Abstract
The G-quadruplex, a non-B DNA motif that forms in certain G-rich sequences, is often located near transcription start sites in growth regulatory genes. Multiple lines of evidence show that reactive oxygen species generated as second messengers during physiologic signaling target specific DNA sequences for oxidative base modifications. Because guanine repeats are uniquely sensitive to oxidative damage, and G4 sequences are known “hot spots” for genetic mutation and DNA translocation, we hypothesized that G4 sequences are targeted for oxidative base modifications in hypoxic signaling. Approximately 25% of hypoxia-regulated genes in pulmonary artery endothelial cells harbored G4 sequences within their promoters. Chromatin immunoprecipitation showed that common base oxidation product 8-oxoguanine was selectively introduced into G4s, in promoters of hypoxia up-, down-, and non-regulated genes. Additionally, base excision DNA repair (BER) enzymes were recruited to, and transient strand breaks formed in these sequences. Transcription factor Sp1, constitutively bound to G4 sequences in normoxia, was evicted as 8-oxoguanine accumulated during hypoxic exposure. Blocking hypoxia-induced oxidant production prevented both base modifications and decreased Sp1 binding. These findings suggest that oxidant stress in hypoxia causes oxidative base modifications, recruitment of BER enzymes, and transient strand breaks in G4 promoter sequences potentially altering G4 integrity and function.
Keywords: G-quadruplex, hypoxia, 8-oxoguanine, DNA base oxidation, base excision repair
Introduction
While the vast majority of genomic DNA resides in the typical Watson and Crick double helical structure (B-form DNA), it has become widely appreciated that certain nucleotide sequences can adopt non-B-form secondary motifs. One such secondary structure is the G-quadruplex, or G4 [1, 2]. G-quadruplexes are formed in certain guanine-rich nucleic acid sequences and consist of multiple stacked, planar guanine tetrads. These tetrads are stabilized by Hoogsteen hydrogen bonding between the guanines and each contains a coordinated monovalent cation, typically K+ or Na+ [3]. The specific conformation of the G4 structure probably depends on multiple factors, including the primary DNA sequence, the local ionic environment, protein-DNA interactions, supercoiling and other determinants [1, 2, 4].
G-quadruplex motifs are fairly common in the genomes of higher eukaryotes, [5–8], and multiple lines of indirect and direct evidence suggest that they serve a transcriptional regulatory function. For example, although scarce within the coding sequences of genes, >40% of human gene promoters may contain one or more potential G4 elements [3, 8–10]. A number of proteins, some of which are documented transcription factors, are known to bind preferentially to G4 sequences [11–14]. It has been postulated that both DNA binding proteins as well as mechanical forces linked to transcription initiation can shift the equilibrium between G4 conformational states [4, 15–18]. Treatment of cells with G4-stabilizing compounds alters expression of many genes harboring such sequences [14, 19–23]. A final line of evidence supporting a key regulatory role for G4 sequences is that mutations in G4 sequences are linked to genomic instability and disease-related phenotypes. In certain B-cell lymphomas, for example, the presence of multiple runs of guanines on the non-template DNA strand is strongly correlated with hypermutation and translocation [24]. Reporter constructs containing telomeric sequences capable of forming G4s display extensive instability [25], and in yeast, insertion of telomeric sequences promoted homologous recombination and formation of DNA strand breaks [26]. The repetitive mini-satellite sequences displaying the highest incidence of instability in vivo are G-rich sequences capable of forming G4s [27]. Most recently it has been demonstrated in a genome-wide survey that there is an enrichment for G-quadruplex sequences in the vicinity of DNA strand-breaks associated with somatic copy number alterations within multiple varieties of cancers [28].
The basis of the genetic instability of guanine-rich promoter sequences is unknown. Studies using oligonucleotide models predict that guanine-rich sequences should be hot spots for oxidative base damage [29–34]. In particular, oligonucleotide models of G-quadruplex-forming sequences show that the external G-tetrads are frequently targeted for oxidative base modification [35] by a mechanism termed charge tunnelling [29]. Importantly, it has not been established whether such motifs are sensitive to oxidative base damage in living cells.
Herein we tested the hypothesis that G4 motifs are sensitive to oxidative base modifications evoked by hypoxia. Hypoxia is of particular interest since it is a central feature of the tumor microenvironment, promoting tumor growth [36]. Hypoxia also increases the mutation rate in cancer cell lines and thereby contributes to the acquisition of an invasive, metastatic, phenotype [37]. Of equal significance, reactive oxygen species (ROS) used as second messengers in hypoxic signalling cause oxidative base damage in G-rich sequences of hypoxic response elements in the VEGF and other hypoxia-inducible promoters [38–41] that do not harbor putative G4 motifs. Our results indicate that putative G-quadruplex forming promoter regions are indeed targets for hypoxia-induced base modifications and offer interesting clues regarding the potential biological significance of such base damage.
Materials and Methods
Overall strategy
To explore the postulated link between hypoxia-induced ROS generation and G-quadruplex-targeted base modifications, the current studies used a responsive cell population – pulmonary artery endothelial cells (PAECs) – to evaluate the association between the hypoxic transcriptome and a genome-wide database of putative G-quadruplex forming sequences. We then applied chromatin immunoprecipitation analyses (ChIP) to G-quadruplex-forming and non-G4 containing (control) sequences in promoters of hypoxia- up-, down-, and non-regulated genes to search for the common base oxidation product, 8-oxoguanine (8-oxoG), and identify those sequences associating with base excision DNA repair (BER) enzymes and with a G4-DNA binding protein, the common transcription factor Sp1.
Pulmonary artery endothelial cell culture, hypoxic exposure, and drug treatments
Rat pulmonary artery endothelial cells (PAECs) isolated and cultured as previously described were used between passages 6 and 16 [40, 41]. Cells were grown in 50/50 DMEM/F-12 (Mediatech, Inc) + 10% fetal bovine serum in 150 mm tissue culture dishes (Corning) at 5% CO2 and 21% O2 until 85–90% confluent. Hypoxia exposure was performed in an environment of 5% CO2 and 2% O2 for the specified durations (1, 3 or 6 hours). In some experiments, PAECs were incubated with myxothiazol (Sigma) at a concentration of 10uM to inhibit ROS production by the mitochondrial complex III [41]. To stabilize the G-quadruplex structures, TMPyP4 (Calbiochem) was added to PAECs at a final concentration of 100 µM one hour prior to hypoxic exposure.
RNA isolation
PAECs were grown as above and exposed to hypoxic (2% O2) or normoxic (21% O2) atmosphere for 3 hours. Cells were washed in ice cold PBS harvested with TRIzol (Invitrogen) following manufacturer instructions.
Microarray and identification of G-quadruplex sequences
RNA from rat PAECs cultured for 3 hours in either hypoxia or normoxia was labeled and hybridized to Rat Gene 1.0 ST Arrays (Affymetrix) following standard manufacturer’s protocols. Each condition was tested in triplicate. The Rat Gene 1.0 ST Array provides full genome coverage and approximately 26 independent probes spread across the full length of each gene. Microarray data were normalized, analyzed and visualized using Partek Genomics Suite v 6.5 (Partek, St. Louis, MO). Hybridization intensities from the Affymetrix arrays were quantified and normalized using GC robust multiarray average probe set summarization. All microarray data met the quality control criteria established by the Tumor Analysis Best Practices Working Group (PMID:14970825). Non-expressed transcripts were eliminated if there were less than two out of a total of 15 calls with a detection p-value < 0.05. The remaining ‘present’ transcripts (23,592 of 27,342 total) were used for all subsequent statistical and visual analysis. A one-way analysis of variance (ANOVA) model was used to test for differentially expressed genes with a false discovery rate (FDR) of less than 10% used to control for multiple testing.
After identifying those genes differentially regulated by hypoxia, we next compared that list to a database enumerating genes in the rat genome containing putative G-quadruplexes in promoter sequences [5]. Hypoxia up-regulated genes selected for additional study are listed in Table 1 and included: Vascular endothelial growth factor (VEGF), Metallothionein 1a (Mt1a), and Nuclear factor of kappa light polypeptide gene enhancer in B cells (Nfκb2). Hypoxia down-regulated genes included arginine vasopressin receptor 1a (Avpr1a) and Breast cancer 2 (Brca2), both of which contain two potential G4 sequences. G-quadruplex-containing genes used for this study whose baseline expression was unaltered in hypoxia were the Dual specificity phosphatase 10 (Dusp10) and Mothers against DPP homolog 6 (Smad6) genes. It should be noted that all three groups of genes are transcribed at some level under both conditions (normoxia and hypoxia). We selected multiple sequences from two genes to serve as negative controls; two are within the Vegf gene, one in the coding region and the other in the 1st intron. Another lies just downstream of the Vegf stop codon. The remaining three negative control sequences are in the promoter and coding region of the α-actin gene, which lacks a putative G-quadruplex. One is in the promoter, one is entirely within intron 2 and the last contains part of exons 3 and 4 and all of intron 3. These control sequences contain a similar GC content despite lacking the G-rich G-quadruplex. The control sequences are summarized in table 2. Both sets of sequences utilized for this study have substantially higher GC content than the Rattus norvegicus genome, which is approximately 42%.
Table 1.
Properties of G-quadruplex regions analyzed for oxidative damage.
| Gene | Regulation by hypoxia |
Distance from TSS |
% GC of amplified fragment |
|---|---|---|---|
| Avpr1a | down | 318 | 59.73 |
| 85 | 55.02 | ||
| Brca2 | down | 998 | 71.56 |
| 263 | 50.57 | ||
| Dusp10 | unchanged | 594 | 68.81 |
| Mt1a | up | 341 | 49.56 |
| Nfkb2 | up | 502 | 62.2 |
| Smad6 | unchanged | 172 | 65.13 |
| Vegf | up | 66 | 63.14 |
Table 2.
Properties of control regions used in ChIP analyses.
| Gene | Location | % GC of amplified fragment |
|---|---|---|
| Actb | Promoter | 66.67 |
| Inton 2 | 45.92 | |
| Exon3-Exon4 | 50.84 | |
| Vegf | Exon1 | 73.59 |
| Intron1 | 50.66 | |
| 3'UTR | 36.69 |
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed using the commercially available ChIP-IT Express kit as recommended by the manufacturer (Active Motif). Briefly, ~2×107 PAECs were fixed with 1% formaldehyde (Sigma) for 10 minutes and washed with ice-cold 1x PBS. The fixation reaction was terminated by addition of Glycine Stop Fix solution and incubation with gentle shaking at room temperature for 5 minutes. PAECs were washed with 1xPBS and collected in Cell Scraping Solution supplemented with 0.5 mM PMSF. Chromatin was sheared to ~300–500 base pair fragments by sonication on ice for ten 20s pulses at 30% amplitude with a Vibracell VCX 130PB (Sonics & Materials). The chromatin was then immunoprecipitated with 2 μg of antibody for 12–14 hours on a rotary mixer at 4°C. Antibodies used were directed at 8-oxoguanine (Millipore), Ogg1 (Abcam), Ref-1/APE1 (Novus Biologicals), Sp1 (Santa Cruz Biotechnology), and IgG (Santa Cruz Biotechnology). The immunoprecipitation and purification steps were performed as per manufacturer’s specifications.
Precipitated DNA and input DNA controls were amplified by PCR using primers specific for target sequence. The following primers were used for ChIP of G4 regions: Brca2; 5’-CGAAGATCTCTTGTTGAATCCA, 5’-TAATTGTCTTATTGAGGAAG-GG; 5’-TCGCAGGAGCGTGAGAAGTGAG, 5’-CTGCAAGGGGCCGGGAGGAG, Avpr1a; 5’-AGTGCAGGGGCAAACCGTGTG, 5’-ATAAGTGAATGTAAAGCCAGGT; 5’-GAATCCAGGAGTCTCAGCTCC, 5’-ATGGGATATTCAAACATGTCC, Mt1a; 5’-ATGGTGGCACATGACTGGATG, 5’-CTGCATTTGGTTCTTCTGCTC, Nfkb2; 5’-GATGAAACGCACATTCCTGTAG, 5’-GAGAATGTGATTCGGGCCGCTG, Smad6; 5’-CGTGTTTGTTAGCGGGTGTGC, 5’-CTGATTGTTGCGCAAACAAGGTT, Dusp10; 5’-AGGAGTGACAGGGGCACAGAAG, 5’-GTCCAACTTTCCCGAGGCCA, and VEGF; 5’-GGCTATGGACCCTGGTAAGGG, 5’-GAGGTTTGAATATCAAATCC. PCR products were separated by agarose gel electrophoresis (2% gel), and integrated density values were obtained using Kodak Gel Logic 1500 (Eastman Kodak). Densitometry values were normalized to input DNA.
Detection of DNA strand breaks
PAECs were fixed at room temperature for 20 minutes with Streck Tissue Fixative (Streck) + 10 mM EDTA. This fixative solution does not damage the DNA. After 2 washes in ice cold 1xPBS, the cells were resuspended in PBS and counted. The PAECs were permeabilized in 100 mM Tris-HCl [pH7.4], 50 mM EDTA, and 1% Triton X-100 for 30 minutes at 4°C. After 2 ice cold PBS washes, the pellet was resuspended in 1x Terminal deoxynucleotidyl transferase (TdT) buffer (Promega) supplemented with 1x hexanucleotide mix (Roche), 1nmol/µl biotin-16-dUTP (Roche), and 2 units of TdT (Promega) and held at 37°C for 30 minutes. Nuclei were washed once with 100 mM Tris-HCl + 150 mM NaCl, resuspended in PBS, sonicated, and ChIP was performed as above using anti-biotin antibody (Novus Biologicals).
Results
Hypoxia-regulated genes harboring promoter G-quadruplex sequences in rat PAECs
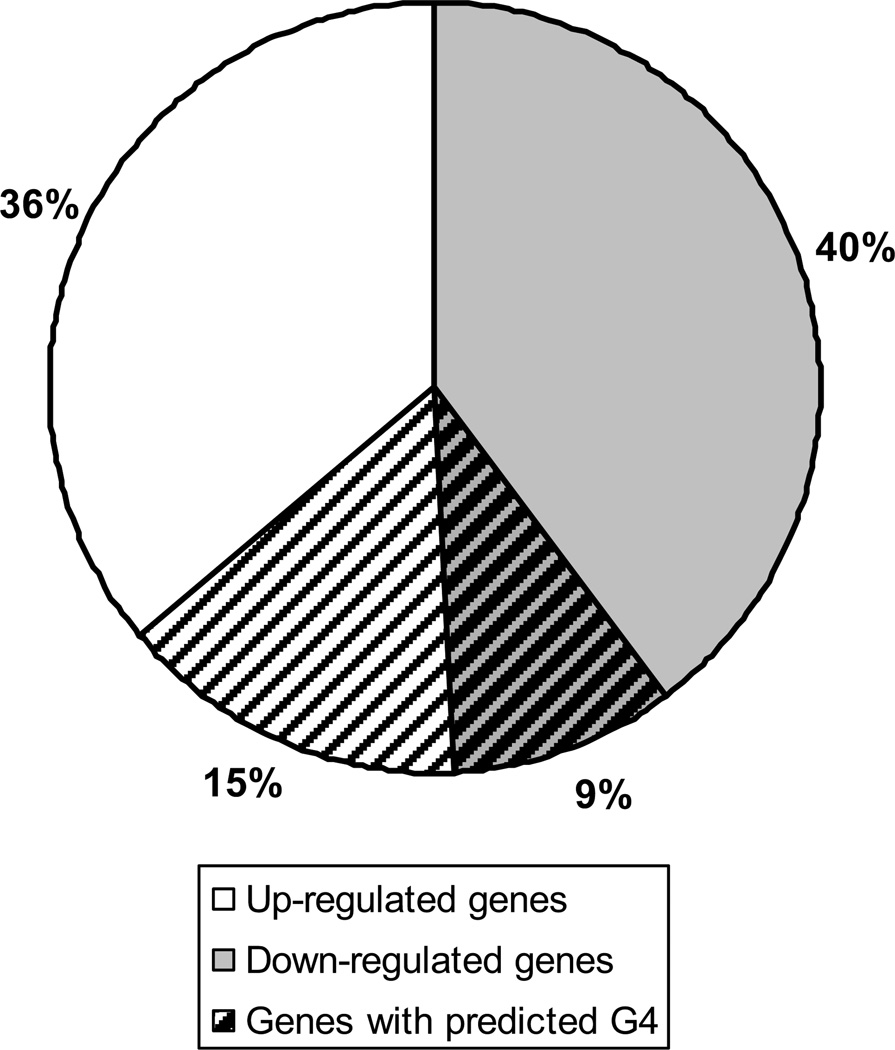
Microarray analysis comparing normoxic rat PAECs and cells exposed to hypoxia for 3 hours yielded 620 transcripts whose expression was differentially altered by hypoxia exposure. Of the 507 hypoxia-responsive transcripts searchable in a G4-predictor database [5], 123, or 24.3%, contained a predicted G4 sequence within the promoter region (Figure 1). The percentage of hypoxia-responsive genes containing a predicted G-quadruplex sequence calculated in this study is very similar to that previously estimated for all genes in the Rattus norvegicus genome (25.69%) [5]. Interestingly, G4 sequences tended to be slightly more common in the promoters of genes that were up-regulated in response to hypoxia (29.4%) in comparison to genes down-regulated in hypoxia (18.4%) in these cells.
Figure 1.
Representation of the 507 hypoxia regulated genes that were able to be analyzed for the presence of a potential promoter G4 sequence. White indicates genes that were found by microarray to be up-regulated in PAECs after 3 hours of hypoxia (258 genes or 51%). Gray represents those genes that were down-regulated (249 genes or 49%). The portion with lines represents those genes predicted to have a G4 within the promoter (123 total genes or 24%). The genes up-regulated in hypoxia were more likely to have a promoter G4 (75 or 61% of G4s).
Impact of hypoxia on 8-oxoG formation in G-quadruplex sequences
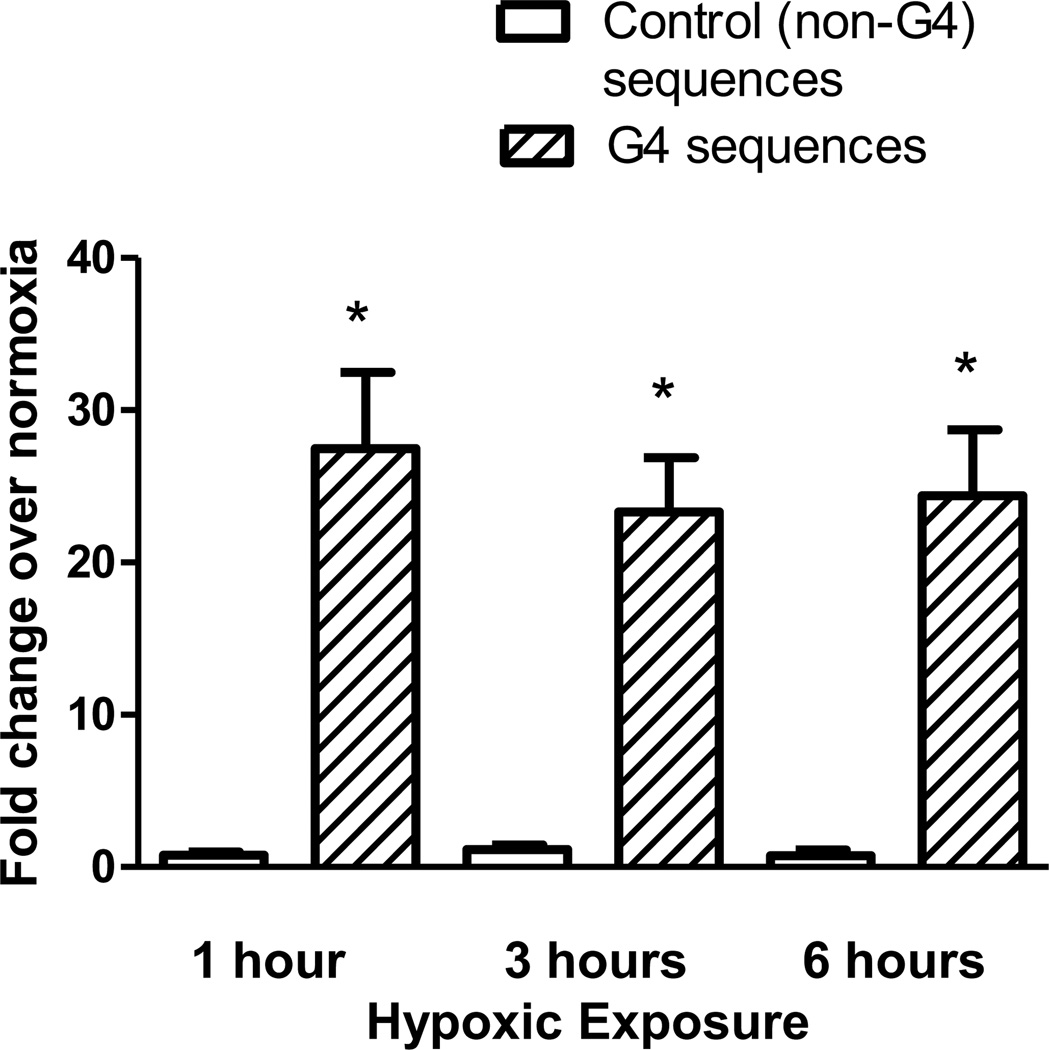
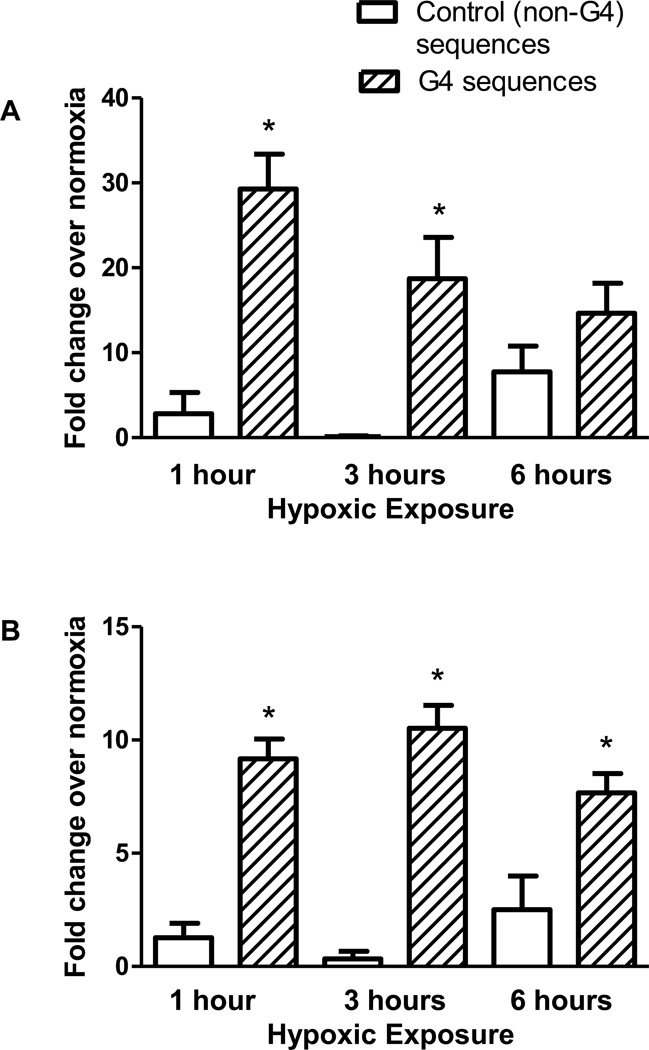
To determine whether G4s are prone to oxidative base modifications within the intact genome of living cells, PAECs were exposed to hypoxic conditions (2% O2) known to generate an oxidant stress directed at functionally-significant promoter sequences in hypoxia-inducible genes [40, 41]. Chromatin immunoprecipitation analysis was utilized to detect the presence of 8-oxoG in DNA sequences containing G-quadruplexes and control sequences that do not contain potential G4s. In order to decrease the possibility that any differences could be due to guanine content of the sequences, both the G4 sequences (mean GC 60.64% ± 2.559 SEM) and the control sequences (mean GC 54.06% ± 5.569 SEM) were selected on the basis that they contain a similar proportion of GC in comparison to G4-containing sequences. Figure 2 demonstrates a substantial increase in 8-oxoG incorporation in the nine G4-containing regions when compared to the six non-G4 control regions at all time points examined.
Figure 2.
Comparison of incorporation of 8-oxoG in G4-containing regions versus control sequences. ChIP was used to compare incorporation of 8-oxoG during hypoxic exposure between G4-containing sequences and non-G4 control sequences which do not contain a predicted G4 sequence. N≥8. The error bars indicate S.E.M. * Significance between G4 and non-G4 sequences (P<0.05).
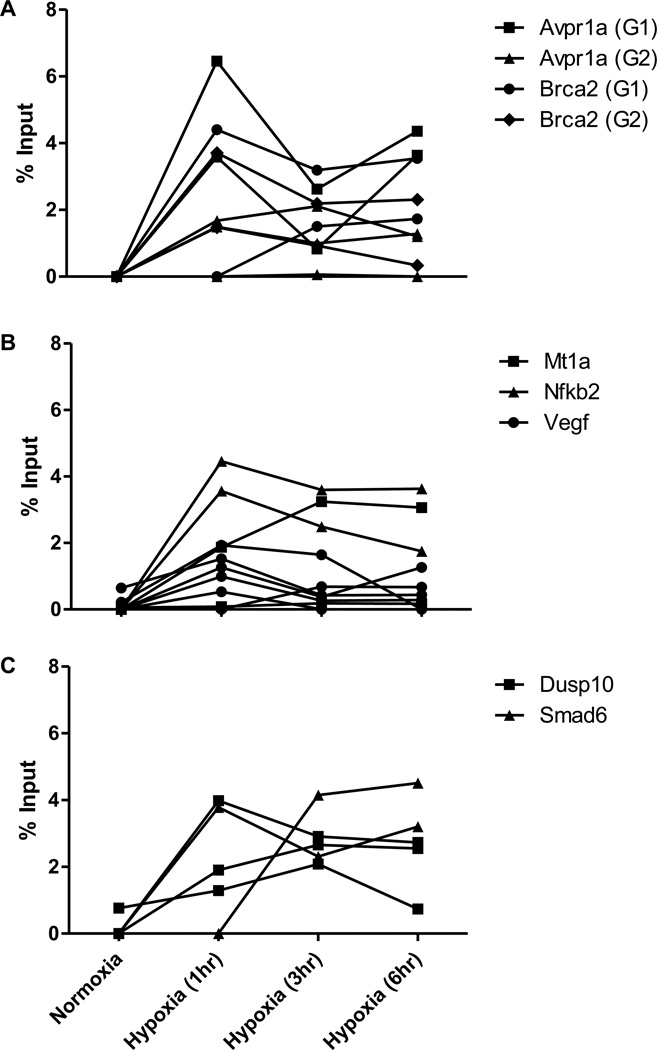
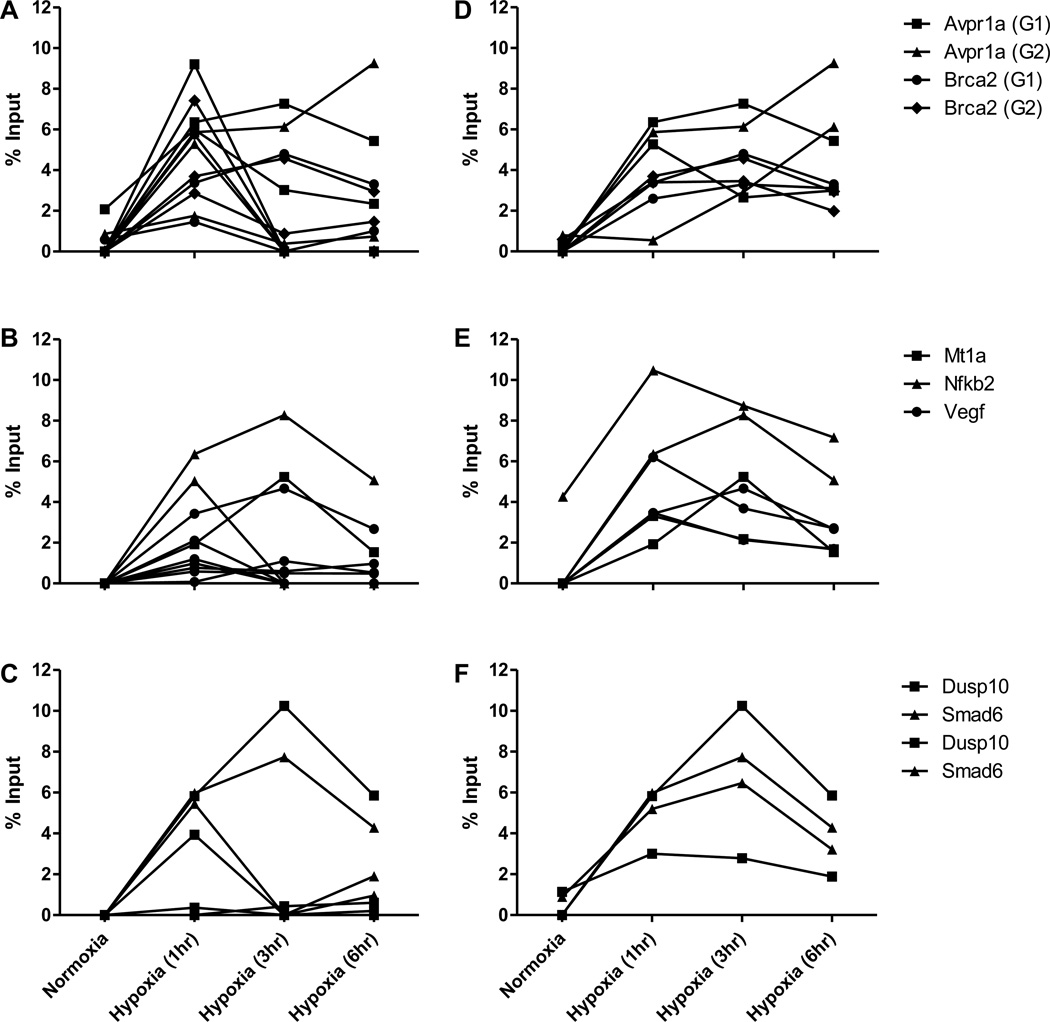
DNA damage at the G4 sequences appears independent of the transcriptional state of the gene in which they reside insofar as all three groups demonstrate similar increases in 8-oxoG content (Figure 3). While there is conspicuous variability in time courses – even for the same G4 – the occurrence of the increased 8-oxoG is consistent in all groups. In many experiments there is a spike of 8-oxoG at one hour that begins to decrease, but often the damage persists or rises over time as the cells are exposed to hypoxia. Potentially noteworthy is the fact that in the two genes whose expression was unaltered by hypoxia (Dusp10 and Smad6), the 8-oxoG tends to persist at a higher level longer than in the two transcriptionally regulated groups (Comparisons shown in Supplementary Table 1).
Figure 3.
Frequency of incorporation of the oxidized nucleotide 8-oxoG in the G4 region of multiple promoters in hypoxia. ChIP was used to detect changes in level of incorporation of 8-oxoG between cells in normoxia and various periods of hypoxia (1, 3, or 6 hours) in the regions of promoters that contain predicted G4 sequences of genes that were determined to be down-regulated (A), up-regulated (B), or transcriptionally unaffected (C) by hypoxia. Each line represents an independent cell culture and chromatin preparation for ChIP.
Impact of the G4-stabilizing compound TMPyP4 on hypoxia-induced base oxidation in promoter G4 sequences
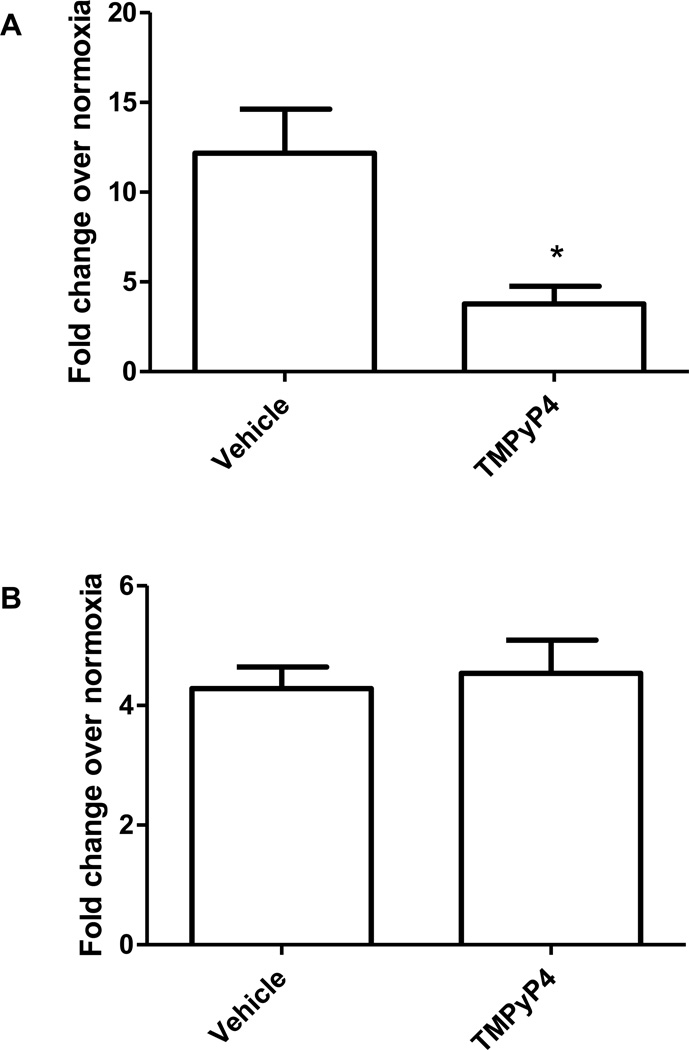
To determine whether the oxidative damage is localized to the G-quadruplexes and not adjacent regions, cells were treated with the cationic porphyrin compound TMPyP4 prior to hypoxic exposure. TMPyP4 has been shown to react with high specificity to G4 motifs, where it intercalates into the DNA and enhances the formation and stability of the G4 tertiary structure [23, 42]. In PAECs pre-treated for one hour with TMPyP4 prior to hypoxic exposure the VEGF G4 was highly protected against 8-oxoG formation (Figure 4A). In contrast, TMPyP4 failed to protect against formation of 8-oxoG in VEGF hypoxic response element (HRE) sequence (Figure 4B), which is known to be prone to base oxidation under these conditions and does not contain a G4 sequence [38, 40, 43]. TMPyP4 also provided a similar protective effect for the other G4 sequences examined (Supplementary Figure 1).
Figure 4.
Effect of G4-stabilizing compound TMPyP4 on 8-oxoG incorporation into VEGF G4 and HRE sequences. Pre-treating PAECs with TMPyP4 prior to exposure to hypoxia for 3 hours protected the VEGF G4 (A) but not the VEGF HRE (B) from oxidation of guanine to 8-oxoG. *Significantly different from vehicle control (P<0.05).
Recruitment of base excision repair enzymes to promoter G4 sequences in hypoxic cells
To test whether the oxidation of G4 guanines leads to recruitment of BER enzymes, ChIP analyses were applied to the DNA glycosylase, Ogg1, and the bi-functional transcriptional co-activator and apurinic/apyrimidinic endonuclease, Ref-1/APE1. Figure 5 shows recruitment of the BER enzymes to G4- and non-G4-containing sequences in hypoxic PAECs. Both Ogg1 and Ref-1/APE1 increased slightly over time in the non-G4-containing sequences, an interesting finding in light of the absence of an hypoxia-induced increase 8-oxoG in these regions (Figure 2). As shown in Figure 6, both of these proteins were present at significantly increased levels in hypoxia. Ogg1 displayed a tendency to spike early and then decrease (Figure 6A–C), whereas Ref-1/APE1 tended to persist at higher levels for the duration of exposure (Figure 6D–F). Curiously, Ogg1 decreased most rapidly from the G4s located in the promoters of the two unregulated genes, which as noted above, displayed more persistent elevations in 8-oxoG.
Figure 5.
Comparison of BER enzyme recruitment in G4-containing regions versus control sequences. ChIP was utilized to compare the association of the BER enzymes Ogg1 (A) and Ref-1/APE1 (B) during hypoxic exposure between G4-containing sequences and non-G4 control sequences. N≥8. The error bars indicate S.E.M. * Significantly different between G4 and non-G4 sequences (P<0.05)
Figure 6.
Frequency of association of BER pathway enzymes with the G4 region of multiple promoters in hypoxia. ChIP was used to analyze the levels of BER enzymes Ogg1 (A–C) and Ref-1/APE1 (D–F) between cells in normoxia and various periods of hypoxia (1, 3, or 6 hours) in the regions of promoters that contain predicted G4 sequences of genes that were determined to be down-regulated (A and D), up-regulated (B and E), or transcriptionally unaffected (C and F) by hypoxia. Each line represents ChIP of an independent chromatin preparation.
Impact of hypoxia on strand break formation in G4-containing promoter sequences
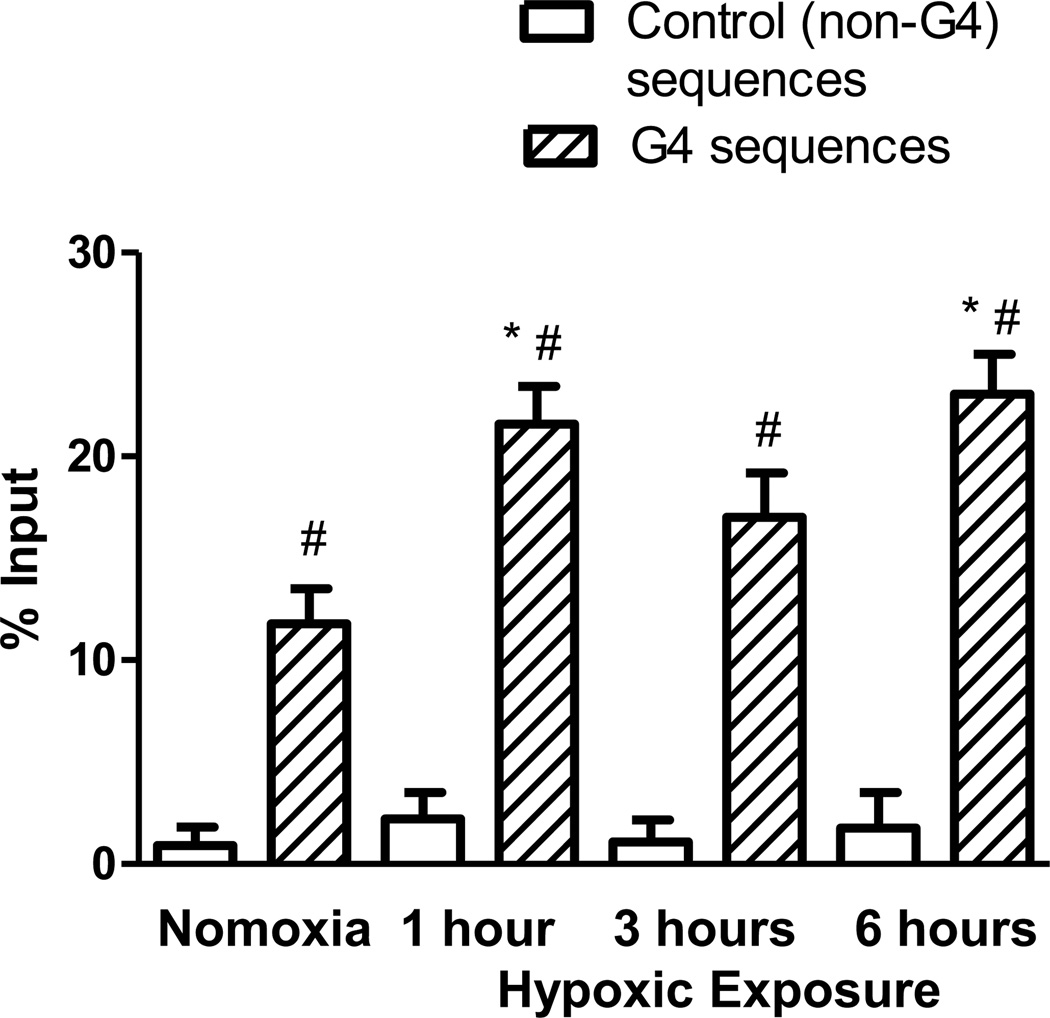
A modified ChIP assay was used to determine if G4 sequences were susceptible to strand breaks during hypoxic exposure. Such breaks could be a direct result of DNA damage or an intermediate in the BER process. As shown in Figure 7, unlike 8-oxoG or the BER enzymes, there was a low but detectable level of strand breaks in cells cultured in normoxic conditions. However, the frequency of strand breaks was increased for cells in hypoxia. The high level of DNA strand breaks was specific to sequences containing G4s, as the non-G4 regions only exhibited a low level of strand breaks in either normoxia or hypoxia.
Figure 7.
Formation of DNA strand breaks in G4 sequences. DNA strand breaks were labeled with dUTP by TdT and detected by ChIP for G4-containing and non-G4 sequences. N≥4. The error bars indicate S.E.M. * Significant change from normoxia (P<0.05). #Significant difference between the G4- and non-G4 signal (P<0.05).
Relation between hypoxia-induced G4 guanine oxidation and Sp1 binding
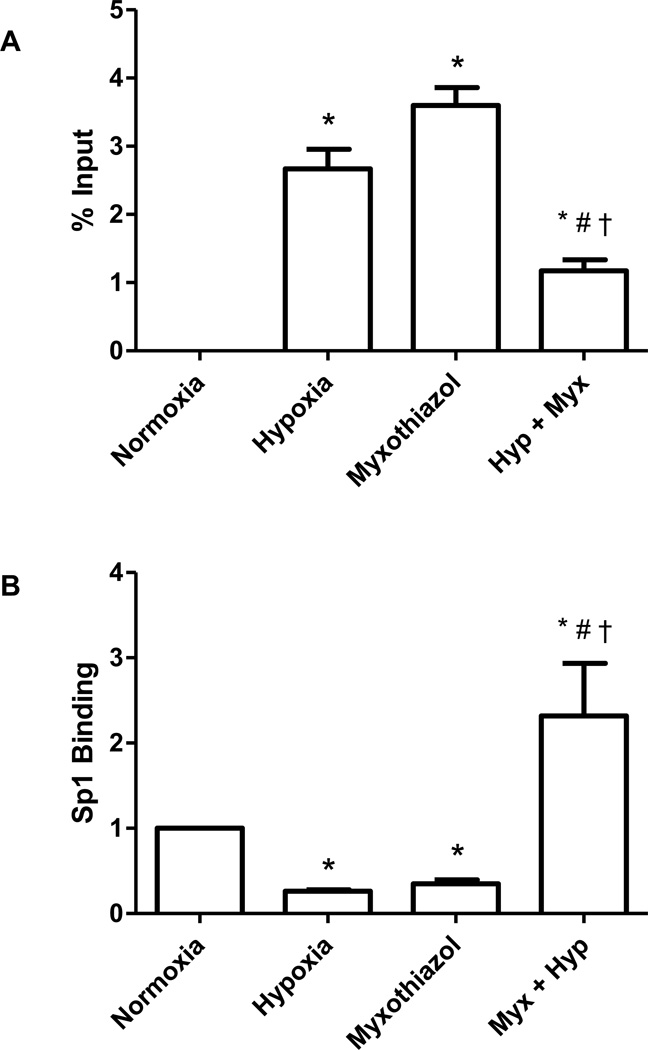
The VEGF promoter contains three Sp1 binding sites which overlap the G4 sequence. Ghosh and Mitchell have previously shown that oxidative damage to the guanines in the Sp1 binding site blocks binding of the transcription factor [44]. Accordingly, we tested the idea that the hypoxia-induced formation of 8-oxoG in the VEGF G4 motif altered Sp1 binding to the sequence using myxothiazol, a mitochondrial complex III inhibitor, to suppress ROS production and base damage, as shown previously [41]. As displayed in Figure 8A, three hours exposure to hypoxia introduced 8-oxoG into a portion of the VEGF promoter G4 sequences. In normoxic PAECs, treatment with myxothiazol mimicked the effect of hypoxia, increasing 8-oxoG-containing VEGF promoter G4 sequences. This finding is not unexpected since it has been reported earlier that myxothiazol, although inhibiting complex III, causes oxidant leakage from other sites in the electron transport chain [45]. In hypoxic cells, however, the complex III inhibitor decreased formation of 8-oxoG in the VEGF promoter G4 sequence by 56%. Sp1 binding to the VEGF promoter G4 sequence demonstrated an inverse relationship to 8-oxoG incorporation. As shown in Figure 8B, Sp1 binding to the VEGF promoter G4 sequence was reduced in hypoxia by approximately 75%. In normoxic cells, myxothiazol decreased Sp1 binding to a similar extent (68%). In contrast, the complex III inhibitor not only blocked the hypoxia-induced eviction of Sp1 from the VEGF promoter G4 but triggered an increase in Sp1-promoter G4 association.
Figure 8.
Effect of the mitochondrial complex III inhibitor, myxothiazol (Myx), on (A) 8-oxoG incorporation into the VEGF promoter G4-containing sequence and (B) Sp1 association with the VEGF G4-containing promoter sequence in normoxic and hypoxic PAECs. N=3–5. *Significantly different from normoxia (P<0.05). * Significantly different from normoxia (P<0.05). # Differs significantly from hypoxia (P<0.05). † Significantly different from myxothiazol alone (P<0.05).
Discussion
Because the concept that G4 motifs are prone to oxidative damage has been derived largely from studies on oligonucleotide models [29–34], herein we tested the hypothesis that promoter G4 sequences are sensitive to oxidative base modifications in the setting of hypoxia-induced oxidant stress. Our microarray analysis of the hypoxia-regulated transcriptome in rat PAECs identified a little more than 600 genes – similar to previous analyses in endothelial cells [46, 47] – of which 24% harbored putative G4 motifs in their promoters, with 15% up-regulated and 9% down-regulated. We concentrated our search for oxidative base modifications in functionally-relevant genes selected from these categories as well as genes that were not regulated by hypoxia.
Hypoxia is a fundamental stimulus in biology and medicine, and considerable effort has been directed towards understanding mechanisms of hypoxia-regulated adaptive gene expression. Though remaining somewhat controversial [48], studies in transformed and non-transformed cells suggest that hypoxia-induced ROS generation, either by mitochondria or by membrane NADPH oxidase, plays a key role in this process. Reactive oxygen species so generated may function to stabilize the critical transcription factor in hypoxia-regulated gene expression, Hif-1 [49–51]. There are also several reports that hypoxia-generated ROS oxidatively modify specific bases in the hypoxic response elements (HREs) of hypoxia-inducible genes [38, 40, 41]. Based on multiple lines of indirect evidence [39, 41, 43], including similarities between hypoxia-induced and ligand-mediated base modifications in response elements of ligand-inducible promoters [52–55], it has been speculated that hypoxia-induced base modifications might contribute to transcriptional regulation, possibly by altering transcription factor binding or local sequence flexibility/topology [56–58]. Against this background, the present study determined if G4 sequences in hypoxia-regulated and non-regulated genes were targeted for oxidative base modifications during hypoxia.
Our data demonstrate that each of the 9 selected genes acquired oxidative base damage in G4 sequences during hypoxic exposure; “control” sequences selected for their similar GC content were unaffected by hypoxia. The oxidative damage appears selectively targeted to the G-quadruplex sequence insofar as the oxidation is largely blocked by TMPyP4, which specifically interacts with G4 motifs [23, 42]. The reason(s) for the protective activity of TMPyP4 is yet to be uncovered. It is possible that the intact G4 conformation, which is enhanced by the drug, is more resistant to oxidative damage. Counter to this, however, is in vitro evidence that intact quadruplexes are more prone to oxidative damage than the same sequence in a typical duplex arrangement [35]. Alternatively, it is conceivable that intercalation of TMPyP4 could simply shield the guanines of the G4 sequence from oxidative attack. Additional studies will be required to define the mechanism by which TMPyP4 suppresses oxidative damage to G4 sequences.
During hypoxic exposure we noted that the selected G4 sequences associated with the BER enzymes, Ogg1 and Ref-1/APE1, to a greater extent than control sequences and that the G4 sequences displayed transient strand breaks. These findings provide direct evidence in living cells that G-quadruplex motifs are unusually sensitive to oxidant stress. That G4 damage is increased in the context of hypoxia, a physiologically and pathologically-relevant stimulus utilizing ROS as second messengers, has implications relating to the mechanisms of spontaneous mutation and in terms of the documented acquisition of highly aggressive and metastatic behavior of tumor cells [24–27].
While there is mounting evidence that G-quadruplexes play a role in gene regulation, very little is known about the nature or mechanism of the regulatory function. Genome-wide studies of G-quadruplexes have shown that stabilizing promoter G4 structures enhances transcription of some genes while inhibiting others (Ref). The precise mechanism by which G4 structures govern expression is further complicated by the discovery that the opposite C-rich DNA strand can, at least in some cases, form a structure called an i-motif which may also function in gene regulation [2, 15]. The most thoroughly studied G-quadruplex resides in the nuclease hypersensitive element III1 (NHEIII1) region of the Myc promoter [15, 17]. In the current model for Myc transcriptional regulation, the G-quadruplex serves as both a silencer element and as a buffer for the negative supercoiling generated during transcription [17]. Because G4 structures are remarkably stable, in order to play such a regulatory role they would require the assistance of proteins in order to destabilize and return to normal B-form DNA. In the aforementioned model, the primary protein that is predicted to serve this function is Topoisomerase I. Upon Myc activation toposiomerase I nicks the DNA releasing the negative supercoiling and prompting disassembly of the G-quadruplex structure [15, 17]. Here we propose that oxidation of the G4 sequence and the attendant repair process could perform an analogous function under conditions of oxidant stress, where transcription of multiple genes must be altered rapidly in order for cell survival and adaptation. Furthermore G4 sequences often contain or overlap putative transcription factor binding sites. Therefore, formation and disassembly of the G-quadruplex structure as well as alteration of the DNA sequence through oxidation could also have a role in regulation of transcription factor binding as suggested previously.
Our model also contends that base oxidation and repair in G-quadruplex sequences is necessary but not sufficient for gene expression. This position derives from the observation that promoter G4 sequences in genes that are not regulated by hypoxia also are targets for base oxidation and repair enzyme recruitment. It follows then that G-quadruplex oxidation could be a relatively indiscriminate process designed to rapidly prime a broad range of stress response genes by removing supercoiling, altering nucleosome placement, and modulating transcription factor binding to the motif, but that the availability of key transcription factors to interact with distinct promoter sites is critically required for transcriptional activation.
Another interesting functional implication of the oxidative base modifications in G4 sequences is their impact on G4-protein interactions. A recent genome-wide analysis demonstrated that zinc finger transcription factors ubiquitously associate with promoter G4 sequences and postulated that such binding interactions may contribute to G4 functional activity [14]. In the present study, we found that Sp1 associated with the G4 sequence located within the VEGF promoter in normoxia, but was evicted during hypoxic exposure. Loss of Sp1 was prevented when the oxidative base modifications in the G4 sequence were suppressed by the mitochondrial complex III inhibitor, myxothiazol. Interestingly, it has previously been shown that Sp1 binding to model oligonucleotide was attenuated when 8-oxoguanine was incorporated into the consensus Sp1 binding site [44]. These findings raise the interesting possibility that transcription factor interactions with G4 sequences might be influenced by the introduction of oxidative base modifications and their repair, and in so doing, impact functional properties of the sequence.
In summary, our data demonstrate that about 25% of hypoxia-regulated genes harbor putative G4 sequences in close proximity to transcription start sites and that these sequences are oxidatively damaged in hypoxia. The findings of this study point to a mechanistic basis for instability and mutation associated with G-quadruplex motifs. The present observations also attest to the probable complexity of G4 involvement in transcriptional regulation. We detected no associations between the impact of hypoxia on transcriptional activity in specific genes, the presence G4 sequences in promoters, and the kinetics of DNA damage and repair in the specific G4 sequence. On the other hand, introduction of oxidative base damage in the sequence impacted transcription factor binding, and the formation of strand breaks would be expected to profoundly alter conformational properties of the sequences. Additional studies will be needed to understand the biological roles of oxidative damage to G4 sequences in hypoxia-induced signaling.
Conclusion
Hypoxia, one of many physiological stimuli using reactive oxygen species as second messengers, causes oxidative base modifications in G-quadruplex sequences that are associated with recruitment of base excision DNA repair enzymes and formation of DNA strand breaks. While the biological significance of the hypoxia-induced base oxidation on G-quadruplex sequences cannot be discerned from the present observations, the finding that base modifications were associated with eviction of the transcription factor Sp1 suggests that base lesions could impact transcriptional regulatory functions of G4 sequences. Finally, given the importance of mutations in G4 sequences to evolution of the cancer genome, the present observation that G4 sequences are hot spots for hypoxia-induced base modifications suggests a mechanistic basis for the apparent high G4 mutational rate.
Supplementary Material
We tested the idea that DNA G-quadruplex sequences are prone to oxidative damage.
G-quadruplex sequences are prone to oxidative damage in hypoxia.
Damage attracts base excision repair enzymes and associates with DNA strand breaks.
The damage affects transcription factor binding in the vicinity.
Acknowledgements
This work was supported by the National Institutes of Health [RO1 HL58234 and RO1 HL073244 to M.N.G., UL1 RR025780 to M.W.G.]; and the American Heart Association [09PRE2260451 to D.W.C.]. Funding for open access charge: National Institutes of Health HL073244.
Abbreviations
- G4
G-quadruplex
- 8-oxoG
8-oxoguanine
- BER
base excision repair
- Avpr1a
arginine vasopressin receptor 1a
- Brca2
breast cancer 2
- Dusp10
dual specificity phosphatase 10
- Smad6
Mothers against DPP homolog 6
- Mt1a
Metallothionein 1a
- Nfkb2
Nuclear factor of kappa light polypeptide gene enhancer in B cells
- Vegf
Vascular endothelial growth factor
- HRE
hypoxic response element
- ROS
reactive oxygen species
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Huppert JL. Structure, location and interactions of g-quadruplexes. Febs J. 2010;277:3452–3458. doi: 10.1111/j.1742-4658.2010.07758.x. [DOI] [PubMed] [Google Scholar]
- 2.Brooks TA, Kendrick S, Hurley L. Making sense of g-quadruplex and i-motif functions in oncogene promoters. Febs J. 2010;277:3459–3469. doi: 10.1111/j.1742-4658.2010.07759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 4.Kendrick S, Hurley LH. The role of g-quadruplex/i-motif secondary structures as cis-acting regulatory elements. Pure Appl Chem. 2010;82:1609–1621. doi: 10.1351/PAC-CON-09-09-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang R, Lin Y, Zhang CT. Greglist: A database listing potential g-quadruplex regulated genes. Nucleic Acids Res. 2008;36:D372–D376. doi: 10.1093/nar/gkm787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yadav VK, Abraham JK, Mani P, Kulshrestha R, Chowdhury S. Quadbase: Genome-wide database of g4 DNA--occurrence and conservation in human, chimpanzee, mouse and rat promoters and 146 microbes. Nucleic Acids Res. 2008;36:D381–D385. doi: 10.1093/nar/gkm781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hammond-Kosack MC, Docherty K. A consensus repeat sequence from the human insulin gene linked polymorphic region adopts multiple quadriplex DNA structures in vitro. FEBS Lett. 1992;301:79–82. doi: 10.1016/0014-5793(92)80214-2. [DOI] [PubMed] [Google Scholar]
- 10.Murchie AI, Lilley DM. Supercoiled DNA and cruciform structures. Methods Enzymol. 1992;211:158–180. doi: 10.1016/0076-6879(92)11010-g. [DOI] [PubMed] [Google Scholar]
- 11.Fry M. Tetraplex DNA and its interacting proteins. Front Biosci. 2007;12:4336–4351. doi: 10.2741/2391. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez V, Guo K, Hurley L, Sun D. Identification and characterization of nucleolin as a c-myc g-quadruplex-binding protein. J Biol Chem. 2009;284:23622–23635. doi: 10.1074/jbc.M109.018028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez V, Hurley LH. The c-myc nhe iii(1): Function and regulation. Annu Rev Pharmacol Toxicol. 2010;50:111–129. doi: 10.1146/annurev.pharmtox.48.113006.094649. [DOI] [PubMed] [Google Scholar]
- 14.Kumar P, Yadav VK, Baral A, Saha D, Chowdhury S. Zinc-finger transcription factors are associated with guanine quadruplex motifs in human, chimpanzee, mouse and rat promoters genome-wide. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brooks TA, Hurley LH. The role of supercoiling in transcriptional control of myc and its importance in molecular therapeutics. Nat Rev Cancer. 2009;9:849–861. doi: 10.1038/nrc2733. [DOI] [PubMed] [Google Scholar]
- 16.Dexheimer TS, Carey SS, Zuohe S, Gokhale VM, Hu X, Murata LB, Maes EM, Weichsel A, Sun D, Meuillet EJ, Montfort WR, Hurley LH. Nm23-h2 may play an indirect role in transcriptional activation of c-myc gene expression but does not cleave the nuclease hypersensitive element iii(1) Mol Cancer Ther. 2009;8:1363–1377. doi: 10.1158/1535-7163.MCT-08-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun D, Hurley LH. The importance of negative superhelicity in inducing the formation of g-quadruplex and i-motif structures in the c-myc promoter: Implications for drug targeting and control of gene expression. J Med Chem. 2009;52:2863–2874. doi: 10.1021/jm900055s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hurley LH. Secondary DNA structures as molecular targets for cancer therapeutics. Biochem Soc Trans. 2001;29:692–696. doi: 10.1042/0300-5127:0290692. [DOI] [PubMed] [Google Scholar]
- 20.Balasubramanian S, Hurley LH, Neidle S. Targeting g-quadruplexes in gene promoters: A novel anticancer strategy? Nat Rev Drug Discov. 2011;10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brooks TA, Hurley LH. Targeting myc expression through g-quadruplexes. Genes Cancer. 2010;1:641–649. doi: 10.1177/1947601910377493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rangan A, Fedoroff OY, Hurley LH. Induction of duplex to g-quadruplex transition in the c-myc promoter region by a small molecule. J Biol Chem. 2001;276:4640–4646. doi: 10.1074/jbc.M005962200. [DOI] [PubMed] [Google Scholar]
- 23.Sun D, Liu WJ, Guo K, Rusche JJ, Ebbinghaus S, Gokhale V, Hurley LH. The proximal promoter region of the human vascular endothelial growth factor gene has a g-quadruplex structure that can be targeted by g-quadruplex-interactive agents. Mol Cancer Ther. 2008;7:880–889. doi: 10.1158/1535-7163.MCT-07-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duquette ML, Huber MD, Maizels N. G-rich proto-oncogenes are targeted for genomic instability in b-cell lymphomas. Cancer Res. 2007;67:2586–2594. doi: 10.1158/0008-5472.CAN-06-2419. [DOI] [PubMed] [Google Scholar]
- 25.Kilburn AE, Shea MJ, Sargent RG, Wilson JH. Insertion of a telomere repeat sequence into a mammalian gene causes chromosome instability. Mol Cell Biol. 2001;21:126–135. doi: 10.1128/MCB.21.1.126-135.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu D, Gruber A, Peterson C, Pisa P. Supression of telomerase activity in hl60 cells after treatment with differentiating agents. Leukemia. 1996;10:1354–1357. [PubMed] [Google Scholar]
- 27.Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Intracellular transcription of g-rich dnas induces formation of g-loops, novel structures containing g4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De S, Michor F. DNA secondary structures and epigenetic determinants of cancer genome evolution. Nat Struct Mol Biol. 2011;18:950–955. doi: 10.1038/nsmb.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merino EJ, Boal AK, Barton JK. Biological contexts for DNA charge transport chemistry. Curr Opin Chem Biol. 2008;12:229–237. doi: 10.1016/j.cbpa.2008.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen JH, Hales CN, Ozanne SE. DNA damage, cellular senescence and organismal ageing: Causal or correlative? Nucleic Acids Res. 2007;35:7417–7428. doi: 10.1093/nar/gkm681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Genereux JC, Boal AK, Barton JK. DNA-mediated charge transport in redox sensing and signaling. J Am Chem Soc. 2010;132:891–905. doi: 10.1021/ja907669c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henle ES, Han Z, Tang N, Rai P, Luo Y, Linn S. Sequence-specific DNA cleavage by fe2+-mediated fenton reactions has possible biological implications. J Biol Chem. 1999;274:962–971. doi: 10.1074/jbc.274.2.962. [DOI] [PubMed] [Google Scholar]
- 33.Limoli CL, Giedzinski E, Morgan WF, Swarts SG, Jones GD, Hyun W. Persistent oxidative stress in chromosomally unstable cells. Cancer Res. 2003;63:3107–3111. [PubMed] [Google Scholar]
- 34.Ohno M, Miura T, Furuichi M, Tominaga Y, Tsuchimoto D, Sakumi K, Nakabeppu Y. A genome-wide distribution of 8-oxoguanine correlates with the preferred regions for recombination and single nucleotide polymorphism in the human genome. Genome Res. 2006;16:567–575. doi: 10.1101/gr.4769606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delaney S, Barton JK. Charge transport in DNA duplex/quadruplex conjugates. Biochemistry. 2003;42:14159–14165. doi: 10.1021/bi0351965. [DOI] [PubMed] [Google Scholar]
- 36.Brown JM. Exploiting the hypoxic cancer cell: Mechanisms and therapeutic strategies. Mol Med Today. 2000;6:157–162. doi: 10.1016/s1357-4310(00)01677-4. [DOI] [PubMed] [Google Scholar]
- 37.Yoo YG, Christensen J, Gu J, Huang LE. Hif-1alpha mediates tumor hypoxia to confer a perpetual mesenchymal phenotype for malignant progression. Sci Signal. 2011;4:pt4. doi: 10.1126/scisignal.2002072. [DOI] [PubMed] [Google Scholar]
- 38.Grishko V, Solomon M, Breit JF, Killilea DW, Ledoux SP, Wilson GL, Gillespie MN. Hypoxia promotes oxidative base modifications in the pulmonary artery endothelial cell vegf gene. Faseb J. 2001;15:1267–1269. doi: 10.1096/fj.00-0755fje. [DOI] [PubMed] [Google Scholar]
- 39.Ziel KA, Grishko V, Campbell CC, Breit JF, Wilson GL, Gillespie MN. Oxidants in signal transduction: Impact on DNA integrity and gene expression. FASEB J. 2005;19:387–394. doi: 10.1096/fj.04-2805com. [DOI] [PubMed] [Google Scholar]
- 40.Pastukh V, Ruchko M, Gorodnya O, Wilson GL, Gillespie MN. Sequence-specific oxidative base modifications in hypoxia-inducible genes. Free Radic Biol Med. 2007;43:1616–1626. doi: 10.1016/j.freeradbiomed.2007.08.027. [DOI] [PubMed] [Google Scholar]
- 41.Ruchko MV, Gorodnya OM, Pastukh VM, Swiger BM, Middleton NS, Wilson GL, Gillespie MN. Hypoxia-induced oxidative base modifications in the vegf hypoxia-response element are associated with transcriptionally active nucleosomes. Free Radic Biol Med. 2009;46:352–359. doi: 10.1016/j.freeradbiomed.2008.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a g-quadruplex in a promoter region and its targeting with a small molecule to repress c-myc transcription. Proc Natl Acad Sci U S A. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Breit JF, Ault-Ziel K, Al-Mehdi AB, Gillespie MN. Nuclear protein-induced bending and flexing of the hypoxic response element of the rat vascular endothelial growth factor promoter. Faseb J. 2008;22:19–29. doi: 10.1096/fj.07-8102com. [DOI] [PubMed] [Google Scholar]
- 44.Ghosh R, Mitchell DL. Effect of oxidative DNA damage in promoter elements on transcription factor binding. Nucleic Acids Res. 1999;27:3213–3218. doi: 10.1093/nar/27.15.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Starkov AA, Fiskum G. Myxothiazol induces h(2)o(2) production from mitochondrial respiratory chain. Biochem Biophys Res Commun. 2001;281:645–650. doi: 10.1006/bbrc.2001.4409. [DOI] [PubMed] [Google Scholar]
- 46.Chi JT, Wang Z, Nuyten DS, Rodriguez EH, Schaner ME, Salim A, Wang Y, Kristensen GB, Helland A, Borresen-Dale AL, Giaccia A, Longaker MT, Hastie T, Yang GP, van de Vijver MJ, Brown PO. Gene expression programs in response to hypoxia: Cell type specificity and prognostic significance in human cancers. PLoS Med. 2006;3:e47. doi: 10.1371/journal.pmed.0030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ning W, Chu TJ, Li CJ, Choi AM, Peters DG. Genome-wide analysis of the endothelial transcriptome under short-term chronic hypoxia. Physiol Genomics. 2004;18:70–78. doi: 10.1152/physiolgenomics.00221.2003. [DOI] [PubMed] [Google Scholar]
- 48.Chua YL, Dufour E, Dassa EP, Rustin P, Jacobs HT, Taylor CT, Hagen T. Stabilization of hypoxia-inducible factor-1alpha protein in hypoxia occurs independently of mitochondrial reactive oxygen species production. J Biol Chem. 2010;285:31277–31284. doi: 10.1074/jbc.M110.158485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Comito G, Calvani M, Giannoni E, Bianchini F, Calorini L, Torre E, Migliore C, Giordano S, Chiarugi P. Hif-1alpha stabilization by mitochondrial ros promotes met-dependent invasive growth and vasculogenic mimicry in melanoma cells. Free Radic Biol Med. 2011;51:893–904. doi: 10.1016/j.freeradbiomed.2011.05.042. [DOI] [PubMed] [Google Scholar]
- 50.Hamanaka RB, Chandel NS. Mitochondrial reactive oxygen species regulate hypoxic signaling. Curr Opin Cell Biol. 2009;21:894–899. doi: 10.1016/j.ceb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Poyton RO, Ball KA, Castello PR. Mitochondrial generation of free radicals and hypoxic signaling. Trends Endocrinol Metab. 2009;20:332–340. doi: 10.1016/j.tem.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 52.Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, Chiariotti L, Malorni A, Abbondanza C, Avvedimento EV. DNA oxidation as triggered by h3k9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319:202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- 53.Amente S, Lania L, Avvedimento EV, Majello B. DNA oxidation drives myc mediated transcription. Cell Cycle. 2010;9:3002–3004. doi: 10.4161/cc.9.15.12499. [DOI] [PubMed] [Google Scholar]
- 54.Amente S, Bertoni A, Morano A, Lania L, Avvedimento EV, Majello B. Lsd1-mediated demethylation of histone h3 lysine 4 triggers myc-induced transcription. Oncogene. 2010;29:3691–3702. doi: 10.1038/onc.2010.120. [DOI] [PubMed] [Google Scholar]
- 55.Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG. A topoisomerase iibeta-mediated dsdna break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 56.Gillespie MN, Pastukh V, Ruchko MV. Oxidative DNA modifications in hypoxic signaling. Ann N Y Acad Sci. 2009;1177:140–150. doi: 10.1111/j.1749-6632.2009.05036.x. [DOI] [PubMed] [Google Scholar]
- 57.Gillespie MN, Pastukh VM, Ruchko MV. Controlled DNA "damage" and repair in hypoxic signaling. Respir Physiol Neurobiol. 2010;174:244–251. doi: 10.1016/j.resp.2010.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gillespie MN, Wilson GL. Bending and breaking the code: Dynamic changes in promoter integrity may underlie a new mechanism regulating gene expression. Am J Physiol Lung Cell Mol Physiol. 2007;292:L1–L3. doi: 10.1152/ajplung.00275.2006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.