SUMMARY
BEAMing is a feasible, accurate and sensitive method for detection of PIK3CA mutations in circulating tumor DNA in blood. Mutation status of PIK3CA may change between primary tumor and recurrence. The results suggest a new approach for non-invasive determination of current mutation status in patients with metastatic disease.
In this issue of Clinical Cancer Research, Higgins et al. report the feasibility and accuracy of PIK3CA mutation detection using free circulating tumor DNA (ctDNA) from peripheral blood samples in a prospective study of metastatic breast cancer patients (1).
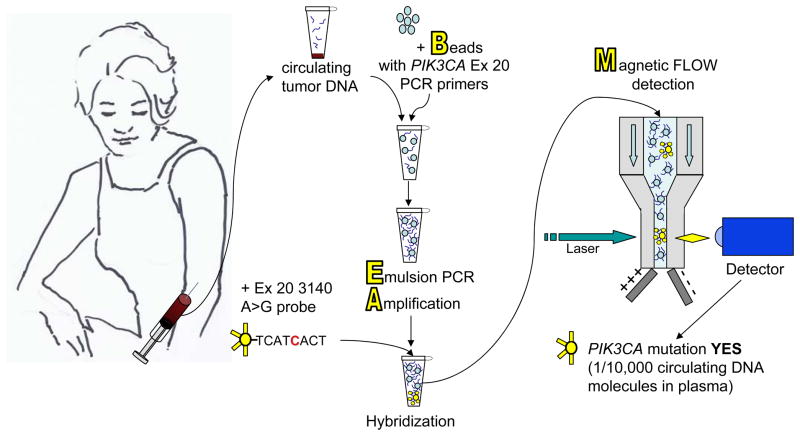
The study uses a recently described mutation detection assay called BEAMing (Beads, Emulsification, Amplification, and Magnetics, Figure 1), reported to be sensitive and capable of detecting mutations present in 1 in 10, 000 DNA molecules (2–4). Higgins and co-authors cite 100% agreement in PIK3CA mutation status determined by BEAMing when compared to standard sequencing performed on the same tumor samples. More important clinically, there was 100% concordance between mutations detected in paired tumor samples and plasma ctDNA when the samples are obtained from the patient concurrently. The technical feasibility of BEAMing on blood ctDNA is supported by the 100% success in determining the presence or absence of a PIK3CA mutation in all patients enrolled in a prospective study. In contrast, only 85% of the patients in this study had adequate and available primary tumor blocks for mutation testing.
Figure 1.
Mutation detection from blood by BEAMing. Blood samples from patients with metastatic cancer contain free circulating tumor DNA (ctDNA) released into the blood stream from tumor growing at primary or metastatic sites. Specific point mutations in targetable genes of interest such as PIK3CA can be detected in ctDNA using BEAMing technology. Circulating tumor DNA molecules are loaded onto magnetic Beads coated with specific polymerase chain reaction primers for the gene of interest. Polymerase chain reaction is performed on the beads in an oil and water Emulsion (Emulsion PCR)to Amplify the DNA. Fluorescent-tagged probes specific for either the wild type sequence or for particular common point mutations are added and hybridize to the amplified DNA. Magnetic flow cytometry of the beads is performed to detect the fluorescent tag and quantify the number of beads containing mutated DNA. The results suggest BEAMing ctDNA in blood plasma is a sensitive and accurate method for relatively non-invasive assessment of current mutation status in patients with metastatic disease.
The number of new targeted anti-cancer drugs under development and entering into human trials has grown dramatically, and many of these agents are specifically aimed at gene products that bear activating mutations(5). This approach has led to a number of recent successes in the treatment solid tumors including drugs targeting mutant B-raf in melanoma, mutant EGFR or translocation-activated ALK in non-small cell lung cancer, to name a few. To ensure new agents are tested appropriately in the right population and any potential efficacy is recognized, many early phase trials attempt to enrich for patients with tumors containing the “presumed” target of the drug being tested. Caution should be taken when deciding the appropriate time during drug development to limit trial entry on the basis of a companion tumor biomarker, including results of a tumor genotype test. In the absence of strong pre-clinical data showing a tight correlation between a particular biomarker and drug sensitivity, trial eligibility should not be restricted too early (6). For instance, in early phase trials of inhibitors of the PIK3CA/mTOR pathway, responses are observed in patients both with and without detected mutations in the presumed target (7). For drugs with a validated role against a targeted mutation, there is need for efficient and cost-effective mutational analysis that informs treatment selection. Many cancer centers are developing gene mutation screening approaches that can be applied to archived FFPE tumor samples (8,9). These centers will undoubtedly encounter the logistical nightmare of identifying which tissue block is most appropriate for tumor DNA analysis, locating the blocks in the pathology archives, transferring the blocks to the correct laboratory for nucleic acid extraction and reporting out mutational analysis in a way that can be used clinically. When primary tumor blocks cannot be located or tumor content is not adequate, an invasive biopsy procedure may be required to obtain tissue from a patient to determine if they are eligible for a particular trial or treatment.
There are potential benefits that BEAMing ctDNA may provide that overcome some of these issues. Obtaining a blood sample is less invasive and risky than biopsy of metastatic lesions. Patients may be monitored for changes in mutation status or mutation burden over time by serial testing of blood during the course of treatment. In the study by Higgins et al, the authors find a difference in PIK3CA mutation status, in both directions, between the archived primary tumor and prospectively collected ctDNA in patients with recurrent metastatic disease in up to 27.5% of the cases. All of the changes in PIK3CA mutation status were seen when the recurrence was more than 3 years after the primary diagnosis. Thus, mutation status in the archived tumor sample may not reflect current mutation status in a patient with recurrent metastatic disease, especially those with a long disease-free interval. The current study reports a non-invasive approach to assess mutation status at the time of tumor recurrence.
With tumor genome sequencing, we are beginning to understand the depth and breadth of intratumor heterogeneity, both within the primary tumor, between primary tumors and distant metastases, and between different metastatic sites (10). Higgins and co-authors report two patients in whom two different PIK3CA mutations were identified in ctDNA. This finding suggests “convergent evolution”, where different tumor subclones acquire different mutations in the same presumably “driver” gene. A recent study sequenced multiple areas of a renal cell carcinoma and several metastatic sites. The investigators identified three different mutations in the gene SETD2 present in different regions of the tumor (11). The co-occurrence of multiple PIK3CA mutations in a breast cancer has also been reported (12). BEAMing mutation detection in circulating free tumor DNA may overcome issues of tumor heterogeneity by detection of rare mutations in minor cancer subclones, or in disparate subclones from different metastatic sites, that might not be identified by sequencing from a single site of disease. However, it is not clear how representative circulating tumor DNA will be of the various subclones of malignancy present throughout the body. Some subclones may release tumor DNA more readily than others. For instance, ctDNA might be more representative of bone metastasis compared to brain metastasis. Answers to these questions will require future clinical trials.
The BEAMing assay is not full gene or even full exon sequencing and will not identify all mutations in a particular gene. The assay is designed to detect specific commonly observed mutations in “hot spot” locations. For PIK3CA , such hot spot mutations account for around 80% of all reported mutations. The current study only tested for four of the most common hot spot mutations in PIK3CA. Therefore, a negative result for a BEAMing test cannot be interpreted as an absence of mutation in the gene. This is a feature that is shared with most of the other currently available mutation detection methods being used for clinical mutation screening. The methodologies we will use in the future for cancer mutation assessment will depend on such factors as cost, breadth of mutation detection, specificity and sensitivity. This study of the BEAMing technology suggests it may be a useful method for sensitive but limited mutation determination from a blood sample. Additional clinical trials are required to confirm the positive and negative value of a BEAMing ctDNA mutation result for prediction of treatment response.
Footnotes
The authors have no conflicts of interest to declare.
References
- 1.Higgins MJ, Jelovac D, Barnathan E, Blair B, Slater S, Powers P, et al. Detection of tumor PIK3CA status in metastatic breast cancer using peripheral blood. Clin Cancer Res. 2012;xx:XXXX. doi: 10.1158/1078-0432.CCR-11-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diehl F, Li M, Dressman D, He Y, Shen D, Szabo S, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci U S A. 2005;102:16368–73. doi: 10.1073/pnas.0507904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diehl F, Li M, He Y, Kinzler KW, Vogelstein B, Dressman D. BEAMing: single molecule PCR on microparticles in water-in-oil emulsions. Nat Methods. 2006;3:551–9. doi: 10.1038/nmeth898. [DOI] [PubMed] [Google Scholar]
- 4.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–90. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maitland ML, Schilsky RL. Clinical trials in the era of personalized oncology. CA Cancer J Clin. 2011;61:368–81. doi: 10.3322/caac.20135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Juric D, Baselga J. Tumor genetic testing for patient selection in phase I clinical trials: the case of PI3K inhibitors. J Clin Oncol. 2012;30:765–766. doi: 10.1200/JCO.2011.39.6390. [DOI] [PubMed] [Google Scholar]
- 7.Baselga J, Semiglazov V, van Dam P, Manikhas A, Bellet M, Mayordomo J, et al. Phase II randomized study of neoadjuvant everolimus plus letrozole compared with placebo plus letrozole in patients with estrogen receptor-positive breast cancer. JClinOncol. 2009;27:2630–2637. doi: 10.1200/JCO.2008.18.8391. [DOI] [PubMed] [Google Scholar]
- 8.MacConaill LE, Campbell CD, Kehoe SM, Bass AJ, Hatton C, Niu L, et al. Profiling critical cancer gene mutations in clinical tumor samples. PLoS One. 2009;4:e7887. doi: 10.1371/journal.pone.0007887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su Z, Dias-Santagata D, Duke M, Hutchinson K, Lin YL, Borger DR, et al. A platform for rapid detection of multiple oncogenic mutations with relevance to targeted therapy in non-small-cell lung cancer. J Mol Diagn. 2011;13:74–84. doi: 10.1016/j.jmoldx.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, et al. Genome remodeling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, Neve RM, Kuo WL, Davies M, et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008;68:6084–91. doi: 10.1158/0008-5472.CAN-07-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]