Abstract
Structural studies have revealed some of the organizing principles and mechanisms involved in the assembly of the COPII coat including the location of the Sec23/24 adapter layer. Previous studies, however, were unable to unambiguously determine the positions of Sec23 and Sec24 in the coat. Here we have determined a cryogenic electron microscopic structure of Sec13/31 together with Sec23. Electron tomography revealed that the binding of Sec23 induces Sec13/31 to form a variety of different geometries including a cuboctahedron, as was previously characterized for Sec13/31 alone. Single particle reconstruction of the Sec13/31-23 cuboctahedra revealed that the binding of Sec23 induces a conformational change in Sec13/31 resulting in a more extended conformation. Docking Sec23 crystal structures into the EM map suggested that Sec24 projects its cargo binding surface out into the large open faces of the coat. These results have implications for the mechanisms by which COPII transports large cargos, cargos with large intracellular domains, and for tethering complexes that must project out of the coat in order to interact with their binding partners. Furthermore, Sec23 binds Sec13/31 at two unique sites in the coat, which suggests that each site may have unique roles in the mechanisms of COPII vesiculation.
Introduction
The COPII proteins Sar1, Sec23/Sec24 (Sec23/24), and Sec13/Sec31 (Sec13/31) are involved in transporting secreted and membrane proteins out of the endoplasmic reticulum (ER) 1-7. The roles of the individual proteins have been worked out through a combination of genetics and biochemistry. Sar1 is a regulatory GTPase that has an amphipathic α-helix at its N-terminus 8,9. Sar1 exchanges GDP for GTP in a reaction that is catalyzed by the ER resident protein Sec12 8,10. In its GTP bound state, Sar1 undergoes a conformational change that exposes its amphipathic helix allowing Sar1 to bind the ER membrane. Once it is bound to the ER, Sar1 recruits Sec23/24 where Sec23 is the Sar1 GTPase activating protein (GAP) and Sec24 is involved in binding cargo proteins. Together Sar1 and Sec23/24 are called the prebudding complex and are thought to be involved in initiating vesicle curvature11. Next, Sec13/31 binds individual prebudding complexes and polymerizes into a cage structure. The collective action of the Sec13/31 and the prebudding complexes are thought to organize the coat into a coherent bud. The final step of budding involves fission of the nascent bud from the ER to form a complete vesicle in a reaction that has been implicated to involve Sar19,12-15.
Though the roles of the individual COPII proteins have been well characterized, the mechanisms by which they perform their activities are largely unknown. Structures of the COPII proteins have yielded many of the mechanistic insights to date. X-ray crystal structures have been determined of each of the COPII proteins and several of their complexes together 11,16-18. These structures have been combined with three-dimensional structures of large COPII complexes determined by cryogenic electron microscopy (cryoEM) to reveal some of the rules dictating the formation of the COPII coat and interactions with cargo. It was shown that Sec13/31 will self-assemble into a cage, and this structure was determined by single particle cryoEM 19. The cryoEM map showed that Sec13/31 assembles into a geometrical cage-like structure with cuboctahedral geometry where Sec13/31 makes up the edges of the cuboctahedron. Four Sec13/31 edges come together to form each of the vertices of the cage. Fitting the crystal structure of Sec13/31 into this map revealed that the assembly of the cage is mediated by contacts between the WD40/β-propeller domains at the N-terminus of Sec31 at the cuboctahedral vertices 17,19,20.
Further insights were gleaned from a cryoEM reconstruction of Sec13/31 assembled in a cage together with Sec23/24 (Sec13/31-23/24)21. This structure formed a 1000 Å cage with icosidodecahedral geometry, thus we will henceforth call it the IDD. The edge structure for this assembly was the same as the cuboctahedron, but the angles between adjacent edges were larger which allowed the cage to assemble with a larger diameter. Sec23/24 in that structure was located underneath the cage vertices, and it was hypothesized that Sec23/24 modulates the diameter of the cage in response to the demands of the cargo. Crystal structures of Sec23/24 were fit into the cryoEM density for the IDD, but the resolution of that reconstruction was too low to unambiguously position the structures in the map. This information is necessary for determining the mechanisms by which the COPII coat interacts with and recruits cargo.
Here we determined the structure of Sec13/31 together with Sec23 (Sec13/31-23) by single particle cryoEM. The specimen showed both conformational and compositional heterogeneity that was minimized by classifying particles by the 1D profiles of their rotational averages. This allowed us to reconstruct the complex with high-enough resolution to identify the density corresponding to Sec23. By combining this information with the reconstruction of Sec13/31-Sec23/24 and the available crystal structures, we constructed a model for the position of Sec23/24 where the cargo binding surfaces of Sec24 face the large open square or pentagonal faces of the COPII coat.
Results
Assembly of the Sec13/31-23 cage
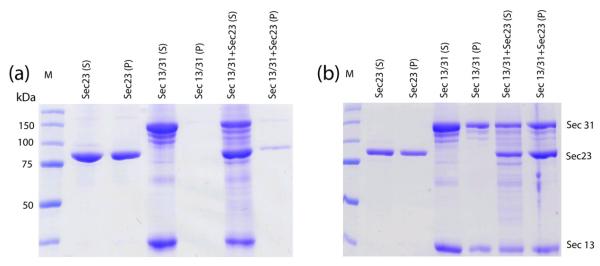
Previously, it was demonstrated that Sec13/31 self-assembles into a cage when the proteins were dialyzed into assembly buffer 19 (see Materials and Methods). Here we assembled Sec13/31 cages together with Sec23 by adding the full-length proteins together in a 1:1 stoichometric ratio and then dialyzing them in assembly buffer. Centrifugation assays were then performed to demonstrate that Sec23 binds to the Sec13/31 cages (Fig. 1). In the absence of Sec13/31, Sec23 precipitated in assembly buffer and was present in the pellet of a low speed spin (Fig. 1a). The presence of Sec13/31, on the other hand, prevented Sec23 from pelleting which suggested an interaction between the proteins. The Sec13/31 cages pellet at 16,000 × g, and pelleting the cages co-precipitates Sec23 (Fig. 1b). Together, these data suggest that Sec23 proteins interact specifically with the assembled Sec13/31 cages.
Figure 1.
Spin down assay. Samples of Sec23 alone, Sec13/31, and Sec13/31-23 were dialyzed into high-salt assembly buffer and then centrifuged at low speed and high speed and pellets and supernatants were run on a PAGE gel. (a) Gel showing supernatant (S) and pellet (P) lanes for samples spun at 500 × g. Lanes are labeled with the sample name for the particular lane. Sec23 pellets in assembly buffer, but is soluble in presence of Sec13/31. (b) Gel showing supernatant and pellet lanes for samples spun at 16,000 × g. The Sec13/31 cages pellet at 16,000 × g, and Sec23 co-precipitates with Sec13/31 cages at this speed.
Compositional and Conformational Heterogeneity in Sec13/31-23
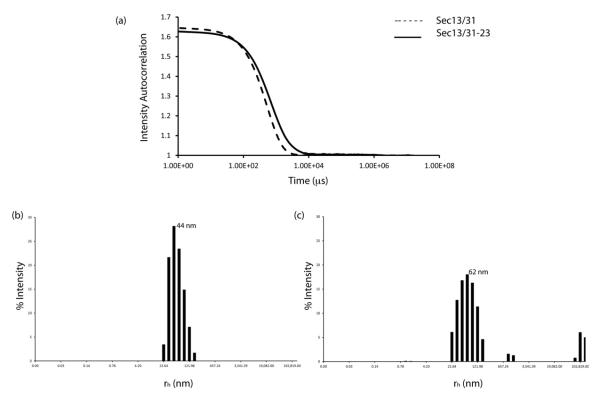
The diameter and polydispersity of the Sec13/31 and Sec13/31-23 samples recovered after the low speed centrifugation were characterized by dynamic light scattering (DLS) and cryoEM. Cumulant analysis of the autocorrelation curves from the DLS (Fig. 2a) indicated that the assembled Sec13/31 particles had a mean hydrodynamic radius (rh) of 440 Å (880 Å diameter) and were 16% polydisperse. Sec13/31-23 particles, on the other hand, were larger and more polydisperse with a mean rh of 620 Å (1240 Å diameter) and 31% polydispersity (Fig. 2b,c). CryoEM confirmed that the Sec13/31-23 particles assembled in a range of sizes with diameters ranging from 500 Å to 800 Å and the majority of the particles displaying a diameter of ~600 Å (Fig. 3a). The discrepancy between the absolute diameters measured by DLS and cryoEM is likely caused by the increased viscosity of the assembly buffer relative to pure water as this would tend to inflate the radii measured by DLS.
Figure 2.
Dynamic light scattering. (a) Autocorrelation curves of fluctuations in light intensity are shown for Sec13/31 (dashed line) and Sec13/31-23 (solid line). (b) Histogram for the size distribution of Sec13/31. Assembled Sec13/31 particles had a polydispersity of 16% with a mean rh of 44 nm (440 Å). (c) Histogram for size distribution of Sec13/31-23. Assembled Sec13/31-23 particles had a polydispersity of 31 % with a mean rh of 62 nm (620) Å.
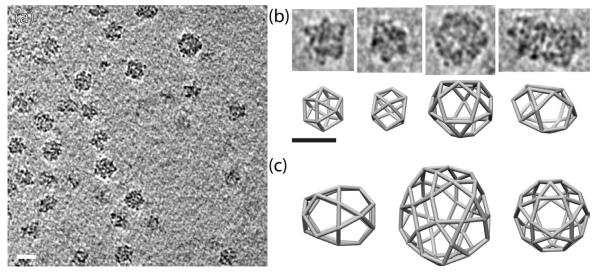
Figure 3.
Tomography of Sec13/31-23 assemblies. (a) Representative tomogram showing Sec13/31-23 assemblies of various sizes. (b) Individual subtomograms of Sec13/13-23 cages (top panel) with corresponding cage geometries that match the size of the individual cages (bottom panel). The two left-most sets of panels in (b) show cuboctahedrons while the two right-most sets of panels show larger more irregular structures. (c) Possible Sec13/31 geometries that form cages with a range of sizes. Scale bar: 600 Å.
We collected electron tomographic tilt series of the sample to distinguish whether the differences in the sizes of the Sec13/31-23 particles were due to particles that assembled with different geometry or cages that were flattened during freezing. When reconstructed into tomograms, the tilt series demonstrated that the 600 Å particles corresponded to the cuboctahedral cages that we have seen previously 22 (Fig. 3a). The particles with diameters > 600 Å were not flattened but instead corresponded to COPII cages with some larger geometry than the cuboctahedrons yet were not large enough to be IDDs that are formed when Sec13/31 is assembled with Sec23/24 21. Instead, they must form with some other geometry that follows the rules determined previously for the formation of COPII cages. In earlier reconstructions, it was shown that the heterotetrameric Sec13/31 edges are off-center such that one end, the so called + end, is closer to a vertex than the other end, the − end. Four Sec13/31 edges must come together to create a vertex, and the α angle (between the + end of one edge and the adjacent edge in the clockwise direction) is 60°, and the β angle (between the − end of an edge and the adjacent edge in the clockwise direction) (Fig. 5a) is 90° to 108° 21. It is possible to create many different types of cage geometries using these rules though these are not spherically circumscribable and are less symmetrical than the cuboctahedron or IDD structures (Fig. 3b,c). Based on these rules, we constructed 3D COPII cage geometries that roughly match the diameter and shape of the cages observed in the tomograms (Fig. 3b). The majority of the cages matched well with a cuboctahedron (Fig. 3b left panels), but there were several larger and irregularly shaped structures. Sec13/31 cages can be constructed to match these shapes (Fig. 3b right panels) but the tomograms were not high enough resolution to resolve the individual Sec13/31 edges in the cages.
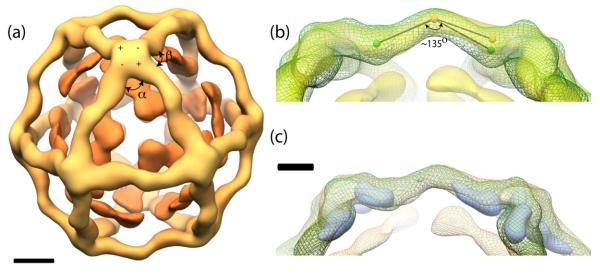
Figure 5.
Single particle reconstruction of Sec13/31-23. (a) An outer Sec13/31 cage structure (light orange) can be observed around inner Sec23 densities (dark orange). The polarity (+ and −) and α and β angles are shown. Scale bar: 100 Å. (b) Superimposition of Sec13/31 (green mesh)19 and Sec13/31-23 (yellow). Approximately 10° increase is shown when an angle is drawn from the center of the edge to a landmark in the Sec13/31 edge structure (spheres). (c) A difference map (blue) derived by subtracting Sec13/31-23 from Sec13/31 demonstrates the conformational change of Sec13/31. Scale bar: 50 Å.
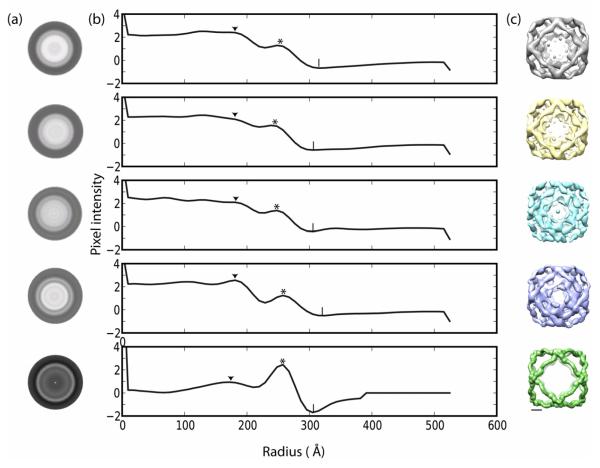
Since the majority of the Sec13/31-23 particles corresponded to cuboctahedral cages, we collected 2850 cryoEM images for single particle reconstruction. 28,000 particles were manually selected, focussing only on the 600 Å cages. The particles were initially characterized by centering, rotationally averaging, and performing multivariate statistical analysis (MSA). Since the vertices of a cuboctahedron are spherically circumscribable, we were able to more accurately classify the particles by their radius and the occupancy of Sec23 by rotationally averaging them (Fig. 4, rows 1-4). The Sec13/31-23 particles were then compared to empty Sec13/31 cages that were used as a control (Fig. 4, row 5). The MSA split the Sec13/31-23 particles into four classes of 7478, 6983, 5528, and 5556 particles in classes 1-4 respectively (Fig. 4a). Radial density plots of the four classes show two prominent features; there is an outer density peak that corresponds to Sec13/31 (* in Fig. 4b) and an inner peak (▼ in Fig. 4b). In the absence of Sec23, the inner peak is small and is less than half the height of the outer peak (Fig. 4b, row 5). The height of the inner peak increases greatly in the presence of Sec23 (Fig. 4b, rows 1-4), thus we attribute increase in peak height to Sec23 binding to Sec13/31. The four Sec13/31-23 classes produced by MSA differ primarily by the height of the inner Sec23 peak, and there also appears to be a difference in the overall radius. In two classes, the Sec23 peak is almost twice the height of the Sec13/31 peak. In class 1 (Fig. 4b, row 1) the Sec23 peak is 1.9 times the Sec13/31 peak, and in class 4 (Fig. 4b, row 4) the Sec23 peak is 2.0 times greater than the Sec13/31 peak. In the other two classes, the Sec23 peak is 1.3 and 1.5 times the Sec13/31 peak respectively (Fig. 4b, rows 2 and 3). The increase in the Sec23 peak correlates with a small increase in radius; the two classes with higher Sec23 peaks have outer radii of ~ 314 Å(| in Fig. 4b, rows 1 and 4) while the other classes have radii of ~ 298 Å (| in Fig. 4b, rows 2 and 3).
Figure 4.
Sub-classification of 600 Å cages by multivariate statistical analysis. (a) Rows 1-4; Rotational average of Sec13/31-23 particles split into four separate classes by MSA; Row 5: Rotational average of Sec13/31 particles. (b) One dimensional plots of pixel intensity vs radius (in Angstrom) of rotationally averaged classes. The radial density shows an outer peak corresponding to Sec13/31(*) and a smaller peak that corresponds to binding of Sec23 to Sec13/31 (▼). Symbol (|) marks the outer radii of the particles. (c) Independent reconstruction of the different classes by single particle refinement. Rows 2 and 3 show structures with artifacts. Scale bar: 100 Å.
Three-dimensional structure of Sec13/31-23
The four classes of Sec13/31-23 particles were independently reconstructed by single particle refinement (Fig. 4c) enforcing octahedral symmetry. Two of the reconstructions converged on structures with features consistent with the COPII proteins, and two generated structures with artifacts (Fig. 4c rows 2 and 3). The latter two structures corresponded to particles with radial class averages that exhibited lower Sec23 occupancy and smaller radii. There are several possible reasons for the artifacts in the reconstructions. For example, Sec23 heterogeneity could cause misclassification, or the particles could be incompletely formed or damaged. The remaining two classes (Fig. 4c row 1 and row 4), had a distinct and well formed inner density from Sec23 and are comparatively larger. Given the similarities between classes 1 and 4, the particles in these classes were combined so that we could achieve a higher-resolution reconstruction. The combined dataset was then refined until it converged, leading to a final reconstruction that had a resolution (FSC0.5) of 38 Å. The final reconstruction shows a cuboctahedral Sec13/31 cage with contours similar to previously determined cage structures and new densities that we attribute to Sec23 underneath the vertices and in the triangular faces (Fig. 5a).
The structure of the Sec13/31 cage is similar to the structure previously observed for Sec13/31 alone 19. The edges of both structures measure approximately 300 Å, have a characteristic curve in the middle of the edge, and show an asymmetry such that one end of a given heterotetramer (the + end) is closer to its vertex than the other end is to its nearest vertex (the − end) 21. A few differences can also be seen. The edges are more extended in the Sec13/31-23 cage as compared to Sec13/31. The original Sec13/31 cage reconstruction shows a ~135° (Fig. 5b) angle in the center of the heterotetramer 19. This angle appears to expand by ~10° in the presence of Sec23 (Fig. 5b), and this is confirmed by a difference map calculated by subtracting Sec13/31-23 from Sec13/31. The difference map (Fig. 5c blue) shows regions of maximum difference to be near the vertices.
Identification of the position of Sec23 in the COPII coat
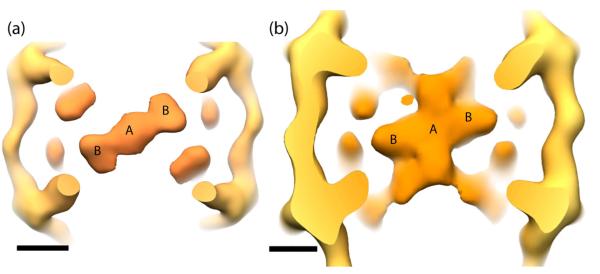
In addition to the outer layer of Sec13/31 density in the 3D reconstruction, there is an inner layer that we interpret to be Sec23 (Fig. 5a). In the previously determined IDD structure of Sec13/31 together with Sec23/24, we were unable to unambiguously assign densities for Sec23 and Sec24 respectively 21. In comparing the IDD with the current map, we see similar features, but there are differences with respect to the IDD densities that run along the edges and extend from the central-most densities (Fig. 6). The new structure determined here allows us to assign Sec23 to the density located underneath the Sec13/31 vertices and in the middle of the triangular faces (Fig. 6a) and allows us build a model for the position of Sec24.
Figure 6.
Comparison of Sec13/31-23 with Sec13/31-23/24 by viewing along the two-fold symmetry axis. (a) The Sec23 density lies just inside the Sec13/31 cage. (b) Similar densities are seen in Sec13/31-23/24. Two locations where Sec23 binds Sec13/31 are marked as ‘A’ and ‘B’. Scale bar: 100 Å.
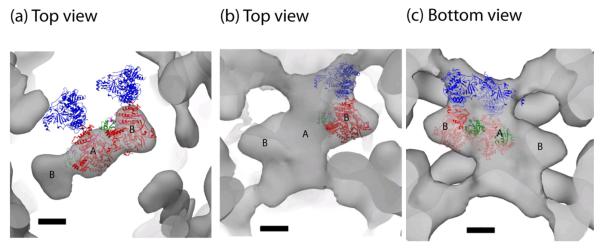
Crystal structures for Sec13/31 and Sec23 are available 11,17,18 and were fit into the Sec13/31-23 map. As seen previously, the crystal structure for the yeast Sec13/31 heterotetramer fits well into the edge of the Sec13/31 cage. The overall topology of the yeast heterotetramer agrees well with the edge of the new cryoEM Sec13/31-23 structure with lobes of density at the ends of the edges corresponding to the WD40/β propeller domains of Sec13 and Sec31. Sec23 also fits well in the cryoEM map. Viewed along the two-fold symmetry axis, the Sec23 density appears as a structure with a central density (position A) and two symmetry-related lobes (positions B) (Fig. 6a). Copies of a Sec23 crystal structure fit in each of the lobes in position B, and one copy fits in the central density in position A (Fig. 7a). Recently, a crystal structure was solved of Sec23 in complex with Sar1 and the Sar1 GAP activating peptide from Sec3118. The position of the Sec31 peptide in that structure determines which side of Sec23 faces Sec13/31 and which faces the membrane. Combining this information with the structure of the IDD suggests that there is only one position that satisfies the previous constraint and positions Sec23 such that Sec24 fills the remaining density in the IDD in both positions A and B (Fig. 7b,c). In these orientations, Sec23/24 in the IDD is nearly orthogonal to our previous model. In the earlier model, Sec24 ran approximately parallel to the Sec13/31 heterotetramers. Based on our new structure, however, we predict that Sec24 is rotated ~90° with respect to the previous model such that it projects out into the large open square faces of the cuboctahedral structure (equivalent to the pentagonal faces of the IDD). A consequence of this docking is that Sar1 in position A is oriented away from the two-fold while Sar1 in position B is oriented towards the two-fold axis (Fig. 7a,c). Another consequence is that the regions of Sec24 that bind cargo are now facing the middle of the large open square faces of the cage, and this position of Sec24 is ideal to interact with potential cargo to form pre-budding complexes.
Figure 7.
COPII protein crystal structure docking into the EM density. (a) Crystal structure of Sec23 (red) fit into the central (position A) and side lobe (position B). Sec24 (blue) and Sar1 (green) are shown in their respective positions based on the Sec23 docking. (b) Top view of a crystal structure of Sec23/24-Sar1 docked into the Sec13/31-23/24 IDD density map. (c) Bottom view showing positions of Sar1. Scale bar: 50 Å.
Discussion
Here we have determined the structure of Sec13/31 together with Sec23. Our cryoEM reconstruction reveals that Sec23 in the COPII coat is located underneath the Sec13/31 vertices and extends toward the triangular faces. Analysis of the structure has revealed some new principles in COPII coat design.
Cage heterogeneity
Addition of stoichiometric amounts of Sec23 and Sec13/31 formed cages of various sizes (500-800 Å). Using rules for cage assembly 19 that four edges come together to form a vertex, we were able to construct cages of similar shape and size of the structures observed in tomograms of the Sec13/31-23 assemblies. This suggests that COPII proteins may form cages of a variety of sizes instead of the discrete symmetrical cuboctahedron and icosidodecahedron observed previously. This observation agrees with observations of complete COPII coated vesicles which display a range of sizes averaging 500-700 Å in diameter 1,23. Among the 600 Å cages, we also observed heterogeneity in the occupancy of Sec23 that correlated with a slight increase in cage diameter. This type of flexibility was predicted by the yeast Sec13/31 crystal structure which is more linear than the curved Sec13/31 that we observe in the EM structures17. Our data shows comparatively larger diameter cages for Sec13/31-23 samples than Sec13/31 alone (Fig. 2, 4), suggesting that part of the Sec13/31 edge curvature exists to accommodate an expansion of the cage upon binding Sec23. Moreover, these properties give COPII coats the ability to adapt to the physical demands of cargo during secretion by flexing and straightening to accommodate cargo of different shapes and sizes.
Sec13/31-23 forms a cuboctahedron instead of an icosidodecahedron
The fact that the Sec13/31-23 cage forms with cuboctahedral geometry or with other geometry smaller than IDD, supports the hypothesis that Sec24 is capable of further influencing the diameter of the cage in response to the needs of cargo21. In our previous reconstruction of Sec13/31-Sec23/24, the structure formed a 1000 Å diameter assembly with icosidodecahedral geometry. The current structure has a diameter of 600 Å with cuboctahedral geometry. This suggests that Sec24 is involved in influencing the diameter of the cage. The mechanism by which this occurs is unclear, but it is likely that Sec24 is in position underneath the vertices to interact sterically with Sec13/31 and influence the diameter of the cage in response to the requirements of the specific cargo with which they are interacting.
Symmetry mismatch
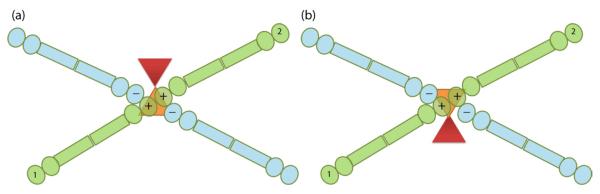
Sec23 appears to have two binding sites called positions A and B (Fig. 6a). The density in position A spans the two-fold symmetry axis and is large enough to accommodate only one Sec23. This suggests that there is a symmetry mismatch at this Sec23 binding site. The symmetry mismatch is made possible by the fact that only one of the two Sec31 + ends at the vertices can interact with Sec23 at a time, given their proximity to the two-fold symmetry axis (Fig. 8). A consequence of this is that in the reconstruction, two Sec23s are rotated 180° with respect to each other and are averaged at that position. At the current resolution, this appears to blur the density so that it takes on a roughly square shape instead of the triangular density expected for a single Sec23 protein.
Figure 8.
Model for symmetry mismatch of Sec23 at position A. In our model, Sec23 at position A can be oriented in one of two ways depending on which Sec13/31 + end it interacts with at the vertex. Panel (a) demonstrates interaction with a given Sec13/31 (1, green), while panel (b) demonstrates interaction with the symmetry related Sec13/31 (2, green). The two positions orient Sec23 180° from each other and consequently orient Sec24 (red) in opposite directions in each position.
Position of Sec24 and cargo binding
The positioning of Sec23 shown here places Sec24 in the middle of the square faces of the Sec13/31 cuboctahedron (and the pentagonal faces for the IDD). This position exposes the cargo binding domain of Sec24 in an open area of the structure. This position gives Sec24 plenty of free space to interact with cargo and allows cargo proteins with large cytosolic domains to extend out of the large open pores in the structure 11. This also has relevance for activities like tethering. For example, the tether complex TRAPPI remains bound to Sec24 until it tethers with its receptor in the ER-Golgi intermediate complex (ERGIC) 24. With the tether complexes extending out of the open faces, they are more available to interact with their binding partners.
Do the different Sec23 binding sites have different roles in vesiculation?
The fact that there are two unique Sec23 binding sites leads us to hypothesize that the two sites may have different roles in vesiculation. One possibility is that position A (below the vertices) is involved in building and maintaining the integrity of the coat vertices in response to the GTP/GDP state of Sar125. This would help explain how nucleotide state of Sar1 is transduced to the rest of the coat to mediate assembly and disassembly. In this model, position B (in the middle of the triangular face) would be more important in cargo or tether binding via Sec24. More experiments are required to test these hypotheses. It is important to note that residues 763-1273 of Sec31 (including the proline-rich region) are not accounted for in our structure or any of the Sec31 structures determined so far. It is assumed that this region of Sec13/31 is highly mobile and so is unresolved with both x-ray crystallography and cryoEM techniques. It is possible, however, that some of the density that we observe here is attributable to the unresolved region of Sec31.
Our results now demonstrate that Sec23 binds to the Sec13/31 cage underneath the vertices and in the triangular faces. This positioning suggests that Sec24 points its cargo binding domain in the middle of the large open faces of the cage. However, many questions remain: how is Sar1 involved in fission, how is curvature initiated, how are cargo positioned in the coat, etc. Clearly much more structural work is necessary to reveal all the mechanisms of COPII vesiculation and ultimately reveal an atomic model of complete COPII coated vesicles.
Materials and Methods
Preparation of Sec13/31, Sec23 COPII Coats
Human Sec23A-His, Sec24C-His, Sec13R, and Sec31L1 (GenBank accession NM_006364.2, NM_004922.2, NM_183352, and NM_014933, respectively) were expressed in Sf9 and Hi5 insect cells using baculovirus and purified as described 19. COPII coats were assembled by incubating equimolar ratios of purified human Sec13/31 and Sec23 on ice or at 4°C for 30 min followed by dialysis against assembly buffer (25 mM HEPES (pH 7.5), 700 mM KOAc, 1 mM MgOAc, 1 mM DTT) overnight. Samples were concentrated to 5-6 mg/ml using Agilent Technologies concentrators spin 10K MWCO.
Dynamic light scattering
DLS analysis were carried out at 20°C using Protein solutions DynaPro instrument with temperature controlled microsampler. Data analysis was done using Dynamics V6.12.0.3 software (Wyatt Technology Corp.). Eighty microlitres of purified Sec13/31 and Sec13/31-23 (~ 1 mg/ml) in assembly buffer were first spun at 16000 g and then transferred to a cuvette and allowed to equilibrate at 20°C before taking ten readings with an acquisition time of 20 s each. This was repeated a minimum of three times for reproducibility. Regularization algorithms in Dynamics were used to calculate size distribution in Fig. 2b and c. Cumulant fits to the autocorrelation curves were used to calculate the polydispersity and the mean hydrodynamic radius (rh).
Cryo-Electron Microscopy
For cryo-EM analysis the COPII coats were preserved in vitreous ice. Four microliters of the Sec13/31-23 sample were placed on a Quantifoil R2/2 400 mesh holey carbon grid that had been plasma-cleaned for 12 s using a Gatan SOLARIS model 950 advanced plasma cleaner. The samples were fixed moments before freezing by adding 3μL of 2.5% glutaraldehyde to the sample already on the grid. The sample was vitrified using an FEI Vitrobot (FEI Company). Data were collected using Titan Krios (FEI) transmission electron microscope operated at 300 keV that was equipped with a Gatan Ultrascan 4k × 4k CCD camera and the Leginon software for automatic data acquisition 26. 2850 exposure pairs at near to focus (1-3μm) and far from focus (−5 μm) were collected at a dose of 30 e−/Å2 and magnification of 37,000.
Tomography
Tilt series were acquired using Leginon’s tomography application. Images were collected at 29,000 × magnification which corresponded to a pixel size of 3.05 Å/pixel at the specimen level and were collected through a range ±68° with an angular step of 2°. The total dose for the series was 200 e−/Å2. The relative shifts between images and the position of the tilt axis was determined using marker-free alignment with the PROTOMO27 software package. 3D volumes were created by weighted backprojection.
Single-Particle Processing
Images were processed using Appion28. Particles were picked using template based matching, and this resulted in a 32,000 particle data set that was then manually edited to keep good particles. This resulted in a final dataset of 28,000 particles. The contrast transfer function (CTF) was estimated using the ACE software package29 integrated into Appion. Phases were corrected for individual particles based on the defocus reported from ACE for the particular micrograph from which the particle was picked. Particles were initially reconstructed using EMAN refinement30. The near-to-focus particles were used initially, but contrast for those particles was insufficient to generate a coherent reconstruction whereas the far-from-focus particles generated reconstructions that had features consistent with COPII cages. Thus, the far-from-focus particles were used for all subsequent processing.
Particle heterogeneity
Conformational and compositional heterogeneity was minimized by classifying the radial density profiles for the individual particles. Particles were first centered and rotationally averaged. SPIDER31 scripts for correspondence analysis and hierarchical ascendant classification were used to split the rotationally averaged particles into four classes that differed in their radii and the occupancy of Sec23. The original particles corresponding to the four classes were refined against an earlier Sec13/31 reconstruction (EMDB ID1232) to convergence using EMAN. The 1st and 4th subclass generated good reconstructions with similar features (Fig. 4c). The particles from these sets were combined to make a relatively homogeneous set of particles for further refinement. These particles were refined for 10 iterations using EMAN projection matching, and heterogeneity was further reduced by performing MSA on each class of projections between iterations. Correspondence analysis with hierarchical ascendant classification was performed generating 2-5 subclasses for each projection. Only the subclass that correlated best with the projection of the model for that iteration was to contribute to the reconstruction for a given iteration. The resolution of the final reconstruction was 38 Å, as measured by the FSC0.5 criterion (Fig 5a). A total of 9571 particles went into the final reconstruction.
Model cage creation
Cage lattices that roughly match the shape and diameters of cages observed in the cryo-tomograms were constructed using the Cage Builder module of UCSF Chimera 32. Subtomograms for individual cages were visualized in Chimera and the cage lattices were built around the individual density maps.
Calculating the bend angle for the Sec13/31 edge
The bend angle at the center of the Sec13/31 edge for both the Sec13/31 cage and the Sec13/31-23 cage was calculated using UCSF Chimera, alpha version 1.6. For each map, a reference point was placed in the center of an edge, and two other points were placed at the center of the densities for Sec13 on either end of the given Sec13/31 heterotetramer. Angles were then calculated between the three points for both the Sec13/31 cage and Sec13/31-23 cage.
Highlights.
The structure of the Sec13/31 cage together with Sec23 was determined.
Binding Sec23 induces heterogeneity in the Sec13/31 cage.
Sec23 binds Sec13/31 at two sites one of which crosses the two-fold symmetry axis.
Docking Sec23/24 in the structure positions Sec24 towards the open faces of the cage.
Acknowledgments
We thank Dr. Beth Stroupe for helpful discussions. The work was supported by grants from the National Institutes of Health (GM086892) and the American Heart Association (#0835300N).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers
The Sec13/31-23 cage structure was deposited in the Electron Microscopy Database under the accession number EMD-5408.
References
- 1.Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach MF, Ravazzola M, Amherdt M, Schekman R. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 1994;77:895–907. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- 2.Shaywitz DA, Espenshade PJ, Gimeno RE, Kaiser CA. COPII subunit interactions in the assembly of the vesicle coat. J Biol Chem. 1997;272:25413–25416. doi: 10.1074/jbc.272.41.25413. [DOI] [PubMed] [Google Scholar]
- 3.Antonny B, Gounon P, Schekman R, Orci L. Self-assembly of minimal COPII cages. EMBO Rep. 2003;4:419–424. doi: 10.1038/sj.embor.embor812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bickford LC, Mossessova E, Goldberg J. A structural view of the COPII vesicle coat. Curr Opin Struct Biol. 2004;14:147–153. doi: 10.1016/j.sbi.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 5.Watson P, Stephens DJ. ER-to-Golgi transport: form and formation of vesicular and tubular carriers. Biochim Biophys Acta. 2005;1744:304–315. doi: 10.1016/j.bbamcr.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Townley AK, Feng Y, Schmidt K, Carter DA, Porter R, Verkade P, Stephens DJ. Efficient coupling of Sec23-Sec24 to Sec13-Sec31 drives COPII-dependent collagen secretion and is essential for normal craniofacial development. J Cell Sci. 2008;121:3025–3034. doi: 10.1242/jcs.031070. [DOI] [PubMed] [Google Scholar]
- 7.Russell C, Stagg SM. New Insights into the Structural Mechanisms of the COPII Coat. Traffic. 2009;11:303–310. doi: 10.1111/j.1600-0854.2009.01026.x. [DOI] [PubMed] [Google Scholar]
- 8.Huang M, Weissman JT, Beraud-Dufour S, Luan P, Wang C, Chen W, Aridor M, Wilson IA, Balch WE. Crystal structure of Sar1-GDP at 1.7 A resolution and the role of the NH2 terminus in ER export. J Cell Biol. 2001;155:937–948. doi: 10.1083/jcb.200106039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee MC, Orci L, Hamamoto S, Futai E, Ravazzola M, Schekman R. Sar1p N-terminal helix initiates membrane curvature and completes the fission of a COPII vesicle. Cell. 2005;122:605–617. doi: 10.1016/j.cell.2005.07.025. [DOI] [PubMed] [Google Scholar]
- 10.Bielli A, Haney CJ, Gabreski G, Watkins SC, Bannykh SI, Aridor M. Regulation of Sar1 NH2 terminus by GTP binding and hydrolysis promotes membrane deformation to control COPII vesicle fission. J Cell Biol. 2005;171:919–924. doi: 10.1083/jcb.200509095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bi X, Corpina RA, Goldberg J. Structure of the Sec23/24-Sar1 pre-budding complex of the COPII vesicle coat. Nature. 2002;419:271–277. doi: 10.1038/nature01040. [DOI] [PubMed] [Google Scholar]
- 12.Yoshihisa T, Barlowe C, Schekman R. Requirement for a GTPase-activating protein in vesicle budding from the endoplasmic reticulum. Science. 1993;259:1466–1468. doi: 10.1126/science.8451644. [DOI] [PubMed] [Google Scholar]
- 13.Haucke V. Vesicle budding: a coat for the COPs. Trends Cell Biol. 2003;13:59–60. doi: 10.1016/s0962-8924(02)00011-9. [DOI] [PubMed] [Google Scholar]
- 14.Sato K, Nakano A. Mechanisms of COPII vesicle formation and protein sorting. FEBS Lett. 2007;581:2076–2082. doi: 10.1016/j.febslet.2007.01.091. [DOI] [PubMed] [Google Scholar]
- 15.Long KR, Yamamoto Y, Baker AL, Watkins SC, Coyne CB, Conway JF, Aridor M. Sar1 assembly regulates membrane constriction and ER export. J Cell Biol. 2010;190:115–128. doi: 10.1083/jcb.201004132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lederkremer GZ, Cheng Y, Petre BM, Vogan E, Springer S, Schekman R, Walz T, Kirchhausen T. Structure of the Sec23p/24p and Sec13p/31p complexes of COPII. Proc Natl Acad Sci U S A. 2001;98:10704–10709. doi: 10.1073/pnas.191359398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fath S, Mancias JD, Bi X, Goldberg J. Structure and organization of coat proteins in the COPII cage. Cell. 2007;129:1325–1336. doi: 10.1016/j.cell.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 18.Bi X, Mancias JD, Goldberg J. Insights into COPII coat nucleation from the structure of Sec23.Sar1 complexed with the active fragment of Sec31. Dev Cell. 2007;13:635–645. doi: 10.1016/j.devcel.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stagg SM, Gurkan C, Fowler DM, LaPointe P, Foss TR, Potter CS, Carragher B, Balch WE. Structure of the Sec13/31 COPII coat cage. Nature. 2006;439:234–238. doi: 10.1038/nature04339. [DOI] [PubMed] [Google Scholar]
- 20.Stagg SM, Lapointe P, Balch WE. Structural design of cage and coat scaffolds that direct membrane traffic. Curr Opin Struct Biol. 2007;17:221–228. doi: 10.1016/j.sbi.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 21.Stagg SM, LaPointe P, Razvi A, Gurkan C, Potter CS, Carragher B, Balch WE. Structural basis for cargo regulation of COPII coat assembly. Cell. 2008;134:474–484. doi: 10.1016/j.cell.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stagg SM, Lander GC, Pulokas J, Fellmann D, Cheng A, Quispe JD, Mallick SP, Avila RM, Carragher B, Potter CS. Automated cryoEM data acquisition and analysis of 284742 particles of GroEL. J Struct Biol. 2006;155:470–481. doi: 10.1016/j.jsb.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 23.Matsuoka K, Schekman R, Orci L, Heuser JE. Surface structure of the COPII-coated vesicle. Proc Natl Acad Sci U S A. 2001;98:13705–13709. doi: 10.1073/pnas.241522198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai H, Yu S, Menon S, Cai Y, Lazarova D, Fu C, Reinisch K, Hay JC, Ferro-Novick S. TRAPPI tethers COPII vesicles by binding the coat subunit Sec23. Nature. 2007;445:941–944. doi: 10.1038/nature05527. [DOI] [PubMed] [Google Scholar]
- 25.Rao Y, Bian C, Yuan C, Li Y, Chen L, Ye X, Huang Z, Huang M. An open conformation of switch I revealed by Sar1-GDP crystal structure at low Mg2+ Biochem Biophys Res Commun. 2006;348:908–915. doi: 10.1016/j.bbrc.2006.07.148. [DOI] [PubMed] [Google Scholar]
- 26.Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 27.Winkler H, Taylor KA. Accurate marker-free alignment with simultaneous geometry determination and reconstruction of tilt series in electron tomography. Ultramicroscopy. 2006;106:240–254. doi: 10.1016/j.ultramic.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 28.Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau PW, Lyumkis D, Potter CS, Carragher B. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mallick SP, Carragher B, Potter CS, Kriegman DJ. ACE: automated CTF estimation. Ultramicroscopy. 2005;104:8–29. doi: 10.1016/j.ultramic.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 30.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 31.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 32.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera - A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]