Abstract
Streptococcus pneumoniae (Spn) is the predominant causative organism of acute otitis media in children. To better understand the genes that are regulated at the onset of AOM caused by Spn infection in the middle ear, the transcriptome profile of peripheral blood mononuclear cells isolated from children prior to and during an AOM event was evaluated by microarray. We found that 1903 (6.2%) of 29,187 genes were differentially regulated greater than 2 fold at the onset of AOM compared to the pre-infection stage of the same children. The ontology of differentially regulated genes was dominated by those involved with the immune response. At onset of infection, genes associated with bacterial defenses were significantly up-regulated, including beta-defensin123, S100 protein A12, Toll-like receptor 5, IL-10, and those involved in the classical and alternative complement pathways. Genes associated with inhibition of bacterial entry through clathrin-dependent endocytosis were also up-regulated. In contrast, genes associated with cell-mediated immune responses were broadly down regulated. The results provide the first human transcriptome data identifying genes differentially regulated at the onset of AOM in children.
Keywords: STREPTOCOCCUS PNEUMONIAE, ACUTE OTITIS MEDIA, TRANSCRIPTOME PROFILE
1 Introduction
Acute otitis media (AOM) is the most frequent reason that children seek medical care for illness and receive antibiotics. AOM is associated with significant middle ear inflammation, mainly caused by bacterial infection. Streptococcus pneumoniae (Spn) is one of the most frequent pathogens isolated from middle ear fluid (MEF) of children with AOM [1]. The pathogenesis of AOM in humans involves nasopharyngeal (NP) colonization by the bacteria achieved by adherence and invasion of host NP epithelial cells. The bacterial inoculum in the NP then refluxes up the eustachian tube near the adenoid tissues and gains entry to the middle ear. In in vitro experiments and an animal model, a robust inflammatory response with significant gene regulation has been described during Spn infection [2, 3] but data on human infection are needed.
Here we describe, for the first time in human children, an evaluation of the gene expression profile of peripheral blood mononuclear cells (PBMCs) associated with onset of AOM caused by Spn. This analysis was made possible during a prospective study on the immune responses of children to Spn during AOM with comparable PBMC samples taken within 4 weeks prior to onset of AOM. Double sampling from the same children reduced the complexities of analysis due to genetic heterogeneity among individuals. Closely timed sampling avoided the influence of changing age on responses in pediatric subjects. Our previous work identified significant changes in blood of a limited number of specific pro-inflammatory molecules during AOM caused by Spn [4,5,6]. However, with transcriptome analysis, we were able to identify a much broader range of significant changes in gene regulation.
Materials and methods
2.1 Subjects
The samples were collected from children (age 10-15 months) as approved by the IRB at Rochester General Hospital. The diagnosis of AOM was based on symptoms of fever, irritability or ear ache, and physical signs of inflammation of the tympanic membrane (bulging) with the presence of middle ear fluid (pus-laden fluid) documented by tympanocentesis. The inclusion criteria for children were: Caucasian children; pre-infection PBMC/serum sample within 4 weeks before the onset of AOM and the collection of an acute stage of illness PBMC/serum sample within 24 hours of onset of AOM; age 10-15 months at onset of AOM; AOM caused by Spn as proven by tympanocentesis; and moderate to severe AOM as judged by the physician. Exclusion criteria were: a history of immunodeficiency; chronic or recurrent AOM; any chronic disease; any other concurrent infectious disease; or receipt of steroids or other immunomodulatory agents within 1 month of any PBMC/serum sample.
2.2 Identification of pathogens
Identification of Spn in MEF was performed based on presence of α-hemolysis and inhibition of optochin and confirmed by a positive slide agglutination test according to established CLSI procedures (BBL™ Pneumoslide™ Kit, BD, Tetax, USA). Nontypeable Haemophilus influenzae (NTHi), Moraxella catarrhalis and other otopathogens were identified as described previously [7]. The culture results were further verified by multiplex PCR as described previously [8].
2.3 RNA isolation
Four to ten milliliters of heparinized peripheral venous blood was collected for isolation of PBMCs and sera. PBMCs were isolated by Ficoll gradient and total RNA was extracted from PBMCs using a QIAamp RNA blood Mini Kit (Qiagen, Gaithersburg, MD) according to the manufacturer's instructions. Total RNA was treated with DNase I to remove contaminating DNA (Qiagen). RNA quality and integrity were determined utilizing an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). Only high quality RNA, having a RIN of >7.0, and an A260/A280 absorbance ratio of >1.8, was utilized for further experimentation.
2.4 Microarray analysis
DNA microarray analysis was performed in a blinded manner by Phalanx Biotech using the Human Whole Genome OneArray™ (Phalanx Biotech, Palo Alto, CA), which included 29,187 human genome probes and 1088 experimental control probes. RNA was converted to double-stranded cDNA and amplified using in vitro transcription that included amino-allyl UTP, and the aRNA product was subsequently conjugated with Cy5™ NHS ester (GEH Life sciences, Piscataway, NJ). Fragmented aRNA was hybridized at 42°C overnight using the HybBag mixing system with 1X OneArray Hybridization Buffer (Phalanx Biotech, Belmont, CA), 0.01 mg/ml sheared salmon sperm DNA (Promega, Madison, WI), at a concentration of 0.025 mg/ml labeled target. After hybridization, the arrays were washed according to the OneArray protocol. For the hybridization, the controls included dye intensity controls, positive controls containing a mixture of probes from 58 robustly expressed housekeeping genes and negative controls containing eight 60-mer oligonucleotide probes that do not cross-hybridize with human genomic probes on the Human OneArray DNA microarray. Gene expression was assessed in three replicates of each PBMC sample.
2.5 Quantitative real-time reverse transcriptase PCR analysis
Real time PCR was performed in our laboratory at the Rochester General Hospital Research Institute using a saved aliquot of the RNA preparation from the microarray experiments as templates. One hundred nanograms of total RNA was reverse transcribed to cDNA using an RT2 First Strand Kit (QIAGEN). Detection and quantification of gene expression in PBMCs was performed using an RT2 Profiler Human Custom Kit according to the manufacturer's instructions (QIAGEN). Quantitative real-time reverse transcriptase PCR was performed using a CFX 96 Thermocycler (Bio-Rad, Fairfield, CA). The threshold and baseline were set automatically using the pcr/array analysis method according to the manufacturer's instructions (SABiosciences). CT data were uploaded into the data analysis template on the manufacturer's website (http://www.sabiosciences.com/pcr/arrayanalysis.php). The relative expression of genes compared with the expression in control samples was calculated on the website using the Δ ΔCT method with five housekeeping genes as controls.
2.6 Proteome assays
The protein levels of S100A12 in the serum and MEF samples were determined from the same children where microarray was performed using the CircuLex S100A12 ELISA Kit according to standard instructions in the protocol. IL-10 levels were determined in the same samples using a Bio-Plex Pro Assay (Bio-Rad Laboratories, Hercules, California, USA) according to manufacturer's instructions and read with a Luminex instrument.
2.7 Data Analysis
Primary data analysis was performed by Phalanx Biotech. Raw intensity signals for each microarray were captured using a Molecular Dynamics™ Axon 4100A scanner, measured using GenePixPro™ Software, and stored in GPR format. The data from all microarrays in each experimental set was then passed to Rosetta Resolver (Rosetta Biosoftware, Cambridge, MA) for analysis. Testing was performed by combining technical replicates (http://www.phalanxbiotech.com) and performing statistical analyses using Rosetta Resolver's proprietary modeling techniques (http://www.rosettabio.com/products/resolver). The gene intensities were adjusted by subtracting the array background, and then normalized to housekeeping genes (four probes per gene were selected from seven robustly expressed housekeeping genes). The data with a consistent and high quality readout verified by quality control at Phalanx Biotech of microarray were analyzed at the Rochester Institute of Technology (L.C.) for biological function groupings. The data from 12 hybridizations of 4 children were pooled and the fold change was calculated by comparing the readout at onset of AOM compared to the pre-infection healthy state. (4 children at 2 stages = 8 data sets and for each stage and triple hybridizations were performed; therefore, each stage -AOM or healthy- had 4 samples × 3 hybridizations yields 12 hybridizations for each stage.) Genes with greater than a two-fold increase or decrease were considered significant and included for further analysis. The biological themes associated with these genes were analyzed using DAVID software [http://david.abcc.ncifcrf.gov]. To identify lexical enrichments, the terminology defined by the Gene Ontology (GO) Consortium [http://www.geneontology.org] was used.
3 Results
3.1 Transcriptome responses in children with AOM due to Spn
1903 (6.2%) of 29,187 genes assessed were differentially expressed 2-fold up or down in all 4 of the studied children at the onset of AOM compared to their pre-infection stage. Of the 1903 genes differentially regulated, 970 transcripts (51% of the total) were significantly up-regulated during AOM compared to the pre-infection state and 933 transcripts (49 % of the total) were significantly down-regulated (Table 1). The complete list of differentially expressed genes is provided in supplementary Table 1 and accession number GSE23140.
Table 1. Comparison of the fraction of the genes differentially expressed in differential disease models.
Spn-AOM: data derived from microarray in 4 children with AOM caused by Spn infection. Nontypeable Haemophilus influenzae (NTHi)-AOM: data derived from microarray in 4 children with AOM caused by NTHi. Spn (mice): data derived from microarray in mice infected by Spn.
| Disease model | Gene differentially expressed | Ratio of total probes (%) | Exp/Control (A/H) | Reference |
|---|---|---|---|---|
| Spn-AOM (Human) | 1903 | 6.2 | >2 fold | This study |
| NTHi-AOM (Human) | 1487 | 4.8 | >2 fold | Our Lab |
| HPV31 virus (Mouse) | 7075 | 2 | >2 fold | Chang, 2000 |
| Spn (Mouse) | 490 | 3.2 | >1.5 fold | Zhang, 2006 |
| THP-1, Spn (in vitro) | 142 | 3.4 | > 2 fold | Rogers, 2003 |
| A549 cell, P. aeruginasa (in vitro) | 22 | 1.5 | >2 fold | Ichikawa, 2000 |
3.2 Gene functional clustering is dominated by the immune response
To understand the biological functions of the genes differentially expressed at the onset of AOM, the 1903 transcripts identified above were analyzed with DAVID v6.7 software. It was found that the top-ranked gene ontology was typical of an immune response to bacterial infection, followed by the clusters of regulation of transcription, cell morphogenesis and cellular homeostasis (Table 2). Further analysis found that there were 127 genes that could be grouped in the host immune defense category which could be further classified into the subgroups of innate immune response, leukocyte activation and differentiation, response to bacteria infection, regulation of apoptosis, kinase activity and biosynthesis.
Table 2. Principal biological annotation and functional terms identified by gene set enrichment analysis.
The genes differentially expressed in 4 children with AOM due to Spn relative to their pre-infection healthy stage were analyzed. Only of genes with a high fold change (< 0.5 and > 2 fold) in expression and enrichment score >1.3 and P< 0.05 occurring in all 4 children are shown.
| Cluster* 1 Enrichment Score: 2.01 | ||||
|---|---|---|---|---|
| Term** | Count | % | P Value | Fold Enrichment |
| GO:0016064, immunoglobulin mediated immune response | 14 | 0.77 | 0.0019 | 2.62 |
| GO:0019724, B cell mediated immunity | 14 | 0.77 | 0.0027 | 2.52 |
| GO:0002460, adaptive immune response based on somatic recombination of immune receptors | 16 | 0.88 | 0.0078 | 2.10 |
| GO:0002250, adaptive immune response | 16 | 0.88 | 0.0078 | 2.10 |
| GO:0002449, lymphocyte mediated immunity | 15 | 0.82 | 0.0079 | 2.16 |
| GO:0002455, humoral immune response mediated by circulating immunoglobulin | 9 | 0.49 | 0.0090 | 2.93 |
| GO:0006958, complement activation, classical pathway | 8 | 0.44 | 0.0205 | 2.78 |
| GO:0002252, immune effector process | 21 | 1.15 | 0.0409 | 1.58 |
| GO:0002443, leukocyte mediated immunity | 15 | 0.82 | 0.0426 | 1.76 |
| Total | 128 | |||
| Cluster 2 Enrichment Score: 1.671 | ||||
| Term | Count | % | P Value | Fold Enrichment |
| GO:0045935, positive regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | 82 | 4.49 | 0.0064 | 1.33 |
| GO:0051173, positive regulation of nitrogen compound metabolic process | 84 | 4.60 | 0.0070 | 1.32 |
| GO:0045941, positive regulation of transcription | 74 | 4.05 | 0.0099 | 1.32 |
| GO:0010557, positive regulation of macromolecule biosynthetic process | 84 | 4.60 | 0.0102 | 1.30 |
| GO:0031328, positive regulation of cellular biosynthetic process | 87 | 4.76 | 0.0118 | 1.28 |
| GO:0009891, positive regulation of biosynthetic process | 88 | 4.82 | 0.0122 | 1.28 |
| GO:0010628, positive regulation of gene expression | 75 | 4.11 | 0.0136 | 1.30 |
| GO:0045944, positive regulation of transcription from RNA polymerase II promoter | 47 | 2.57 | 0.0635 | 1.28 |
| Total | 621 | |||
| Cluster 3 Enrichment Score: 1.54 | ||||
| Term | Count | % | P Value | Fold Enrichment |
| GO:0000902, cell morphogenesis | 51 | 2.79 | 0.0070 | 1.45 |
| GO:0000904, cell morphogenesis involved in differentiation | 37 | 2.03 | 0.0097 | 1.53 |
| GO:0048667, cell morphogenesis involved in neuron differentiation | 32 | 1.75 | 0.0146 | 1.54 |
| GO:0032989, cellular component morphogenesis | 54 | 2.96 | 0.0147 | 1.37 |
| GO:0048858, cell projection morphogenesis | 36 | 1.97 | 0.0171 | 1.48 |
| GO:0032990, cell part morphogenesis | 36 | 1.97 | 0.0308 | 1.42 |
| GO:0030030, cell projection organization | 48 | 2.63 | 0.0404 | 1.32 |
| GO:0007409, axonogenesis | 28 | 1.53 | 0.0415 | 1.46 |
| Total | 322 | |||
| Cluster 4 Enrichment Score: 1.31 | ||||
| Term | Count | % | P Value | Fold Enrichment |
| GO:0055074, calcium ion homeostasis | 30 | 1.64 | 0.0106 | 1.61 |
| GO:0055066, di-, tri-valent inorganic cation homeostasis | 36 | 1.97 | 0.0120 | 1.52 |
| GO:0006874, cellular calcium ion homeostasis | 29 | 1.59 | 0.0132 | 1.60 |
| GO:0030005, cellular di-, tri-valent inorganic cation homeostasis | 34 | 1.86 | 0.0159 | 1.51 |
| GO:0055065, metal ion homeostasis | 31 | 1.70 | 0.0191 | 1.53 |
| GO:0006875, cellular metal ion homeostasis | 29 | 1.59 | 0.0304 | 1.49 |
| Total | 189 |
Number of functional annotation cluster (enrichment score >1.3).
Number of functional annotation terms(enrichment 0f >1.3 and P<0.051.These genes were analyzed using DAVID, software [http://david.abcc.ncifcrf.gov].
3.3 Differentially expressed genes related to innate immunity
Since the understanding of the immune response mechanism in children is still incomplete, our analysis of the transcriptome profile was focused on the 127 transcripts of host immune defense responses. We found that 19 genes were classified to innate immune responses which included genes for the classical and alternative complement pathway, toll-like receptors and interleukins.
Of the genes involved in the classical complement pathway, the highest up-regulated gene was C1QA and C1QB. In addition, C1S was down regulated significantly and CFD, a component of the alternative complement pathway with function similar to C1S in the classical pathway, was up regulated (Table 3).
Table 3. Fold change of host immune defense response in children with AOM due to Spn.
Genes are grouped functionally based on ontologies. RefSeq identifier: Entrez gene ID. The fold change of gene expression was derived by comparison of the gene expression intensity at the onset of AOM due to Spn with the pre-infection healthy stage of the same children. The data from the four children are presented as a mean.
| Entrez gene ID | Gene Symbol | Gene description | Fold change |
|---|---|---|---|
| Genes associated with complement activation and TLR | |||
| NM_134470 | IL1RAP | interleukin 1 receptor accessory protein | 6.69 |
| NM_015991 | C1QA | complement component 1, q subcomponent, A chain | 6.33 |
| NM_000491 | C1QB | complement component 1, q subcomponent, B chain | 4.46 |
| NM_000877 | IL1R1 | interleukin 1 receptor, type I | 4.35 |
| NM_022059 | CXCL16 | chemokine (C-X-C motif) ligand 16 | 2.48 |
| NM_003268 | TLR5 | toll-like receptor 5 | 2.31 |
| NM_001928 | CFD | complement factor D (adipsin) | 2.25 |
| NM_007268 | VSIG4 | V-set and immunoglobulin domain containing 4 | 2.23 |
| NM_145343 | APOL1 | apolipoprotein L, 1 | 2.09 |
| NM_183236 | RAB27A | RAB27A, member RAS oncogene family | 2.01 |
| NM_001733 | C1R | complement component 1, r subcomponent | -2 |
| NM_000562 | C8A | complement component 8, alpha polypeptide | -2.17 |
| NM_000715 | C4BPA | complement component 4 binding protein, alpha | -2.36 |
| NM_005582 | CD180 | CD180 molecule | -2.48 |
| NM_201442 | C1S | complement component 1, s subcomponent | -2.6 |
| NM_001145805 | IRGM | immunity-related GTPase family, M | -3.13 |
| NM_001877 | CR2 | complement component receptor 2 | -3.32 |
| NM_001032295 | SERPING1 | serpin peptidase inhibitor, member 1 | -3.76 |
| NM_001017388 | TLR10 | toll-like receptor 10 | -3.97 |
| Genes associated with antibacterial activity | |||
| NM_153324 | DEFB123 | Beta-defensin 123 | 4.78 |
| NM_005621 | S100A12 | S100 calcium-binding protein A12 | 4.52 |
| NM_021058 | HIST1H2BJ | Histone H2B type 1-J | 3.59 |
| NM-002965 | S100A9 | S100 calcium binding protein A9 | 2.71 |
| NM_001725 | BPI | Bactericidal permeability-increasing protein | 2.59 |
| NM_005091 | PGLYRP1 | Peptidoglycan recognition protein | -2.06 |
| NM_207469 | DEFB132 | Beta-defensin 132 | -3.07 |
| NM_002343 | LTF | Lactotransferrin | -3.98 |
| Cytokine genes | |||
| NM_002981 | CCL1 | chemokine (C-C motif) ligand 1 | 4.77 |
| NM_148672 | CCL28 | chemokine (C-C motif) ligand 28 | 4.6 |
| NM_002985 | CCL5 | chemokine (C-C motif) ligand 5 | 3.92 |
| NM_014438 | IL1F8 | interleukin 1 family, member 8 (eta) | 2.93 |
| NM_000572 | IL10 | interleukin 10 | 2.93 |
| NM_022059 | CXCL16 | chemokine (C-X-C motif) ligand 16 | 2.48 |
| NM_172088 | TNFSF12, 13 | tumor necrosis factor, member 12, 13 | 2.23 |
| NM_006573 | TNFSF13B | tumor necrosis factor superfamily, member 13b | 2.17 |
| NM_003810 | TNFSF10 | tumor necrosis factor superfamily, member 10 | -2.05 |
| NM_000586 | IL2 | interleukin 2 | -2.12 |
| NM_000576 | IL1B | interleukin 1, beta | -2.38 |
| NM_172140 | IL29 | interleukin 29 (interferon, lambda 1) | -3.17 |
| NM_002620 | PF4V1 | platelet factor 4 variant 1 | -6.18 |
| NM_005623 | CCL8 | chemokine (C-C motif) ligand 8 | -6.31 |
| NM_001565 | CXCL10 | chemokine (C-X-C motif) ligand 10 | -9.3 |
| NM_004591 | CCL20 | chemokine (C-C motif) ligand 20 | -9.77 |
| NM_002982 | CCL2 | chemokine (C-C motif) ligand 2 | -11.13 |
| Genes for lymphocyte activation and differentiation | |||
| NM_001765 | CD1C | T-cell surface glycoprotein CD1c | -2.13 |
| NM_001033667 | LY9 | T-lymphocyte surface antigen Ly-9 | -2.16 |
| NM_181780 | BTLA | B- and T-lymphocyte attenuator | -2.34 |
| NM_001039933 | CD79B | B-cell Ag receptor complex-associated protein beta-chain | -2.39 |
| NM_000073 | CD3G | T-cell surface glycoprotein CD3 gamma chain | -2.49 |
| NM_001783 | CD79A | B-cell Ag receptor complex-associated protein alpha-chain | -2.53 |
| NM_001770 | CD19 | B-lymphocyte antigen CD19 | -2.81 |
| NM_005191 | CD80 | T-lymphocyte activation antigen CD80 | -3.69 |
| NM_001025159 | CD74 | HLA class II histocompatibility antigen gamma chain | -3.39 |
| NM-152866 | CD20 | membrane-spanning 4-domains, subfamily A | -3.41 |
| NM_015055 | SWAP70 | SWAP switching B-cell complex 70kDa subunit | -2.64 |
| NM_013314 | BLNK | B-cell linker protein | -2.33 |
| NM_000586.3 | IL2 | interleukin 2 | -2.12 |
| NM_032682 | FOXP1 | forkhead box P1 | -2 |
| NM_145912 | NFAM1 | NFAT activating protein with ITAM motif 1 | 2 |
| NM_183236 | RAB27A | RAB27A, member RAS oncogene family | 2.01 |
| NM_005178 | BCL3 | B-cell CLL/lymphoma 3 | 2.19 |
| NM_007268 | VSIG4 | V-set and immunoglobulin domain containing 4 | 2.23 |
| NM_000676 | ADORA2B | Adenosine A2b receptor | 2.53 |
| NM_005442 | EOMES | eomesodermin homolog | 2.55 |
| NM_002485 | NBN | nibrin | 2.8 |
| NM_000572 | IL10 | interleukin 10 | 2.93 |
Surprisingly, there were not many pathogen-specific-receptors differentially expressed. TLR2, a common receptor up-regulated in Spn infection in rodent models, did not show differential expression at onset of Spn infection in the middle ear in the children. However, TLR5 was significantly up-regulated and TLR10 showed significant down-regulation (Table 3).
3.4 Differentially expressed genes related to anti-bacterial response
Among the 127 genes related to host immune defense responses, 8 genes associated with anti-bacterial activity were differentially expressed at onset of AOM. The highest up-regulated genes were beta-defensin 123 (DEFB123), S100 proteins S100A12 and S100A9, and HIST1H2BJ (a broad antibacterial molecule) (Table 3). Other genes related to bactericidal activity towards Gram-positive bacteria were down-regulated such as DEFB132 and lactotransferrin (LTF).
3.5 Differentially expressed cytokine genes
Genes associated with 16 cytokines were differentially expressed at onset of AOM, including 5 interleukins and 8 chemokines (Table 3). Among the 8 up-regulated genes, the highest expression was IL-10, which plays an important role in inhibition of CD4 T-cell activation (accept its antibacterial activity). CCL5, associated with chemotaxis of T cells was up-regulated 3.9 fold. CCL1, with chemotactic activity for monocytes but not for neutrophils, was up-regulated 4.8 fold. CCL28, chemotactic for resting CD4 or CD8 T cells and eosinophils, was up-regulated 4.6 fold. IL1F8, involved in the inflammatory response, was up-regulated 2.9 fold. Down-regulated genes included CCL20, a chemotactic factor for lymphocytes, neutrophils and dendritic cells towards an infection site, IL-2, required for T-cell proliferation and other activities crucial to regulation of the immune response, and IL-1β, involved in inflammatory reactions. In short, the regulated genes associated with cytokines were consistent with enhancement of antibacterial activity and inhibition of cell mediated immune responses.
3.6 Differentially expressed genes related to cellular immunity
22 differentially expressed genes were grouped to lymphocyte activation and differentiation. Fifteen of them showed down regulation at the onset of Spn AOM. The most predominant down regulated genes were CD80, which is involved in antigen presentation and T-lymphocyte activation, and CD74, which plays a critical role in MHC class II antigen processing. Among 7 up regulated genes there were 3, including IL10, ADORA2B and VSIG4 that play an important role in inhibition of lymphocyte activation and proliferation (Table 3). In addition, among the genes associated with B-cells activation and proliferation, CD19, SWAP70, CD79A, BLNK, and FOXP1 were all down-regulated.
3.7 Differentially expressed genes indicative of endocytosis
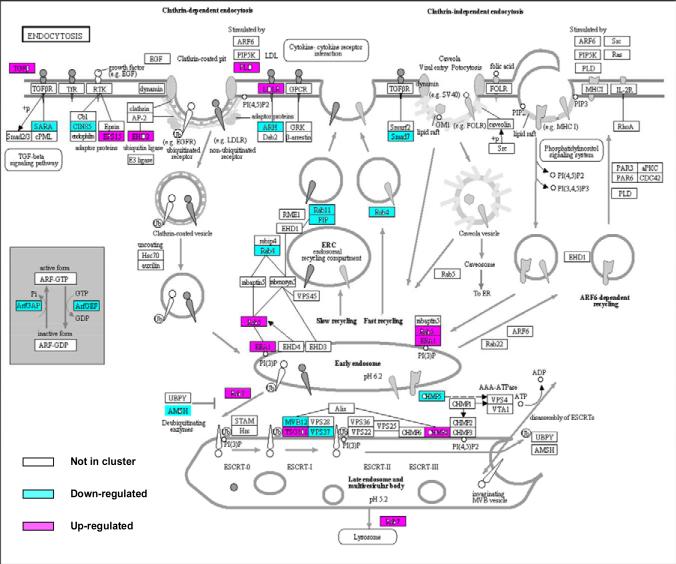
We found many genes involved in the clathrin-mediated endocytosis cascade were differentially regulated, including down regulation of CD2 associated protein (CD2AP) and Cb1-interacting protein of 85 kDa (CIN85), consistent with an effect whereby the entrance of Spn into host cells was inhibited. Other differentially expressed genes in this cascade included PLD, LDLR, ARH, FIP, RAB11, RAB4, RAB5, AMSH, EEA1 and MVB12. In contrast, no genes involved in the clathrin-independent endocytic machinery were differentially regulated (Fig. 1).
Fig. 1. Gene expression in 4 children with AOM due to Spn is enriched in genes involved in clathrin-dependent endocytosis.
Fold change in gene expression from children with AOM due to Spn versus their pre-infection healthy stage is shown on the KEGG pathway map (modified from map04144, at the Kyoto Encyclopedia of Genes and Genomes [KEGG] website http://www.genome.jp/kegg/kegg1.html). The genes are colored according to the transcript with the fold change in expression. Red color indicates up-regulation at onset of Spn infection, green down-regulation.
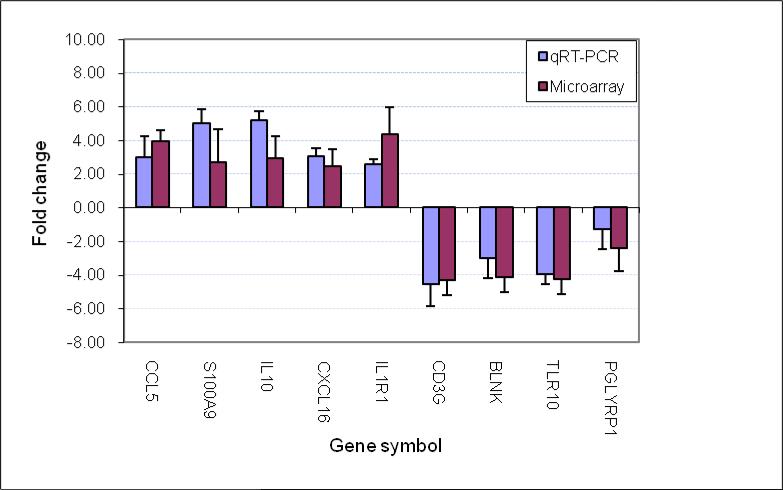
3.8 Data verification
To support the microarray data, we performed qRT-PCR analysis for a subset of genes with different biofunctional categories, including IL-1 receptors (IL-1R1), cytokines (IL-10), chemokines (CCL5), antibacterial molecules (S100A9, PGLYRP1), innate immune response (TLR10), B-cell activation (BLNK) and T-cell activation (CD3g). Our qRT-PCR data were consistent with the data generated with microarray (Fig. 2).
Fig. 2. Comparison of genes regulated as measured by microarray and qRT-PCR.
The fold change of gene expression was derived by comparison of the gene expression at the onset of AOM due to Spn (AOM) with the pre-infection healthy stage (Preinf) in the same children with the same samples. The data from the 4 children are presented as a mean+/-SD. In all the genes studied there was no difference for the fold change between qRT-PCR and microarray (p>0.05).
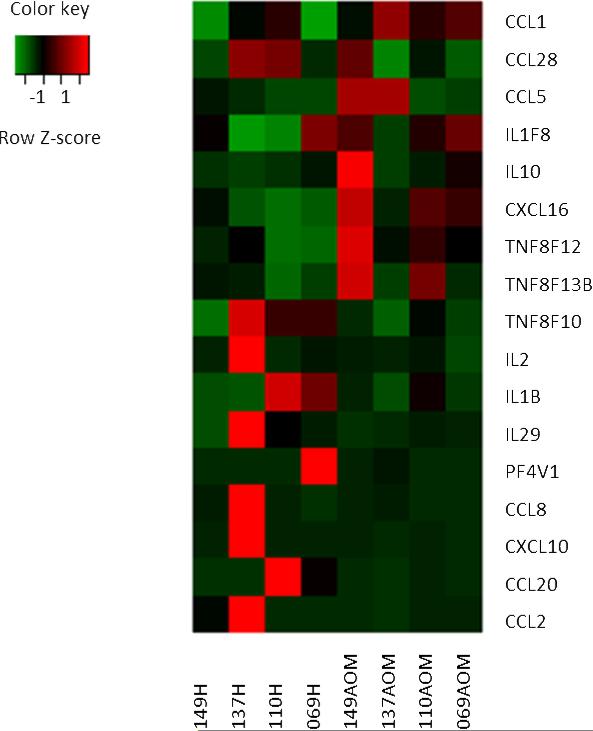
3.9 Individual sample analysis
Although the timing of PBMC samples and the study children were carefully selected to allow pooling of data, sources of variability not strictly genetically controlled may have confounded the results. To further strengthen our array data, the transcriptional changes in 17 genes for each child were analyzed (Fig. 3). Similar levels of gene up-regulation and down-regulation were seen with no differences in the functional clusters. These data are in agreement with our array data and further support the transcriptome changes we found associated with the onset of AOM compared to their healthy pre-infection stages.
Fig. 3. Variability of gene expression among 4 children with AOM due to Spn.
Statistical package software R was used for the identification of differentially expressed genes. The profile includes 17 genes selected from the 127 immune defense response gene transcripts of PBMCs, which were >2 fold change at the comparison of the onset of AOM due to Spn and their pre-infection healthy stage. Shown are normalized expression (AOM stage) greater than (red), near (black), or less than (green) the mean of controls (healthy stage). Each column represents one child of healthy or AOM stage. Genes or transcripts are represented in rows.
3.10 Proteome analysis
To confirm that the transcriptome results were reflected in the proteome, we evaluated concentrations of S100A12 and IL-10 proteins in the sera and MEF of the study children. Similar to the transcriptome results, the levels of S100A12 in the serum samples of the 4 children at onset of AOM (32.5 ± 4.5 ng/ml) were significantly higher (p= 0.04) compared to when the children were healthy (11.3 ± 2.5 ng/ml). Interestingly, the levels of S100A12 in the MEF, at the site of infection, were about 450-fold higher (14677 ± 1435 ng/ml) than serum in the AOM stage. Similarly, the levels of IL-10 in serum samples of children at onset of AOM (4.8 ± 0.9 pg/ml) were significantly higher (p= 0.04) compared to when the children were healthy (2.9 ± 0.3 pg/ml). IL-10 in the MEF was higher (314 ± 194 pg/ml) at the site of infection in the AOM children over serum during infection.
4. Discussion
To our knowledge, this is the first report on transcriptome changes associated with Spn infection of the middle ear in children. The genes associated with the immune response ontology were dominant and could be further classified into the subgroups of innate immune response, leukocyte activation and differentiation, and response to bacteria infection. To our surprise, genes associated with cell mediated immune response were significantly down-regulated in the studied children. In addition, genes associated with the inhibition of bacterial entry to host cells through clathrin-dependent endocytosis were down-regulated.
4.1. Number of genes mobilized during AOM due to Spn in young children
Overall, we found that 6.2% of 29,187 genes evaluated were differentially expressed greater than 2 fold in all four children with AOM caused by Spn compared to their healthy stage. The percentage of genes regulated in AOM children upon Spn infection is consistent with results from children with other infections and models of infection. For example in a study of children infected with influenza virus, 1.3% of the studied genes were differentially expressed in the acute phase of infection [9]. In another study by our group involving other children with AOM caused by NTHi, 4.8% of the studied genes showed a > 2 fold change compared to the pre-infection healthy stage [10]. Using an in vitro model, the transcriptional response of RAW 264.7 cell's to Salmonella typhimurium infection involved 5.8% (34/588) of genes studied using a 4-fold or greater change as a cut off [11]. Using a microarray of the human genome, 1.5% (22/1506) of host genes were identified as induced by Pseudomonas aeruginosa infection of A549 cells [12].
4.2. Up-regulation of antibacterial response genes predominate in children with AOM due to Spn infection
We found the genes related to antibacterial activity, such as S100 proteins, defensins, cytokines/chemokines, complement and Toll like receptors were particularly up-regulated at onset of AOM due to Spn. We confirmed that the transcriptome signature was reflected in the proteome for S100A12 in serum and at the site of infection in the middle ear. In agreement, we had previously observed an increase in S100A12 in serum of children at onset of AOM and a return to pre-infection levels with recovery from AOM [5]. However the up-regulation of DEFB123 and BPI at onset of the AOM stage in young children is a new finding and indicates that these additional specific antibacterial responses are mobilized as part of the innate defense at onset of the infection of Spn in the middle ear. It is known that defensins are a family of secreted antimicrobial peptides directly interfering with bacterial membranes in the innate immune response. DEFB123 exerts antimicrobial activity against Gram-positive bacteria [13]. Administration of a derivative of bactericidal permeability-increasing protein (BPI) provided protection against Spn infection in a mouse model [14], and S100A12 had a direct antimicrobial effect on bacteria [15].
The role of complement in eradication of Spn is a well-known immune defense mechanism. Therefore, finding that genes for the classical complement pathway were up-regulated in young children at onset of AOM due to Spn was expected. Using strains of mice with genetic deficiencies of complement components, Brown et al found that the classical pathway is the dominant pathway for activation of the complement system during innate immunity to Spn [16]. Our study is consistent with the mouse model observation. However, surprisingly, we also found that complement factor D gene (CFD), encoding a protein in the alternative complement pathway, was also up-regulated at the onset of AOM, suggesting activation of the alternative pathway as well. The result suggests the need for further study of the role of the alternative pathway in the innate response to Spn.
Toll-like receptors are transmembrane proteins expressed by cells of the innate immune system that recognize invading microbes and activate signaling pathways that launch immune and inflammatory responses. The TLR2 receptor has been classically described as the main receptor activated by Spn. However, recently, Muñoz et al [17] reported that flagellin of Spn could locally activate innate immunity against Spn as flagellin signals through TLR5. In the current study, we found that TLR5 was up-regulated significantly in children with AOM caused by Spn. Unexpectedly, we found that TLR10 was down regulated. Human TLR10 is an orphan member of the TLR family and its exact function is not known. However, TLR signaling negatively regulated by various proteins has been reported previously [18]. Further study of the role of TLR5 and TLR10 during Spn infection may be warranted.
4.3. Down regulation of cell-mediated immune response in young children at onset of AOM caused by Spn
The current study indicated that the genes associated with the cellular immune response to infection were down regulated at onset of AOM as demonstrated by the indirect cytokine gene regulation and direct cell proliferation and activation regulation. Specifically,cytokine IL-2 and IL-1β were down-regulated. It is known that IL-2 is required for T-cell proliferation as well as other activities crucial to the regulation of the immune response. IL-1β is an important mediator of the inflammatory response, and is involved in a variety of cellular activities, including cell proliferation and differentiation. The observed down-regulation of IL-1β at onset of AOM is consistent with the weak cellular immune response in young children that was observed [19]. We confirmed that the transcriptome signature was reflected in the proteome for IL-10 in serum and at the site of infection in the middle ear.
4.4. Endocytosis of bacteria by host cells in young children
Endocytosis is a process used by cells to engulf bacteria [20]. We found that many genes associated with endocytosis were differentially regulated in children with AOM due to Spn. The genes all belonged to the clathrin-mediated endocytosis group. The platelet-activating factor receptor (PAFR) gene was up-regulated and PAFR has been shown to be a receptor mediating entry of Spn into cells [21]. Data from studies using drugs that inhibit endocytosis support a role for clathrin-dependent endocytosis in the entry of Spn into cells [21,22]. In one of those studies, uptake of the Spn vacuole involved clathrin, and half thebacteria proceeded into vacuoles marked by Rab5 and Rab7 [21]. Our study showed significant regulation of genes encoding Rab5 and Rab7 at onset of AOM due to Spn infection.
We also found that genes for both CD2 associated protein (CD2AP) and Cb1-interacting protein of 85 kDa (CIN85) were significantly down-regulated. Those genes encode proteins necessary for ligand-induced endocytosis of receptor tyrosine kinases during bacterial entry. A previous study found that decreased expression of these proteins strongly inhibited Listeria monocytogenes entry into host cells [23]. Therefore, the down-regulation of these genes in the current study indicates that host cells in children with AOM may have defects in antigen presentation by antigen presenting cells.
4.5. General conclusions, limitations of study results and future directions
The remarkable power of microarray analysis is demonstrated in this study of gene regulation associated with a common pediatric infection caused by respiratory bacteria. With a focus on the immune response genes, we found genes related to antibacterial activity, such as S100 proteins, defensins, cytokines/chemokines, complement and Toll like receptors were particularly up-regulated as expected. The regulation of genes related to DEFB123 and BPI, the alternative complement pathway, TLR5 and TLR10, 5 interleukins and 8 chemokines with functions of enhanced antibacterial activity and inhibition of lymphocyte activation and differentiation and the clathrin-mediated endocytosis cascade are new findings.
Limitations of our study include those inherent in clinical studies in humans of gene regulation where genetic heterogeneity, timing of exposure to pathogens, and dose and strain of the pathogen. To reduce the heterogeneity of the samples, we selected samples from children of very similar age, of the same race, medical history record, pathogen in the middle ear and severity of disease. In addition, to avoid the technical error and subjectivity in microarray assay interpretation, the data was blinded and compiled by an outside laboratory (Phalanx Biotech). We used PBMCs as the source of clinical material because blood is the reservoir for immune cells that migrate to the middle ear during AOM. Leukocytes isolated from the peripheral blood constitute an accessible source of clinically relevant information from which a comprehensive molecular phenotype can be obtained by microarray analysis [24-29].
Future directions of our work include analysis of the gene regulation and protein products involved in the innate and adaptive immune response to Spn infection and the paradoxical results regarding the cell mediated immune response. The regulation of genes involved in the clathrin-dependent endocytosis cascade also should be further understood. Thus, the current results provide valuable information for further identifying new targets for study to understand the AOM disease process and direct us to development of intervention and therapeutic agents.
Supplementary Material
Supplementary Table 1. Genes differentially expressed in 4 children with AOM due to Spn versus their pre-infection healthy stage. Genes whose relative transcription levels changed by >2 fold were considered as differentially regulated. The fold change of gene expression was derived by comparison of the normalized intensity of the signals at the time of onset of AOM due to Spn with the pre-infection healthy stage in the same children. The data from the 4 children are presented as a mean.
Acknowledgments
Work supported by R01DC08671 and the Thrasher Research Fund (to MEP). This study would not have been possible without the help and dedication of Dr. Janet Casey at Legacy Pediatrics and her study coordinator Sally Thomas, LPN, CCRC. We also thank the collaborating pediatricians from Long Pond Pediatrics, Genesis Pediatrics, Rainbow Pediatrics and Lewis Pediatrics, and the parents who consented to this challenging study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authorship Contributions and Disclosure of Conflicts of Interest
The authors declare no competing commercial and financial interests. KL designed the study and performed all transcriptome experiments, analyzed the data and wrote the first draft of the paper. LC analyzed the data. RK performed the proteome experiments. MEP contributed to the design of the research, analyzed data and finalized writing of the paper.
References
- 1.Pichichero ME, Casey JR. Emergence of a Multiresistant Serotype 19A Pneumococcal Strain Not Included in the 7-Valent Conjugate Vaccine as an Otopathogen in Children. JAMA. 2007:1772–1778. doi: 10.1001/jama.298.15.1772. [DOI] [PubMed] [Google Scholar]
- 2.Rogers PD, Thornton J, Barker KS, McDaniel DO, Sacks GS, Swiatlo E, Daniel McL.S. Pneumolysin-dependent and –independent gene expression identified by cDNA microarray analysis of THP-1 human mononuclear cells stimulated by Streptococcus pneumonia. Infect. Immun. 2003;71:2087–2094. doi: 10.1128/IAI.71.4.2087-2094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen A, Li HS, Hebda PA, Zeevi A, Swarts JD. Gene expression profiles of early pneumococcal otitis media in the rat. Int. J. Pediatr. Otorhinolaryngol. 2005;69:1383–1393. doi: 10.1016/j.ijporl.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 4.Liu K, Casey J, Pichichero ME. Serum Intercellular Adhesion Molecule 1 Variations in Young Children During Acute Otitis Media. Clin Vaccine Immunol. 2010;17:1909–16. doi: 10.1128/CVI.00194-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu K, Pichichero ME. S100A12 is a biomarker of acute otitis media caused by Streprococcus pneumoniae in children.. 10th International Otitis Media Symposium; New Orleans. June 5-9, 2011. [Google Scholar]
- 6.Liu K, Pichichero ME. Does IL-10 regulate Intercellular Cell-Adhesion Molecule-1 Differently in otitis prone children. 10th International Otitis Media Symposium; New Orleans. June 5-9, 2011. [Google Scholar]
- 7.Casey JR, Adlowitz DG, Pichichero ME. New Patterns in the otopathogens causing acute otitis media six to eight years after introduction of pneumococcal conjugate vaccine. Pediatr Infect Dis J. 2010;29:304–9. doi: 10.1097/INF.0b013e3181c1bc48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaur R, Adlowitz DG, Casey JR, Zeng M, Pichichero ME. Simultaneous Assay for Four Bacterial Species Including Alloiococcus otitidis Using Multiplex-PCR in Children With Culture Negative Acute Otitis Media. Pediatric Infectious Disease Journal. 2010;29:741–745. doi: 10.1097/INF.0b013e3181d9e639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawada J, Kimura H, Kamachi Y, Nishikawa K, Taniguchi M, Nagaoka K, Kurahashi H, Kojima S, Morishima T. Analysis of gene-expression profiles by oligonucleotide microarray in children with influenza. J Gen Virol. 2006;87:1677–83. doi: 10.1099/vir.0.81670-0. [DOI] [PubMed] [Google Scholar]
- 10.Liu K, Chen L, Pichichero ME. Nontypeable Haemophilus influenzae induces weak defense responses in young children with acute otitis media. Infection, Genetics and Evolution. 2011 Under Review. [Google Scholar]
- 11.Rosenberger CM, Scott MG, Gold MR, Hancock RE, Finlay BB. Salmonella typhimurium infection and lipopolysaccharide stimulation induce similar changes in macrophage gene expression. J. Immunol. 2000;164:5894–5904. doi: 10.4049/jimmunol.164.11.5894. [DOI] [PubMed] [Google Scholar]
- 12.Ichikawa JK, Norris A, Bangera G, Geiss GK, van't Wout AB, Bumgarner RE, Lory S. Interaction of Pseudomonas aeruginosa with epithelial cells: Identification of differentially regulated genes by expression microarray analysis of human cDNAs. Proc. Natl. Acad. Sci. USA. 2000;97:9659–9664. doi: 10.1073/pnas.160140297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Motzkus D, Schulz-Maronde S, Heitland A, Schulz A, Forssmann WG, Jübner M. Martin, Maronde E. The novel β-defensin DEFB123 prevents lipopolysaccharide-mediated effects in vitro and in vivo. The FASEB Journal. 2006;20:1701–1702. doi: 10.1096/fj.05-4970fje. [DOI] [PubMed] [Google Scholar]
- 14.Srivastava A, Casey H, Johnson N, Levy O, Malley R. Recombinant bactericidal/permeability-increasing protein rBPI21 protects against pneumococcal disease. Infect. Immun. 2007;75:342–349. doi: 10.1128/IAI.01089-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lutzow YC, Donaldson L, Gray CP, Vuocolo T, Pearson RD, Reverter A, Byrne KA, Sheehy PA, Windon R, Tellam RL RL. Identification of immune genes and proteins involved in the response of bovine mammary tissue to Staphylococcus aureus infection. BMC Vet Res. 2008;4:18. doi: 10.1186/1746-6148-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown JS, Hussell T, Gilliland SM, Holden DW, Paton JC, Ehrenstein MR, Walport MJ, Botto M. The classical pathway is the dominant complement pathway required for innate immunity to Streptococcus pneumoniae infection in mice. Proc Natl Acad Sci U S A. 2002;99:16969–74. doi: 10.1073/pnas.012669199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muñoz N, Maele LV, Marqués JM, Rial A, Sirard JC, José A. Chabalgoity. Mucosal Administration of Flagellin Protects Mice from Streptococcus pneumoniae. Lung Infection Infection and Immunity. 2010;78:4226–4233. doi: 10.1128/IAI.00224-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gopal R, Birdsell D, Monroy FP. Regulation of toll like receptors in intestinal epithelial cells by stress and Toxoplasma gondii infection. Parasite Immunol. 2008;30:563–576. doi: 10.1111/j.1365-3024.2008.01055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma SK, Casey JR, Pichichero ME. Reduced memory CD4+ T-cell generation in the circulation of young children may contribute to the otitis-prone condition. J Infect Dis. 204(2011):645–53. doi: 10.1093/infdis/jir340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marsh M. Endocytosis. Oxford University Press; 2001. p. vii. ISBN 978-0-19-963851-2. [Google Scholar]
- 21.Cundell DR, Gerard NP, Gerard C. Ilona Idanpaan-Heikkila & Elaine I. Tuomanen. Streptococcus pneumoniae anchor to activated human cells by the receptor for platelet-activating factor. Nature. 2002;377:435–438. doi: 10.1038/377435a0. [DOI] [PubMed] [Google Scholar]
- 22.Radin JN, Orihuela CJ, Murti G, Guglielmo C, Murray PJ, Elaine I. Tuomanen. β-Arrestin 1 Participates in Platelet-Activating Factor Receptor-Mediated Endocytosis of Streptococcus pneumonia. Infection and Immunity. 2005;73:7827–7835. doi: 10.1128/IAI.73.12.7827-7835.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Veiga E, Cossart P. Listeria hijacks the clathrin-dependent endocytic machinery to invade mammalian cells. Nat. Cell Biol. 2005;7:894–900. doi: 10.1038/ncb1292. [DOI] [PubMed] [Google Scholar]
- 24.Alizadeh AA, Eisen MB, Davis RE, Ma C, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 25.Golub TR, Slonim DK, Tamayo P, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 26.Rubins KH, Hensley LE, Jahrling PB. The host response to smallpox: analysis of the gene expression program in peripheral blood cells in a nonhuman primate model. Proc Natl Acad Sci USA. 2004;101:15190–15195. doi: 10.1073/pnas.0405759101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bennett L, Palucka AK, Arce E, Cantrell V, Borvak J, Banchereau J. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med. 2003;197:711–23. doi: 10.1084/jem.20021553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baechler EC, Batliwalla FM, Karypis G. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci USA. 2003;100:2610–2615. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salzman MB, Sood SK, Slavin ML, Rubin LG. Ocular manifestations of Mycoplasma pneumoniae infection. Clin Infect Dis. 1992;14:1137–9. doi: 10.1093/clinids/14.5.1137. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Genes differentially expressed in 4 children with AOM due to Spn versus their pre-infection healthy stage. Genes whose relative transcription levels changed by >2 fold were considered as differentially regulated. The fold change of gene expression was derived by comparison of the normalized intensity of the signals at the time of onset of AOM due to Spn with the pre-infection healthy stage in the same children. The data from the 4 children are presented as a mean.