds to the proline-rich domain in the N-terminus of zyxin. Since VASP is a critical regulator of actin dynamics, we analyzed VASP displacement in the tandem LIM protein expressing cells.
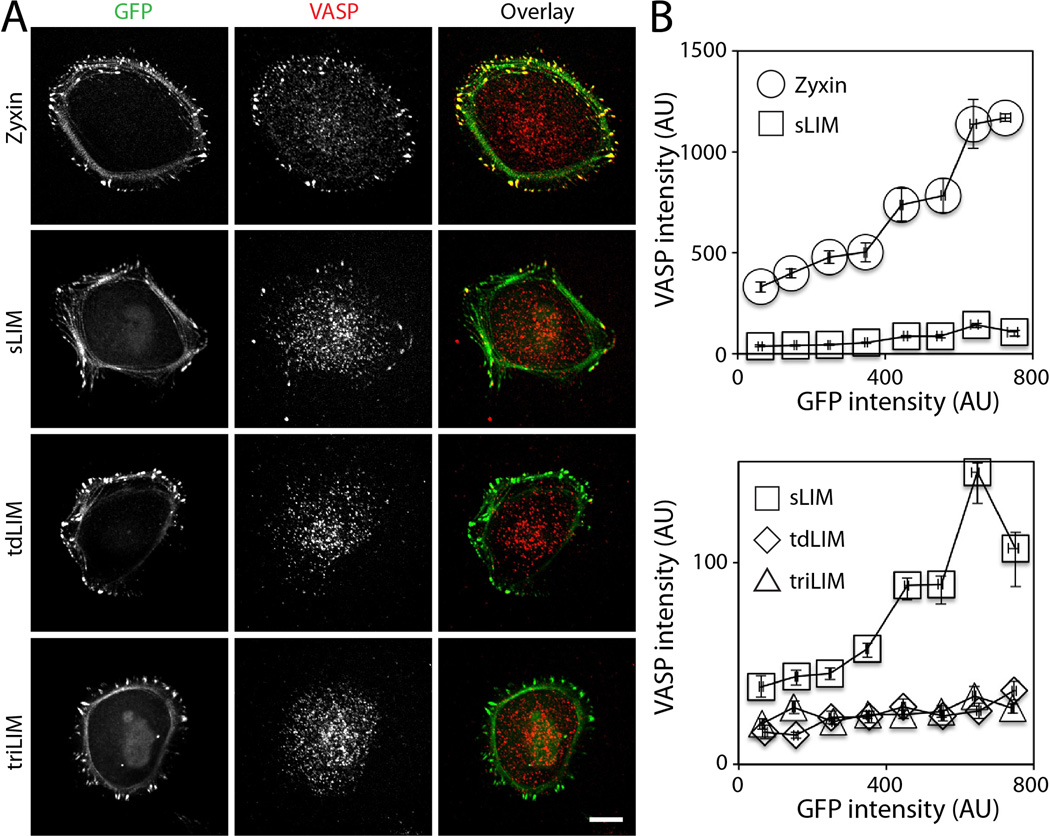
In the full-length zyxin expressing cells, VASP localized at focal adhesions (Figure 2A), whereas significantly less VASP was localized at focal adhesions in the single LIM domain expressing cells (Figure 2A). Quantitative analyses of VASP and zyxin intensities show that, when the GFP-zyxin signal is used to define the area of focal adhesion, VASP increasingly accumulated with increased zyxin (Figure 2B, top). This observation is consistent with the notion that zyxin recruits VASP to focal adhesions. In contrast, only a few VASP at focal adhesions were observed in the single LIM domain expressing cells (Figure 2A and B).
Figure 2. The LIM proteins displace VASP from focal adhesions.
(A) Immuno-flourescence labeling of VASP using GFP-tagged full-length zyxin, single LIM (sLIM), tandem dimer LIM (tdLIM), and tandem trimer LIM (triLIM). Scale bar 10 µm. (B) Quantification of GFP and VASP signal at focal adhesions. Error bars are standard error of the mean.
In the tandem dimer or trimer LIM expressing cells, VASP was not localized to focal adhesions (Figure 2A). The VASP intensity in both tandem LIM expressing cells was lower than the single LIM expressing cells (Figure 2B, bottom). The extra LIM domain is most likely preventing endogenous zyxin accumulation at focal adhesions and thereby, also preventing VASP from accumulating at focal adhesions. The similar levels of VASP in the tandem dimer and trimer LIM expressing cells suggest that one extra LIM domain are sufficient to completely remove VASP from focal adhesions in these cells.
Similar force sensitive accumulation of LIM variants
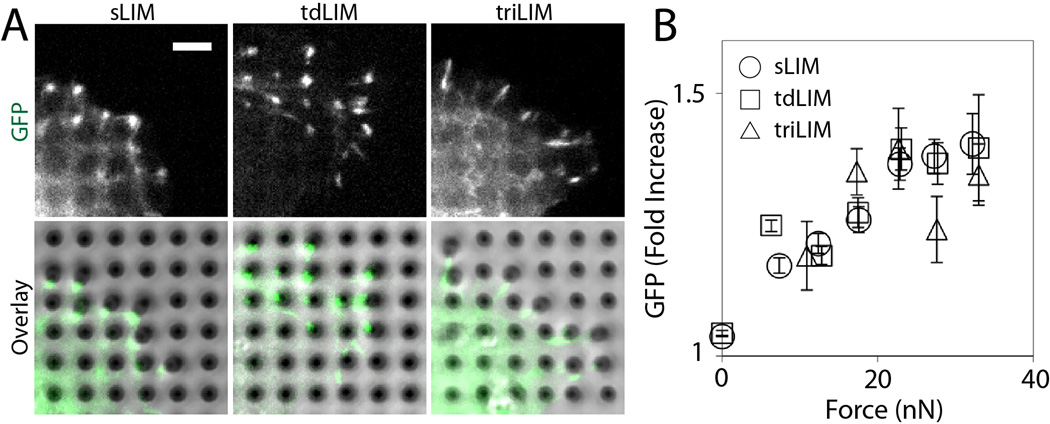
Using a micropillar array as a traction force sensor, we analyzed the relative accumulation of LIM variants at force-bearing sites. The ability of zyxin to accumulate at force-bearing sites is solely due to the LIM domain [4]. The GFP-tagged LIM domain accumulated at the bending pillars (Figure 3A) and the GFP intensity of LIM domain increased with the increasing amount of traction forces (Figure 3B). When tandem LIM expressing cells were plated onto the force-sensing pillars, both tandem dimer and trimer LIM expressing cells spread out and pulled on pillars (Figure 3A). Both tandem dimer and trimer LIM proteins accumulated at force-bearing sites and the GFP intensities increased as traction forces increased (Figure 3B). The GFP accumulation was similar in single, tandem dimer and trimer LIM expressing cells (Figure 3B), suggesting that the force sensitivities of the LIM variants are similar and that there are no observable benefits with the presence of extra LIM sequences.
Figure 3. Force-dependent accumulation of LIM proteins.
(A) MDCK cells expressing single (sLIM), tandem dimer (tdLIM), or tandem trimer (triLIM) LIM constructs adhere to micro-fabricated pillar arrays. Scale bar 5 µm. (B) Quantification of traction forces exerted at the pillar tips and GFP signals at the deflecting pillars. Error bars are standard error of the mean.
We hypothesized that additional LIM sequence provides increased force sensitivity. We reasoned that the presence of additional binding sites will increase the binding probability or affinity to force-bearing regions, therefore, allowing the tandem LIM proteins to be more force-sensitive than single LIM proteins. Yet, the single LIM proteins were just as sensitive to force as the tandem LIM variants (Figure 3B). Since VASP accumulation at focal adhesions was lower in tandem LIM expressing cells than single LIM expressing cells (Figure 2), the extra LIM domains are functionally competent to displace endogenous zyxin, and therefore VASP from focal adhesions. While the VASP displacement by tandem LIM protein demonstrated that the extra LIM domains are functional, the single LIM domain is sufficient for the maximum force sensitivity.
One explanation for the similar force sensitivities observed between the LIM protein variants is that the affinity to force-bearing sites does not improve with the presence of multiple LIM domains. Since individual LIM motifs or two consecutive LIM motifs within the LIM domain do not accumulate to the force-bearing sites [4], three LIM motifs or the complete single LIM domain is required for the maximum force sensitivity. This in turn suggests that the binding partner of zyxin at force-bearing sites interacts with all three LIM motifs in the zyxin LIM domain, and is unlikely to interact with multiple LIM domains. Another possible explanation for the similar force sensitivities observed between the LIM protein variants may lie in the accessibility of additional LIM binding partners to the tandem dimer and trimer sequences once an initial LIM binding partner attaches. Steric hindrance may prevent further LIM binding partner interactions, resulting in similar force sensitivities observed between the variants. It also remains possible that the difference between the force-sensitivity of single and tandem LIM domains are too small to detect with our force sensors. Nevertheless, the difference in the force sensitivity, if any, of tandem LIM variants is not significant compared to the single LIM domain. Thus, the additional LIM domains do not enhance force sensitivity.
The binding partner(s) of zyxin LIM domain at force-bearing sites remains unclear. While VASP and α-actinin have been identified as binding partners of zyxin, VASP and α-actinin interact with the N-terminus of zyxin and not the LIM domain. Currently, there are four known zyxin LIM domain binding proteins: Cell Cycle and Apoptosis Regulator-1 (CARP-1) [11], Cysteine-Rich Protein (CRP) [12], p130Cas [13], and synemin [14]. While CRP and p130Cas proteins localize to focal adhesions, CARP-1 and synemin do not and are, therefore, unlikely to recruit zyxin to focal adhesions.
Interestingly, p130Cas and CRP have reduced affinities toward truncated zyxin LIM domains [12,13]. Since truncated zyxin LIM domains are less likely to accumulate at focal adhesions [4,6] and force-bearing sites [4] than the full-length LIM domain, p130Cas and CRP may be responsible for zyxin recruitment at focal adhesions or force-bearing sites. However, in p130Cas deficient cells, zyxin proteins remain localized at focal adhesions, but not at focal complexes (nascent adhesions at the tips of lamellipodia). Furthermore, the first LIM motif of zyxin LIM domain is sufficient to interact with CRP, yet, the first zyxin LIM motif is not sufficient for focal adhesion localization [4,6]. These two results suggest that the recruitment of zyxin to focal adhesions or force-bearing sites may not be due to p130Cas or CRP, but rather through another, as of yet unidentified zyxin binding partner. Since the force-sensing mechanism is the critical for mechano-transduction, identifying molecular components of zyxin complex at force-bearing contacts will aid the understanding of how cells sense mechanical signals.
Highlights.
We hypothesize that multiple zyxin LIM domains may enhance force sensitivity
We analyze the accumulation of tandem dimer or trimer LIM domain and traction force
Tandem LIM variants reduces VASP accumulation at focal adhesions
Force sensitivities are similar among single and tandem LIM variants
Single LIM domain of zyxin has the optimal force sensitivity
ACKNOWLEGEMENTS
We thank Dr. Juergen Wehland (Gesellschaft für Biotechnologische Forschung, Germany) for the zyxin-GFP plasmid, and Dr. Hiroaki Hirata (Nagoya University) for the zyxin-LIM plasmid. This work was supported by a Beckman Young Investigator Award, a Hellman Family New Faculty Award, a NIH EUREKA GM094798, and the funds from the University of California Cancer Research Coordinating Committee.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Vogel V, Sheetz M. Local force and geometry sensing regulate cell functions. Nat Rev Mol Cell Biol. 2006;7:265–275. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- 2.Colombelli J, Besser A, Kress H, Reynaud EG, Girard P, Caussinus E, Haselmann U, Small JV, Schwarz US, Stelzer EH. Mechanosensing in actin stress fibers revealed by a close correlation between force and protein localization. J Cell Sci. 2009;122:1665–1679. doi: 10.1242/jcs.042986. [DOI] [PubMed] [Google Scholar]
- 3.Smith MA, Blankman E, Gardel ML, Luettjohann L, Waterman CM, Beckerle MC. A zyxin-mediated mechanism for actin stress fiber maintenance and repair. Dev Cell. 2010;19:365–376. doi: 10.1016/j.devcel.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uemura A, Nguyen TN, Steele AN, Yamada S. The LIM domain of zyxin is sufficient for force-induced accumulation of zyxin during cell migration. Biophys J. 2011;101:1069–1075. doi: 10.1016/j.bpj.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kadrmas JL, Beckerle MC. The LIM domain: from the cytoskeleton to the nucleus. Nat Rev Mol Cell Biol. 2004;5:920–931. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- 6.Nix DA, Fradelizi J, Bockholt S, Menichi B, Louvard D, Friederich E, Beckerle MC. Targeting of zyxin to sites of actin membrane interaction and to the nucleus. J Biol Chem. 2001;276:34759–34767. doi: 10.1074/jbc.M102820200. [DOI] [PubMed] [Google Scholar]
- 7.Nguyen TN, Uemura A, Shih W, Yamada S. Zyxin-mediated actin assembly is required for efficient wound closure. J Biol Chem. 2010;285:35439–35445. doi: 10.1074/jbc.M110.119487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drees B, Friederich E, Fradelizi J, Louvard D, Beckerle MC, Golsteyn RM. Characterization of the interaction between zyxin and members of the Ena/vasodilator-stimulated phosphoprotein family of proteins. J Biol Chem. 2000;275:22503–22511. doi: 10.1074/jbc.M001698200. [DOI] [PubMed] [Google Scholar]
- 9.Hirata H, Tatsumi H, Sokabe M. Mechanical forces facilitate actin polymerization at focal adhesions in a zyxin-dependent manner. J Cell Sci. 2008;121:2795–2804. doi: 10.1242/jcs.030320. [DOI] [PubMed] [Google Scholar]
- 10.Hoffman LM, Jensen CC, Kloeker S, Wang CL, Yoshigi M, Beckerle MC. Genetic ablation of zyxin causes Mena/VASP mislocalization, increased motility, and deficits in actin remodeling. J Cell Biol. 2006;172:771–782. doi: 10.1083/jcb.200512115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hervy M, Hoffman LM, Jensen CC, Smith M, Beckerle MC. The LIM Protein Zyxin Binds CARP-1 and Promotes Apoptosis. Genes Cancer. 2010;1:506–515. doi: 10.1177/1947601910376192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmeichel KL, Beckerle MC. The LIM domain is a modular protein-binding interface. Cell. 1994;79:211–219. doi: 10.1016/0092-8674(94)90191-0. [DOI] [PubMed] [Google Scholar]
- 13.Yi J, Kloeker S, Jensen CC, Bockholt S, Honda H, Hirai H, Beckerle MC. Members of the Zyxin family of LIM proteins interact with members of the p130Cas family of signal transducers. J Biol Chem. 2002;277:9580–9589. doi: 10.1074/jbc.M106922200. [DOI] [PubMed] [Google Scholar]
- 14.Sun N, Huiatt TW, Paulin D, Li Z, Robson RM. Synemin interacts with the LIM domain protein zyxin and is essential for cell adhesion and migration. Exp Cell Res. 2010;316:491–505. doi: 10.1016/j.yexcr.2009.10.015. [DOI] [PubMed] [Google Scholar]