Summary
Many viruses induce hepatitis in humans, highlighting the need to understand the underlying mechanisms of virus-induced liver pathology. The murine coronavirus, mouse hepatitis virus (MHV), causes acute hepatitis in its natural host and provides a useful model for understanding virus interaction with liver cells. The MHV accessory protein, ns2, antagonizes the type I interferon response and promotes hepatitis. We show that ns2 has 2′,5′-phosphodiesterase activity, which blocks the interferon inducible 2′,5′-oligoadenylate synthetase (OAS)-RNase L pathway to facilitate hepatitis development. Ns2 cleaves 2′,5′-oligoadenylate, the product of OAS, to prevent activation of the cellular endoribonuclease RNase L and consequently block viral RNA degradation. An ns2 mutant virus was unable to replicate in the liver or induce hepatitis in wild-type mice, but was highly pathogenic in RNase L deficient mice. Thus, RNase L is a critical cellular factor for protection against viral infection of the liver and the resulting hepatitis.
Graphical Abstract
Highlights
► Mouse hepatitis virus ns2 protein inhibits the IFN-induced OAS-RNase L pathway ► ns2 reduces the intracellular level of 2′,5′-oligoadenylate (2-5A) ► ns2 has a 2′,5′-phosphodiesterase activity to directly cleave 2-5A ► ns2 mutant virus recovers wild-type virulence in RNase L-deficient mice
Introduction
Many viral infections induce acute and/or chronic liver pathology in humans, for which there are often no known effective therapies or vaccines. There is little known about the early virus-host interactions that determine whether an infection is established, highlighting the need to understand mechanisms regulating the pathogenesis of viral hepatitis. The liver is constantly exposed to intestinal derived antigens present in the blood circulating through the liver sinusoids and, therefore, has evolved a unique immune environment that is relatively tolerant to foreign antigens and permissive to chronic infections (Crispe, 2009, Neumann-Haefelin et al., 2007). Thus, the innate immune response is a particularly important defense against pathogen invasion of the liver. Type I interferon (IFN) is the first line of defense available to restrict viral infection including infections with human hepatitis viruses (García-Sastre and Biron, 2006, Qu and Lemon, 2010). Kupffer cells (liver macrophages) found in the liver sinusoids are among the first cells to encounter viruses and as such, along with endothelial cells present a barrier that must be breached in order to access hepatocytes in the liver parenchyma (Crispe, 2009). Indeed, it has been reported that IFN signaling in macrophages is crucial for control of hepatitis induced by mouse hepatitis virus (Cervantes-Barragán et al., 2009) and lymphocytic choriomeningitis virus (Lang et al., 2010).
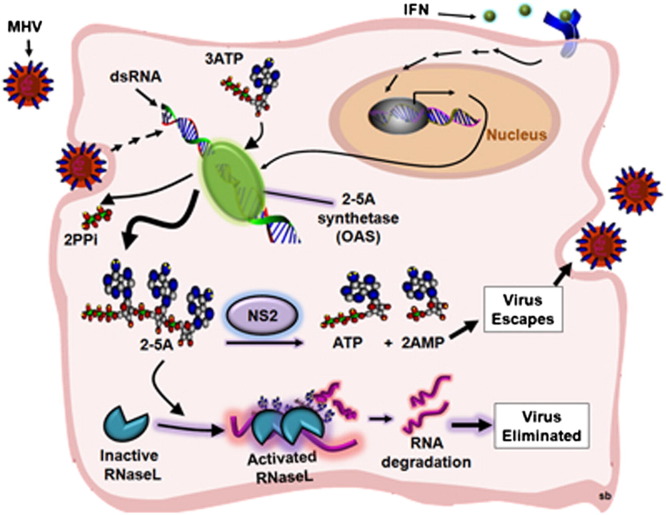
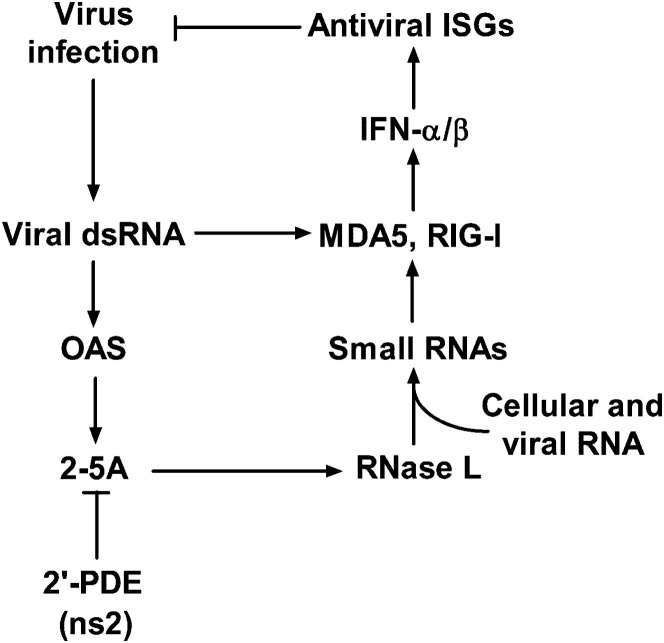
Type I IFN, primarily IFN-α/β, produced by virus-infected cells, induces expression of over four hundred interferon-stimulated genes (ISGs), whose products cooperate to induce an antiviral state. One potent antiviral IFN induced activity is the 2′,5′-oligoadenylate synthetase-ribonuclease L (OAS-RNase L) pathway (Sadler and Williams, 2008) (Figure 1 ). After infection, IFNs induce a group of OAS genes whose products are activated by viral double-stranded RNA (dsRNA). OAS uses ATP to generate the 2′,5′-linked oligoadenylates with the structures [px5′A(2′p5′A)n; x = 1–3; n ≥ 2 (2-5A)]. 2-5A binds to the ubiquitous cellular endoribonuclease RNase L, causing inactive RNase L monomers to form activated dimers (Dong and Silverman, 1995). RNase L cleaves single-stranded regions of both viral and cellular messenger RNA (mRNA), leading to inhibition of viral replication and protein synthesis (Silverman, 2007). In addition, detection of the newly generated short RNAs by cellular pattern recognition receptors, MDA5 and RIG-I, further enhances IFN production and the ensuing antiviral activities (Malathi et al., 2007). The concentration of 2-5A is believed to be the primary factor controlling RNase L activation. The cellular enzyme 2′-phosphodiesterase (2′-PDE, also known as PDE12) has been proposed to reduce or prevent 2-5A-mediated activation of RNase L by degrading 2-5A, presumably to prevent excessive tissue damage caused by the effects of RNase L (Kubota et al., 2004). The antiviral activity of the OAS-RNase L pathway has been demonstrated during infections of mice with numerous RNA and DNA viruses (Flodström-Tullberg et al., 2005, Li et al., 1998, Samuel et al., 2006, Scherbik et al., 2006, Washenberger et al., 2007, Xiang et al., 2002, Zhou et al., 1997).
Figure 1.
The Interferon-Induced OAS-RNase L Pathway
After infection, viral RNA is detected by pattern recognition receptors RIG-I and MDA5, resulting in the induction of IFN-α/β, which in turn induces ISGs, including OAS. OAS is activated by dsRNA to produce 2-5A, which activates RNase L. RNase L degrades cellular and viral RNA producing more RNA that is recognized by MDA5 and RIG-I, resulting in enhanced IFN induction. 2′-PDE cleaves 2-5A and inhibits the activation of RNase L. MHV ns2, like the cellular enzyme 2′-PDE, is a 2′,5′-phosphodiesterase. OAS, 2′-5′-oligoadenylate synthetase; 2-5A, 2′,5′ oligoadenylate; 2′-PDE, 2′-phosphodiesterase.
Hepatitis caused by the murine coronavirus, mouse hepatitis virus (MHV), is an acute disease induced in the natural host and provides a convenient and valuable model for understanding the interaction of virus with liver cells (Weiss and Leibowitz, 2011). Previous studies using reverse genetics and MHV strains with different tropisms demonstrated that events that occur after viral entry play a crucial role in the likelihood of liver invasion and the development of hepatitis (Navas and Weiss, 2003). Indeed, we recently reported that MHV accessory protein ns2 confers the ability to antagonize the type I IFN antiviral response in macrophages and that this is required for efficient viral replication in the liver and the development of hepatitis (Zhao et al., 2011). Based on sequence homology to cellular phosphodiesterases, ns2 had been predicted to be a cyclic phosphodiesterase (Mazumder et al., 2002, Snijder et al., 2003). We show here that ns2 has in fact a 2′,5′-phosphodiesterase activity that can downregulate intracellular levels of 2-5A, thereby preventing the activation and antiviral activity of RNase L. Our results underscore the importance of the OAS-RNase L pathway in vivo for protection of the host from developing viral hepatitis.
Results
MHV A59 Resists RNase L Activity
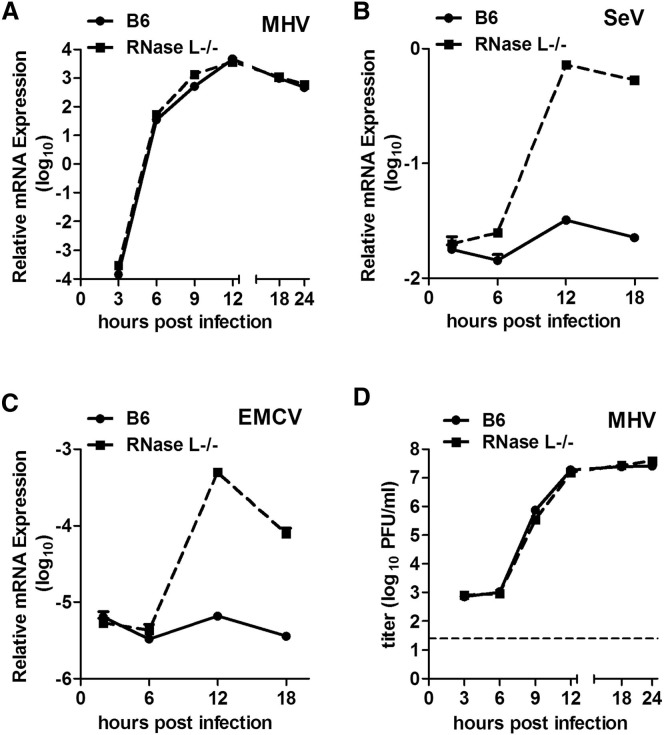
MHV differs from many other viruses in that it stimulates type I IFN production in only limited cell types, primarily in macrophages (Roth-Cross et al., 2008) and plasmacytoid dendritic cells (Cervantes-Barragan et al., 2007). Furthermore MHV displays cell type-specific sensitivity to the antiviral effects of IFN treatment, with macrophage lineage cells being most successful in restricting MHV replication (Rose and Weiss, 2009, Zhao et al., 2011). We also observed that MHV was unable to prevent IFN induction by a coinfecting virus and was also unable to protect a coinfecting virus from the effects of IFN (Roth-Cross et al., 2007). Thus, we reasoned that MHV is unable to subvert major IFN signaling pathways, but rather may interfere with one or more IFN-induced antiviral activities. Indeed, a previous study showed that MHV-A59 infection does not promote RNase L-mediated degradation of 18S and 28S ribosomal RNA (rRNA) in HeLa cells (Ye et al., 2007), suggesting that the OAS-RNase L pathway may be downregulated. To more directly evaluate the role of RNase L in MHV replication, we compared the replication of wild-type A59 in bone-marrow derived macrophages (BMMs) from wild-type C57BL/6 (B6) mice or from mice genetically ablated for RNase L expression [RNase L−/− (B6)]. The kinetics of accumulation of A59 genome RNA were indistinguishable in the two cell types (Figure 2A), as was the extent of infectious virus replication (Figure 2D). In contrast, the genome RNA levels of Sendai virus (SeV) and encephalomyocarditis virus (EMCV) were approximately 16-fold and 64-fold higher at 12 hr after infection in BMMs derived from RNase L−/− mice, compared with B6 (Figures 2B and 2C, respectively). These results indicate that RNase L activity restricts the replication of both SeV and EMCV but not that of MHV A59.
Figure 2.
MHV Replication Is Not Inhibited by RNase L Signaling
(A–C) BMMs from B6 or RNase L−/− mice were infected with MHV-A59, SeV, or EMCV (1 PFU/cell). At the indicated time points, RNA was isolated from cell lysates and viral mRNA expression quantified by qRT-PCR (n = 3). Relative viral mRNA expression levels were expressed as 2−ΔCT, relative to β-actin mRNA [ΔCT = CT(gene of interest) – CT(β-actin)].
(D) Cells infected with MHV-A59 were lysed by freeze-thawing and titers determined by plaque assay (n = 3).
See also Table S1.
Ns2 Specifically Inhibits the OAS-RNase L Pathway
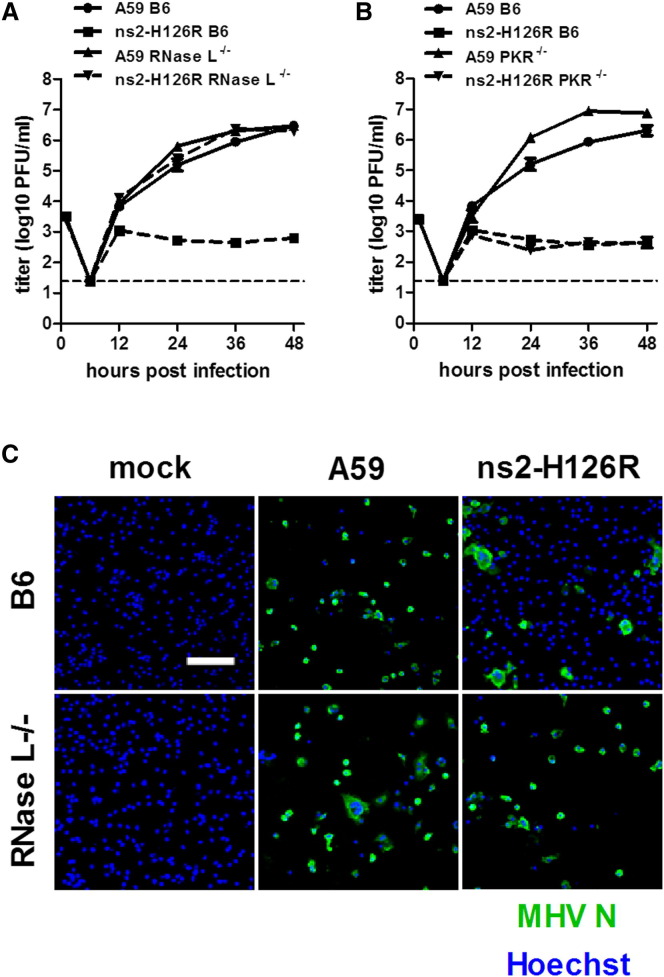
We showed previously that A59 ns2 antagonizes the IFN-signaling pathway in macrophage lineage cells (Zhao et al., 2011). Furthermore the A59 mutant virus, ns2-H126R, carrying a single amino acid substitution in a predicted phosphodiesterase catalytic His residue (Mazumder et al., 2002, Roth-Cross et al., 2009, Snijder et al., 2003), is attenuated for replication in macrophages (Zhao et al., 2011). Thus, we hypothesized that ns2 could protect the viral genome RNA from RNase L-mediated degradation through its phosphodiesterase activity, which might cleave 2-5A, thereby reducing the level of inducer of RNase L activity. To test this hypothesis, we first compared the replication kinetics of A59 and ns2-H126R in BMMs derived from B6 and RNase L−/− mice. Along with robust replication of A59 in B6 BMMs (Figure 3A), we observed extensive lysis of most of the cells by 48 hr postinfection (p.i.), while most of ns2-H126R-infected BMMs remained intact (Figure 3C). The few cells left intact after A59 infection expressed viral antigen, while there were small numbers of cells expressing viral antigen after ns2-H126R infection. In contrast, in RNase L−/− BMMs, ns2-H126R recovered the ability to replicate efficiently, with similar kinetics (Figure 3A) and induced similar wide spread cell lysis as A59 (Figure 3C). Similar results were obtained for two other mutants carrying single amino acid substitutions in ns2, H46A, and L94P (Figure S1 available online). These data suggest that ns2 confers the ability to antagonize the antiviral activities of RNase L.
Figure 3.
The MHV ns2 Mutant Replicates to the Same Extent as WT Virus in BMM from RNase L−/− but Not PKR−/− mice
(A and B) BMMs, derived from RNase L−/− or PKR−/− mice were infected (0.01 PFU/cell). At the indicated times, titers of viruses in the cell lysates combined with supernatants were determined by plaque assay (n = 3).
(C) BMMs derived from B6 or RNase L−/− mice were infected (MOI = 0.01 PFU/cell). At 48 hr p.i., the cells were stained with anti-MHV N antibody and Hoechst. The scale bar represents 100 μm.
See also Figure S1.
To gain further insight into the mechanism of ns2 action, we compared the replication of A59 and ns2-H126R in mice genetically ablated for expression of PKR (protein kinase R), a potent antiviral mediator that like OAS is bound and activated by dsRNA (Sadler and Williams, 2008). A59 replicated to 10-fold higher titer in PKR−/− as compared to B6 BMMs, indicating that MHV is sensitive to the antiviral effects of PKR. However, the replication kinetics of ns2-H126R in PKR−/− BMMs were the same as in B6 BMMs (Figure 3B), indicating that ns2 does not antagonize PKR, but rather specifically inhibits the OAS-RNase L pathway. Together with the results obtained in the RNase L−/− BMMs (Figure 3A), these data imply that ns2 acts through a mechanism other than that employed by other viral proteins, including vaccinia E3L (Chang et al., 1992, Xiang et al., 2002) and influenza ns1 (Lu et al., 1995, Min and Krug, 2006), which inhibit both OAS and PKR activity by binding and sequestering dsRNA.
Ns2 Inhibits RNase L Cleavage by Decreasing 2-5A Levels
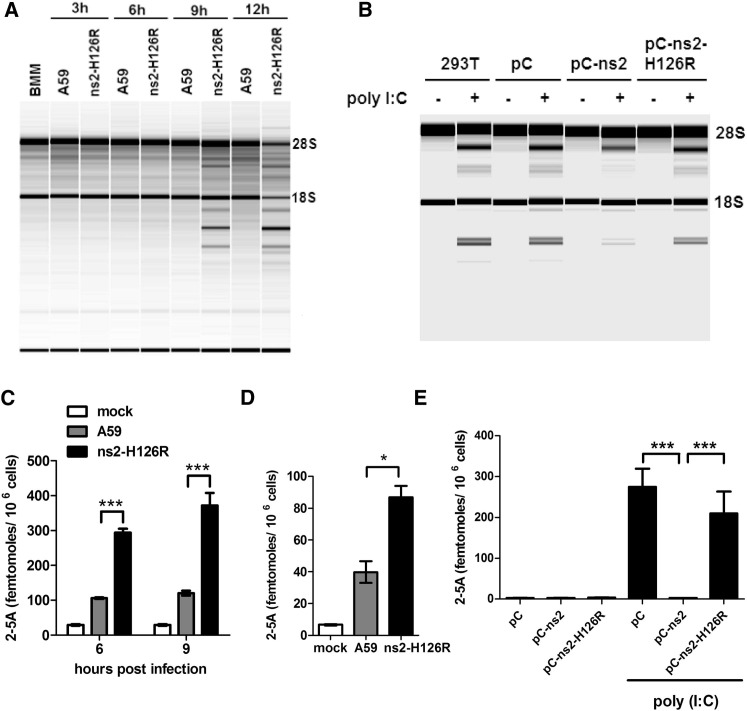
In order to compare the extent of RNaseL activity during A59 and ns2-H126R infection, we compared the state of cleavage of rRNA in B6 BMMs infected with either ns2-H126R or A59. RNA chip analysis of infected BMMs showed that ns2-H126R induced obvious rRNA cleavage at 9 and 12 hr p.i. In contrast, A59 did not induce rRNA cleavage above background even at the later time points, indicating that ns2 efficiently inhibits rRNA cleavage (Figure 4A). As expected, neither virus induced rRNA cleavage in RNase L−/− BMMs (Figure S2A), confirming the cleavage was RNase L mediated.
Figure 4.
MHV ns2 Protein Inhibits Ribosomal RNA Degradation as well as Accumulation of 2-5A in Infected BMMs and in 293T Cells Induced with Poly(I:C)
(A) B6 BMMs were infected (1 PFU/cell). At the indicated times, RNA was purified and analyzed on a Bioanalyzer. The positions of 18S and 28S rRNA are indicated.
(B) 293T cells were transfected with pCAGGS (pC), pC-ns2, or pC-ns2-H126R and 24 hr later were transfected with 10 μg/ml poly(I:C). Four hours later, RNA was purified and analyzed on a Bioanalyzer.
(C and D) 2-5A was isolated from virus-infected BMMs described in (A) derived from B6 mice at 6 and 9 hr p.i (C) or RNase L−/− mice at 9 hr p.i. (D) and analyzed by FRET. Error bars represent the SEM (n = 3). Asterisks indicate that differences are statistically significant (∗p < 0.05 and∗∗∗p < 0.001).
(E) 2-5A from transfected 293T cells described in (B) was isolated and quantified by FRET. Error bars represent the SEM (n = 3). Asterisks indicate that differences are statistically significant (∗∗∗p < 0.001).
See also Figure S2.
Latent RNase L is activated by 2′,5′-linked oligoadenylate (2-5A), which is synthesized by OAS upon activation with dsRNA (Silverman, 2007). To further investigate whether ns2 directly inhibits accumulation of 2-5A, through its predicted phosphodiesterase activity (Mazumder et al., 2002), we measured intracellular levels of 2-5A with A59 or ns2-H126R in BMMs derived from B6 (Figure 4C) and RNase L−/− (Figure 4D) mice. In both B6 and RNase L−/− BMMs, the concentration of 2-5A in ns2-H126R-infected cells was significantly higher than that in A59-infected cells. It should be noted that the 2-5A concentration in RNase L−/− BMMs, as measured by activation of RNase L in a fluorescence resonance energy transfer (FRET)-based assay (see the Experimental Procedures) after infection with either viral strain was much lower than that in virus-infected B6 BMMs (Figure 4D), probably because induction of optimal OAS levels is dependent on an active OAS-RNase L pathway (Malathi et al., 2007).
To directly assess the effects of ns2 on the level of 2-5A, we transfected ns2- and ns2-H126R-expressing plasmids separately into HEK293T cells. Staining with anti-ns2 monoclonal antibody (obtained from Dr. Stuart Siddell, University of Bristol) demonstrated similar expression levels of WT and H126R mutant ns2 (Figure S2B). The OAS-RNase L pathway was subsequently activated in ns2-expressing cells by treatment with IFN and transfection of poly(rI):poly(rC) [poly(I:C)], which mimics naturally occurring dsRNA in activating OAS activity, and the level of RNase L mediated cleavage of rRNA assessed. The expression of ns2 but not ns2-H126R mutant protein in 293T cells decreased the extent of rRNA cleavage as compared with empty vector-transfected cells (Figure 4B). In addition, the level of 2-5A in ns2-transfected cells was significant lower (p < 0.001) than that in empty vector- or ns2-H126R mutant-transfected cells (Figure 4E). Taken together, these results demonstrate that ns2 inhibits RNase L-induced cleavage by limiting the amount of available 2-5A.
Ns2 Degrades 2-5A into ATP and AMP
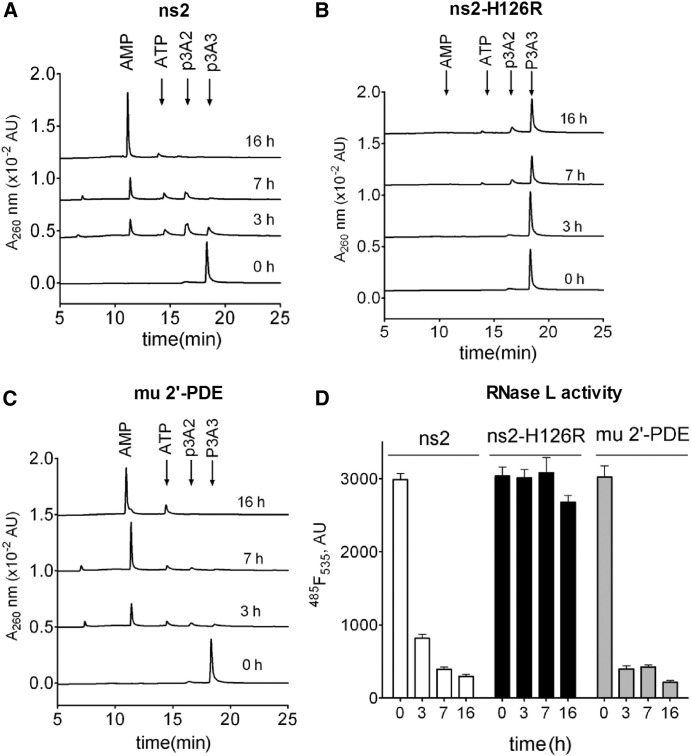
Previous studies reported that the cellular protein 2′-phosphodiesterase (2′-PDE) degrades 2-5A into AMP and ATP and was presumed to regulate the level of RNase L activity (Kubota et al., 2004). Since ns2 was predicted to have phosphodiesterase activity, we hypothesized that ns2 could display a similar enzymatic activity to 2′-PDE and directly cleave 2-5A, thus reducing the activation of RNase L. To test this hypothesis, we expressed ns2, ns2-H126R, and murine 2′-PDE in E. coli and purified the recombinant proteins as described in the Supplemental Experimental Procedures. Each protein was incubated with trimer 2-5A [(2′-5′)p3A3], and at several time points the degradation products were monitored by a high-performance liquid chromatography (HPLC) assay. ns2 and murine 2′-PDE, but not ns2-H126R, degraded 2-5A to the final products of AMP and ATP (Figures 5A–5C). As confirmed by FRET assays (Thakur et al., 2005), the quantity of (2′-5′)p3A3 remaining was significantly decreased after incubation with both ns2 and murine 2′-PDE but not ns2-H126R inactive protein (Figure 5D). These findings demonstrated that ns2 has specificity for 2′,5′-phosphodiester bonds and can directly degrade 2-5A with similar kinetics as 2′-PDE.
Figure 5.
MHV ns2 Cleaves (2′-5′)p3A3 into AMP and ATP In Vitro
(A–C) Purified (2′-5′)p3A3 was incubated with 10 μg/ml of purified MHV ns2, ns2-H126R, or murine 2′-PDE. At the times indicated, the reactions were stopped; (2′-5′)p3A3 and its degradation products were analyzed by HPLC.
(D) The amount of uncleaved (2′-5′)p3A3 remaining was determined by FRET. Error bars represent the SEM (n = 3).
Inhibition of the OAS-RNase L Pathway Is Required for MHV Replication and Spread In Vivo
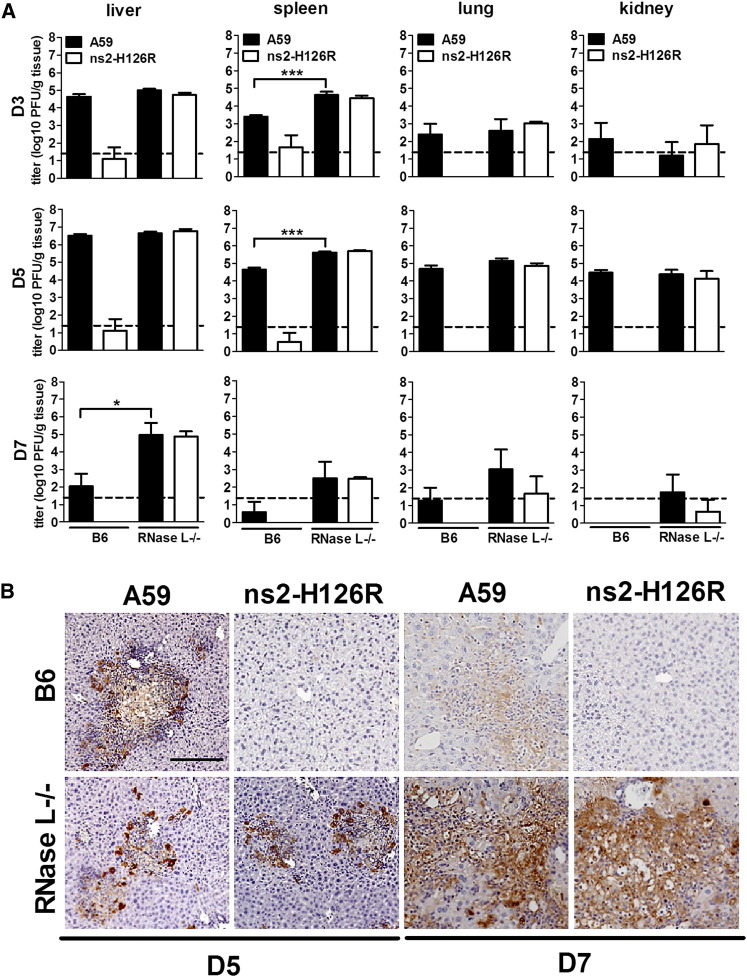
Our previous studies demonstrated that after intrahepatic (IH) inoculation of A59, viral replication is robust in the liver by 3 days postinfection (d.p.i.), peaks at 5 d.p.i., and is mostly cleared by 7 d.p.i. (Navas et al., 2001, Roth-Cross et al., 2009). In contrast, ns2-H126R replicates minimally in the liver (Roth-Cross et al., 2009, Zhao et al., 2011). However, in mice depleted of macrophages prior to infection, ns2-H126R replication and induction of hepatitis were significantly enhanced, approaching that of A59 (Zhao et al., 2011). Taking these in vivo data in combination with the finding that ns2-H126R replicates minimally in B6 BMMs (Figure 3A) (Zhao et al., 2011), we speculated that the ability to replicate in Kupffer cells (liver macrophages) was a prerequisite for spread of virus into the liver parenchyma and for the consequent development of hepatitis. Since ns2-H126 recovers the ability to replicate in RNase L−/− BMMs (Figure 3A), we predicted that the ns2 mutant virus would also replicate and spread in the absence of RNase L activity in vivo. For testing of this hypothesis, both B6 and RNase L−/− mice were inoculated IH with 200 PFU of either A59 or ns2-H126R and virus titers in the liver were quantified. In B6 mice, A59 replicated to high titers peaking at 5 d.p.i., whereas ns2-H126R replicated near the limit of detection. In contrast, in RNase L−/− mice, ns2-H126R reached titers, comparable to those attained by A59 (Figure 6A), reflecting that wild-type ns2 expression is not necessary for efficient replication in the absence of RNase L expression. Notably, there was no significant difference in A59 titers between livers from B6 mice and RNase L−/− mice, indicating that ns2 is efficient at antagonizing RNase L activation in B6 mice. Surprisingly, however, by 7 d.p.i., there were striking differences in viral clearance between B6 and RNase L−/− mice. While both viruses were nearly cleared from the liver of B6 mice at 7 d.p.i., there were still considerable titers of both A59 and ns2-H126R in the livers of RNase L−/− mice (Figure 6A).
Figure 6.
MHV ns2 Enhances Virus Replication In Vivo by Inhibiting RNase L Signaling
(A) Four-week-old B6 or RNase L−/− mice were inoculated intrahepatically with A59 or ns2-H126R (200 PFU/mouse). At 3, 5, and 7 d.p.i., organs were harvested and homogenized, and virus titers were determined. The dashed line represents the limit of detection and the error bars represent the SEM (n = 5). Asterisks indicate that differences are statistically significant (∗p < 0.05 and ∗∗∗p < 0.001).
(B) Liver sections from mice sacrificed at 5 d.p.i. were stained with anti-MHV N monoclonal antibody. The scale bar represents 200 μm. Sections are representative of two sections from each of three animals from each group.
See also Figure S3.
Consistent with viral titers, in B6 mice, A59 expresses abundant viral antigen at 5 d.p.i., which is mostly cleared at 7 d.p.i., while ns2-H126R expresses minimal if any viral antigen. In contrast, in RNaseL−/− mice, both viruses express viral antigen at 5 d.p.i. to a similar extent as in B6 mice (Figure 6B). However instead of clearing virus at 7 d.p.i., both viruses expressed increase viral antigen compared to 5 d.p.i. These data indicate that ns2-mediated antagonism of RNase L activation confers the ability to replicate in the liver, but complete genetic ablation of RNase L expression in mice has additional effects on viral clearance.
The patterns of replication of A59 and ns2-H126R in the spleen, lung, and kidney of B6 mice were similar to those in the liver; ns2-H126R replicated minimally, while A59 replicated efficiently, although not to as high titers as in the liver (Figure 6A). There were no significant differences between A59 and ns2-H126R replication in the spleen, lung, and kidney of RNase L−/− mice at all times tested (Figure 6A), and viral antigen expression in the spleen confirmed that the viral replication and spread for ns2-H126R were similar to those of A59 in the RNase L−/− mice (Figure S3A).
As in comparison of replication in the liver, B6 and RNase L−/− mice showed no difference in infectious A59 titers in the lung and kidney at 3 and 5 d.p.i. However, the A59 titer in the spleen at both 3 and 5 d.p.i of RNase L−/− mice was 10-fold higher than that in B6. This may be at least in part due to the previously reported reduced level of apoptosis in the spleen of RNase L−/− mice (Zhou et al., 1997). Though not statistically significant, the low, but detectable titers of virus at 7 d.p.i. in the spleen, lung, and kidney of RNase L−/− mice were all higher than in B6 mice, perhaps resulting from the delayed clearance in the liver.
Inhibition of the OAS-RNase L Pathway Is Required for the Induction of Hepatitis
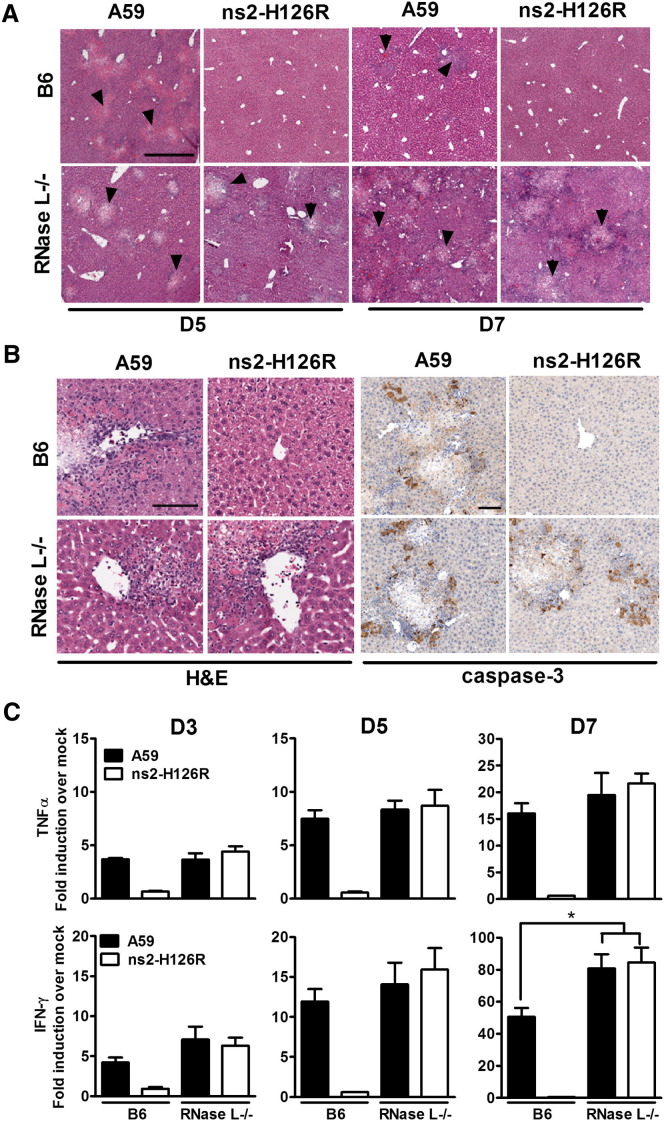
Viral replication in the liver is associated with induction of hepatitis. Indeed, necrotic foci and parenchymal inflammation were observed in liver sections from A59 infected B6 mice sacrificed at 5 d.p.i. (Figures 7A and 7B). Similar pathology was observed in sections from A59- or ns2-H126-infected RNase L−/− mice at 5 d.p.i., while liver sections from ns2-H126R-infected B6 mice, exhibited no obvious pathological changes (Figure 7A). However, as might be predicted by the lack of viral clearance from the livers of RNase L−/− mice, there was extensive necrosis and inflammation in A59- or ns2-H126R-infected livers from RNase L−/− mice compared with the few necrotic foci in the A59-infected livers and the absence of pathology in ns2-H126R-infected livers from B6 mice at 7 d.p.i. (Figure 7A). Consistent with the pathology, staining with anti-caspase-3 antibody demonstrated extensive apoptosis around necrotic foci in A59-infected livers from both B6 and RNase L−/− mice, but only in livers of RNase L−/− mice infected with ns2-H126R (Figure 7B).
Figure 7.
MHV ns2 Facilitates the Induction of Viral Hepatitis in vivo
(A) Livers sections from mock or virus-infected mice sacrificed at 5 and 7 d.p.i. were stained with hematoxylin and eosin (H&E). Arrows show areas of necrosis. The scale bar represents 500 μm. Sections are representative of two sections from three animals from each group.
(B) Liver sections from mice sacrificed at 5 d.p.i. were stained with H&E for detection of inflammation or with anti-caspase-3 antibody for detection of apoptosis. The scale bars represent 100 μm. Sections are representative of two sections from three animals from each group.
(C) Livers were harvested from mock- or virus-infected mice (n = 5) at 3, 5, and 7 d.p.i., and RNA was isolated. TNF-α and IFN-γ mRNAs were quantified by qRT-PCR, normalized to β-actin mRNA level, and expressed as the fold change relative to mock infected with the formula 2−Δ[ΔCT].The error bars represents the SEM. Asterisks indicate that differences are statistically significant (∗p < 0.05).
See also Table S1.
Inflammation in the liver is associated with the local production of inflammatory cytokines, especially TNF-α and IFN-γ (Das et al., 2009). The patterns of cytokine mRNAs induced by A59 and ns2-H126R were consistent with the levels of necrosis and inflammation observed above. There were no differences in the induction of TNF-α and IFN-γ mRNAs in the livers of A59-infected B6 and RNase L−/− mice at 3 and 5 d.p.i., while ns2-H126R induced only minimal levels of both cytokines in livers of B6 mice. A59 and ns2-H126R induced similar levels of TNF-α and IFN-γ mRNAs in the livers of RNase L−/− mice (Figure 7C), indicating, as expected, that ns2-H126R induced the same extent of hepatitis as A59, in the absence of expression of the OAS-RNase L pathway. However, there was a significantly higher (p = 0.0308) level of expression of IFN-γ mRNA in the liver of RNase L−/− mice compared with B6 mice at 7 d.p.i., consistent with the increased virus titers (Figure 6A) and necrosis (Figure 7A).
Discussion
We present evidence for a molecular mechanism of subversion of the RNase L pathway in macrophages that regulates acute hepatitis during MHV infection. MHV and other closely related betacoronaviruses (de Groot et al., 2012) encode ns2 proteins that together with related proteins of toroviruses and human rotavirus form a protein family (known also as LigT-like or family II) with cellular proteins of diverse origins, some of which possess cyclophosphodiesterase (CPD) activity. The family is part of a larger superfamily of 2H phosphodiesterases characterized by the presence of a pair of conserved His-x-Thr/Ser motifs (Mazumder et al., 2002, Snijder et al., 2003). We were unable to demonstrate that ns2 has a CPD activity using as substrates 2′,3′ cAMP, 3′,5′ cAMP, and ADP-ribose 1″,2″ cyclic phosphate (unpublished data), but instead found that ns2 is a 2′,5′-phosphodiesterase cleaving, and thus eliminating, 2-5A, the activator of RNase L. In this respect, ns2 resembles the E. coli 2′-5′-RNA ligase, also within the 2H phosphodiesterase superfamily, which can ligate 2′,5′ linkages of the noncognate tRNA splicing intermediates and is also able to carry out the reverse 2′,5′-phosphodiesterase reaction in vitro (Arn and Abelson, 1996). Before this study, 2′-PDE (PDE12) of the different exonuclease-endonuclease-phosphatase family (Goldstrohm and Wickens, 2008) was the only mammalian enzyme known that cleaves the 2′,5′ bond in 2-5A. Accordingly it was proposed to be a key regulator of the OAS/RNase L system (Kubota et al., 2004). Thus, two proteins, 2′-PDE and ns2, both of which can cleave 2-5A, evolved independently. It remains to be seen whether other incompletely characterized homologs of ns2 of the viral and cellular origins have similar substrate specificity.
Substitution of the catalytic His residue (H126R) of ns2 abrogated phosphodiesterase activity in vitro (Figure 5) as well as virus replication in macrophages and in the livers of B6 mice (Zhao et al., 2011), demonstrating that the enzymatic activity of ns2 was necessary for the type I IFN antagonism and virulence conferred by ns2. Two additional ns2 mutant viruses, one with H46A substitution of the second predicted catalytic His residue (Roth-Cross et al., 2009), and the other with L94P substitution (Sperry et al., 2005), displayed similar phenotypes to ns2-H126R (Figure S1). These data confirm the crucial roles of His126 and His46, each within conserved His-x-Thr/Ser motifs in enzymatic activity.
Viruses have evolved multiple mechanisms for inhibiting RNase L. Thus, the susceptibility to RNase L in vitro and in mice varies among viruses and cell types (Silverman, 2007, Zhou et al., 1997). HIV (Martinand et al., 1999) and EMCV (Martinand et al., 1998) induce expression of an RNase L inhibitor protein, RLI (a.k.a. ABCE1). However, RLI is apparently less efficient than ns2 in the control of RNase L activity during EMCV infections (Figure 2). The group C enterovirus genome contains a conserved RNA structure that inhibits the activity of RNase L (Townsend et al., 2008). Herpes simplex virus I (Cayley et al., 1984) and SV40 (Hersh et al., 1984) induce synthesis of 2-5A derivatives that antagonize or fail to activate RNase L. Vaccinia E3L protein antagonizes OAS by reducing the level of dsRNA available and successfully escapes the effects of RNase L (Xiang et al., 2002). Based on the finding that the A59 nucleocapsid (N) protein could functionally replace the E3L protein in rescuing vaccinia virus ΔE3L from the antiviral effects of IFN it was suggested that N could also bind dsRNA, thus interfering with the activation of OAS (Ye et al., 2007). However, expression of N protein, in the context of MHV infection, is clearly not potent enough to shut down OAS-RNase L pathway as ns2-H126R induces RNase L-mediated cleavage of rRNA in BMMs (Figure 4A). In contrast, rRNA cleavage was not detectable in A59 infected BMMs, providing further evidence that ns2 is a potent antagonist of the OAS-RNase L pathway. The comparable level of A59 replication in B6 and RNase L−/− BMMs and in the infected liver at least through 5 d.p.i. suggests that ns2 is able to completely antagonize RNase L activity, although there may be cell types in which this is not complete. Our findings demonstrate a mechanism by which the OAS-RNase L pathway can be antagonized, that is by blocking the activation of RNase L through the ns2-mediated cleavage of 2-5A.
Interestingly, while there were no significant differences in viral titers or pathology in the livers of A59-infected B6 and RNase L−/− mice during early infection, by 7 d.p.i., a time at which virus is nearly cleared from B6 mice, the livers of infected RNase L−/− mice still had significant titers of virus, antigen expression and pathology (Figures 6 and 7), implying that RNase L−/− mice are defective in viral clearance. One possible reason for the delay in clearance is that RNase L−/− mice have lower levels of apoptosis and in addition are relatively resistant to apoptosis, resulting in increased numbers of cells in the thymus and spleen (Zhou et al., 1997). Indeed, we observed increased virus titers in the spleen of infected RNase L−/− mice compared to B6 mice even at 3 d.p.i. (Figure 6A). This could potentially confer defects in adaptive immune responses; the T cell response, responsible for viral clearance, peaks at 7 d.p.i., the time at which differences in RNase L−/− and B6 mice infections are detectable. RNase L−/− mice were, however, eventually able to clear virus; there was no detectable infectious virus, viral RNA, N protein or pathology in the liver by 30 d.p.i. (Figure S3B). The requirement for basal RNase L activity in the timely clearance of MHV is under investigation.
The replication of ns2-H126R was attenuated in macrophage lineage cells, but not in other primary cells or transformed cell lines (Zhao et al., 2011), indicating that the OAS-RNase L pathway is targeted by ns2 specifically in macrophages. This correlates with basal levels of ISGs and MHV-induced IFN production, which differs among cell types, with the highest expression in macrophages (Ida-Hosonuma et al., 2005, Zhou et al., 1997). Accordingly, we found that the basal levels of expression of mRNAs for genes encoding active OAS proteins (Kakuta et al., 2002), OAS1a, OAS2, and OAS3, were also higher in macrophages as compared with primary hepatocytes (data not shown) (Asada-Kubota et al., 1998). We hypothesize that Kupffer cells serve as a gateway to restrict entrance from the sinusoids into the liver parenchyma. Indeed, it has been reported that IFN signaling in macrophages is crucial for control of hepatitis induced by MHV (Cervantes-Barragán et al., 2009) as well as by LCMV (Lang et al., 2010). We propose that upon infection of Kupffer cells, the IFN response, most importantly the OAS-RNaseL pathway, creates an antiviral state restricting infection and limiting spread of virus to the parenchymal hepatocytes, thus preventing hepatitis. However, ns2 facilitates viral replication in Kupffer cells by antagonizing the OAS-RNase L pathway, thus downregulating the IFN response and enhancing entry into the liver parenchyma. Depletion of macrophages confers susceptibility to ns2-H126R as well as increased susceptibility to A59 (Zhao et al., 2011), supporting the notion that the Kupffer cells provide protection against invasion of the liver parenchyma.
After intracranial inoculation, ns2 mutants replicate to the same titers as A59 in the brains of B6 (Roth-Cross et al., 2009) and RNase L−/− mice (data not shown), implying that antagonism of RNase L is not required for efficient MHV replication in the brain. However, we found that clearance is delayed in the brains (data not shown) as in the livers (Figure 6A) of RNase L−/− mice. Similarly, the MHV-JHM v-2.2 variant replicated to a similar titer in the CNS during acute infection of B6 and RNaseL−/− mice; however, v-2.2 spread into microglia/macrophages within the gray matter of the spinal cord and induced more demyelination in RNase L−/− mice than in B6 mice (Ireland et al., 2009). (While there are no published data on ns2 expression by v-2.2, it is very likely to express an active ns2 as the closely related JHM.SD encodes an ns2 capable of inhibiting RNase L in vitro [data not shown].) These findings support our previous conclusion that the requirement for ns2 expression for MHV virulence is organ specific (Zhao et al., 2011); they also show that basal levels of RNase L expression are required for efficient MHV clearance in the brain as well as the liver (Figure 6).
Our results underscore the important role of RNase L in host restriction of viral infection of the liver. The success of IFN-α treatment of chronic hepatitis C virus (HCV) may depend in part on the susceptibility of viral RNA genome to degradation by RNase L, which cleaves single-stranded viral RNA on the 3′ sides of UU and UA dinucleotides. Cleavage of HCV genome RNA by RNase L releases a small RNA that is recognized by RIG-I and thus serves to enhance IFN induction (Malathi et al., 2010). Indeed, the strains of HCV that are most susceptible to IFN-α treatment are those strains with genomes predicted to be most vulnerable to RNase L-mediated degradation (Han et al., 2004, Washenberger et al., 2007). Thus, enhancement of the activity of the OAS-RNase L pathway may have potential for antiviral therapy for liver infections. A possible approach to enhancing RNase L mediated viral RNA genome degradation might be to limit activities of enzymes that degrade 2-5A; treatment with an inhibitor for 2′-PDE was shown to decrease vaccinia virus replication in vitro (Kubota et al., 2004). In this regard, inhibitors of virus-encoded phosphodiesterases that degrade 2′,5′-oligoadenylates could provide an avenue for development of targeted antiviral drugs.
Experimental Procedures
Cell Lines, Viruses, and Mice
Murine L2 fibroblast and HEK293T cells were cultured as described previously (Roth-Cross et al., 2007). Plaque assays were performed on murine L2 cell monolayers (Hingley et al., 1994). Recombinant coronaviruses inf-MHV-A59 (A59) and inf-ns2-H126R (ns2-H126R) have been described previously (Roth-Cross et al., 2009, Schwarz et al., 1990). (See the Supplemental Experimental Procedures for other ns2 mutants.) EMCV (Babu et al., 1985) was obtained from Dr. Sally Huber (University of Vermont), and Sendai virus (SeV), strain 52 (López et al., 2006), was obtained from Dr. Carolina Lopez (University of Pennsylvania). C57BL/6 (B6) mice were purchased from Jackson Laboratory. RNase L−/− mice (B6, ten generations of backcrossing) (Zhou et al., 1997) were bred in the University of Pennsylvania animal facility. PKR−/− mice (B6 background) (Yang et al., 1995) were a gift from Bryan R.G. Williams (Monash Institute, Melbourne).
Plasmid Constructs
The ns2 and ns2-H126R open reading frames were ligated into mammalian expression vector pCAGGS, resulting in pC-ns2 and pC-ns2-H126R. The ns2 and ns2-H126R open reading frames (ORFs) were inserted into E. coli expression vector pMAL-c2, resulting in pMAL-ns2 and pMAL-ns2-H126R. The murine 2′-PDE-ORF cloned into pET-SUMO was obtained from Reinhard Seidlmeier, Ingenium Pharmaceuticals AG, Martinsried, Germany.
BMM Isolation and Infection
Primary BMMs were generated from the hind limbs of B6 or RNase L−/− mice as described previously (Caamaño et al., 1999) and cultured in DMEM supplemented with 10% FBS and 20% L929 cell-conditioned medium for 6 days. BMMs were either mock infected or infected with 0.01 or 1 plaque forming units (PFU)/cell with A59 or ns2-H126R and harvested at time points indicated.
Real-Time Quantitative RT-PCR
RNA was isolated with an RNeasy mini kit (QIAGEN). Quantitative RT-PCR (qRT-PCR) was performed with an iQ5 iCycler (Bio-Rad), and cycle threshold (CT) values were recorded as described previously (Rose et al., 2010, Roth-Cross et al., 2007). Viral mRNA levels were expressed as relative to β-actin mRNA with the formula 2−ΔCT (ΔCT = CT (gene of interest) – CT (β-actin)). Cytokine mRNA expression levels were expressed as the fold change relative to mock-infected levels with the formula 2−Δ(ΔCT). The primers used in qRT-PCR are listed in Table S1.
RNase L-Mediated Ribosomal RNA Cleavage in Intact Cells
For quantification of rRNA cleavage, total RNA from virus-infected BMMs or transfected 293T cells (see the Supplemental Experimental Procedures) was isolated with the RNeasy kit (QIAGEN) and quantified by Nanodrop analyzer. Equal quantities of RNA (500 ng) were separated on RNA chips and analyzed with an Agilent Bioanalyzer 2100 (Agilent Technologies) (Xiang et al., 2003).
Quantification of 2-5A
2-5A was quantified by an indirect assay in which activation of RNase L activity is measured (Elbahesh et al., 2011). In brief, cells were lysed in preheated (95°C) buffer containing 1% Nonidet-P40 and clarified by centrifugation, and molecules >3 kDa were removed by centrifugation in Microcon filters (Millipore). RNase L activity was determined with a FRET assay, with recombinant human RNase L and a cleavable substrate consisting of a 36 nucleotide synthetic oligoribonucleotide [5′(6-FAMUUAUCAAAUUCUUAUUUGCCCCAUUUUUUUGGUUUA-BHQ-1)-3′] derived from respiratory syncytial virus (Thakur et al., 2005). A standard curve was generated with authentic trimer 2-5A, (2′-5′)p3A3 (95% purity), and fluorescence was measured with a Wallac 1420 fluorimeter (Perkin-Elmer LAS).
2′-PDE Activity Assay
2′-PDE activity was quantified by monitoring of the degradation of purified (2′-5′)p3A3 with HPLC (Molinaro et al., 2006) or the FRET-based RNase L activity assay, as above (Thakur et al., 2005) (see the Supplemental Experimental Procedures). For the 2-5A degradation assay, 100 μM purified (2′-5′)p3A3 was incubated for the indicated time with 10 μg/ml purified ns2, ns2-H126R, or 2′-PDE protein and then centrifuged in microcon centrifugal filter MWCO 3kDa (Millipore). The filtrate was diluted 1:5 and loaded onto a Dionex P100 analytical HPLC column, interfaced with Beckman System Gold HPLC system connected to a 32 Karat workstation. The chromatograms were analyzed with 32 Karat software (Beckman-Coulter).
Infection of Mice
Four- to five-week-old B6 or RNase L−/− mice were anesthetized with isoflurane (IsoFlo, Abott Laboratories) and inoculated intrahepatically with A59 or ns2-H126R (200 PFU) in 50 μl PBS containing 0.75% BSA. At 3, 5, and 7 d.p.i., animals were sacrificed, livers, spleens, lungs, and kidneys were harvested, and viral titers in lysates were determined by plaque assay (Gombold et al., 1993). Alternatively, organs were used for RNA isolation or immunohistochemistry.
This study was carried out in strict accordance with the Guide for the Care and Use of Laboratory Animals and the Public Health Service Policy on Humane Care and Use of Laboratory Animals. The protocol was approved by the University of Pennsylvania's Institutional Animal Care and Use Committee (permit number A3079-01).
Histology and Immunohistochemistry
Livers were fixed overnight in 4% paraformaldehyde, embedded in paraffin and sectioned. Sections were stained with hematoxylin and eosin or alternatively blocked with 10% normal donkey serum and immunostained with a 1:20 dilution of a monoclonal antibody against MHV nucleocapsid (N) protein or a 1:1000 dilution of polyclonal antibody against caspase-3 antibody. Staining was developed using avidin-biotin-immunoperoxidase (Vector Laboratories).
Statistical Analysis
An unpaired two-tailed t test was used to determine statistical significance. Data were analyzed with GraphPad Prism software (GraphPad Software).
Acknowledgments
We thank Drs. Kristine Rose, Serge Fuchs, Katalin Kariko, Rahul Kohli, Carolina Lopez, and Beth Schachter for discussion and editing. We thank Dr. Stuart Siddell, University of Bristol, England, for inf-MHV-A59 and inf-ns2-H126R and inf-ns2-H46A, Dr. Reinhard Sedlmeier, Ingenium Pharmaceuticals GmbH for the 2′-PDE cDNA clone, and Dr. Bryan R.G. Williams, Monash Institute, Melbourne, Australia, for the PKR−/− mice. A.E.G. acknowledges his prior collaboration with Dr. E. Snijder on the anti-2-5A activity of ns2. This work was supported by NIH grants R21-AI-080797, R56-AI-095285, and RO1-NS-054695 (S.R.W.) and RO1-CA044059 (R.H.S.). A.E.G. was supported in part by the Leiden University Fund. R.H.S. is a consultant and inventor on patents relating to RNase L licensed to Alios BioPharma.
Published: June 13, 2012
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, three figures, and one table and can be found with this article online at doi:10.1016/j.chom.2012.04.011.
Supplemental Information
References
- Arn E.A., Abelson J.N. The 2′-5′ RNA ligase of Escherichia coli. Purification, cloning, and genomic disruption. J. Biol. Chem. 1996;271:31145–31153. doi: 10.1074/jbc.271.49.31145. [DOI] [PubMed] [Google Scholar]
- Asada-Kubota M., Tatsumi R., Ueda T., Kobayashi M., Hamada K., Maekawa S., Sokawa Y. The target cells of injected type I interferons in mouse liver. J. Interferon Cytokine Res. 1998;18:71–74. doi: 10.1089/jir.1998.18.71. [DOI] [PubMed] [Google Scholar]
- Babu P.G., Huber S., Sriram S., Craighead J.E. Genetic control of multisystem autoimmune disease in encephalomyocarditis virus infected BALB/cCUM and BALB/cBYJ mice. Curr. Top. Microbiol. Immunol. 1985;122:154–161. doi: 10.1007/978-3-642-70740-7_23. [DOI] [PubMed] [Google Scholar]
- Caamaño J., Alexander J., Craig L., Bravo R., Hunter C.A. The NF-kappa B family member RelB is required for innate and adaptive immunity to Toxoplasma gondii. J. Immunol. 1999;163:4453–4461. [PubMed] [Google Scholar]
- Cayley P.J., Davies J.A., McCullagh K.G., Kerr I.M. Activation of the ppp(A2'p)nA system in interferon-treated, herpes simplex virus-infected cells and evidence for novel inhibitors of the ppp(A2'p)nA-dependent RNase. Eur. J. Biochem. 1984;143:165–174. doi: 10.1111/j.1432-1033.1984.tb08355.x. [DOI] [PubMed] [Google Scholar]
- Cervantes-Barragan L., Züst R., Weber F., Spiegel M., Lang K.S., Akira S., Thiel V., Ludewig B. Control of coronavirus infection through plasmacytoid dendritic-cell-derived type I interferon. Blood. 2007;109:1131–1137. doi: 10.1182/blood-2006-05-023770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Barragán L., Kalinke U., Züst R., König M., Reizis B., López-Macías C., Thiel V., Ludewig B. Type I IFN-mediated protection of macrophages and dendritic cells secures control of murine coronavirus infection. J. Immunol. 2009;182:1099–1106. doi: 10.4049/jimmunol.182.2.1099. [DOI] [PubMed] [Google Scholar]
- Chang H.W., Watson J.C., Jacobs B.L. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc. Natl. Acad. Sci. USA. 1992;89:4825–4829. doi: 10.1073/pnas.89.11.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispe I.N. The liver as a lymphoid organ. Annu. Rev. Immunol. 2009;27:147–163. doi: 10.1146/annurev.immunol.021908.132629. [DOI] [PubMed] [Google Scholar]
- Das M., Sabio G., Jiang F., Rincón M., Flavell R.A., Davis R.J. Induction of hepatitis by JNK-mediated expression of TNF-alpha. Cell. 2009;136:249–260. doi: 10.1016/j.cell.2008.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R.J., Baker S.C., Baric R., Enjuanes L., Gorbalenya A.E., Holmes K.V., Perlman S., Poon L., Rottier P.J.M., Talbot P.J. Family Coronaviridae. In: King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J., editors. Virus Taxonomy, Classification and Nomenclature of Viruses, Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier; Oxford: 2012. pp. 806–828. [Google Scholar]
- Dong B., Silverman R.H. 2-5A-dependent RNase molecules dimerize during activation by 2-5A. J. Biol. Chem. 1995;270:4133–4137. doi: 10.1074/jbc.270.8.4133. [DOI] [PubMed] [Google Scholar]
- Elbahesh H., Jha B.K., Silverman R.H., Scherbik S.V., Brinton M.A. The Flvr-encoded murine oligoadenylate synthetase 1b (Oas1b) suppresses 2-5A synthesis in intact cells. Virology. 2011;409:262–270. doi: 10.1016/j.virol.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flodström-Tullberg M., Hultcrantz M., Stotland A., Maday A., Tsai D., Fine C., Williams B., Silverman R., Sarvetnick N. RNase L and double-stranded RNA-dependent protein kinase exert complementary roles in islet cell defense during coxsackievirus infection. J. Immunol. 2005;174:1171–1177. doi: 10.4049/jimmunol.174.3.1171. [DOI] [PubMed] [Google Scholar]
- García-Sastre A., Biron C.A. Type 1 interferons and the virus-host relationship: a lesson in détente. Science. 2006;312:879–882. doi: 10.1126/science.1125676. [DOI] [PubMed] [Google Scholar]
- Goldstrohm A.C., Wickens M. Multifunctional deadenylase complexes diversify mRNA control. Nat. Rev. Mol. Cell Biol. 2008;9:337–344. doi: 10.1038/nrm2370. [DOI] [PubMed] [Google Scholar]
- Gombold J.L., Hingley S.T., Weiss S.R. Fusion-defective mutants of mouse hepatitis virus A59 contain a mutation in the spike protein cleavage signal. J. Virol. 1993;67:4504–4512. doi: 10.1128/jvi.67.8.4504-4512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J.Q., Wroblewski G., Xu Z., Silverman R.H., Barton D.J. Sensitivity of hepatitis C virus RNA to the antiviral enzyme ribonuclease L is determined by a subset of efficient cleavage sites. J. Interferon Cytokine Res. 2004;24:664–676. doi: 10.1089/jir.2004.24.664. [DOI] [PubMed] [Google Scholar]
- Hersh C.L., Brown R.E., Roberts W.K., Swyryd E.A., Kerr I.M., Stark G.R. Simian virus 40-infected, interferon-treated cells contain 2′,5′-oligoadenylates which do not activate cleavage of RNA. J. Biol. Chem. 1984;259:1731–1737. [PubMed] [Google Scholar]
- Hingley S.T., Gombold J.L., Lavi E., Weiss S.R. MHV-A59 fusion mutants are attenuated and display altered hepatotropism. Virology. 1994;200:1–10. doi: 10.1006/viro.1994.1156. [DOI] [PubMed] [Google Scholar]
- Ida-Hosonuma M., Iwasaki T., Yoshikawa T., Nagata N., Sato Y., Sata T., Yoneyama M., Fujita T., Taya C., Yonekawa H., Koike S. The alpha/beta interferon response controls tissue tropism and pathogenicity of poliovirus. J. Virol. 2005;79:4460–4469. doi: 10.1128/JVI.79.7.4460-4469.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireland D.D., Stohlman S.A., Hinton D.R., Kapil P., Silverman R.H., Atkinson R.A., Bergmann C.C. RNase L mediated protection from virus induced demyelination. PLoS Pathog. 2009;5:e1000602. doi: 10.1371/journal.ppat.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakuta S., Shibata S., Iwakura Y. Genomic structure of the mouse 2′,5′-oligoadenylate synthetase gene family. J. Interferon Cytokine Res. 2002;22:981–993. doi: 10.1089/10799900260286696. [DOI] [PubMed] [Google Scholar]
- Kubota K., Nakahara K., Ohtsuka T., Yoshida S., Kawaguchi J., Fujita Y., Ozeki Y., Hara A., Yoshimura C., Furukawa H. Identification of 2′-phosphodiesterase, which plays a role in the 2-5A system regulated by interferon. J. Biol. Chem. 2004;279:37832–37841. doi: 10.1074/jbc.M400089200. [DOI] [PubMed] [Google Scholar]
- Lang P.A., Recher M., Honke N., Scheu S., Borkens S., Gailus N., Krings C., Meryk A., Kulawik A., Cervantes-Barragan L. Tissue macrophages suppress viral replication and prevent severe immunopathology in an interferon-I-dependent manner in mice. Hepatology. 2010;52:25–32. doi: 10.1002/hep.23640. [DOI] [PubMed] [Google Scholar]
- Li X.L., Blackford J.A., Hassel B.A. RNase L mediates the antiviral effect of interferon through a selective reduction in viral RNA during encephalomyocarditis virus infection. J. Virol. 1998;72:2752–2759. doi: 10.1128/jvi.72.4.2752-2759.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López C.B., Yount J.S., Hermesh T., Moran T.M. Sendai virus infection induces efficient adaptive immunity independently of type I interferons. J. Virol. 2006;80:4538–4545. doi: 10.1128/JVI.80.9.4538-4545.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Wambach M., Katze M.G., Krug R.M. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the elF-2 translation initiation factor. Virology. 1995;214:222–228. doi: 10.1006/viro.1995.9937. [DOI] [PubMed] [Google Scholar]
- Malathi K., Dong B., Gale M., Jr., Silverman R.H. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448:816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malathi K., Saito T., Crochet N., Barton D.J., Gale M., Jr., Silverman R.H. RNase L releases a small RNA from HCV RNA that refolds into a potent PAMP. RNA. 2010;16:2108–2119. doi: 10.1261/rna.2244210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinand C., Salehzada T., Silhol M., Lebleu B., Bisbal C. RNase L inhibitor (RLI) antisense constructions block partially the down regulation of the 2-5A/RNase L pathway in encephalomyocarditis-virus-(EMCV)-infected cells. Eur. J. Biochem. 1998;254:248–255. doi: 10.1046/j.1432-1327.1998.2540248.x. [DOI] [PubMed] [Google Scholar]
- Martinand C., Montavon C., Salehzada T., Silhol M., Lebleu B., Bisbal C. RNase L inhibitor is induced during human immunodeficiency virus type 1 infection and down regulates the 2-5A/RNase L pathway in human T cells. J. Virol. 1999;73:290–296. doi: 10.1128/jvi.73.1.290-296.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder R., Iyer L.M., Vasudevan S., Aravind L. Detection of novel members, structure-function analysis and evolutionary classification of the 2H phosphoesterase superfamily. Nucleic Acids Res. 2002;30:5229–5243. doi: 10.1093/nar/gkf645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J.Y., Krug R.M. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: Inhibiting the 2′-5′ oligo (A) synthetase/RNase L pathway. Proc. Natl. Acad. Sci. USA. 2006;103:7100–7105. doi: 10.1073/pnas.0602184103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinaro R.J., Jha B.K., Malathi K., Varambally S., Chinnaiyan A.M., Silverman R.H. Selection and cloning of poly(rC)-binding protein 2 and Raf kinase inhibitor protein RNA activators of 2′,5′-oligoadenylate synthetase from prostate cancer cells. Nucleic Acids Res. 2006;34:6684–6695. doi: 10.1093/nar/gkl968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navas S., Weiss S.R. Murine coronavirus-induced hepatitis: JHM genetic background eliminates A59 spike-determined hepatotropism. J. Virol. 2003;77:4972–4978. doi: 10.1128/JVI.77.8.4972-4978.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navas S., Seo S.H., Chua M.M., Das Sarma J., Lavi E., Hingley S.T., Weiss S.R. Murine coronavirus spike protein determines the ability of the virus to replicate in the liver and cause hepatitis. J. Virol. 2001;75:2452–2457. doi: 10.1128/JVI.75.5.2452-2457.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann-Haefelin C., Spangenberg H.C., Blum H.E., Thimme R. Host and viral factors contributing to CD8+ T cell failure in hepatitis C virus infection. World J. Gastroenterol. 2007;13:4839–4847. doi: 10.3748/wjg.v13.i36.4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu L., Lemon S.M. Hepatitis A and hepatitis C viruses: divergent infection outcomes marked by similarities in induction and evasion of interferon responses. Semin. Liver Dis. 2010;30:319–332. doi: 10.1055/s-0030-1267534. [DOI] [PubMed] [Google Scholar]
- Rose K.M., Weiss S.R. Murine Coronavirus Cell Type Dependent Interaction with the Type I Interferon Response. Viruses. 2009;1:689–712. doi: 10.3390/v1030689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose K.M., Elliott R., Martínez-Sobrido L., García-Sastre A., Weiss S.R. Murine coronavirus delays expression of a subset of interferon-stimulated genes. J. Virol. 2010;84:5656–5669. doi: 10.1128/JVI.00211-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth-Cross J.K., Martínez-Sobrido L., Scott E.P., García-Sastre A., Weiss S.R. Inhibition of the alpha/beta interferon response by mouse hepatitis virus at multiple levels. J. Virol. 2007;81:7189–7199. doi: 10.1128/JVI.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth-Cross J.K., Bender S.J., Weiss S.R. Murine coronavirus mouse hepatitis virus is recognized by MDA5 and induces type I interferon in brain macrophages/microglia. J. Virol. 2008;82:9829–9838. doi: 10.1128/JVI.01199-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth-Cross J.K., Stokes H., Chang G., Chua M.M., Thiel V., Weiss S.R., Gorbalenya A.E., Siddell S.G. Organ-specific attenuation of murine hepatitis virus strain A59 by replacement of catalytic residues in the putative viral cyclic phosphodiesterase ns2. J. Virol. 2009;83:3743–3753. doi: 10.1128/JVI.02203-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler A.J., Williams B.R. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel M.A., Whitby K., Keller B.C., Marri A., Barchet W., Williams B.R., Silverman R.H., Gale M., Jr., Diamond M.S. PKR and RNase L contribute to protection against lethal West Nile Virus infection by controlling early viral spread in the periphery and replication in neurons. J. Virol. 2006;80:7009–7019. doi: 10.1128/JVI.00489-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherbik S.V., Paranjape J.M., Stockman B.M., Silverman R.H., Brinton M.A. RNase L plays a role in the antiviral response to West Nile virus. J. Virol. 2006;80:2987–2999. doi: 10.1128/JVI.80.6.2987-2999.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz B., Routledge E., Siddell S.G. Murine coronavirus nonstructural protein ns2 is not essential for virus replication in transformed cells. J. Virol. 1990;64:4784–4791. doi: 10.1128/jvi.64.10.4784-4791.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman R.H. Viral encounters with 2′,5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. J. Virol. 2007;81:12720–12729. doi: 10.1128/JVI.01471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L., Guan Y., Rozanov M., Spaan W.J., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperry S.M., Kazi L., Graham R.L., Baric R.S., Weiss S.R., Denison M.R. Single-amino-acid substitutions in open reading frame (ORF) 1b-nsp14 and ORF 2a proteins of the coronavirus mouse hepatitis virus are attenuating in mice. J. Virol. 2005;79:3391–3400. doi: 10.1128/JVI.79.6.3391-3400.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur C.S., Xu Z., Wang Z., Novince Z., Silverman R.H. A convenient and sensitive fluorescence resonance energy transfer assay for RNase L and 2′,5′ oligoadenylates. Methods Mol. Med. 2005;116:103–113. doi: 10.1385/1-59259-939-7:103. [DOI] [PubMed] [Google Scholar]
- Townsend H.L., Jha B.K., Han J.Q., Maluf N.K., Silverman R.H., Barton D.J. A viral RNA competitively inhibits the antiviral endoribonuclease domain of RNase L. RNA. 2008;14:1026–1036. doi: 10.1261/rna.958908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washenberger C.L., Han J.Q., Kechris K.J., Jha B.K., Silverman R.H., Barton D.J. Hepatitis C virus RNA: dinucleotide frequencies and cleavage by RNase L. Virus Res. 2007;130:85–95. doi: 10.1016/j.virusres.2007.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S.R., Leibowitz J.L. Coronavirus pathogenesis. In: Maramorosch K., Shatkin A.J., Murphy F.A., editors. Volume 81. Elsevier; London: 2011. (Advances in Virus Research). pp. 85–173. [Google Scholar]
- Xiang Y., Condit R.C., Vijaysri S., Jacobs B., Williams B.R., Silverman R.H. Blockade of interferon induction and action by the E3L double-stranded RNA binding proteins of vaccinia virus. J. Virol. 2002;76:5251–5259. doi: 10.1128/JVI.76.10.5251-5259.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y., Wang Z., Murakami J., Plummer S., Klein E.A., Carpten J.D., Trent J.M., Isaacs W.B., Casey G., Silverman R.H. Effects of RNase L mutations associated with prostate cancer on apoptosis induced by 2′,5′-oligoadenylates. Cancer Res. 2003;63:6795–6801. [PubMed] [Google Scholar]
- Yang Y.L., Reis L.F., Pavlovic J., Aguzzi A., Schäfer R., Kumar A., Williams B.R., Aguet M., Weissmann C. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J. 1995;14:6095–6106. doi: 10.1002/j.1460-2075.1995.tb00300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Hauns K., Langland J.O., Jacobs B.L., Hogue B.G. Mouse hepatitis coronavirus A59 nucleocapsid protein is a type I interferon antagonist. J. Virol. 2007;81:2554–2563. doi: 10.1128/JVI.01634-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Rose K.M., Elliott R., Van Rooijen N., Weiss S.R. Cell-type-specific type I interferon antagonism influences organ tropism of murine coronavirus. J. Virol. 2011;85:10058–10068. doi: 10.1128/JVI.05075-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou A., Paranjape J., Brown T.L., Nie H., Naik S., Dong B., Chang A., Trapp B., Fairchild R., Colmenares C., Silverman R.H. Interferon action and apoptosis are defective in mice devoid of 2′,5′-oligoadenylate-dependent RNase L. EMBO J. 1997;16:6355–6363. doi: 10.1093/emboj/16.21.6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.