SUMMARY
Systemic bacterial infection is characterized by a robust whole organism inflammatory response. Analysis of the immune response to infection involves technologies that typically focus on single organ systems and lack spatial information. Additionally, the analysis of individual inflammatory proteins requires antibodies specific to the protein of interest, limiting the panel of proteins that can be analyzed. Herein we describe the application of matrix-assisted laser desorption/ionization imaging mass spectrometry (MALDI IMS) to mice systemically infected with Staphylococcus aureus to identify inflammatory protein masses that respond to infection throughout an entire infected animal. Integrating the resolution afforded by magnetic resonance imaging (MRI) with the sensitivity of MALDI IMS provides three-dimensional spatially resolved information regarding the distribution of innate immune proteins during systemic infection, allowing comparisons to in vivo structural information and soft tissue contrast via MRI. Thus, integrating MALDI IMS with MRI provides a systems biology approach to study inflammation during infection.
INTRODUCTION
The inflammatory response to infection is characterized by the robust recruitment of innate immune cells to affected tissue. A properly controlled inflammatory response ensures pathogen clearance while a dysregulated immune response can lead to significant tissue destruction and an inability to control infection. Understanding the kinetics and anatomical distribution of this response is paramount to the ability to monitor and control inflammation.
Staphylococcus aureus is a bacterial pathogen that is capable of causing a diverse array of diseases ranging from superficial skin complications to systemic infections with a high mortality rate (DeLeo et al., 2010; Lowy, 1998). A hallmark of S. aureus infection is the formation of tissue abscesses that are visible representations of the inflammatory response to infection. Abscesses are composed of leukocytes of the innate immune system, and the most prevalent cell type within the abscess is the neutrophil (Cheng et al., 2009). Despite the appreciation of abscesses as the primary tissue lesion associated with staphylococcal infection, little is known regarding the dynamics of abscess formation and maintenance.
The inflammatory response to infection can be monitored using a variety of techniques. Quantification of immune cell subsets using fluorescence activated cell sorting provides a snapshot of the cell populations that respond to infection. In addition, monitoring cytokine abundance through expression analyses reveals the immune signaling molecules that are expressed in infected animals. However, these technologies do not provide spatial information regarding the immune response, nor do they allow investigators to monitor the inflammatory response to infection in real time. To obtain spatial information regarding the immune response to infection, researchers traditionally have relied on immunohistochemistry, which requires the availability of antibodies or stains for proteins and cell types of interest. More recently, imaging modalities have permitted the real-time non-invasive monitoring of infection progression. Magnetic resonance imaging (MRI) is the primary method to image inflammation based on its high soft tissue contrast and increased spatial resolution in comparison to other imaging technologies (Hertlein et al., 2011). However, MRI does not provide information on the immune cell subsets or innate immune proteins that are recruited in response to infection. A technological advance is needed that permits the analysis of immune proteins that are expressed in response to infection while preserving spatial information regarding the expression pattern of these proteins throughout an infected animal.
Matrix-assisted laser desorption/ionization imaging mass spectrometry (MALDI IMS) has emerged as a powerful tool for imaging the distribution of macromolecules in tissue sections. A raster of the tissue section by a UV laser in a two-dimensional array enables the collection of mass spectral data at predefined points within the section (McDonnell and Heeren, 2007; Seeley and Caprioli, 2008; Seeley et al., 2011). The collected data can then be converted to a color representation of protein expression and superimposed on an optical image of the animal. The end result is a two-dimensional map of molecular abundance within the tissue section under study (Khatib-Shahidi et al., 2006). The application of MALDI IMS to a murine model of S. aureus infection identified the neutrophil protein calprotectin as being abundantly recruited to staphylococcal kidney abscesses (Corbin et al., 2008). This work led to the discovery of proteins that are expressed within kidneys of infected mice; however, it did not provide a whole-animal view of the host response to infection. Moreover, applying MALDI IMS analysis to an individual tissue section leads to the loss of three-dimensional anatomic information regarding the inflammatory response to infection. During systemic infection of mice, S. aureus colonizes virtually every organ, and monitoring protein expression across the entire animal would undoubtedly provide significant insight into the whole body response to systemic infection.
We have developed a method for using MALDI IMS to image the inflammatory response to infection across an entire infected animal. By integrating MALDI IMS and MRI, it is possible to combine the significant resolution afforded by MRI with the tremendous sensitivity of mass spectrometry to image changes in protein abundance in three dimensions in response to infection (Sinha et al., 2008).
RESULTS
Manipulating the inflammatory response to S. aureus infection
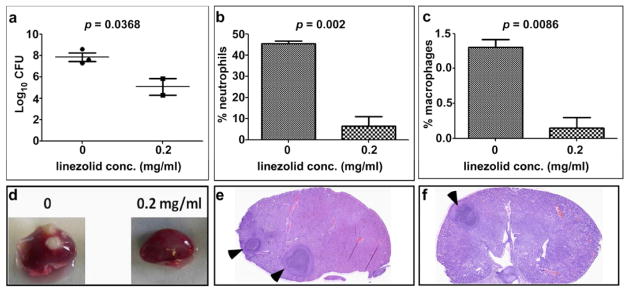
To establish a model that allows for monitoring and manipulating the inflammatory response to infection, we first intravenously infected mice with S. aureus. Ninety-six hours following infection, mice were sacrificed and bacterial burden was quantified by tissue homogenization followed by enumeration of bacterial colony forming units (CFU). The kidneys of mice infected with S. aureus harbored approximately 1 × 107 CFU at the termination of this experiment (data not shown). To reduce staphylococcal colonization and the associated inflammatory response, mice that were infected for 96 hours were treated with linezolid and the infection was allowed to proceed for an additional 96 hours. Linezolid is a clinically relevant antibiotic that exhibits potent anti-inflammatory properties (Bernardo et al., 2004; Garcia-Roca et al., 2006; Sandberg et al., 2010). As expected, linezolid decreased the bacterial burden within infected tissue (Figure 1A). Moreover, linezolid significantly reduced the numbers of neutrophils and macrophages that responded to infected kidneys following staphylococcal challenge (Figure 1B and C). This reduction in bacterial burden and inflammation decreased the number of detectable abscesses within the kidneys (Figure 1D). In addition, in cases where abscesses were visualized in the linezolid treated mice, these lesions were smaller and more organized as compared to untreated animals (Figure 1E and F). Taken together, these data establish a model that mimics the clinical condition of systemic staphylococcal infection followed by linezolid treatment and enables the experimental manipulation of the inflammatory response to infection.
Figure 1. Linezolid reduces staphylococcal abscess formation and inflammation in systemically infected mice.
Groups of three animals were infected with S. aureus for 96 hours at which point they were either treated with 0.2 mg/ml linezolid or left untreated and the infection was allowed to proceed for an additional 96 hours. (A) Bacterial burdens in the kidneys of the infected mice were estimated by tissue homogenization and plating. (B and C) The percentage of neutrophils (B) and macrophages (C) in the kidneys of the two groups of mice as determined by fluorescently activated cell sorting. Error bars in panels A–C represent +/− standard deviation. (D) Photographic image of representative kidneys from the treated and control mice. (E and F) Hematoxylin and eosin stain of kidney sections from control (E) and treated (F) mice 8 days following infection. Black arrows denote abscesses.
Monitoring abscess formation and dynamics using magnetic resonance imaging
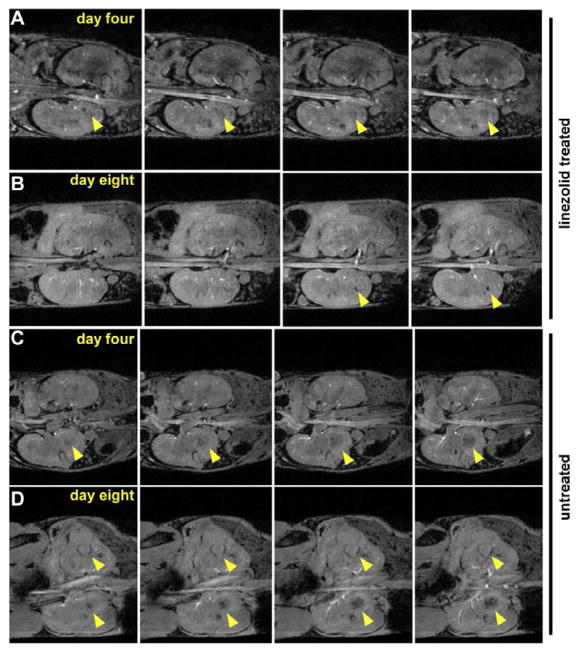
To visualize abscess formation within living animals in real-time, mice were intravenously infected with S. aureus. Four days following infection, mice were either treated with linezolid or left untreated and the infection was allowed to proceed for an additional four days. Mice were imaged using MRI at four and eight days following infection. These experiments revealed that abscess formation was visible by four days following infection in both experimental groups, as expected (Figure 2A and C). Eight days following infection, untreated animals displayed significant kidney abscess formation characterized by severe organ deformation (Figure 2D). In contrast, organ architecture was largely maintained in the linezolid treated mice at this time point, and smaller spherical abscesses were visible (Figure 2B). These data emphasize the value of MRI for imaging the inflammatory response to S. aureus infection and underscore the therapeutic benefit of linezolid through reduced bacterial burden and decreased tissue abscess formation.
Figure 2. MRI of S. aureus infected mice that have been treated with linezolid or left untreated.
Four contiguous slices depicting the kidneys in the coronal plane for one treated and one untreated animal, acquired using a 3D gradient echo imaging sequence at 9.4T in approximately 33 minutes. Panels (A) and (B) show one infected animal on day 4 (just prior to treatment with linezolid) and day 8, respectively. Abscesses are marked with arrows. Panels (C) and (D) show an untreated animal on day 4 and day 8, respectively.
Imaging the inflammatory response to infection across an entire animal
Conventional assays to measure inflammation are focused on individual organs or lack anatomical information due to the requirement for tissue homogenization or blood sampling procedures. This technical barrier is particularly cumbersome when studying systemic bacterial infections that invariably involve a whole body response to infection. Mice intravenously infected with S. aureus exhibit significant abscess formation in the kidneys with visible lesions rarely present in the heart and liver, however bacteria can be cultured from virtually every murine organ in this infection model (data not shown). Therefore, the ability to monitor the inflammatory response to infection across an entire infected animal would provide significant insight into how the host responds to systemic challenge by an infectious agent.
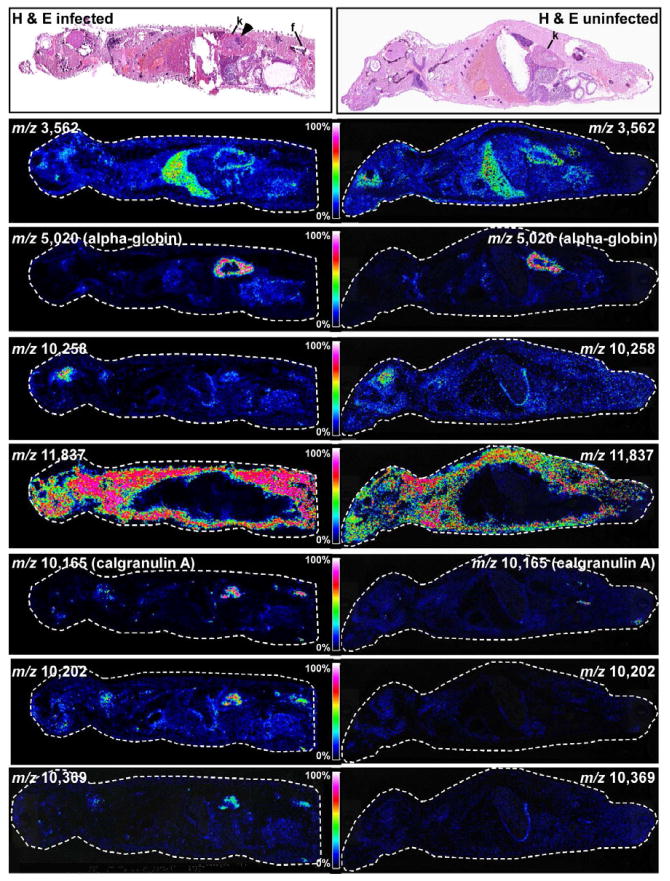
To image protein expression in response to infection across an entire infected animal, mice were intravenously infected with S. aureus as described above. Four days following infection, mice were sacrificed and the entire animal was sectioned through the sagittal plane. An uninfected mouse (injected with 100 μl PBS) was included as a control in this analysis. Sections were either stained with hematoxylin and eosin or imaged for protein expression using MALDI IMS (Figure 3). Consistent with previous experiments, abscesses were visible in the kidneys of systemically infected mice. Figure 3 shows a series of representative proteins whose abundance is either unaffected or affected by the infection status of the animal. Mass signals that are abundant in the liver (m/z 3,562), kidney (m/z 5,020), and brain (m/z 10,258) were detected in both infected and uninfected mice. Also, we detected a mass signal that was abundant extra-peritoneally but was not detected within the peritoneal cavity (m/z 11,837) of both infected and uninfected animals. Based on the significant tissue pathology visible in the kidneys of infected animals, we sought to identify the kidney-specific protein that was detected at m/z 5,020. These experiments revealed that the signal in the kidney cortex at m/z 5,020 is a truncated form of alpha-globin (residues 2–47, gi 122441).
Figure 3. Whole animal MALDI IMS of the systemic response to S. aureus infection.
(Top panels) Hematoxylin and eosin stained sections of entire mice that were either infected with S. aureus or left uninfected. Abscesses are marked by an arrow. Kidneys are marked with “k” and femur is marked with “f”. Masses corresponding to proteins that are abundant in the liver (m/z 3,562), kidney (m/z 5,020), brain (m/z 10,258), or systemically (m/z 11,837) in both infected and uninfected mice are shown. In addition, masses corresponding to proteins that are only expressed in infected animals are shown (m/z 10,165, 10,202, 10,369). The color scale correlates to 0–100% full scale on the intensity of each ion.
In addition to imaging the distribution of proteins that did not change abundance in infected animals, a series of proteins were detected that increase abundance in response to infection. The mass signal corresponding to the calgranulin A subunit of calprotectin (m/z 10,165) was most abundant at the sites of kidney abscesses (Figure 3). This finding is consistent with previously published reports describing calprotectin as an abundant abscess protein and a marker of neutrophil deposition (Steinbakk et al., 1990; Urban et al., 2009). Other proteins were detected that also increased abundance within the abscess (m/z 10,202, m/z 10,369), potentially representing additional immune proteins that respond to staphylococcal infection. In keeping with this, all three of these abscess-specific proteins were also detected in the femur of infected mice, suggesting that MALDI IMS enables the visualization of immune cell development in the bone marrow in response to infection. These results provide a whole animal view of inflammation, and establish whole animal IMS as a technique to study the inflammatory response to systemic infections.
Imaging inflammation through integrated IMS and MRI
The generation of three-dimensional maps of protein expression within infected animals was accomplished by combining proteomic information obtained by MALDI IMS with spatial information obtained by MRI. Mice were infected as described above and imaged using MRI. The kidney is prone to deformation when the mouse is moved between modalities complicating co-registration of kidney images. Therefore, we rigidly fixed the mouse’s body using wooden splints prior to MR imaging. Following MRI, mice were sacrificed and flash frozen in a mix of dry ice and hexane and then embedded in an ice block to minimize protein degradation and distortion during sectioning. The frozen tissue was sagittally sectioned at 15 μm on a cryomacrotome in the cranial-caudal direction and images were acquired every 30 μm of the blockface section using a digital camera. Images were then reconstructed into a continuous blockface volume through serial registration of each optical image. These blockface images serve as an intermediary for registration of the MALDI data to the previously acquired MR images. The 3D blockface volume can accurately be coregistered to the MR volume and the MALDI data are then inherently registered to the blockface images via rigid section collection using the tape transfer system.
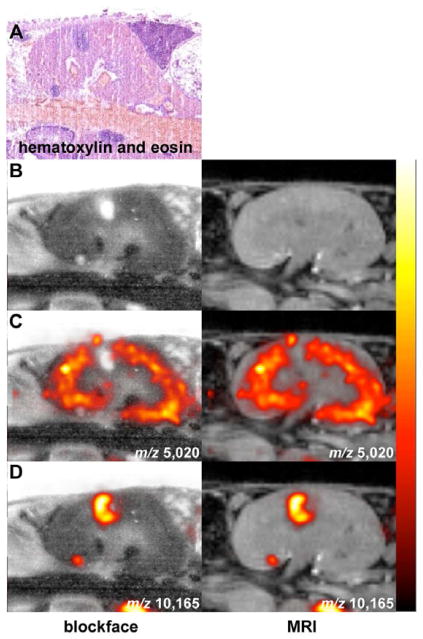
During sectioning, individual sections were collected and prepared for MALDI IMS. Approximately 40 sections were collected, every 210 μm, in the anterior to posterior direction encompassing the abscessed kidney. Tissue samples were prepared using previously described methods and acquired on a MALDI time-of-flight mass spectrometer (Reyzer et al., 2010). Following image acquisition, the mass spectral data were processed for spectral smoothing and base-line correction in MATLAB software (Andersson et al., 2008). Image overlays revealed that calprotectin (m/z 10,165) was localized precisely at the site of abscess formation as visualized using hematoxylin and eosin, blockface, or MRI (Figure 4A, B, D). Conversely, truncated alpha-globin (m/z 5,020), a kidney cortex protein that is not impacted by infection status (Figure 3), was excluded from the abscess (Figure 4C). These data establish the integration of MRI and MALDI IMS as a strategy for imaging inflammatory protein expression at sites of infection.
Figure 4. Co-registered MALDI IMS and MRI reveals proteins that exhibit disease state distribution patterns.
(A) Hematoxylin and eosin stained section showing the presence of kidney abscesses. (B–D) Blockface image (left) and MRI (right) co-registered with MALDI IMS signals at (C) m/z 5,020 (alpha-globin), or (D) m/z 10,165 (calgranulin A). The data are presented as arbitrary units of intensity from 0 (dark red) to 1 (white).
The inflammatory response to infection imaged in three dimensions
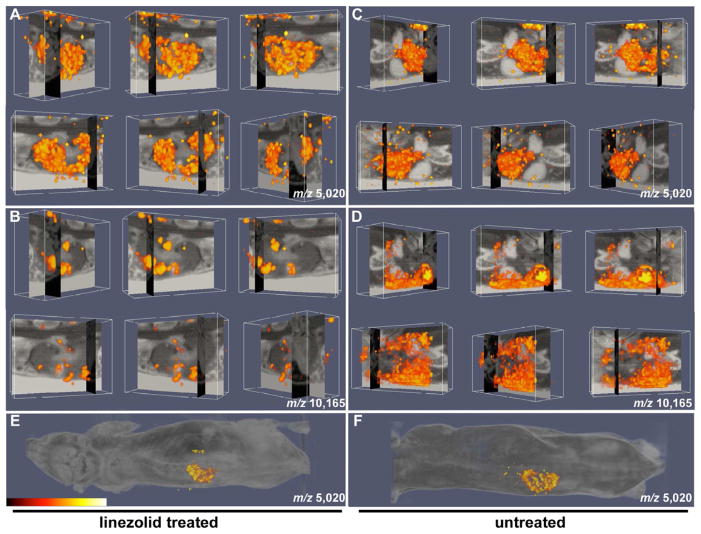
To recreate the spatially resolved three-dimensional volume reconstruction of MALDI IMS images, a series of postprocessing steps was carried out. To cross-correlate the three dimensional MALDI IMS and MRI images, we rigidly co-registered the blockface volume to the magnetic resonance volume of the mouse’s midsection. We generated ion volumes for two proteins, calprotectin (m/z 10,165) and truncated alpha-globin (m/z 5,020), in S. aureus infected mice. We created images for both the untreated and linezolid treated animals in an effort to determine how modulation of inflammation impacts protein abundance and distribution. In these experiments, a concentration of linezolid was utilized that reduces bacterial load and inflammation while still allowing for the formation of visible abscesses. Following linezolid treatment, we found substantial overlap between the sites of kidney abscess formation as determined by MRI, and the abundance of calprotectin as determined by MALDI IMS (Figure 5B and Movie S1). In fact, calprotectin was only detectable within kidney abscesses, and not in the surrounding healthy tissue. In the untreated infected animals, kidney morphology was severely deformed and abscess architecture was difficult to discern. However, significant calprotectin signal could be detected through the infected organ at this time point suggesting the presence of a robust inflammatory response (Figure 5D and Movie S2). Consistent with the reduction of inflammation imparted by linezolid, more calprotectin was detected in the kidneys of untreated animals as compared to treated animals. Additionally, the signal associated with truncated alpha-globin (m/z 5,020) was confined to the cortex and not detectable within the abscess in either the linezolid-treated or untreated mice (Figure 5A and C and Movies S3 and S4). Figures 5E and F show the signal for truncated alpha-globin (m/z 5,020) from linezolid treated and untreated mice superimposed over a whole animal image to provide anatomical orientation. Together, these findings establish three-dimensional co-registration of MALDI IMS and MRI data as a valuable tool for the study of the inflammatory response to infection.
Figure 5. Three-dimensional integration of MALDI IMS and MRI for imaging the inflammatory response to infection.
(A–B) Orthogonal blockface and MRI slice data of linezolid-treated mouse with overlaid (A) alpha-globin protein density (m/z 5,020) and (B) calgranulin A protein density (m/z 10,165) volume renderings. (C–D) Orthogonal blockface and MRI slice data of untreated mouse with overlaid (C) alpha-globin protein density (m/z 5,020) and (D) calgranulin A protein density (m/z 10,165) volume renderings. (E and F) Protein density (m/z 5,020) from (E) linezolid-treated and (F) untreated mice superimposed on whole mouse image. The data in all panels are presented as arbitrary units of intensity from 0 (dark red) to 1 (white). See also Supplemental Movies S1–4.
DISCUSSION
To our knowledge, no methods currently exist to study the whole animal inflammatory response to infection without loss of spatial information. Systemic pathogens such as S. aureus are capable of colonizing virtually every organ, necessitating an organism-wide response to infection. Although the immune response to infection can reach every major organ system, organ-specific defense strategies are critical to protect against microbial challenge. Cellular and cytokine analyses have provided valuable information regarding the immune factors that respond to infection; however, these techniques typically are done on single organ systems and require tissue homogenization, which results in loss of spatial information. Similarly, histological analysis of tissue sections is useful for two-dimensional analysis of infected tissue, but no three-dimensional information is obtained. Finally, the analysis of individual inflammatory proteins requires the availability of antibodies specific for the protein of interest, severely limiting the panel of proteins that can be analyzed. Simply stated, one has to know that a protein exists and have reagents that recognize it before one can study it. We have circumvented each of these shortcomings through the application of integrated MALDI IMS and MRI to an animal model of infectious disease. Although we applied this method to a murine model of S. aureus infection, this technique should be widely applicable to numerous inflammatory diseases with both infectious and non-infectious etiology.
Using this approach, we have revealed multiple protein masses with altered abundance in response to systemic staphylococcal infection in an organ specific manner. While the identities of many of these proteins remain unknown, they may represent important factors involved in the immune response to infection. Proteins that dramatically change abundance in response to infection may play critical roles in the host-pathogen interaction and hence make valuable targets for the development of therapeutics that manipulate the host response. This discovery-based application has the potential to uncover proteins involved in defense against infection. Conversely, the technique described here is equally powerful when studying the distribution of known proteins. The whole animal distribution of calprotectin in systemically infected mice establishes this protein as an excellent marker of both abscess formation and neutrophil development in the bone marrow. Calprotectin is currently used as a diagnostic marker of inflammatory bowel disease (Lewis, 2011). Our experiments extend the potential utility of calprotectin as a marker of infectious disease and inflammation to sites beyond the gastrointestinal tract. Together, these results demonstrate the significant potential that integrated MALDI IMS has to infectious disease research by enabling a systems biology approach to the study of inflammation.
EXPERIMENTAL PROCEDURES
Animal infections
All animal experiments were approved by the Vanderbilt University Institutional Animal Care and Use Committee. Six to eight-weeks old BALB/c mice were infected retro-orbitally with approximately 1×107 CFU of S. aureus wildtype strain Newman. Ninety-six hours post-infection, mice were moved into new cages and divided into two groups; one group was supplied with plain distilled water for drinking and the other group was supplied with distilled water to which linezolid was added to a final concentration of 0.2 mg/ml. The water bottles were wrapped with aluminum foil in order to protect linezolid from light and the bottles were changed every 48 hours. Mice were monitored for another 96 hours and their food was sprinkled each day with either plain water or the linezolid solution to ensure continuous administration of the antibiotic. For IMS analysis, mice were imaged with MRI at day 8 post infection, the mouse was euthanized, and the whole mouse was then flash frozen in dry ice/hexane mixture and kept at −80°C until IMS analysis. For histological analysis, infected kidneys were formalin fixed and paraffin-embedded by the Vanderbilt immunohistopathology core, and stained with H&E in the mass spectrometry research center.
Fluorescently activated cell sorting (FACS)
For the FACS experiments, at day 8 post-infection mice were euthanized with CO2, kidneys were removed, imaged, homogenized, and pushed through a 70 μm cell strainer. RBCs were lysed and the kidney cells were washed and re-suspended in FACS staining buffer (0.01% BSA in PBS, 20% mouse serum). Cells were then stained with anti-CD11b- PerCB, Ly6c (anti-Gr1)-FITC, anti-B220 (B cell marker)-PE, and anti-F4/80 (macrophage marker)-APC. Cells were immediately analyzed by flow cytometry. Forward and side scatters were used to exclude dead and clumped cells. Cells positively stained with both anti-CD11b and anti-Gr1 were marked as neutrophils while those positively stained with anti-CD11b and anti-F4/80 were marked as macrophages.
Preparation of mice for MALDI IMS
Following euthanasia and freezing in a mix of dry ice and hexane, mice were positioned on a block of paraffin wax which was placed on a large chuck. A freezing frame was applied and the setup was placed in a −80°C freezer. After approximately thirty minutes, chilled water was added to the frame to fully submerge the animal and the block was left to freeze for 24 hours. This embedding allowed for more rigid section collection free from distortions. The freezing frame was removed and the ice block was placed in the cryomacrotome for 24 hours to warm to approximately −20°C.
The mouse was sectioned sagittally on a Leica 3600 Cryomacrotome at 15 μm thickness. Pictures were taken of the blockface every 30 μm using a Cannon EOS 20D digital camera mounted on a frame above the crytotome. Two serial sections were collected every 210 μm using a macro-tape transfer system (Instrumedics) and transferred onto indium-tin oxide (ITO) coated glass slides for imaging mass spectrometry (IMS) and standard histology slides for hematoxylin and eosin staining. Approximately 40 sections were collected from each animal. Each glass histology slide was stained with hematoxylin and eosin using standard protocols. The slides were scanned using a MiraxScan Digital Slide Scanner (Zeiss, Budapest, Hungary) at 0.23 μm pixel resolution and the images were exported to JPEG format for future reference.
The ITO slides were washed with 70%, 90%, and 95% ethanol to remove salts and lipids, and excess transfer polymer in order to produce better signal during MALDI IMS analysis. The regions on each MALDI target surrounding the kidneys were spotted with a 20 mg/mL sinapinic acid matrix solution (in 50:50 water and acetonitrile with 0.1% TFA added) using a Portrait 630 (Labcyte, Sunnyvale, CA) Acoustic Robotic Microspotter. Matrix was applied in flyby mode in a block pattern (68 columns and 48 rows) at 210 μm resolution consisting of six passes of five drops for a total of 30 drops.
Mass spectrometry
Spotted regions of tissue were imaged using a Bruker Autoflex Speed Mass Spectrometer (Billerica, MA). The mass spectrometer was run in linear positive ion mode with a detection voltage of 2.745 kV. Spectra were collected from each matrix spot as a sum of 400 laser shots, in 50 shot steps, with a laser repetition rate of 1000 Hz, ion source voltages of 19.5 kV (1) and 18.1 kV (2), a delay time of 200 nsec, and a lens voltage of 6 kV. The mass range measured was from 4,000 to 20,000 Da. The spectra were analyzed using FlexImaging software in order to establish the location of prominent proteins, including calprotectin, within the tissue.
Magnetic Resonance Imaging
Six female mice were anesthetized via inhalation of 2%/98% isoflurane/oxygen. Animals were secured in a prone position in a 38-mm inner diameter radiofrequency (RF) coil and placed in a Varian 9.4T horizontal bore imaging system (Varian Inc, Palo Alto, CA) for data collection. Respiration rate and internal body temperature were continuously monitored. A constant body temperature of 37°C was maintained using heated air flow.
On each day of imaging, multislice scout images were collected in all three imaging planes for localization of the kidneys, using a gradient echo sequence with repetition time (TR) = 75 ms, echo time (TE) = 5 ms, slice thickness = 2 mm, flip angle = 35°, and an average of 4 acquisitions. Additional parameters include field of view (FOV) = 32 mm × 32 mm and data matrix = 128 × 128.
Following localization of the kidneys, three-dimensional gradient echo imaging data was acquired with FOV = 25.6 mm × 25.6 mm × 25.6 mm, TR = 15 ms, TE = 7.5 ms, data matrix = 128×128×128, with 8 acquisitions per phase encode step, for a total acquisition time of approximately 33 minutes per animal. The acquired data were zero padded and reconstructed using Matlab 2010a (The Mathworks, Inc., Natick, MA) at 256×256×256 matrix using an inverse Fourier transform, resulting in a nominal isotropic resolution of 100 μm.
Blockface Reconstruction
Collecting tissue sections on the macrotome while acquiring slice-by-slice blockface data does not provide intrinsically registered blockface images. After these data are collected, the misalignment between consecutive images must be corrected using an inter-slice image registration. For the registration process, we implemented a method optimizing a normalized mutual information metric through a 2D rigid-body transformation (Viola, 1997). As the central blockface images contained the largest section of mouse, we began the registration process using the median slice. Moving back toward the first slice, the successive image was aligned to the median image. The resulting registered image was used as the reference for the next successive image. This process continued iteratively until the first image was aligned. The procedure was repeated for the second half of the image beginning at the median slice and moving toward the last image. Once all individual blockface images were aligned they were combined into one three-dimensional reconstructed blockface image. These data were coregistered with the MR data, aligning the MR, blockface, and MALDI data in a single coordinated system.
MALDI data processing
A series of post-processing steps were taken to create three-dimensional spatially resolved MALDI data with a known transform to the blockface data coordinate system. Raw MALDI data were processed for baseline correction and spectral smoothing using the MATLAB Bioinformatic toolbox (The Mathworks, Inc., Natick, MA). Since the data were collected in a regular grid (210 μm × 210 μm spacing), two-dimensional spatially resolved MALDI data were immediately available. To create three-dimensional spatially resolved MALDI data each section of data were registered to their corresponding blockface slice. The transform was calculated by manually adjusting translation and in-plane rotation values to align MALDI data with their sampling locations using the section’s spotting image as a reference. These three-dimensional spatially resolved MALDI data allow the creation of ion volumes for specified protein(s).
MR Registration
Since the mice were rigidly fixed between MRI acquisitions and freezing, a 6 degrees-of-freedom rigid body normalized mutual information based registration was used to align the MR data to blockface data. After alignment, the MR data, blockface data, and MALDI data are known in a single coordinate system allowing data to be compared on a voxel-by-voxel basis.
Tissue Homogenization for Protein Identification
Frozen uninfected control kidney was dissected to obtain only cortex. Approximately 70 mg of tissue was collected and transferred to a small glass tissue homogenizer. To this, 1 mL of T-PER (Pierce, Rockford, IL) was added and the tissue was homogenized. The homogenate was transferred to a Falcon tube and further disrupted with 5 pulses of a sonic dismembrator (Thermo Fisher Scientific, Suwanee, GA). The homogenate was centrifuged for 10 minutes at 16,000 g to pellet cellular debris and the supernatant collected in a clean tube. A Bradford Assay for protein concentration was conducted using a SpectraMax M2e Spectrophotometer (Molecular Devices, Sunnyvale, CA) and found to be 3559 μg/mL.
HPLC Separation for Protein Identification
A 300 μL aliquot of the protein extract was diluted to 900 μL using 98:2 water:ACN with 0.1% TFA and spun for 5 minutes at 16,000 g to pellet any precipitate. Chromatography was carried out using a Waters Alliance HPLC system (Milford, MA) configured with a 2690 separations module and a 2487 dual wavelength absorbance detector. Samples were fractionated on a Vydac (Hesperia, CA) 208TP5315 C8 reversed-phase column (5 μm particle size, 3.2 mm × 150 mm) at 40 °C. The analytical column was fitted with a Vydac 208GD52 C8 reversed phase guard column (5 μm, 2.1 mm × 10 mm). Mobile phase solvents A and B were 0.1% TFA (v/v) in water and 0.1% TFA (v/v) in acetonitrile, respectively. Proteins present in the homogenate were fractionated at a flow rate of 0.5 mL/min using linear gradients and the following program: 5% B for 10min, 5 to 25% B over 5 min, 25 to 60% B over 50 min, 60 to 95% B over 10 min and hold for 5 min. The mobile phase was ramped back to the initial conditions and the column allowed to re-equilibrate. Chromatographic effluent was monitored using ultraviolet (UV) detection at both 214 and 280 nm. Fractions were collected at 1-minute intervals (0.5 mL each) into a 96-well format microplate and stored at −80 °C until use. Fractions were evaporated to dryness using a speedvac (Thermo Fisher Scientific).
Mass Spectral Analysis for Protein Identification
Dried HPLC fractions were reconstituted using 30 μL of 40:60 acetonitrile:water containing 0.1% TFA and spotted to a MALDI target plate with an equal volume of 20 mg/mL sinapinic acid (SA) in 40% ACN with 0.1% TFA. Spectra were collected in linear positive ion mode on a Bruker Autoflex Speed mass spectrometer (Billerica, MA) to determine the fraction(s) containing masses of interest. A mixture of standard proteins containing bovine insulin (Mr 5733.6), cytochrome C (horse heart, Mr 12360.2), apomyoglobin (horse, Mr 16951.5) and trypsinogen (bovine pancreas, calculated Mr 23981) along with SA matrix was spotted onto the MALDI target for external mass calibration. Spectra were evaluated using FlexAnalysis (Bruker) and 3 different fractions were found to contain peaks within 5 Da of [M+H]+ 5020. To determine which of these corresponded to the imaged protein of interest, accurate masses were obtained from a Bruker SolariX 9.4T FTMS for the fractions as well as a tissue section. The accurate monoisotopic mass of the protein from the tissue was found to be 5016.49 Da. The HPLC fraction (32 min.) corresponding to this mass was further processed.
1D Gel Analysis for Protein Identification
The fraction of interest from 2 HPLC runs were combined and taken to dryness and reconstituted in water. The fraction was separated on a NuPage 4–12% Bis-Tris (Invitrogen, Carlsbad, CA) gel and bands corresponding to the mass range 3–6 kDa were cut out. Gel bands were subjected to in-gel tryptic digestion, and the tryptic peptides were extracted from the gel with 60% ACN with 0.1% formic acid. The extract was taken to dryness in a speedvac and reconstituted in 15 μL of 0.1% formic acid. Peptides were analyzed via LC-MS/MS, similar to that described below (in the following experimental section), except that collision-induced dissociation (CID) was used for peptide fragmentation. A total of 7 proteins were identified, which consisted of beta-globin, cntm_P13645, fructose-bisphosphate aldolase, alpha-globin, ATP synthase subunit epsilon, 10kDa heat shock protein, alcohol dehydrogenase, and myotrophin. None of these had a molecular weight of 5016.49 Da and therefore could not be identified as the protein of interest therefore the following top-down approach was performed.
Identification of Truncated Alpha-Globin (residues 2–47)
The remainder of the enriched fraction was then speedvac concentrated, and reconstituted in 0.1% formic acid. A 50%-aliquot of the reconstituted fraction was loaded onto a reversed-phase capillary trap column using a helium-pressurized cell (pressure bomb). The trap column (360μm × 100μm ID) was fitted with a filter end-fitting (IDEX Health & Science, Oak Harbor, WA), and was packed with 4 cm of Jupiter C18 material (5 μm, 300Å, Phenomenex, Torrance, CA). Once the sample was loaded, the trap column was connected using an M-520 microfilter union (IDEX Health & Science) to an analytical column (360μm × 100μm ID), equipped with a laser-pulled emitter tip and packed with 18cm of C18 reversed-phase material (Jupiter, 3μm beads, 300Å, Phenomenex). Using an Eksigent NanoLC Ultra HPLC, peptides were gradient-eluted at a flow rate of 400 nL/min, and the mobile phase solvents consisted of 0.1% formic acid, 99.9% water (solvent A) and 0.1% formic acid, 99.9% acetonitrile (solvent B). The gradient consisted of 2–35 %B in 40 min, followed by 35–90 %B in 10 min. Upon gradient-elution, proteins were mass analyzed on a LTQ Orbitrap Velos mass spectrometer, equipped with a nanoelectrospray ionization source (Thermo Scientific, San Jose, CA). The instrument was operated using targeted methods to enable analysis of low abundant proteins of interest, corresponding to protein species imaged in IMS experiments. For all LC-MS/MS analyses, full scan (m/z 400–2000) spectra were acquired with the Orbitrap as the mass analyzer (resolution 60,000) as the initial scan event. Subsequent scan events consisted of targeted MS/MS of specific m/z values corresponding to the protein of interest. Electron transfer dissociation (ETD) was used to fragment the intact protein species, and ETD MS/MS spectra were acquired consecutively using the ion trap followed by the Orbitrap as the mass analyzer. An isolation width of 2 m/z and an activation time of 70 ms were used for ETD MS/MS. The MSn AGC target value in the ion trap was set to 2e4, with a maximum injection time of 125 ms. For MS/MS spectra collected in the Orbitrap, the MSn AGC target was 8e5, the maximum injection time was set to 750 ms, and spectra were collected with resolving powers of either 7,500 or 30,000. The ETD reagent ion AGC target was 3e6 with a maximum inject time of 500 ms. To identify the truncated alpha-globin protein (residues 2–47, MW 5016, gi 122441), the [M+6H]+6 ion with m/z 837.43 was targeted for ETD MS/MS, and the resulting spectra were de novo sequenced. These data were in agreement with the identification of alpha-globin from the 1D gel band.
Supplementary Material
HIGHLIGHTS.
IMS provides a whole-animal view of the inflammatory response to infection.
IMS identifies protein masses that are abundant at sites of inflammation.
Anatomic information at the site of infection can be generated by MRI.
Integrated IMS and MRI enable a 3-D view of the host-pathogen interaction.
Acknowledgments
We would like to thank the members of the Skaar laboratory for critical reading of the manuscript. We also thank Jeffrey Spraggins for help determining accurate mass on the FT-ICR MS as well as David Friedman, Sarah Stuart and Hayes McDonald for identification of proteins from in-gel digestion. This research was supported by the United States Public Health Service Grants AI69233, AI091771 and AI073843 from the National Institute of Allergy and Infectious Diseases, GM 58008 and EB000214 from the National Institute of General Medical Sciences, and a Pfizer 2009 ASPIRE research award. EPS is a Burroughs Wellcome Fellow in the Pathogenesis of Infectious Diseases.
Footnotes
The authors have no conflicts of interest to report.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersson M, Groseclose MR, Deutch AY, Caprioli RM. Imaging mass spectrometry of proteins and peptides: 3D volume reconstruction. Nature Methods. 2008;5:101–108. doi: 10.1038/nmeth1145. [DOI] [PubMed] [Google Scholar]
- Bernardo K, Pakulat N, Fleer S, Schnaith A, Utermohlen O, Krut O, Muller S, Kronke M. Subinhibitory concentrations of linezolid reduce Staphylococcus aureus virulence factor expression. Antimicrob Agents Chemother. 2004;48:546–555. doi: 10.1128/AAC.48.2.546-555.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AG, Kim HK, Burts ML, Krausz T, Schneewind O, Missiakas DM. Genetic requirements for Staphylococcus aureus abscess formation and persistence in host tissues. FASEB J. 2009;23:3393–3404. doi: 10.1096/fj.09-135467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, Torres VJ, Anderson KL, Dattilo BM, Dunman PM, Gerads R, et al. Metal Chelation and Inhibition of Bacterial Growth in Tissue Abscesses. Science. 2008;319:962–965. doi: 10.1126/science.1152449. [DOI] [PubMed] [Google Scholar]
- DeLeo FR, Otto M, Kreiswirth BN, Chambers HF. Community-associated meticillin-resistant Staphylococcus aureus. The Lancet. 2010;375:1557–1568. doi: 10.1016/S0140-6736(09)61999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Roca P, Mancilla-Ramirez J, Santos-Segura A, Fernandez-Aviles M, Calderon-Jaimes E. Linezolid diminishes inflammatory cytokine production from human peripheral blood mononuclear cells. Arch Med Res. 2006;37:31–35. doi: 10.1016/j.arcmed.2005.05.022. [DOI] [PubMed] [Google Scholar]
- Hertlein T, Sturm V, Kircher S, Basse-Lusebrink T, Haddad D, Ohlsen K, Jakob P. Visualization of abscess formation in a murine thigh infection model of Staphylococcus aureus by 19F-magnetic resonance imaging (MRI) PLoS ONE. 2011;6:e18246. doi: 10.1371/journal.pone.0018246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatib-Shahidi S, Andersson M, Herman JL, Gillespie TA, Caprioli RM. Direct molecular analysis of whole-body animal tissue sections by imaging MALDI mass spectrometry. Anal Chem. 2006;78:6448–6456. doi: 10.1021/ac060788p. [DOI] [PubMed] [Google Scholar]
- Lewis JD. The utility of biomarkers in the diagnosis and therapy of inflammatory bowel disease. Gastroenterology. 2011;140:1817–1826. e1812. doi: 10.1053/j.gastro.2010.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- McDonnell LA, Heeren RM. Imaging mass spectrometry. Mass Spectrom Rev. 2007;26:606–643. doi: 10.1002/mas.20124. [DOI] [PubMed] [Google Scholar]
- Reyzer ML, Chaurand P, Angel PM, Caprioli RM. Direct molecular analysis of whole-body animal tissue sections by MALDI imaging mass spectrometry. Methods Mol Biol. 2010;656:285–301. doi: 10.1007/978-1-60761-746-4_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg A, Jensen KS, Baudoux P, Van Bambeke F, Tulkens PM, Frimodt-Moller N. Intra- and extracellular activity of linezolid against Staphylococcus aureus in vivo and in vitro. J Antimicrob Chemother. 2010;65:962–973. doi: 10.1093/jac/dkq052. [DOI] [PubMed] [Google Scholar]
- Seeley EH, Caprioli RM. Molecular imaging of proteins in tissues by mass spectrometry. Proc Natl Acad Sci U S A. 2008;105:18126–18131. doi: 10.1073/pnas.0801374105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley EH, Schwamborn K, Caprioli RM. Imaging of intact tissue sections: moving beyond the microscope. J Biol Chem. 2011;286:25459–25466. doi: 10.1074/jbc.R111.225854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha TK, Khatib-Shahidi S, Yankeelov TE, Mapara K, Ehtesham M, Cornett DS, Dawant BM, Caprioli RM, Gore JC. Integrating spatially resolved three-dimensional MALDI IMS with in vivo magnetic resonance imaging. Nature Methods. 2008;5:57–59. doi: 10.1038/nmeth1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbakk M, Naess-Andresen CF, Lingaas E, Dale I, Brandtzaeg P, Fagerhol MK. Antimicrobial actions of calcium binding leucocyte L1 protein, calprotectin. Lancet. 1990;336:763–765. doi: 10.1016/0140-6736(90)93237-j. [DOI] [PubMed] [Google Scholar]
- Urban CF, Ermert D, Schmid M, Abu-Abed U, Goosmann C, Nacken W, Brinkmann V, Jungblut PR, Zychlinsky A. Neutrophil extracellular traps contain calprotectin, a cytosolic protein complex involved in host defense against Candida albicans. PLoS Pathog. 2009;5:e1000639. doi: 10.1371/journal.ppat.1000639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viola PaWIW. Alignment by maximization of mutual information. International Journal of Computer Vision. 1997;24:137–154. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.