Abstract
KP772 is a new lanthanum complex containing three 1,10-phenathroline molecules. Recently, we have demonstrated that the promising in vitro and in vivo anticancer properties of KP772 are based on p53-independent G0/G1 arrest and apoptosis induction. A National Cancer Institute (NCI) screen revealed significant correlation of KP772 activity with that of the ribonucleotide reductase (RR) inhibitor hydroxyurea (HU). Consequently, this study aimed to investigate whether KP772 targets DNA synthesis in tumor cells by RR inhibition. Indeed, KP772 treatment led to significant reduction of cytidine incorporation paralleled by a decrease of deoxynucleoside triphosphate (dNTP) pools. This strongly indicates disruption of RR activity. Moreover, KP772 protected against oxidative stress, suggesting that this drug might interfere with RR by interaction with the tyrosyl radical in subunit R2. Additionally, several observations (e.g. increase of transferrin receptor expression and protective effect of iron preloading) indicate that KP772 interferes with cellular iron homeostasis. Accordingly, co-incubation of Fe(II) with KP772 led to generation of a coloured iron complex (Fe-KP772) in cell free systems. In electron paramagnetic resonance (EPR) measurements of mouse R2 subunits, KP772 disrupted the tyrosyl radical while Fe-KP772 had no significant effects. Moreover, coincubation of KP772 with iron-loaded R2 led to formation of Fe-KP772 suggesting chelation of RR-bound Fe(II). Summarizing, our data prove that KP772 inhibits RR by targeting the iron centre of the R2 subunit. As also Fe-KP772 as well as free lanthanum exert significant -though less pronounced- cytotoxic/static activities, additional mechanisms are likely to synergise with RR inhibition in the promising anticancer activity of KP772.
Keywords: Lanthanum; ribonucleotide reductase; 1,10-phenanthroline; cell cycle arrest; anticancer activity; KP772
INTRODUCTION
[Tris(1,10-phenanthroline)lanthanum(III)]trithiocyanate (KP772) is a new lanthanum compound containing three 1,10-phenathroline (1,10-phen) molecules. This complex exerts potent cytotoxic activity against a wide range of tumor cell lines in vitro and a colon carcinoma xenograft model in vivo [1]. Recent investigations revealed that multidrug-resistant cancer cells are especially sensitive to KP772, which indicates that this lanthanum drug might be suitable for the second line treatment of patients after failure of standard chemotherapy [2]. The anticancer activity of KP772 is characterized by apoptosis induction and potent cell cycle arrest in G0/G1 phase not based on radical- or intercalation-induced DNA damage [1].
Additionally, the antiproliferative effects of KP772 were evaluated against a panel of 60 cell lines as part of the in vitro anticancer-screening services provided by the NCI [1]. Only moderate correlations were observed comparing the cytotoxicity profile of KP772 with that of other drugs. Notably, several antimetabolic compounds including HU and methotrexate (MTX) were among those reaching significant correlations (Table 1). Both, HU and MTX are known to exert their antineoplastic effects through nucleotide pool depletion [3-5]. MTX, a well-known structural analogue of folic acid, is a potent inhibitor of the dihydrofolate reductase and, consequently, the de novo thymidylate and purine nucleotide synthesis [4]. In contrast, HU blocks DNA synthesis through inhibition of the RR. This enzyme, which is rate-limiting for synthesis of dNTPs, is an α2β2 complex composed of two subunits [6]. The R1 subunit has a α2 homodimeric structure with substrate and allosteric effective sites that control enzyme activity and substrate specificity. The small R2 subunit, a β2 homodimer, forms two dinuclear iron centers each stabilizing a tyrosyl radical. Additionally, a p53-inducible R2-homologoue (p53R2) has been described [5]. Expression of this subunit is induced by DNA damage and it has been reported that p53R2 supplies dNTPs for DNA repair in G0/G1 cells in a p53-dependent manner. Moreover, recent studies revealed that p53R2 activity is crucial for mitochondrial DNA replication [7]. With regard to drug-induced RR inhibition, HU is believed to destabilize R2 iron centers causing thereby destruction of the radical essential for enzyme activity [4, 5]. Additionally, several iron chelating agents including desferrioxamine (DFO) [5, 8] and several thiosemicarbazones (e.g. Triapine) [3, 5] were shown to interact with the iron-containing R2 subunit of RR. Also the rigid planar 1,10-phen, contained in the KP772 complex, is a known metal chelator [9] but there are no reports on RR inhibition or any other impacts on the cellular nucleotide pools for 1,10-phen so far. The structurally-related Fe(II) chelator bathophenanthroline was reported to have even at very high concentrations (1 mM) only very minor effects on the tyrosyl radical of mouse R2 [10].
Table 1. GI50 Compare Analysis for KP772 (NSC632737).
| rank | CI | NSC Number | Compound |
|---|---|---|---|
| 1 | 0.379 | NSC32065 | hydroxyurea |
| 2 | 0.372 | NSC153353 | L-alanosine |
| 3 | 0.339 | NSC752 | thioguanine |
| 4 | 0.338 | NSC6396 | thio-tepa |
| 5 | 0.334 | NSC135758 | piperazinedione |
| 6 | 0.333 | NSC740 | methotrexate |
| 7 | 0.331 | NSC339638 | fostriecin |
| 8 | 0.328 | NSC132313 | dianhydrogalatitol |
| 9 | 0.32 | NSC750 | busulfan |
| 10 | 0.317 | NSC145668 | cyclocytidine |
In this study, we demonstrate that KP772 treatment leads to dNTP pool depletion and functional inhibition of the RR. Moreover, our data indicate that, based on its iron- chelating properties, KP772 interacts with the R2 subunit of this essential cellular enzyme.
MATERIAL AND METHODS
Drugs
KP772 (La(phen)3(SCN)3) was prepared as described previously [1]. For all experiments, the compound was dissolved in water and diluted into culture media at the concentrations indicated. All other substances were purchased from Sigma-Aldrich (St. Louis, USA).
Cell Culture
The following human cell lines were used in this study [1]: the cervix carcinoma Hela-derivative KB-3-1, the promyelocytic leukaemia cell line HL60, and the non-small lung cancer cell line A549 (from American Type Culture Collection, Manassas, VA), as well as the colon carcinoma cell line HCT116 generously donated by B. Vogelstein, John Hopkins University, Baltimore. All cells were grown in RMPI 1640 medium supplemented with 10% fetal bovine serum with the exception of HCT116 which were grown in McCoy’s culture medium. Cultures were regularly checked for Mycoplasma contamination.
Cytotoxicity Assays
Cells were plated (2×103 cells in 100 μl/well) in 96-well plates and allowed to recover for 24 h. Drugs were added in another 100 μl growth medium and cells exposed for 72 h. The proportion of viable cells was determined by MTT assay following the manufacturer’s recommendations (EZ4U, Biomedica, Vienna, Austria). Cytotoxicity was expressed as IC50 values calculated from full dose-response curves (drug concentrations leading to a 50% reduction of cell survival in comparison to the control cultured without drugs). Synergism was determined by calculating the combination index (CI) according to Chou and Talalay [11] with CalcuSyn software (Biosoft, Ferguson, MO USA). CI<1, CI=1 or CI>1 represent synergism, additive effects and antagonism of the two investigated substances, respectively.
Incorporation of 3H-Labelled Cytidine into DNA
To indirectly analyze the effect of KP772 on RR activity, incorporation 3H-cytidine assay [12] was performed. For this purpose, 5×106 HL60 cells were incubated with the test substances for 24 h. After the incubation period, the cells were pulsed with 3H-cytidine (0.3125 μCi, 5 nM) for 30 min at 37 °C. Then cells were collected, washed and total DNA was extracted. Radioactivity was determined as described in [1].
Determination of dNTP Pools
HL60 cells were incubated with KP772 for 24 h. Afterwards, 5×107 cells were separated for the extraction of dNTPs according to the method described by Garrett and Santi [13]. Briefly, cells were washed in phosphate buffered saline (PBS) and lysed by 30 min incubation with trichloroacetic acid. After centrifugation supernatant was collected and neutralized by adding 1.1 vol of freon containing 0.5 M tri-n-octylamin. Aliquots of 100 μl were periodated by adding 30 μl of 4 M methylamine and 10 μl sodium periodate (100 g/l). After 30 min incubation at 37 °C, the reaction was stopped by adding 5 μl of 1 M rhamnose solution. The extracted dNTPs were measured using a Merck ”La Chrom” HPLC system equipped with L-7200 autosampler, L-7100 pump, L-7400 UV detector, and D-7000 interface. Samples were eluted with a 3.2 M ammonium phosphate buffer, pH 3.4 (pH adjusted by addition of 0.32 mmol/l H3PO4), containing 20 mM acetonitrile using a 4.6 × 250 mm Partisil 10 SAX analytical column (Whatman, Kent, UK). Separation was performed at constant ambient temperature with a flow rate of 2 ml/min. The concentration of dNTPs was calculated as percent of total area under the curve for each sample.
Western Blot Analysis
Cell fractionation, protein separation and Western blotting were performed as previously described [2]. The following antibodies: anti-RR subunit R1 polyclonal goat T-16, anti-RR subunit R2 polyclonal goat I-15; anti-RR subunit p53R2 polyclonal goat N-16 were purchased from Santa Cruz Biotechnology and used in a 1:200 dilution. Anti-ß-actin monoclonal mouse antibody AC-15 (Sigma at a 1:1000 dilution) was used to detect ß-actin as loading control. All secondary, peroxidase-labelled antibodies from Pierce (Rockford, IL, USA) were used at working dilutions of 1:10000.
Alkaline Comet Assay
The induction of DNA strand breaks was determined using the alkaline comet assay as described [1]. Each microscope slide was precoated with a layer of 1.5% normal melting point agarose and dried at room temperature. KB-3-1 cells were pretreated for 30 min with KP772 or NAC followed by 1 h H2O2 treatment. After PBS washing, cells were mixed with 0.5% low melting point agarose at 37 °C and dropped on top of the first layer. The agarose was allowed to solidify on a cooled tray and then immersed in ice-cold lysing solution (2.5 M NaCl; 100 mM Na2EDTA, 10 mM Tris–HCl, 1% Triton X-100; pH 10) for 1 h. After lysis, the slides were placed in a horizontal gel electrophoresis chamber and DNA was allowed to unwind for 1 h in the electrophoresis buffer (300 mM NaOH, 1 mM Na2EDTA, pH 12.5). Electrophoresis was conducted for 20 min at 25 mV and 300 mA in a chamber cooled on ice. Then the slides were washed with neutralisation buffer (0.4 M Trizma Base, pH 7.5) and water, fixed in 100% ice cold methanol, and stained with ethidium bromide. The comet tails were analysed using a fluorescent microscope (Nikon) with an automated image analysis system based on the public domain program NIH image analysis system. DNA migration was expressed as comet tail length in micrometers. Per experimental group, two slides were prepared and from each at least 50 cells were analysed. Cell viability at the time of assay was in all cases >90%.
Measurement of Intracellular Oxidants
2′,7′-dichlorofluorescin diacetate (DCF-DA) was used to detect the intracellular production of free radicals [14]. Stock solutions of 33.4 mM in DMSO were stored at −20 °C. KB-3-1 cells (2.5×105 cells per sample in phenol-free Hanks balanced salt solutions) were treated with the test compounds for 1 h at 37 °C, followed by 1 h exposure to DCF-DA. Mean fluorescence intensity was measured by flow cytometry using a fluorescence-activated cell sorting (FACS) Calibur (Becton Dickinson, Palo Alto, CA). The resulting DNA histograms were quantified using the ModeFit software (BD).
Expression of Transferrin Receptor (TfR)
Expression of TfR in HL60 cells was analysed by immunostaining and FACS analysis using the monoclonal mouse antibody VIP-1 (generously donated by Dr. Majdic, Medical University of Vienna, Austria). Briefly, 5×105 cells were washed with PBS containing 1% BSA and incubated for 30 min with 20 μg/ml primary antibody at 4 °C. Bound antibody was stained for 30 min at 4 °C with an anti-mouse IgG (Fab specific) FITC conjugate (Sigma) at a 1:150 working dilution.
Cell Cycle Analysis
HL60 or KB-3-1 cells (106 per well) seeded into 6-well plates and cultured for 24 h were treated for another 24 h with the test substance. Then cells were collected, washed with PBS, fixed in 70% ethanol and stored at −20 °C. To determine the cell cycle distribution, cells were transferred into PBS, incubated with RNAse (10 μg/ml) for 30 min at 37 °C, treated with 5 μg/ml propidium iodide (PI) for 30 min and then analyzed by FACS.
UV-Vis Spectra
Optical absorption spectra were recorded at room temperature using a Synergy HT multi-detection microplate reader (Bio-Tek, Vermont, USA) or a V-560 UV/VIS spectrophotometer (Jasco Essex, UK). All spectra were baseline corrected.
Electron Paramagnetic Resonance (EPR) Spectroscopy
Quenching of the tyrosyl radical in mouse R2 subunit by KP772 was monitored kinetically using EPR spectroscopy in comparison with Fe-KP772 and 1,10-phen. Mouse R2 protein was expressed in Rosetta 2(DE3)pLys bacteria as described [15]. Protein was reconstituted with an anaerobic solution of Fe(H4N)2(SO4)2 at a ratio of 10 Fe(II) ions per R2 monomer. Excess iron was removed by gel filtration. The protein contained 0.33 tyrosyl radicals per monomer. Final R2 iron loading was 1.3 ± 0.1 Fe/polypeptide (maximum is 2 Fe/polypeptide). EPR spectra were recorded on a Bruker ESP 300 X-band (9.5 GHz) spectrometer with an Oxford Instruments ESR9 helium cryostat at 40 K, 3.2 mW microwave power and 10 G modulation amplitude. The concentration of the tyrosyl radical was determined by double integration of EPR spectra recorded at non-saturating microwave power levels, and compared with a standard solution of 1 mM CuSO4 in 50 mM EDTA. The calculated radical concentration was normalized and expressed in percent. Experiments were repeated 5 times, which gave an estimate of the uncertainty of each measurement of about 5%.
RESULTS
Effects of KP772 on RR Activity and dNTP Pools
As a first approach the cytotoxic activity of KP772 was compared with the well investigated RR inhibitors Triapine and HU at identical conditions. As shown in Fig. (1), both KP772 and Triapine were significantly cytotoxic at μM levels with IC50 values of 1.4 μM and 16.3 μM, respectively. In contrast, HU was found to be active only at mM levels (IC50 of 1.12 mM), which is in accordance to other published data [16-18]. To measure the impact of KP772 on the enzymatic activity of RR, the incorporation of 3H-labeled cytidine into DNA of HL60 cells was determined. After 24 h treatment with 2.5 μM KP772, 3H-cytidine incorporation into DNA was significantly decreased to 25% of control values (Fig. (2A)). Treatment with lower drug concentrations had no significant effect on cytidine incorporation.
Fig. (1). Cytostatic activity of KP772 in comparison with Triapine and HU.

A549 cells were treated for 72 h with the indicated concentrations of the respective drug. Vitality was evaluated by MTT assay. Values given are means ± standard deviations of one representative experiment performed in triplicates.
Fig. (2). Impact of KP772 on RR activity and cellular dNTP pools.
(A) Influence of KP772 on 3H-cytidine incorporation was measured in HL60 cells after 24 h drug incubation. DNA was isolated and radioactivity determined as described under Material and Methods. Values given are means ± standard deviation of one representative experiment out of three performed in triplets. ** Significantly different from the control cells (p < 0.01, by Student’s t-test) (B) HL60 cells were incubated for 24 h with the indicated concentrations of KP772. dNTPs were isolated and measured by HPLC as described under Material an Methods. Values given are means ± standard deviation of 3 determinations out of one representative experiment. Significant dose-concentration response * p < 0.05, ** p < 0.01, p < 0.001 by one-way ANOVA (C) Expression levels of RR subunits were determined by Western blotting in the indicated cell lines after 24 h drug treatment. Antibodies used are described under Material and Methods.
Next, the impact of KP772 on the cellular dNTP pools was determined using HPLC [13]. To this end, HL60 cells were incubated with 2.5, 5, and 10 μM KP772 for 24 h. Intracellular concentrations of dNTPs in untreated control cells were 1.02 μM, 2.48 μM, and 11.32 μM for dCTP, dTTP, and dATP, respectively. As shown in Fig. (2B), KP772 treatment caused imbalance in the dNTP pools cells leading to a dose-dependent decrease of cellular dCTP. In contrast, dATP and dTTP levels were enhanced at 2.5 μM (up to 190% and 135% in case of dATP and dTTP, respectively) followed by a dose-dependent decrease of all measured dNTPs at higher KP772 concentrations.
To determine whether KP772 has an impact on RR protein expression, amounts of R1, R2 and p53R2 subunits were analysed in several cancer cell lines (Fig. (2C)). In all tested models (HCT116, A549, HL60, and KB-3-1) treatment with 2.5 μM KP772 increased R2 subunit expression, while at higher drug concentrations frequently a decrease of this protein subunit was observable (except KB-3-1 cells). R1 and P53R2 subunit expressions followed a similar pattern in HL60 cells, while they remained widely unchanged in the HCT116 and A549 cell lines. In accordance to the report of Lembo et al. [19], no p53R2 expression in the p53-negative Hela derivative KB-3-1 was detected. Additionally, P53R2 expression in HL60 cells was notably low correlating with the p53 (−/−) status of these cells [20].
Synergism of KP772 with HU
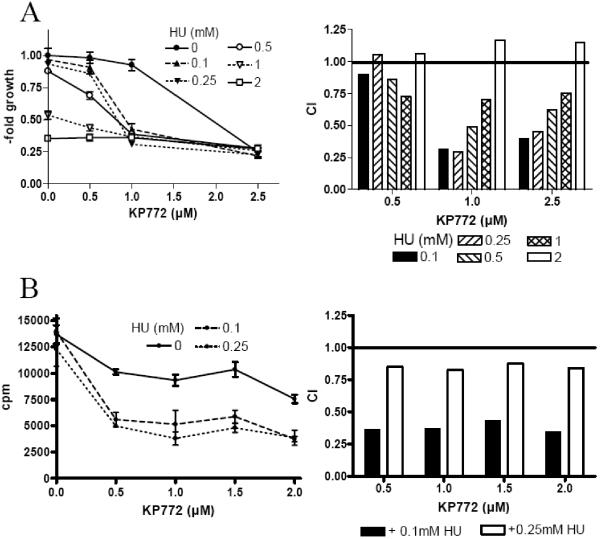
Combinations of diverse RR inhibitors [21, 22] are known to act synergistically and to enhance cytostatic activities of nucleoside analogues due to the RR inhibition-dependent dNTP pool depletion [23-26]. KP772 coadministration had no impact on the effects of 5-FU, DFO, AraC and vinblastine (data not shown). In contrast, the lanthanum drug acted distinctly synergistic with HU in A549 cells (Fig. (2)). As indicated by the combination index (CI, compare Material and Methods section) blot in Fig. (3A) (right graph), maximal synergistic effects (CI between 0.25 and 0.5 μM) were observed by combining 1 and 2.5 μM KP772 with 100 and 250 μM HU. The synergism between KP772 and HU was based on enhanced inhibition of DNA synthesis as measured by reduced 3H-thymidine incorporation (Fig. (3B)). For all KP772 concentrations tested (0.5-2 μM), CI values with 100 and 250 μM HU were <0.45 and <0.88, respectively. Comparable data were achieved using KB-3-1 cells (data not shown).
Fig. (3). Impact of KP772 coincubation on cytotoxic activity of antimetabolic drugs.
Synergistic effect of KP772 and HU on (A) vitality and (B) 3H-thymidine incorporation in A549 cells are depicted. Cell viability was measured using MTT assay. Values given are means ± standard deviations of one representative experiment performed in triplicates. Combination Index (CI) was calculated using CalcuSyn software.
Radical Scavenging Properties of KP772
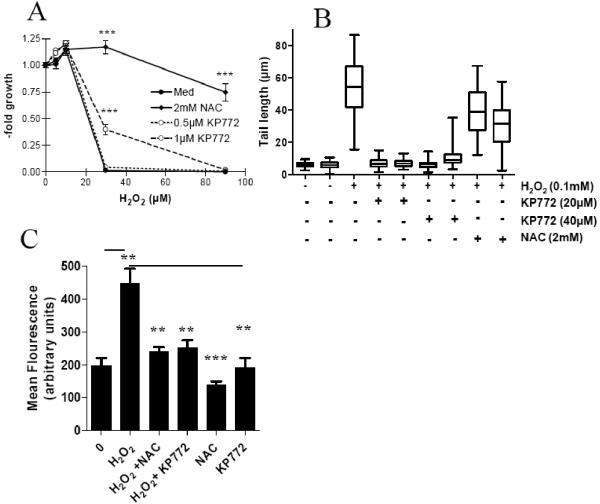
Several RR inhibitors including HU [27], DFO [27], resveratrol [26], and trimidox [28] are all known to scavenge diverse radical species. To test whether KP772 can protect cells from reactive oxygen species (ROS), KB-3-1 cells were treated in presence of subtoxic concentrations of KP772 (0.5 and 1 μM) with various concentrations of H2O2. After 72 h, 1 μM KP772 displayed a significant protective effect against 30 μM H2O2 (Fig. (4A)). To evaluate this effect at higher concentrations of KP772 (inducing cytotoxic effects in long-term exposure assays), the alkaline comet assay allowing short-term exposure times (1 h) was used. We have already reported that up to 300 μM KP772 did not induce any DNA strand breaks as determined by this assay [1]. As shown in Fig. (4B), 30 min pretreatment with 20 μM and 40 μM KP772 was able to completely protect cells from H2O2-induced DNA damage after 1 h co-incubation. In comparison, 2 mM of the radical scavenger NAC only reduced H2O2-induced comet tail length to ~65%. Corroborating results were achieved using the cell permeable dye DCF-DA whose metabolite becomes highly fluorescent after intracellular oxidation by ROS. KP772 widely inhibited H2O2-mediated ROS formation confirming the radical scavenging properties for KP772 (Fig. (4C)).
Fig. (4). Radical scavenging properties of KP772.
(A) After 30 min preincubation with KP772 (0.5 μM and 1 μM) or NAC (2 mM), KB-3-1 cells were treated for 72 h with the indicated concentrations of H2O2. Vitality was determined using MTT assay. Values given are means ± standard deviation of 3 determinations out of 3 experiments. Statistical significance was determined by unpaired Student’s t test (*** p<0.001). (B) Impact of KP772 and NAC on DNA damaging activity of H2O2 was measured by Comet assay as described under Material and Methods. (C) Influence of 30 μM KP772 or 2 mM NAC on the intracellular ROS production in KB-3-1 cells after 1 h incubation with H2O2 (0.1 mM) was determined using the ROS indicator DCF-DA. Fluorescence was measured by flow cytometry. One representative experiment out of three delivering comparable results is shown. Statistical significance in comparison to H2O2 treatment was determined by unpaired Student’s t test; ** p< 0.01, *** p<0.001.
Impact of KP772 on Intracellular Iron Homeostasis
Several iron chelating agents [5, 27] were shown to interact with the iron containing R2 subunit of the RR. Since 1,10-phen is a known metal chelator [9], it was of interest whether KP772 also affects intracellular iron pools. As under low iron conditions regulatory mechanisms rapidly stimulate TfR synthesis and expression [29, 30], changes in TfR expression of HL60 cells were determined as indication for intracellular iron deficiency. After 24 h drug treatment, both compounds, KP772 and the ligand 1,10-phen, induced a dose-dependent increase of membrane-located TfR (Fig. (5A)). Unexpectedly this increase was found only in subtoxic drug concentrations. No changes in TfR expression were observable when cytotoxic effects of both drugs occurred as indicated by appearance of a subG0/G1 peak in PI-staining and FACS analysis (Fig. (5B)). In accordance to our previous reports [1], a profound G0/G1 arrest was detectable after KP772 as well as 1,10-phen treatment. Moreover, KP772 displayed also in this setting an enhanced cytotoxic activity as compared to its ligand [1]. Perhaps due to this reduced toxicity, 1,10-phen was more potent in TfR induction than KP772 (6-fold at 5 μM vs. 4-fold at 2.5 μM). Accordingly, enhanced R1 and R2 subunit expression in HL60 cells was observed in case of 1,10-phen at 5 μM (Fig. (5C)) comparable to the ones induced by KP772 at 2.5 μM (compare Fig. (2C)).
Fig. (5). Interaction of KP772 with intracellular iron.
To assess the impact of KP772 on TfR expression and cell cycle distribution, HL60 cells samples were incubated for 24 h with the indicated KP772 concentrations and separated into 2 sample groups. (A) TfR expression of viable cells in the first group was determined as described under Material and Methods. (B) Changes in cell cycle distribution were determined in the second sample group by PI-staining and FACS analysis of ethanol fixed cells. (C) Impact of 1,10-phen on R1 and R2 subunit expression was determined in HL60 cells by Western blotting. (D) KB-3-1 cells were loaded with FeCl3 followed by treatment with the indicated KP772 concentrations. Values given are means ± standard deviation of 3 experiments performed in triplicates.
To assess whether the cytotoxic activity of KP772 is dependent on cellular iron depletion, cells were preincubated with epimolar concentrations of iron followed by 72 h KP772 treatment. Since 1,10-phen has strong preferences for Fe(II), Fe(III)Cl3, which is converted into Fe(II) during cellular uptake [30], was used in these experiments to minimize extracellular interactions. At all concentrations tested, preincubation with FeCl3 had profound protective effects against KP772 (Fig. (5C)).
In vitro Formation of an Iron Complex
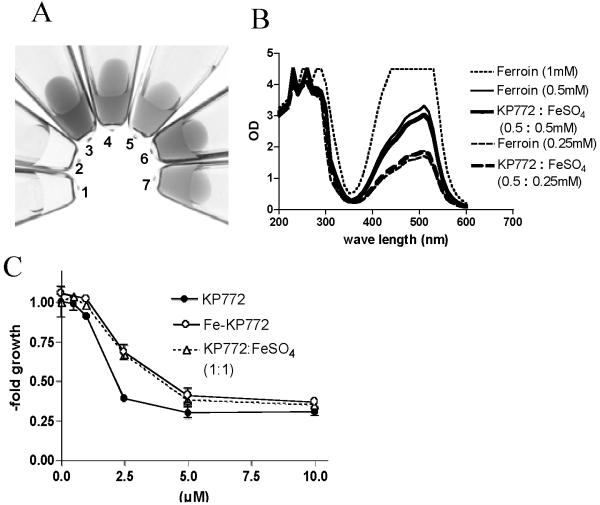
The reaction of 1,10-phen with Fe(II) leading to the formation of an iron complex, called ferrous phen or ferroin, is a well known process and has been used for the analytical determination of iron levels in blood plasma for several decades [31]. To test, whether also KP772 reacts with Fe(II), the colorless lanthanum drug was mixed in aqueous solution with FeSO4 resulting in immediate appearance of orange-red color (Fig. (6A)). Comparison with the UV-Vis spectrum of pure ferroin (Fig. (6B)) suggested that the change in color (broad absorption peak with a maximum between 450 and 550 nm) is based on the formation in a comparable ferrous phen complex (here termed Fe-KP772). Interestingly, this reaction was strongly diminished when performed in cell culture medium containing 10% fetal calf serum (data not shown).
Fig. (6). Formation of Fe-KP772 by coincubation of KP772 with Fe(II)SO4.
(A) Photographs of aqueous solutions of (1) 1 mM KP772, (2) 1 mM FeSO4, (3) 1 mM Ferroin, (4) 0.5 mM Ferroin, (5) 0.25 mM Ferroin, (6) 0.5 mM KP772 mixed with 0.5 mM Fe(II)SO4 or (7) 0.25 mM KP772 with 0.25 mM Fe(II)SO4 were taken immediately after preparation at room temperature. (B) UV-Vis spectra (200-600 nm) of the indicated aqueous solutions were measured immediately after preparation. (C) KB-3-1 cells were exposed to the indicated KP772 concentrations alone, Fe-KP772, or a 1:1 mixture with FeSO4 (mixed in H2O). Values given are means ± standard deviation of 3 experiments performed in triplicates.
For DFO [32] it has been reported that the biological activity of this compound is solely dependent on its iron binding properties. Thus co-incubation with iron completely inhibited the effects of this drug. As shown in Fig. (5C), this is not the case for KP772. Although significantly diminished (1.7-fold enhanced IC50), KP772 had still anticancer activity against KB-3-1 cells after preincubation with equimolar concentrations of Fe(II)SO4. Also, pre-prepared Fe-KP772 exerted comparably diminished (1.5-fold enhanced IC50 as compared to KP772) antiproliferative properties.
Inhibition of RR by KP772-Mediated Iron Chelation
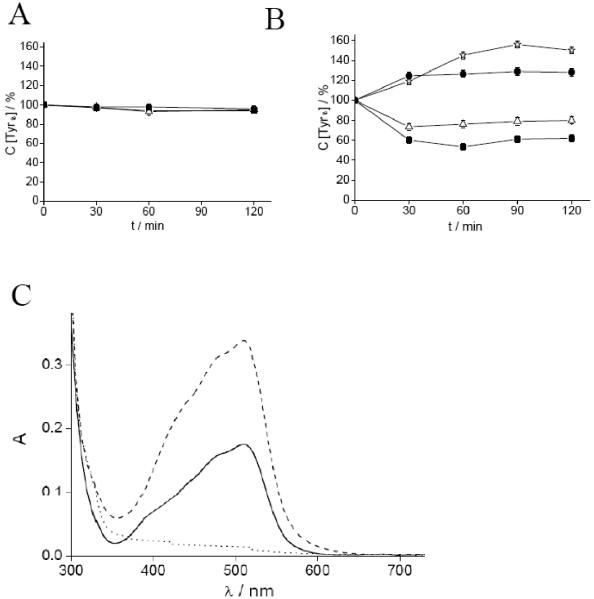
To test whether the RR inhibition observed after treatment with KP772 is based on radical scavenging or iron chelation, EPR measurements of the RR tyrosyl radical were performed. For all EPR measurements, excess iron in R2 protein samples was removed by gel filtration to avoid reactions of KP772 with free, not-R2-bound iron. Fig. (7A) shows the effects on tyrosyl radical content after incubation of mouse recombinant R2 subunit with KP772, Fe-KP772, or 1,10-phen under non-reducing conditions. None of the compounds displayed significant effect in this setting. As a next step, similar experiments were performed in the presence of the reductant dithiothreitol (DTT). In these experiments, 1 mM DTT alone led to a small but significant increase in radical content, probably due to a continued radical reconstitution process. As shown in Fig. (7B), treatment with 20 μM KP772 significantly reduced the R2 EPR signal. Notably, also treatment with 1,10-phen induced disruption of the tyrosyl radical (although to a weaker extent). In contrast, Fe-KP772 had no RR inhibitory potential. DTT alone or with Fe-KP772 led to a small but significant increase in the amount of radical, again suggesting a continued radical reconstitution process. To evaluate whether incubation of KP772 with activated R2 subunits of RR results in the formation of an iron complex, UV-Vis spectra of the samples used for EPR measurements were compared with the spectra of pure KP772 and Fe-KP772 (Fig. (7C)). At these settings, coincubation of R2 protein with KP772 led to generation of a clearly visible Fe-KP772-like spectrum. Together those data indicate that under reducing conditions KP772 is able to chelate the iron of protein R2 sufficiently to destroy the free radical and hence enzyme activity.
Fig. (7). Mouse R2 tyrosyl radical measurements by EPR.
Time course of the relative concentration of the tyrosyl radical signal in mouse R2 protein in presence of Fe-KP772, 1,10-phen and KP772 without (A) and with (B) the addition of DTT. The samples contained: 10 μM mouse R2 protein, 20 μM (●) Fe-KP772, (Δ) 1,10-phen, and (■) KP772. A control sample (◊) contained 10 μM mouse R2 protein and 1 mM DTT. The samples were incubated for indicated times and quickly frozen in cold isopentane. The same samples were used for repeated incubations at room temperature. The natural decay of the tyrosyl radical in the control sample due to freezing/thawing and incubation for each time point (4%, 10%, 24% and 30% at 30, 60, 90 and 120 min incubation, respectively) was subtracted. (C) Light absorption spectra of (—) 30 μM KP772 that was incubated with 10 μM mouse R2 protein for 60 min at room temperature (the contribution from mouse R2 has been removed by subtraction of the 10 μM mouse R2 protein spectrum), (– – –) 30 μM Fe-KP772, and (...) 30 μM KP772 alone. All samples contained 5 mM DTT.
DISCUSSION
RR catalyzes the reduction of ribonucleotides to the corresponding deoxyribonucleotides and thus is essential for DNA synthesis and cell proliferation [5]. Consequently, RR has been in the focus of interest for several decades as target for the treatment of several diseases including malaria, HIV and cancer. Currently, there are several RR inhibiting drugs approved (e.g. HU, gemcitabine) for oncological treatment or under (pre)clinical development (e.g. Triapine, DFO) [33]. Recently, we have demonstrated promising anticancer activity of the 1,10-phen lanthanum complex KP772 both in cell culture and in xenografts models of human tumors [1, 2]. In this study, we have addressed the molecular mechanisms underlying these cytostatic/cytotoxic effects and identified KP772 as a novel and potent RR inhibitory drug.
As canonical for RR inhibitors, treatment with KP772 led to distinct dNTP pool depletion. Regarding the different dNTPs, KP772 most prominently reduced dCTP levels while dATP and dTTP were increased at low and decreased at elevated KP772 concentrations. Similar reports exist for the nucleoside analogue Cl-F-ara-ATP in HL60 cells [34] and for the deoxycytidine analogue gemcitabine in CCRF-CEM cells [35]. This is in contrast to HU, which targets in cell culture [36, 37] as well as in patients [38] primarily dATP. Also the resveratrol-analog M8 reduced dTTP and dATP but increased dCTP levels in Hl60 cells [39]. For Triapine a decline in dATP and dGTP pools of circulating leukemia blasts has been observed during chemotherapy [40]. The reasons underlying these differences are unknown so far but seem to depend on the interaction mode of the inhibitors with the RR enzyme.
According to their targets (R1, R2 or p53R2) and the molecular mechanisms of interaction (e.g. nucleoside analogs, radical scavengers, iron chelators) several categories of small molecule RR inhibitors have been defined [5]. Our data suggest that KP772 almost completely inhibits DNA damage induced by ROS (derived from H2O2 exposure) and, additionally, significantly protects from the respective cytotoxic activity. Moreover, KP772 has a strong iron-chelating activity as also known for its ligand 1,10-phen [9]. H2O2-mediated cytotoxicity is at least partly executed by hydroxyl radicals derived from the Fe(II)-involving Fenton reaction [41]. Consequently, our data suggest that at least part of the antioxidant activity of KP772 is based on its potent Fe(II) chelating properties. Whether KP772 additionally exerts iron-independent radical scavenging activities is unclear and matter of ongoing investigations.
However, with respect to RR our data suggest that KP772 targets the R2 subunit primarily by iron chelation thus preventing the regeneration of the iron center necessary for stabilisation of the essential tyrosine radical [42]. Most importantly, the iron complex Fe-KP772 did not disrupt the R2 tyrosyl radical. Additionally, the reaction of KP772 with the Fe(II) centre of the R2 subunit led to the formation of this iron complex. Notably, these inhibitory effects of KP772 on the R2 tyrosyl radical were observed only under reductive assay conditions. This is in accordance with earlier reports demonstrating that the structurally-related bathophenanthroline had only negligible effects on mouse R2 under aerobic conditions [10] and might be explained by the stronger affinity of 1,10-phen (the KP772 ligand) for Fe(II). These observations place KP772 into the group of iron-chelating RR inhibitors [5] comparable to DFO and Triapine. However, several differences exist in comparison to these iron chelators. In case of the thiosemicarbazone compound Triapine, it has been shown that presence of iron is required for effective RR inhibition and, additionally, that complexation with Fe(II) strongly enhances activity. In sharp contrast, iron preloading protected against KP772-mediated cytotoxicity and the respective Fe(II) complex did not disrupt the R2 tyrosyl radical in EPR experiments. These characteristics of KP772 are shared with the clinically iron chelator DFO [32]. However, in that case several other differences exist. First, KP772 is readily taken up by cells [1] while DFO is characterised by limited membrane permeability [32]. Second, DFO exclusively chelates Fe(III) present mainly in the extracellular compartment, while KP772 formed the coloured Fe-KP772 complex only in presence of the (predominantly intracellular and tightly regulated) Fe(II). Third, the Fe(III) complex of DFO is widely inactive [32], while Fe-KP772 still has significant anticancer activity although lacking impact on RR. These data indicate that KP772 represents an RR inhibitor with a unique mode of action.
These differences also concern the emergence of therapy resistance. Acquired resistance of cancer cells against RR inhibitors including HU [16, 43-45] and gemcitabine [46] based on enhanced RR activity due to R1 and/or R2 subunit overexpression has been frequently reported. In contrast, a HU-resistant KB-3-1 subline overexpressing R2 [16] did not exhibit cross-resistance against KP772 (data not shown). Additionally, despite multiple attempts, selection of a KP772-resistant subline turned out to be impossible so far [2].
These data indicate that KP772 might in parallel to RR target other cellular mechanisms and molecules. Accordingly, in the NCI’s COMPARE analysis (Table 1) only the anticancer activity of HU (Pearson’s correlation coefficient CI 0.379) but of no other RR inhibitor and iron chelators correlated significantly with that of KP772 [1]. In contrast, moderate correlations (CI ≤ 0.380) to several anti-metabolic drugs including alanosine (CI 0.372), MTX (CI 0.333), 6-Thioguanine (CI 0.339), or cyclocytidine (CI 0.317) were observed. This suggests that KP772 may target cell proliferation also by mechanisms independent of RR inhibition. This is in accordance with several other observations as for example that cell cycle changes after KP772 treatment also differed from those induced by other RR inhibitors. In case of HU, cancer cells are arrested on the border from G1 to S phase accompanied by cyclin D1 destabilisation [47, 48]. In contrast, KP772 induced a distinct G0/G1 phase arrest already after an exposure time of around 12 h and predominantly characterised by loss of cyclin B1 [1]. Since R2 expression is reported to be lowest in G0/G1 phase [49], the observed reduction in R2 subunit expression after KP772 treatment might at least partly result from this early cell cycle arrest. This is in accordance with the observation that expression of the cell cycle-independent subunits R1 and p53R2 [50] is widely stable following KP772 treatment. As the p53R2/R1 complex is not only involved in DNA-damage repair but also crucial for the dNTP pool maintenance required for mitochondrial DNA synthesis [50], this indicates that the risk for unwanted side effects during KP772 treatment due to mitochondrial damage of healthy tissue might be low.
Additionally, the stop of DNA synthesis in G0/G1 phase due to KP772 treatment might explains why - in contrast to the reported synergism of RR inhibitors like HU [23, 24], Triapine [25], resveratrol [26] with antimetabolic drugs - no synergistic activity of KP772 with (deoxy)nucleoside analogues AraC or 5-FU was found. Even sequential administration of AraC and KP772 in diverse settings did not reveal any synergistic effect of AraC with KP772 (data not shown).
Furthermore, several reports indicate that HU treatment activates the ATM/ATR pathway similar to DNA-damaging drugs [51]. In contrast, no phosphorylation of the ATM/ATR target proteins CHK1 or CHK2 has been observed in KB-3-1 cells after KP772 treatment (data not shown). Accordingly, ATM kinase inhibition by caffeine cotreatment had only a minor sensitising effect towards the lanthanum drug (data not shown).
Which molecular mechanisms might be responsible for the RR-independent activities of KP772 is speculative so far. On the one hand, KP772 might target besides RR also other iron-dependent proteins such as the mitochondrial respiratory chain or interact with other Fe-S-cluster-containing proteins.
On the other hand, 1,10-phen has affinity not only for Fe(II) but also for other divalent ions including Cu(II) [52, 53] or Zn(II) [54]. In fact there are several reports on the anticancer activity of cupreous 1,10-phen complexes mainly caused by ROS-induced DNA damage [52, 53]. However, we found no indications that either oxidative stress or DNA damage is induced by KP772 [1, 2] arguing against significant formation of Cu(II)-KP772 complexes. In contrast, no generation of ROS is reported for 1,10-phen complexes with Zn(II) [55]. Additionally, the activity of Zn(II)-phenanthroline complexes seem to be rather low [55], arguing against Zn-KP772 as active species exerting the observed anticancer effects. Whether disruption of Zn(II)-containing proteins by chelation is underlying at least some effects of KP772 anticancer activity is unknown and matter of ongoing studies.
Taken together, our data demonstrate that KP772 is a novel RR inhibitor disrupting the essential tyrosyl radical of R2 by iron chelation. In addition, this lanthanum compound is believed to exert other cytostatic/toxic effects, yet to define, probably synergising with RR inhibition. These multifaceted characteristics might explain the broad anticancer activity, the lack of acquired resistance and the high susceptibility of drug-resistant tumor cells against KP772 [2]. In conclusion our data suggest that KP772 should be considered for further clinical development especially against therapy refractory tumors.
ACKNOWLEDGMENTS
We are indebted to Vera Bachinger for the skilful handling of cell culture, Elisabeth Rabensteiner, Rosa-Maria Weiss, and Christian Balcarek for competent technical assistance, Paul Breit for preparing photomicrographs and Irene Herbacek for FACS analysis. Many thanks to Christian Kowol for inspiring discussions.
This work was supported by the Austrian Science Fond (FWF) grant L212-B11 (to WB) and the Medizinischer-Wissenschaftlicher Fonds des Bürgermeisters der Bundeshauptstadt Wien, grant 2460 (to MM).
ABBREVIATIONS
- 1,10-phen
1,10-phenanthroline
- 5-FU
5-fluorouracil
- AraC
cytarabine
- DFO
desferrioxamine
- dNTP
deoxyribonucleoside triphosphate
- DTT
dithiothreitol
- EPR
electron paramagnetic resonance
- FACS
fluorescence-activated cell sorting
- HPLC
high-performance liquid chromatography
- HU
hydroxyurea
- KP772
[Tris(1,10-phenanthroline)lanthanum(III)]trithiocyanate
- MTX
methotrexate
- NCI
National Cancer Institute
- p53R2
p53-inducible RR subunit R2
- PI
propidium iodide
- ROS
reactive oxygen species
- RR
ribonucleotide reductase
- TfR
transferrin receptor
REFERENCES
- [1].Heffeter P, Jakupec MA, Korner W, Wild S, von Keyserlingk NG, Elbling L, Zorbas H, Korynevska A, Knasmuller S, Sutterluty H, Micksche M, Keppler BK, Berger W. Anticancer activity of the lanthanum compound [tris(1,10-phenanthroline)lanthanum(III)]trithiocyanate (KP772; FFC24) Biochem. Pharmacol. 2006;71:426–440. doi: 10.1016/j.bcp.2005.11.009. [DOI] [PubMed] [Google Scholar]
- [2].Heffeter P, Jakupec MA, Korner W, Chiba P, Pirker C, Dornetshuber R, Elbling L, Sutterluty H, Micksche M, Keppler BK, Berger W. Multidrug-resistant cancer cells are preferential targets of the new antineoplastic lanthanum compound KP772 (FFC24) Biochem. Pharmacol. 2007;73:1873–1886. doi: 10.1016/j.bcp.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Tsimberidou AM, Alvarado Y, Giles FJ. Evolving role of ribonucleoside reductase inhibitors in hematologic malignancies. Expert Rev. Anticancer Ther. 2002;2:437–448. doi: 10.1586/14737140.2.4.437. [DOI] [PubMed] [Google Scholar]
- [4].Hatse S, De Clercq E, Balzarini J. Role of antimetabolites of purine and pyrimidine nucleotide metabolism in tumor cell differentiation. Biochem. Pharmacol. 1999;58:539–555. doi: 10.1016/s0006-2952(99)00035-0. [DOI] [PubMed] [Google Scholar]
- [5].Shao J, Zhou B, Chu B, Yen Y. Ribonucleotide reductase inhibitors and future drug design. Curr Cancer Drug Targets. 2006;6:409–431. doi: 10.2174/156800906777723949. [DOI] [PubMed] [Google Scholar]
- [6].Kolberg M, Strand KR, Graff P, Andersson KK. Structure, function, and mechanism of ribonucleotide reductases. Biochim. Biophys. Acta. 2004;1699:1–34. doi: 10.1016/j.bbapap.2004.02.007. [DOI] [PubMed] [Google Scholar]
- [7].Bourdon A, Minai L, Serre V, Jais JP, Sarzi E, Aubert S, Chretien D, de Lonlay P, Paquis-Flucklinger V, Arakawa H, Nakamura Y, Munnich A, Rotig A. Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat. Genet. 2007;39:776–780. doi: 10.1038/ng2040. [DOI] [PubMed] [Google Scholar]
- [8].Green DA, Antholine WE, Wong SJ, Richardson DR, Chitambar CR. Inhibition of malignant cell growth by 311, a novel iron chelator of the pyridoxal isonicotinoyl hydrazone class: effect on the R2 subunit of ribonucleotide reductase. Clin. Cancer Res. 2001;7:3574–3579. [PubMed] [Google Scholar]
- [9].Sun Y, Bian J, Wang Y, Jacobs C. Activation of p53 transcriptional activity by 1,10-phenanthroline, a metal chelator and redox sensitive compound. Oncogene. 1997;14:385–393. doi: 10.1038/sj.onc.1200834. [DOI] [PubMed] [Google Scholar]
- [10].Nyholm S, Thelander L, Graslund A. Reduction and loss of the iron center in the reaction of the small subunit of mouse ribonucleotide reductase with hydroxyurea. Biochemistry. 1993;32:11569–11574. doi: 10.1021/bi00094a013. [DOI] [PubMed] [Google Scholar]
- [11].Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv. Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- [12].Szekeres T, Gharehbaghi K, Fritzer M, Woody M, Srivastava A, van’t Riet B, Jayaram HN, Elford HL. Biochemical and antitumor activity of trimidox, a new inhibitor of ribonucleotide reductase. Cancer Chemother. Pharmacol. 1994;34:63–66. doi: 10.1007/BF00686113. [DOI] [PubMed] [Google Scholar]
- [13].Garrett C, Santi DV. A rapid and sensitive high pressure liquid chromatography assay for deoxyribonucleoside triphosphates in cell extracts. Anal. Biochem. 1979;99:268–273. doi: 10.1016/s0003-2697(79)80005-6. [DOI] [PubMed] [Google Scholar]
- [14].Heffeter P, Pongratz M, Steiner E, Chiba P, Jakupec MA, Elbling L, Marian B, Korner W, Sevelda F, Micksche M, Keppler BK, Berger W. Intrinsic and acquired forms of resistance against the anticancer ruthenium compound KP1019 [indazolium trans-[tetrachlorobis(1H-indazole)ruthenate (III)](FFC14A) J. Pharmacol. Exp. Ther. 2005;312:281–289. doi: 10.1124/jpet.104.073395. [DOI] [PubMed] [Google Scholar]
- [15].Mann GJ, Graslund A, Ochiai E, Ingemarson R, Thelander L. Purification and characterization of recombinant mouse and herpes simplex virus ribonucleotide reductase R2 subunit. Biochemistry. 1991;30:1939–1947. doi: 10.1021/bi00221a030. [DOI] [PubMed] [Google Scholar]
- [16].Yen Y, Grill SP, Dutschman GE, Chang CN, Zhou BS, Cheng YC. Characterization of a hydroxyurea-resistant human KB cell line with supersensitivity to 6-thioguanine. Cancer Res. 1994;54:3686–3691. [PubMed] [Google Scholar]
- [17].Ask A, Persson L, Rehnholm A, Frostesjo L, Holm I, Heby O. Development of resistance to hydroxyurea during treatment of human myelogenous leukemia K562 cells with alpha-difluoromethylornithine as a result of coamplification of genes for ornithine decarboxylase and ribonucleotide reductase R2 subunit. Cancer Res. 1993;53:5262–5268. [PubMed] [Google Scholar]
- [18].Flanagan SA, Robinson BW, Krokosky CM, Shewach DS. Mismatched nucleotides as the lesions responsible for radiosensitization with gemcitabine: a new paradigm for antimetabolite radiosensitizers. Mol. Cancer Ther. 2007;6:1858–1868. doi: 10.1158/1535-7163.MCT-07-0068. [DOI] [PubMed] [Google Scholar]
- [19].Lembo D, Donalisio M, Cornaglia M, Azzimonti B, Demurtas A, Landolfo S. Effect of high-risk human papillomavirus oncoproteins on p53R2 gene expression after DNA damage. Virus Res. 2006;122:189–193. doi: 10.1016/j.virusres.2006.06.011. [DOI] [PubMed] [Google Scholar]
- [20].Smardova J, Pavlova S, Svitakova M, Grochova D, Ravcukova B. Analysis of p53 status in human cell lines using a functional assay in yeast: detection of new non-sense p53 mutation in codon 124. Oncol. Rep. 2005;14:901–907. [PubMed] [Google Scholar]
- [21].Zhu C, Johansson M, Karlsson A. The subcellular location of nucleoside analog phosphorylation is a determinant of synergistic effects of hydroxyurea. Biochem. Biophys. Res. Commun. 2000;276:179–182. doi: 10.1006/bbrc.2000.3441. [DOI] [PubMed] [Google Scholar]
- [22].Myette MS, Elford HL, Chitambar CR. Interaction of gallium nitrate with other inhibitors of ribonucleotide reductase: effects on the proliferation of human leukemic cells. Cancer Lett. 1998;129:199–204. doi: 10.1016/s0304-3835(98)00104-9. [DOI] [PubMed] [Google Scholar]
- [23].Tanaka M, Kimura K, Yoshida S. Mechanism of synergistic cell killing by hydroxyurea and cytosine arabinoside. Jpn. J. Cancer Res. 1985;76:729–735. [PubMed] [Google Scholar]
- [24].Walsh CT, Craig RW, Agarwal RP. Increased activation of 1-beta-D-arabinofuranosylcytosine by hydroxyurea in L1210 cells. Cancer Res. 1980;40:3286–3292. [PubMed] [Google Scholar]
- [25].Karp JE, Giles FJ, Gojo I, Morris L, Greer J, Johnson B, Thein M, Sznol M, Low J. A phase I study of the novel ribonucleotide reductase inhibitor 3-aminopyridine-2-carboxaldehyde thiosemicarbazone (3-AP, Triapine) in combination with the nucleoside analog fludarabine for patients with refractory acute leukemias and aggressive myeloproliferative disorders. Leuk. Res. 2008;32:71–77. doi: 10.1016/j.leukres.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Horvath Z, Saiko P, Illmer C, Madlener S, Hoechtl T, Bauer W, Erker T, Jaeger W, Fritzer-Szekeres M, Szekeres T. Synergistic action of resveratrol, an ingredient of wine, with Ara-C and tiazofurin in HL-60 human promyelocytic leukemia cells. Exp. Hematol. 2005;33:329–335. doi: 10.1016/j.exphem.2004.11.009. [DOI] [PubMed] [Google Scholar]
- [27].Kayyali R, Pannala AS, Khodr H, Hider RC. Comparative radical scavenging ability of bidentate iron (III) chelators. Biochem. Pharmacol. 1998;55:1327–1332. doi: 10.1016/s0006-2952(97)00602-3. [DOI] [PubMed] [Google Scholar]
- [28].Kanno S, Kakuta M, Kitajima Y, Osanai Y, Kurauchi K, Ohtake T, Ujibe M, Uwai K, Takeshita M, Ishikawa M. Preventive effect of trimidox on oxidative stress in U937 cell line. Biol. Pharm. Bull. 2007;30:994–998. doi: 10.1248/bpb.30.994. [DOI] [PubMed] [Google Scholar]
- [29].Aisen P, Enns C, Wessling-Resnick M. Chemistry and biology of eukaryotic iron metabolism. Int. J. Biochem. Cell Biol. 2001;33:940–959. doi: 10.1016/s1357-2725(01)00063-2. [DOI] [PubMed] [Google Scholar]
- [30].Chua AC, Graham RM, Trinder D, Olynyk JK. The regulation of cellular iron metabolism. Crit. Rev. Clin. Lab. Sci. 2007;44:413–459. doi: 10.1080/10408360701428257. [DOI] [PubMed] [Google Scholar]
- [31].Hoyer G. Absorption-photometric determination of serum iron with 1,10-phenanthroline. Scand. J. Clin. Lab. Invest. 1950;2:125–132. doi: 10.3109/00365515009051847. [DOI] [PubMed] [Google Scholar]
- [32].Chaston TB, Lovejoy DB, Watts RN, Richardson DR. Examination of the antiproliferative activity of iron chelators: multiple cellular targets and the different mechanism of action of triapine compared with desferrioxamine and the potent pyridoxal isonicotinoyl hydrazone analogue 311. Clin. Cancer Res. 2003;9:402–414. [PubMed] [Google Scholar]
- [33].Cerqueira NM, Pereira S, Fernandes PA, Ramos MJ. Overview of ribonucleotide reductase inhibitors: an appealing target in anti-tumour therapy. Curr. Med. Chem. 2005;12:1283–1294. doi: 10.2174/0929867054020981. [DOI] [PubMed] [Google Scholar]
- [34].Xie KC, Plunkett W. Deoxynucleotide pool depletion and sustained inhibition of ribonucleotide reductase and DNA synthesis after treatment of human lymphoblastoid cells with 2-chloro-9-(2-deoxy-2-fluoro-beta-D-arabinofuranosyl) adenine. Cancer Res. 1996;56:3030–3037. [PubMed] [Google Scholar]
- [35].Heinemann V, Xu YZ, Chubb S, Sen A, Hertel LW, Grindey GB, Plunkett W. Inhibition of ribonucleotide reduction in CCRF-CEM cells by 2′,2′-difluorodeoxycytidine. Mol. Pharmacol. 1990;38:567–572. [PubMed] [Google Scholar]
- [36].Fox RM. Changes in deoxynucleoside triphosphate pools induced by inhibitors and modulators of ribonucleotide reductase. Pharmacol. Ther. 1985;30:31–42. doi: 10.1016/0163-7258(85)90046-4. [DOI] [PubMed] [Google Scholar]
- [37].Roy B, Guittet O, Beuneu C, Lemaire G, Lepoivre M. Depletion of deoxyribonucleoside triphosphate pools in tumor cells by nitric oxide. Free Radic. Biol. Med. 2004;36:507–516. doi: 10.1016/j.freeradbiomed.2003.11.028. [DOI] [PubMed] [Google Scholar]
- [38].Gandhi V, Plunkett W, Kantarjian H, Talpaz M, Robertson LE, O’Brien S. Cellular pharmacodynamics and plasma pharmacokinetics of parenterally infused hydroxyurea during a phase I clinical trial in chronic myelogenous leukemia. J. Clin. Oncol. 1998;16:2321–2331. doi: 10.1200/JCO.1998.16.7.2321. [DOI] [PubMed] [Google Scholar]
- [39].Horvath Z, Murias M, Saiko P, Erker T, Handler N, Madlener S, Jaeger W, Grusch M, Fritzer-Szekeres M, Krupitza G, Szekeres T. Cytotoxic and biochemical effects of 3,3′,4,4′,5,5′-hexahydroxystilbene, a novel resveratrol analog in HL-60 human promyelocytic leukemia cells. Exp. Hematol. 2006;34:1377–1384. doi: 10.1016/j.exphem.2006.05.018. [DOI] [PubMed] [Google Scholar]
- [40].Giles FJ, Fracasso PM, Kantarjian HM, Cortes JE, Brown RA, Verstovsek S, Alvarado Y, Thomas DA, Faderl S, Garcia-Manero G, Wright LP, Samson T, Cahill A, Lambert P, Plunkett W, Sznol M, DiPersio JF, Gandhi V. Phase I and pharmacodynamic study of Triapine, a novel ribonucleotide reductase inhibitor, in patients with advanced leukemia. Leuk. Res. 2003;27:1077–1083. doi: 10.1016/s0145-2126(03)00118-8. [DOI] [PubMed] [Google Scholar]
- [41].Yu TW, Anderson D. Reactive oxygen species-induced DNA damage and its modification: a chemical investigation. Mutat. Res. 1997;379:201–210. doi: 10.1016/s0027-5107(97)00141-3. [DOI] [PubMed] [Google Scholar]
- [42].Licht S, Gerfen GJ, Stubbe J. Thiyl radicals in ribonucleotide reductases. Science. 1996;271:477–481. doi: 10.1126/science.271.5248.477. [DOI] [PubMed] [Google Scholar]
- [43].Jiang R, Zhang JL, Satoh Y, Sairenji T. Mechanism for induction of hydroxyurea resistance and loss of latent EBV genome in hydroxyurea-treated Burkitt’s lymphoma cell line Raji. J. Med. Virol. 2004;73:589–595. doi: 10.1002/jmv.20130. [DOI] [PubMed] [Google Scholar]
- [44].Wong SJ, Myette MS, Wereley JP, Chitambar CR. Increased sensitivity of hydroxyurea-resistant leukemic cells to gemcitabine. Clin. Cancer Res. 1999;5:439–443. [PubMed] [Google Scholar]
- [45].Carter GL, Cory JG. Cross-resistance patterns in hydroxyurea-resistant leukemia L1210 cells. Cancer Res. 1988;48:5796–5799. [PubMed] [Google Scholar]
- [46].Bergman AM, Eijk PP, Ruiz van Haperen VW, Smid K, Veerman G, Hubeek I, van den Ijssel P, Ylstra B, Peters GJ. In vivo induction of resistance to gemcitabine results in increased expression of ribonucleotide reductase subunit M1 as the major determinant. Cancer Res. 2005;65:9510–9516. doi: 10.1158/0008-5472.CAN-05-0989. [DOI] [PubMed] [Google Scholar]
- [47].Kramer OH, Knauer SK, Zimmermann D, Stauber RH, Heinzel T. Histone deacetylase inhibitors and hydroxyurea modulate the cell cycle and cooperatively induce apoptosis. Oncogene. 2008;27:732–740. doi: 10.1038/sj.onc.1210677. [DOI] [PubMed] [Google Scholar]
- [48].Mukherji A, Janbandhu VC, Kumar V. GSK-3beta-dependent destabilization of cyclin D1 mediates replicational stress-induced arrest of cell cycle. FEBS Lett. 2008;582:1111–1116. doi: 10.1016/j.febslet.2008.02.068. [DOI] [PubMed] [Google Scholar]
- [49].Hakansson P, Hofer A, Thelander L. Regulation of mammalian ribonucleotide reduction and dNTP pools after DNA damage and in resting cells. J. Biol. Chem. 2006;281:7834–7841. doi: 10.1074/jbc.M512894200. [DOI] [PubMed] [Google Scholar]
- [50].Pontarin G, Fijolek A, Pizzo P, Ferraro P, Rampazzo C, Pozzan T, Thelander L, Reichard PA, Bianchi V. Ribonucleotide reduction is a cytosolic process in mammalian cells independently of DNA damage. Proc. Natl. Acad. Sci. USA. 2008;105:17801–17806. doi: 10.1073/pnas.0808198105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Tanaka T, Huang X, Halicka HD, Zhao H, Traganos F, Albino AP, Dai W, Darzynkiewicz Z. Cytometry of ATM activation and histone H2AX phosphorylation to estimate extent of DNA damage induced by exogenous agents. Cytometry A. 2007;71:648–661. doi: 10.1002/cyto.a.20426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Yang J, Wong RN, Yang MS. Protective mechanism of metallothionein against copper-1, 10-phenanthroline induced DNA cleavage. Chem. Biol. Interact. 2000;125:221–232. doi: 10.1016/s0009-2797(00)00148-4. [DOI] [PubMed] [Google Scholar]
- [53].Cai X, Pan N, Zou G. Copper-1,10-phenanthroline-induced apoptosis in liver carcinoma Bel-7402 cells associates with copper overload, reactive oxygen species production, glutathione depletion and oxidative DNA damage. Biometals. 2007;20:1–11. doi: 10.1007/s10534-006-9008-0. [DOI] [PubMed] [Google Scholar]
- [54].Shomron N, Malca H, Vig I, Ast G. Reversible inhibition of the second step of splicing suggests a possible role of zinc in the second step of splicing. Nucleic Acids Res. 2002;30:4127–4137. doi: 10.1093/nar/gkf553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].McCann M, Geraghty M, Devereux M, O’Shea D, Mason J, O’Sullivan L. Insights into the mode of action of the anti-Candida activity of 1,10-phenanthroline and its metal chelates. Met. Based Drugs. 2000;7:185–193. doi: 10.1155/MBD.2000.185. [DOI] [PMC free article] [PubMed] [Google Scholar]