Since the discovery of the DNA double helix, the notion that the two strands might be pulled apart like the two sides of a zipper has fired the scientific imagination (1–3). Now, Essevaz-Roulet et al. (4) have achieved precisely this feat by using the recently developed tool of single DNA micromanipulation, and they even have been able to measure the differing stickiness of GC-rich and AT-rich sequences. Unzipping occurs for applied forces of roughly 10 piconewtons (pN), a very reasonable scale of force given that the resulting work done per base pair unzipped is about 1.5 kcal/mol (note 1 kcal/mol = 6.6⋅10−21 J, and 1 pN⋅1 nm = 10−21 J), comparable to the base-pairing free energy (5).
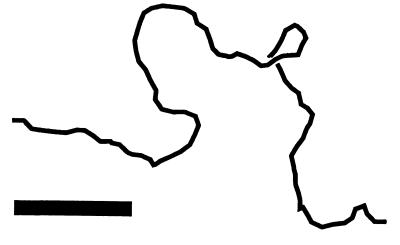
This is the latest in a remarkable series of physical manipulation experiments on DNA (6) carried out over the last five years in several laboratories, starting with force vs. extension measurements for single DNAs by Smith, Finzi, and Bustamante (7). That pioneering experiment showed that the random thermal flopping about of a 100-kb double helix led to an “entropic elasticity” consistent over a thousand-fold range of force with that of the “worm-like chain” model familiar to polymer scientists (8–10). Smith et al. found that it took about 0.1 pN to pull the ends of a DNA apart a distance of half its contour length. This 0.1 pN force scale comes from the energy associated with a thermally excited degree of freedom (at room temperature, 0.6 kcal/mol = 4⋅10−21 J, a.k.a. kBT) divided by the DNA persistence length (50 nm), the contour length that a single appreciable bend occurs over. The sort of shape fluctuations that contribute to this entropic elasticity are shown in Fig. 1, a conformation of a 3-kb DNA obtained from a computer simulation. In the absence of thermal fluctuations, the simulated chain would be perfectly straight; for forces below 0.1 pN thermal fluctuations crumple the molecule. Higher forces reduce fluctuations, straightening out the molecule. A 5-pN force, roughly that generated by single kinesin or myosin motors, stretches a DNA out to nearly its full contour length.
Figure 1.
Sample conformation of a 6-kb dsDNA being stretched by a 0.1-pN force in room temperature aqueous solution (courtesy of A. V. Vologodskii, New York University). The molecule about half stretched. Higher forces of a few pN will fully extend the chain. Bar indicates 100 nm, double the DNA persistence length.
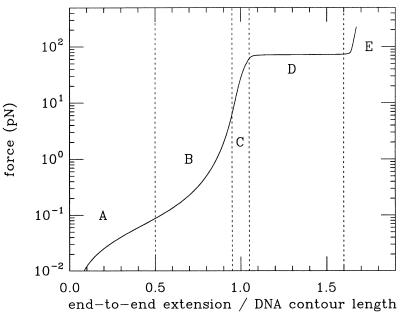
The experiments of Smith et al. (7) were also important because they introduced an elegant technique for manipulating single DNAs. The specificity of base pairing was used to anchor one end of a DNA to a glass slide, and the other end to a micron-sized magnetic bead, to which forces in the range 0.1 to 10 pN could be applied with a centimeter-sized bar magnet (forces also were applied in those original experiments using flow). This basic technique has been the basis of subsequent micromanipulation experiments. For example, this basic method was used by two labs to study “overstretched” DNA, a double-helix structure 1.7 times the B-form length (11–13). A remarkably sharp transition to this new state was observed at a force of roughly 70 pN (see force “plateau” in Fig. 2). The plateau indicates that B-DNA and overstretched DNA are distinct conformations, or phases of the double helix, with the force vs. distance plateau analogous to the constant pressure encountered during a liquid-gas phase transition.
Figure 2.
Sketch of extension (end-to-end distance as a fraction of B-form contour length) vs. force for dsDNA. A, for forces <0.1 pN, the thermal random-walk fluctuations of Fig. 1 give rise to a weak entropic elasticity. B, larger forces (0.1 pN < 10 pN) supress thermal bending and stretch the molecule to near B-form length. C, for forces between 10 and 60 pN simple linear elasticity of DNA is observed, as the double helix starts to be lengthened. D, at about 70 pN, a force plateau is observed, signaling conversion of B-form to overstretched DNA. E, finally, above 70 pN, DNA is entirely converted to its overstretched form.
The structure of overstretched DNA is as yet unknown; in particular the helix repeat (i.e., the amount of untwisting accompanying overstretching) begs to be measured. Molecular model calculations suggest that overstretched DNA still is base-paired, but has highly tilted bases to compromise between stretching and maintaining stacking (14, 15). Recalling that the double helix stretches to 1.5 times B-form length and untwists to have an 18-bp helix repeat instead of the B-form 10.5-bp repeat (16, 17) when coated with RecA suggests a connection between RecA–double-stranded DNA (dsDNA) and force-overstretched DNA (12, 13) (however, note that in RecA-dsDNA the bases are untilted and unstacked). Fig. 2 summarizes these force regimes, from the weak entropic elasticity of random thermal bends (<0.1 pN), to intrinsic elasticity associated with gradual deformation of the double helix itself (10–60 pN), and finally the sharp overstretching transition (70 pN).
Another front was opened by Strick et al. (18), who developed a technique to simultaneously twist and pull DNAs (multiple attachments to one strand were used to make nonswiveling anchors) to study the elasticity of supercoiled DNA. In addition to the expected gradual stiffening of the entropic elasticity with twisting, again force plateaus appeared for roughly 4% underwinding, but at forces of only about 4 pN. So, underwinding DNA by a few percent (roughly the underwinding generated by the balance of DNA gyrase and topoisomerase I on bacterial plasmids and chromosome domains) drops the force needed to disturb double-helix secondary structure down from 70 pN to a few pN. There is no paradox because the total work done during twisting and pulling is, in fact, comparable with ΔG for DNA melting (19).
The precise nature of the DNA state implied by the force plateaus of Strick et al. (18) has not been determined, but it is plausibly melted (strand-separated) because single-stranded regions appear in supercoiled plasmids beyond roughly 7% underwinding (20). The full phase diagram of DNA states as a function of force and twisting awaits exploration: for intermediate forces around 10 pN there might be first overstretching, followed by strand separation as the DNA is progressively underwound.
To these two approaches to physically changing DNA secondary structure—by (over)stretching and by twisting—Essevaz-Roulet et al. (4) have pioneered an important third approach, to literally pull the two strands apart. As this experiment localized a replication-like fork, strand separation occurred progressively down their 50-kb double helix, allowing observation that the GC-rich regions required 15 pN to separate whereas AT-rich regions required only 10 pN. A sequence map thus was obtained from force measurements. Although sequencing by unzipping does not appear possible with the current experiment (the resolution was a few hundred bp, and thermal fluctuations have been argued to set a fundamental resolution limit; ref. 3), mapping by unzipping may be useful when combined with conventional sequencing techniques (4). But another important use of this technique will be precise study of how sequence variation affects DNA structural changes, especially helix opening during DNA replication.
These studies have made the study of single-molecule polymer elasticity an experimental reality. Considered as a polymer, DNA offers a combination of well characterized stretching elasticity, twisting elasticity (a novel feature in the normally single-bond-backboned polymer world), and of course through molecular-biological techniques a structural control unmatched elsewhere in polymer science (where polymerization polydispersities of 1% are difficult to achieve). Already DNA micromanipulation has led to precise physical studies of single-polymer dynamics (21, 22). But even more exciting science lies in a different direction.
Essevaz-Roulet et al. (4) note that their setup gives them insight into the forces applied to DNA by helicases, which act at replication forks to enzymatically separate the strands of the double helix. Why not use their experiment to monitor the actual action of helicases? This is in the spirit of two recent experiments aimed at revealing the structural-energetic activity of DNA-acting factors: a study of DNA looping by lac repressor by Finzi and Gelles (23), and remarkable measurements by Yin et al. (24) of the force generated by RNA polymerase during transcription.
The lac-repressor experiment amounted to direct observation of the thermal hopscotch of DNA loop binding and unbinding. The 1,200-bp substrate provided the tool to do this. Loop formation displaced an attached micron-sized bead a distance large enough to be observed via light microscopy. Force might be included in this kind of experiment to modulate loop binding, thus providing a new probe of local structure and thermodynamics of protein–DNA interactions. It would be instructive to study chromatin fiber by this kind of approach. Given the known binding affinity of histones for DNA, one expects liberation of the 146-bp loop for forces of about 2 pN.
The RNA polymerase study of Yin et al. (24), in addition to giving us movies of the processive action of the enzyme, revealed that RNA polymerase is a champion among molecular motors. It generated up to 20 pN, easily beating out myosin (5 pN) and kinesin (6 pN). RNA polymerase’s fortitude comes from its low gear. Its base-pair step length of 0.3 nm is an order of magnitude less than those other motors, whereas the energy used per step (from NTP hydrolysis) is about the same as for myosin and kinesin. Because force is energy divided by length, the small step length for RNA polymerase translates to a large force. The brutishness of RNA polymerase might well be what is needed to not only denature DNA, but to ensure transcription through chromosomal tangles and anchors, and may lead it to be a determinant of chromosome architecture (25).
The study of helicases on the Y-shaped templates is one of a number of interesting enzymological micromanipulation studies that now can be considered. It will not be long before DNA replication, recombination, topology change, transcription regulation, and chromatin assembly are studied using micromanipulation. Similar methods can be applied to other large biomolecules as evidenced by recent experiments studying denaturation of the protein titin by applied force (26–28); one can imagine using the technique of Essevaz-Roulet et al. (4) to study helix–loop structure of large RNAs (3). Revealing a unique combination of structural and energetic information, this molecular chiropracty is a sharp tool for ultrastructural study of the biochemical transactions of life.
Acknowledgments
I thank J.-F. Allemand, D. Bensimon, C. Bustamante, D. Chatenay, P. Cluzel, V. Croquette, A. Libchaber, T. Perkins, G. Shivashankar, E. D. Siggia, D. Smith, S. Smith, T. Strick, J.-L. Viovy, and A. V. Vologodskii for many enlightening discussions. Acknowledgement is made to the Meyer Foundation and to the donors of The Petroleum Research Fund, administered by the American Chemical Society, for their support.
References
- 1.Levinthal C, Crane H R. Proc Natl Acad Sci USA. 1956;42:436–438. doi: 10.1073/pnas.42.7.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Viovy J-L, Heller C, Caron F, Cluzel P, Chatenay D. C R Acad Sci. 1994;317:795–800. [PubMed] [Google Scholar]
- 3.Thompson R E, Siggia E D. Europhys Lett. 1995;31:335–340. [Google Scholar]
- 4.Essevaz-Roulet B, Bockelmann U, Heslot F. Proc Natl Acad Sci USA. 1997;94:11935–11940. doi: 10.1073/pnas.94.22.11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breslauer K J, Frank R, Blocker H, Marky L A. Proc Natl Acad Sci USA. 1986;83:3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bensimon D. Structure. 1996;4:885–889. doi: 10.1016/s0969-2126(96)00095-0. [DOI] [PubMed] [Google Scholar]
- 7.Smith S B, Finzi L, Bustamante C. Science. 1992;258:1122–1126. doi: 10.1126/science.1439819. [DOI] [PubMed] [Google Scholar]
- 8.Bustamante C, Marko J F, Smith S B, Siggia E D. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 9.Vologodskii A. Macromolecules. 1994;27:5623–5625. [Google Scholar]
- 10.Marko J F, Siggia E D. Macromolecules. 1995;28:8759–8770. [Google Scholar]
- 11.Smith S B, Cui Y, Hausrath A C, Bustamante C. Biophys J. 1995;68:A250. [Google Scholar]
- 12.Cluzel P, Lebrun A, Heller C, Lavery R, Viovy J-L, Chatenay D, Caron F. Science. 1996;271:792–794. doi: 10.1126/science.271.5250.792. [DOI] [PubMed] [Google Scholar]
- 13.Smith S B, Cui Y, Bustamante C. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 14.Lebrun A, Lavery R. Nucleic Acids Res. 1996;24:2260–2267. doi: 10.1093/nar/24.12.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Konrad M W, Bolonick J I. J Am Chem Soc. 1996;118:10989–10994. [Google Scholar]
- 16.Dunn K, Chrysogelos S, Griffith J. Cell. 1982;28:757–765. doi: 10.1016/0092-8674(82)90055-1. [DOI] [PubMed] [Google Scholar]
- 17.Stasiak A, Di Capua E. Nature (London) 1982;299:185–186. doi: 10.1038/299185a0. [DOI] [PubMed] [Google Scholar]
- 18.Strick T R, Allemand J-F, Bensimon D, Bensimon A, Croquette V. Science. 1996;271:1835–1837. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 19.Marko J F, Siggia E D. Science. 1994;265:506–508. doi: 10.1126/science.8036491. [DOI] [PubMed] [Google Scholar]
- 20.Vologodskii A V, Lukashin A V, Anshelevich V V, Frank-Kamenetskii M D. Nucleic Acids Res. 1979;6:967–982. doi: 10.1093/nar/6.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perkins T T, Quake S R, Smith D E, Chu S. Science. 1994;264:822–826. doi: 10.1126/science.8171336. [DOI] [PubMed] [Google Scholar]
- 22.Perkins T T, Smith D E, Larson R G, Chu S. Science. 1995;268:83–87. doi: 10.1126/science.7701345. [DOI] [PubMed] [Google Scholar]
- 23.Finzi L, Gelles J. Science. 1995;267:378–380. doi: 10.1126/science.7824935. [DOI] [PubMed] [Google Scholar]
- 24.Yin H, Wang M D, Svoboda K, Landick R, Block S M, Gelles J. Science. 1995;270:1653–1657. doi: 10.1126/science.270.5242.1653. [DOI] [PubMed] [Google Scholar]
- 25.Cook P. BioEssays. 1994;16:425–430. doi: 10.1002/bies.950160611. [DOI] [PubMed] [Google Scholar]
- 26.Rief M, Gautel M, Oesterhelt F, Fernandez J F, Gaub H E. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 27.Kellermayer M S, Smith S B, Granzier H L, Bustamante C. Science. 1997;276:1112–1116. doi: 10.1126/science.276.5315.1112. [DOI] [PubMed] [Google Scholar]
- 28.Tskhovrebova L, Trinick J, Sleep J A, Simmons R M. Nature (London) 1997;387:308–312. doi: 10.1038/387308a0. [DOI] [PubMed] [Google Scholar]