Abstract
Listeria monocytogenes is a human, food-borne pathogen. Genomic comparisons between L. monocytogenes and Listeria innocua, a closely related non-pathogenic species, were pivotal in the identification of protein-coding genes essential for virulence. However, no comprehensive comparison has focused on the non-coding genome. We used strand-specific cDNA sequencing to produce genome-wide transcription start site maps for both organisms, and developed a publicly available integrative browser to visualize and analyze both transcriptomes in different growth conditions and genetic backgrounds. Our data revealed conservation across most transcripts, but significant divergence between the species in a subset of non-coding RNAs. In L. monocytogenes, we identified 113 small RNAs (33 novel) and 70 antisense RNAs (53 novel), significantly increasing the repertoire of ncRNAs in this species. Remarkably, we identified a class of long antisense transcripts (lasRNAs) that overlap one gene while also serving as the 5′ UTR of the adjacent divergent gene. Experimental evidence suggests that lasRNAs transcription inhibits expression of one operon while activating the expression of another. Such a lasRNA/operon structure, that we named ‘excludon’, might represent a novel form of regulation in bacteria.
Keywords: comparative genomics, Listeria monocytogenes , RNA-seq, transcriptome, TSS map
Introduction
Listeria monocytogenes is a ubiquitous food-borne pathogen that can be silently present in the gastro-intestinal tract of healthy humans. However, in immunocompromised individuals, newborns, elderly, or pregnant women, it may lead to a severe disease with clinical features varying from septicemia to meningitis, meningo-encephalitis, and abortion. Due to its remarkable abilities to exploit basic cellular functions, evade host defense mechanisms, and cross host barriers, this organism has become a model for studying host–pathogen interactions (Hamon et al, 2006; Pizarro-Cerda and Cossart, 2006; Cossart and Toledo-Arana, 2008; Cossart, 2011). The highly sophisticated virulence arsenal of L. monocytogenes consists of a relatively small number of critical genes, and it was at first estimated that merely 1.6% of the L. monocytogenes genes contributes to pathogenicity (Glaser et al, 2001; Doumith et al, 2004; Yang et al, 2008). However, recent and ongoing studies demonstrate that this number is much higher, with increasing discoveries of novel genes that contribute at different levels to the virulence process and survival within the host.
Listeria innocua, a close relative of L. monocytogenes, is present in the same environmental niches as L. monocytogenes, but is non-pathogenic. This organism shares 2523 orthologous genes with L. monocytogenes, representing 88.4% of L. monocytogenes protein-coding genes (Glaser et al, 2001). The striking similarity between the genomes and lifestyles of these two species enabled comparative genomics approaches to investigate L. monocytogenes-specific genes that might be involved in virulence (Glaser et al, 2001; Doumith et al, 2004; Hain et al, 2006a, 2006b). Such comparisons led to the discovery of key virulence-related genes in L. monocytogenes (Cossart and Toledo-Arana, 2008; Dussurget, 2008; Cossart, 2011).
Regulatory RNAs (small RNAs and antisense RNAs; sRNAs and asRNAs, respectively) are now established as key regulators of virulence in diverse bacteria (Heroven et al, 2008; Toledo-Arana et al, 2009; Papenfort and Vogel, 2010; Felden et al, 2011) but, in contrast to the significant advances in identifying protein virulence factors, relatively little is known about the role of regulatory RNAs in L. monocytogenes pathogenesis (Mellin and Cossart, 2012). Valuable information on the L. monocytogenes non-coding genome was revealed by three recent transcriptomic studies, including tiling-array analyses of the strain EGD-e in saprophytic and pathogenic conditions (Toledo-Arana et al, 2009), Illumina high-throughput transcriptome sequencing of strain 10403S (Oliver et al, 2009), and a 454 RNA sequencing study of L. monocytogenes EGD-e grown in macrophages (Mraheil et al, 2011). To date, 101 sRNAs have been identified and annotated in the L. monocytogenes genome, and several have been found to be involved in virulence. These include the three sRNAs Rli31, Rli33-2, and Rli50 that, when deleted, led to an attenuated virulence phenotype in murine macrophage infection studies as well as in mouse and butterfly larvae infection models (Mraheil et al, 2011). A deletion mutant of Rli38 resulted in an attenuated virulence phenotype in orally inoculated mice, demonstrating a role for Rli38 in infection (Toledo-Arana et al, 2009). In addition, an asRNA that overlaps three flagellar biosynthesis genes was found to regulate flagellar biosynthesis (Toledo-Arana et al, 2009). Overall, the Listeria non-coding genome is gaining recognition as an important contributor to the regulation of virulence.
In contrast to the approach that identified L. monocytogenes-specific protein-coding genes by direct genomic comparisons, identification of species-specific sRNAs using purely computational predictions is challenging (Livny and Waldor, 2007; Pichon and Felden, 2008). First, the evolutionary patterns of sRNAs differ from those of protein-coding genes, impeding our ability to estimate conservation using sequence alignments (Gottesman, 2005). Second, conservation of asRNAs that overlap protein-coding genes is not trivial to detect computationally (Kulkarni and Kulkarni, 2007). Finally, even if a regulatory RNA is conserved at the DNA sequence level, its expression pattern might not be conserved.
To better study the Listeria non-coding genomes, we have undertaken a comparative transcriptomic approach that compares the expression of non-coding RNAs in L. monocytogenes and L. innocua. As part of this effort, we have globally mapped the transcription start sites (TSSs) of L. monocytogenes and L. innocua in a genome-wide manner for cells grown under an assortment of conditions. Furthermore, we have developed a publicly available genome browser to analyze and visualize our results with the data of three additional studies (Oliver et al, 2009; Toledo-Arana et al, 2009; Mraheil et al, 2011). We have then validated several of our observations and interpretations by northern blot or qRT–PCR.
Our study is the first to compare two bacterial transcriptomes at a single-base resolution. It has led to the discovery of 33 new sRNAs and 53 new asRNAs in L. monocytogenes. Strikingly, some of the sRNAs and asRNAs were found to be conserved between the two genomes at the DNA level, but not expressed in one species or the other, at least in the experimental conditions used in this study, highlighting an additional layer of divergence between these two organisms. Interestingly, we found new asRNAs that are extremely long (lasRNA) encompassing the coding sequence of the neighboring divergent genes that might represent a novel class of regulatory RNAs, where an RNA downregulates expression of one set of genes and increases the expression of another, which has an opposite or mutually exclusive function, suggesting a new mode of gene regulation in bacteria.
Results
General strategy
Although several recent studies have provided valuable transcriptomic information on L. monocytogenes (Toledo-Arana et al, 2009; Mraheil et al, 2011), no study to date has examined the non-coding genome of L. innocua and no study has undertaken a comparison of the two species to determine conservation and divergence of the virulence-related non-coding genome. Furthermore, genome-wide TSS maps, which allow interrogation of transcriptional activity with single-nucleotide resolution (Wurtzel et al, 2010b), have not been established for either organism.
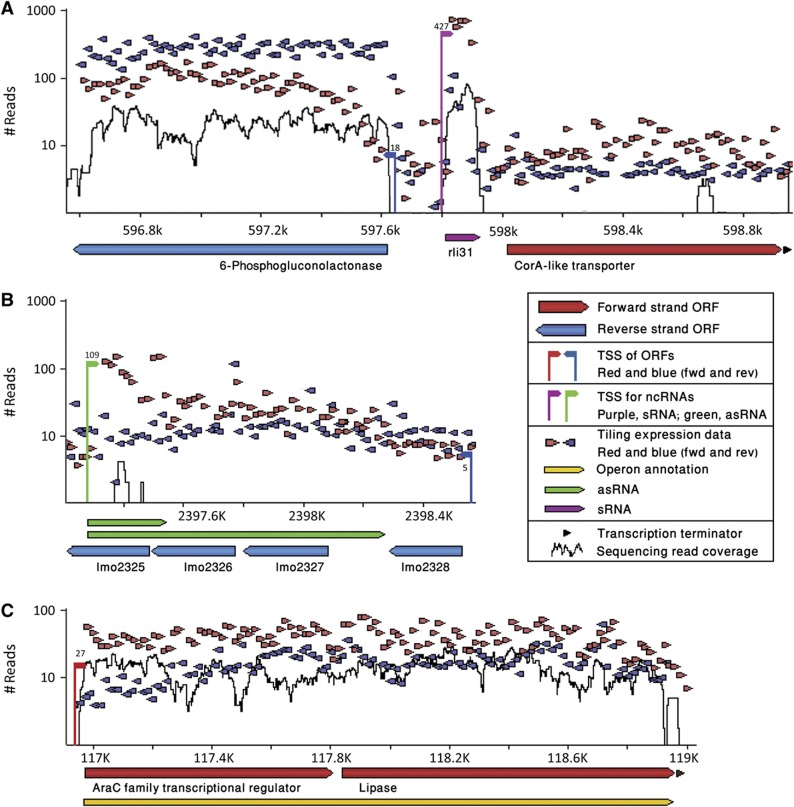
In this study, we first produced genome-wide TSS maps for both organisms. We then centralized all available Listeria transcriptome data, by generating a single, unified web repository that visualizes transcriptome information, the Listeria transcriptome browser. This browser displays a multilayered view of both tiling-array (Toledo-Arana et al, 2009) and deep sequencing transcriptomic data (Oliver et al, 2009; Mraheil et al, 2011), including the TSS maps generated in this study, along with genome annotations (Glaser et al, 2001), RNA-family data (RFAM; Gardner et al, 2011), and terminators predictions (Kingsford et al, 2007) for both organisms (Figure 1; available at http://www.weizmann.ac.il/molgen/Sorek/listeria_browser/). Furthermore, the transcriptome browser allows navigating between the two transcriptomes in synchrony, visually exposing differences and similarities between the two genomes and their transcriptional activity.
Figure 1.
The Listeria transcriptome browser. (A–C) Represent different windows of the genome in the unified browser. X axis represents the position on the genome; y axis the number of cDNA sequences mapped to the genome (log scale). The browser unifies TSS mapping (vertical lines being 5′ ends of genes, with numbers indicating the amount of supporting 5′ end sequencing reads), total RNA sequencing (black line), tiling-array data (red and blue small triangles represent probes on the forward and reverse strands, respectively), genome annotation (arrows underneath the axis; red and blue representing genes on the forward and reverse strands, respectively), operon annotation (yellow arrow), rho-independent terminators prediction (small black arrows), small RNAs (purple), asRNAs (green), as well as additional information (Materials and methods). The browser is available at http://www.weizmann.ac.il/molgen/Sorek/listeria_browser/.
Genome-wide mapping of TSSs in L. monocytogenes and L. innocua
To produce TSS maps for L. monocytogenes and L. innocua, we first isolated RNA from both organisms grown to mid-log and stationary phase at 37°C. Additionally, we studied RNA from L. monocytogenes grown at 30°C and under hypoxic conditions, as well as from L. monocytogenes ΔsigB, and ΔprfA mutants, which are deleted for the two major regulators of stress response and virulence (Cossart, 2011). TSS maps were generated using a sequencing method that relies on selective Tobacco Acid Pyrophosphatase (TAP) treatment and adapter ligation to differentiate between primary transcripts and processed RNAs (Materials and methods; Supplementary Figure S1). We were able to define a total of 2018 TSSs across the L. monocytogenes transcriptome representing 88% of the transcriptional units in this organism (Supplementary Table S1; Materials and methods). These TSSs were inferred from sequencing over 32 million short 5′-end sequencing reads for L. monocytogenes (Supplementary Table S2). Comparison with a set of nine TSSs previously determined by other studies (Loh et al, 2006) validated the accuracy of our TSS determination (Supplementary Table S3). Further analysis of sequences upstream to the determined TSSs revealed the existence of sigma-like consensus promoter motifs in the expected upstream positions (Supplementary Figure S2).
To examine global patterns of conservation between TSSs in L. monocytogenes and L. innocua, we further sequenced 14 million short 5′-end sequencing reads for the L. innocua transcriptome, grown under log and stationary conditions, generating a total of 1670 TSSs (Supplementary Table S1). For the interspecies comparison, we considered only L. monocytogenes TSSs that were assigned to genes conserved in L. innocua, expressed in either log or stationary phases, and transcribed from the same strand as the gene. Out of 1754 such TSSs in L. monocytogenes, 1333 (76%) were also detected in L. innocua. In general, the primary TSSs of genes, represented by TSSs with the highest number of supporting reads preceding defined operons, were highly conserved (981/1114, 88%), while internal TSSs (iTSSs; Mitschke et al, 2011) and predicted alternative TSSs (aTSSs) were much less conserved (352/640, 55%). This might reflect a weaker evolutionary pressure to preserve iTSSs and aTSSs as compared with the dominant TSSs. We note that some of the differences we observed might stem from the fact that we had two-fold less sequencing data for L. innocua; and therefore, the actual conservation in TSSs might be higher. Together, the TSS data reveal strikingly similar expression patterns in L. monocytogenes and L. innocua, albeit with differences which are discussed below.
5′ UTRs in L. monocytogenes and L. innocua
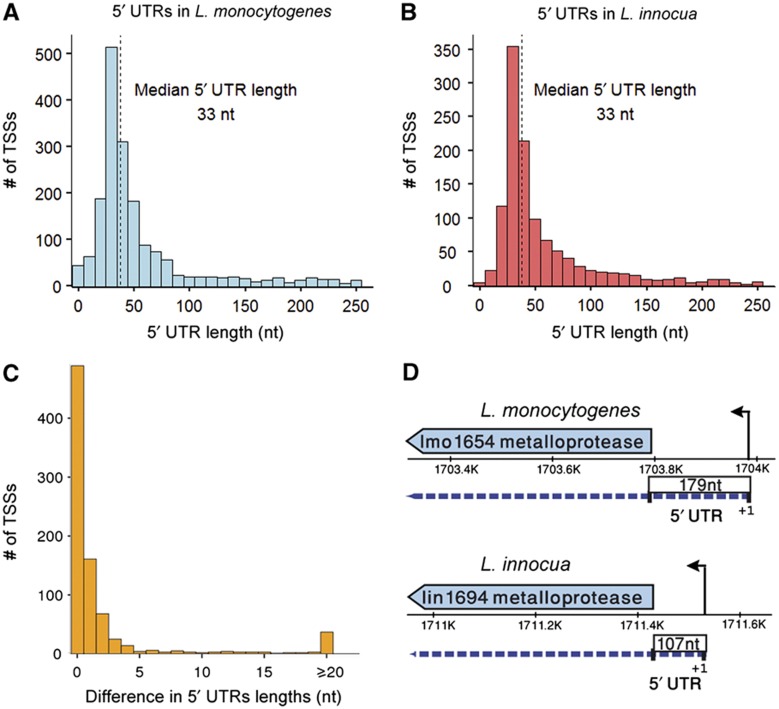
Our mapping of TSSs in L. monocytogenes and L. innocua allowed interrogation of 5′ UTR evolution with a high resolution. As 5′ UTRs can determine translational efficiency of an mRNA as well as a number of post-transcriptional regulatory outcomes (Ingolia et al, 2009; Sharma et al, 2010), we analyzed the similarities and differences between 5′ UTRs lengths in L. monocytogenes and L. innocua. By examining the agreement in primary TSS position relative to the ATG for genes in which a TSS was detected both for L. monocytogenes and for L innocua (Figure 2). In 90.5% of these genes, the TSS was located at the same distance from the ATG in both organisms (±5 nt). However, rare cases (overall 80 genes) of significant differences in 5′ UTR length were recorded (Figure 2C and D), which might lead to changes the regulation of these genes (Loh et al, 2006). For example, the 5′ UTR of L. monocytogenes gene lmo1654, which was recently characterized as a secreted metalloprotease (Desvaux et al, 2010), is preceded by a 179 nt 5′ UTR, while its homolog in L. innocua, lin1694, has a much shorter, 107 nt 5′ UTR (Supplementary Figure S3).
Figure 2.
5′ UTRs in L. monocytogenes and L. innocua. The length distributions of the 5′ UTRs in L. monocytogenes (A) and L. innocua (B) are almost identical, with a median length of 33 nt. The distributions are skewed to the right, representing long 5′ UTRs that include known riboswitches. (C) Divergence of 5′ UTR lengths between L. monocytogenes and L. innocua: the lengths of the 5′ UTRs of genes that have expressed homologs in both organisms were compared. (D) An example for a significant difference in 5′ UTR lengths between homologous genes. The lmo1654 gene is a predicted secreted metalloprotease of L. monocytogenes and has a 179 nt long 5′ UTR. Its homolog in L. innocua, lin1694, has a shorter 5′ UTR of 107 nt.
The distributions of 5′ UTRs lengths were very similar in both organisms, with median length of 33 nucleotides (Figure 2A and B). For the majority of genes in both organisms (87%), 5′ UTRs were shorter than 100 nt. However, the distributions of 5′ UTR lengths showed a subgroup of 101 genes with long 5′ UTRs (>100 nt) that, as expected, included most of the genes with known riboswitches, as well as 10 virulence genes in L. monocytogenes (Toledo-Arana et al, 2009; Figure 2; Supplementary Table S1). This is reminiscent of a similar distribution of 5′ UTR lengths recently described in the related model organism Bacillus subtilis (Irnov et al, 2010).
sRNAs in L. monocytogenes and L. innocua
TSSs in L. monocytogenes intergenic regions were analyzed together with the tiling-array data obtained previously in our laboratory (Toledo-Arana et al, 2009) to detect known and new trans-encoded sRNAs in L. monocytogenes. The criteria used for the novel sRNA annotation were at least 10 reads supporting the transcript presence or reproducibility in at least two of six conditions. In total, 113 intergenic transcripts expressed under the conditions tested were identified in the L. monocytogenes genome. These represented 80 known and 33 new sRNA transcripts, which varied in size from 46 to 596 nt (Table I; Supplementary Table S4). Twenty-one previously identified sRNAs were not detected in this study, possibly due to the differences in growth conditions and techniques used here as compared with previous studies, thus the total number of sRNAs identified in L. monocytogenes so far is 134. Supplementary Figure S4 presents an expression heatmap that includes all new and known sRNAs detected in this study in all tested conditions.
Table 1. Summary of sRNAs and asRNAs found in this study.
| L. monocytogenes | Presence in L. innocua |
|||
|---|---|---|---|---|
| DNA sequence conserved and expressed | DNA sequence conserved but not expressed | DNA sequence absent | ||
| sRNAs | 113a | 74 | 14 | 25 |
| asRNAs | 70b | 36 | 27 | 7 |
aTwenty-one previously identified sRNAs were not detected in this study; thus, the total number of sRNAs in L. monocytogenes is 134.
bSixteen previously identified asRNAs were not detected in this study; thus, the total number of asRNAs in L. monocytogenes is 86.
Interestingly, seven sRNAs, four of which are new, were expressed in the WT strain but not in the ΔsigB strain, suggesting they are directly regulated by SigB (Supplementary Table S4). The SigB dependence of these sRNAs was further supported by the detection of a sigB binding site in the promoters of all seven sRNAs. SigB is a regulator of stress tolerance and has been implicated in virulence (Kazmierczak et al, 2003), suggesting that SigB-regulated sRNAs may contribute to the infectious process.
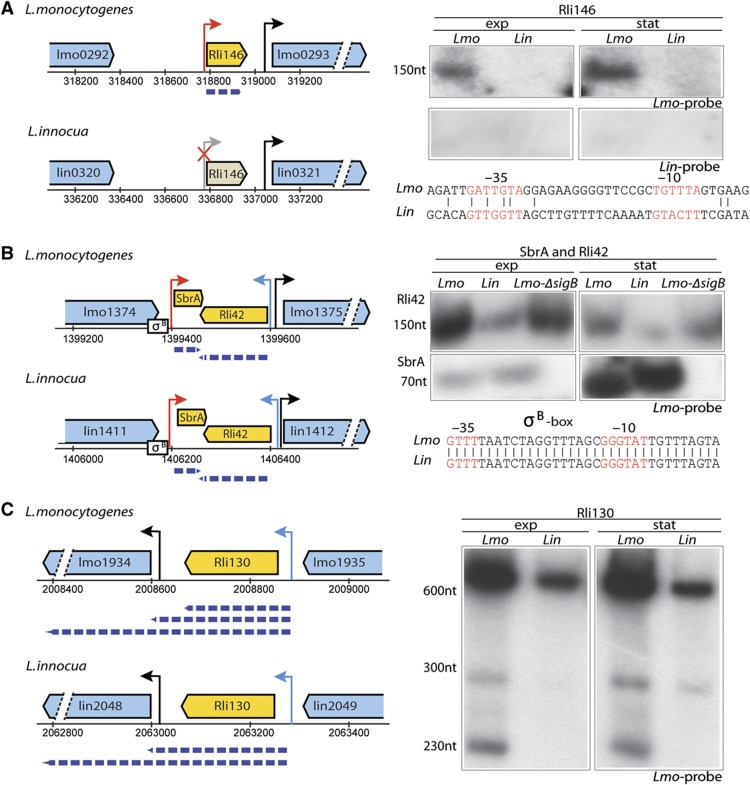
Among the 113 sRNAs identified in L. monocytogenes, 88 sRNAs were transcribed from regions conserved in L. monocytogenes and L. innocua, and 25 were transcribed from regions specific to L. monocytogenes (Table I; Supplementary Table S4). Of the 88 RNAs, 74 were expressed in both species, while the expression of 14 was restricted to L. monocytogenes, possibly reflecting a hidden layer of divergence between the species. Northern blot analyses on two of these sRNAs (Rli146 and Rli87) confirmed their monocytogenes-specific expression patterns. Sequence analysis of their promoter regions showed significant divergence in the Rli146 promoter that might explain its lack of expression in L. innocua (Figure 3A), while only minor differences were identified in the promoter of Rli87 (Supplementary Figure S5A). These results indicate significant divergence of sRNAs between the species, possibly contributing to the differences in pathogenicity between L. monocytogenes and L. innocua (Supplementary Table S4).
Figure 3.
sRNAs in L. monocytogenes and L. innocua. Small RNAs (yellow box-arrows) encoded in the intergenic regions of L. monocytogenes (Lmo) and L. innocua (Lin) genes (blue box-arrows) are defined by an arrow corresponding to the TSS (red arrows on the plus strand; blue arrows on the minus strand, crossed gray arrow if the transcript is not expressed). Size of the transcript (dashed blue arrow) was estimated from the northern blot (right side of the figure) performed using the RNA extracted from L. monocytogenes and L. innocua in the exponential (exp) and stationary phase (stat) with the labeled probeset generated on either the L. monocytogenes (Lmo probe) or L. innocua genome (Lin probe). (A) Rli146 is a small RNA conserved in both genomes but not detected in L. innocua by either RNA sequencing or northern blot. Probes designed specifically to either L. monocytogenes or L. innocua successfully hybridized only in L. monocytogenes validating that the sRNA is not expressed in L. innocua. Differences in the promoter regions that might explain the differences in expression between the species are shown. (B) SbrA and Rli42 are 70 and 150 nt small RNAs conserved and expressed in both species. The SigB-dependent transcription of SbrA is supported by the presence of SigB box (σ-B) within its promoter region and the inability to detect a transcript in the ΔsigB mutant by northern blot. The conserved SigB box preceding SbrA in both L. monocytogenes and L. innocua is shown. Rli42 is a small RNA transcribed opposite to SbrA. (C) Rli130 is a novel sRNA detected in this study that is conserved and expressed in both L. monocytogenes and L. innocua. Northern blot suggests that Rli130 might exist as part of the lmo1934 mRNA as a band of 600 nt is observed. In L. innocua, the shortest form of Rli130 (230 nt) is missing, possibly due to sequence divergence between the two organisms (11 differences between positions 179 and 230 of that gene).
Expression of four of the conserved sRNAs (Rli11/SbrA, Rli42, Rli81, and Rli130) (Figure 3; Supplementary Figure S5B) was also validated by northern blot. Two of these sRNAs, Rli42 and the previously identified Rli11/SbrA (Nielsen et al, 2008), are encoded opposite to each other on the chromosome. Rli42 expression appears higher than that of Rli11/SbrA during exponential growth phase, while the SigB-regulated Rli11/SbrA is more highly expressed during stationary phase indicating an opposite regulation. Rli130 is a new sRNA identified in this study, transcribed upstream of lmo1934 (Figure 3C). Both Rli130 and lmo1934 have their own TSSs and there is a predicted terminator structure between them; however, northern blots indicated that Rli130 transcription appears to continue into the coding sequence of lmo1934. Thus, Rli130 may constitute both an independent trans-acting sRNA and cis-acting regulatory element (e.g., a riboswitch) exerting some control over the expression of the lmo1934 gene. We detected other examples of previously annotated and new sRNAs whose transcription might be associated with the transcription of the downstream gene (Supplementary Table S3).
asRNAs in L. monocytogenes and L. innocua
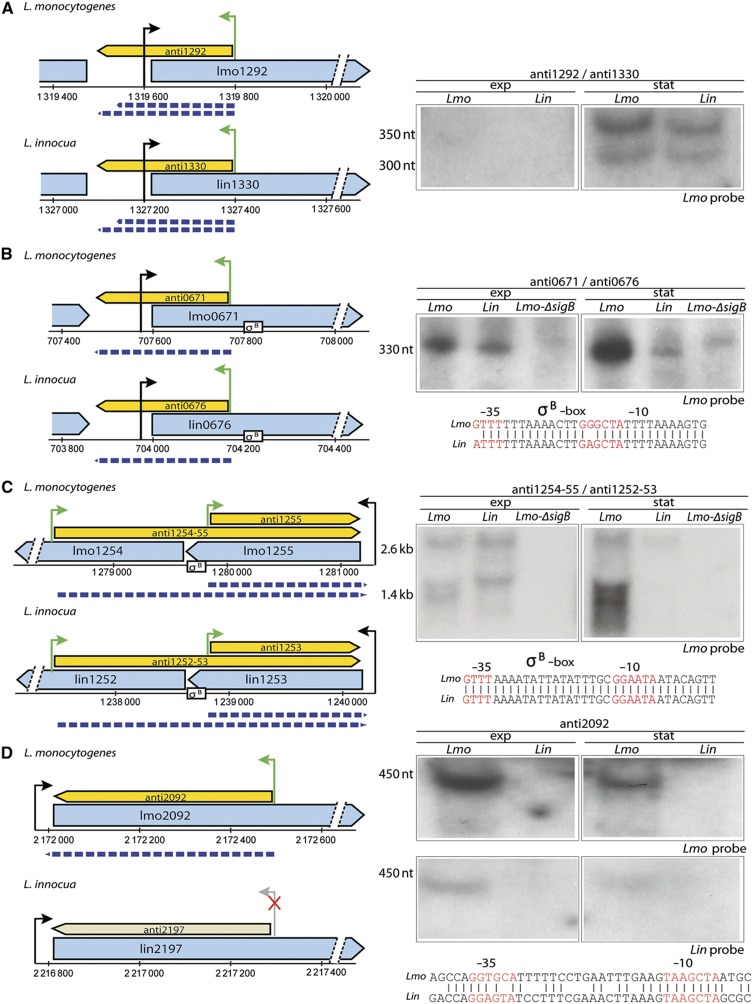
Recent studies of bacterial transcriptomes have revealed a high occurrence of asRNA transcription (Georg et al, 2009; Guell et al, 2009; Sharma et al, 2010; Wurtzel et al, 2010b; Lasa et al, 2011; Mitschke et al, 2011). In L. monocytogenes, 33 asRNAs have been previously reported (Toledo-Arana et al, 2009; Mraheil et al, 2011). However, these previous studies were not ideally designed to detect asRNAs. We took advantage of our approach that can accurately identify TSSs on both strands of the DNA. The higher sensitivity relative to the tiling-array approach, and also the strand specificity of the TSS mapping, allowed us to expand the number of known asRNAs in L. monocytogenes by over two-fold (using similar detection criteria as for the sRNAs). In total, we detected 70 asRNAs, including 17 of the previously annotated asRNAs in L. monocytogenes (Table I; Supplementary Table S5). It should be noted that we observed many additional antisense TSSs albeit with a small number of corresponding reads that did not meet our minimum criteria (>10 reads) and were not included in Supplementary Table S4. We selected 9 of these 70 transcripts for northern blot analysis, and in all 9 cases the presence of asRNAs was verified (Figures 4, 5, 6; Supplementary Figure S6).
Figure 4.
Small antisense RNAs in L. monocytogenes and L. innocua. Small antisense RNAs (yellow box-arrows) transcribed opposite to L. monocytogenes (Lmo) and L. innocua (Lin) genes (blue box-arrows) are defined by green arrows corresponding to TSSs (crossed gray arrow if the transcript was not detected). The size of each transcript (dashed blue arrow) was estimated from the northern blot. Northern blots (right side of the figure) were performed using the RNA extracted from both organisms in the exponential (exp) and stationary phase (stat) with a probe designed against either the L. monocytogenes (Lmo probe) or L. innocua transcript (Lin probe). Where probes cross-hybridized with transcripts from both species and generated identical hybridization patterns, a blot with only one probe is shown. (A) anti1292 is a conserved and expressed sasRNA in both L. monocytogenes and L. innocua. Its transcription starts within the coding region of lmo1292 and terminates near the TSS of the same gene, generating a 350 nt long product that is presumably processed to 300 nt long product or terminated earlier, as observed by the northern blot. (B) anti0671 is an sasRNA conserved and expressed in both L. monocytogenes and L. innocua. Its promoter region contains a SigB box (σ-B), and SigB regulation was further confirmed by northern blot analysis which did not detect an anti0671 transcript in the ΔsigB mutant. The SigB box is not completely conserved in L. innocua. (C) anti1255 is an antisense RNA conserved in both species. It is transcribed from two promoters, one generating a 1.4-kb transcript, and second promoter generating a 2.6-kb transcript. Both transcripts are absent in the ΔsigB mutant, however, only the first promoter generating the 1.4-kb product contains a well-defined SigB box (shown), whereas the putative SigB box of the second promoter slightly differs in sequence (not shown). (D) anti2092 is a 450 nt long asRNA transcribed opposite to the lmo2092 encoding BetL, an osmolyte transporter mediating bile tolerance in L. monocytogenes. Although a BetL homolog is present in L. innocua (lin2197) no antisense transcript was detected in L. innocua, possibly due to differences between the presumed promoter regions as shown.
Figure 5.
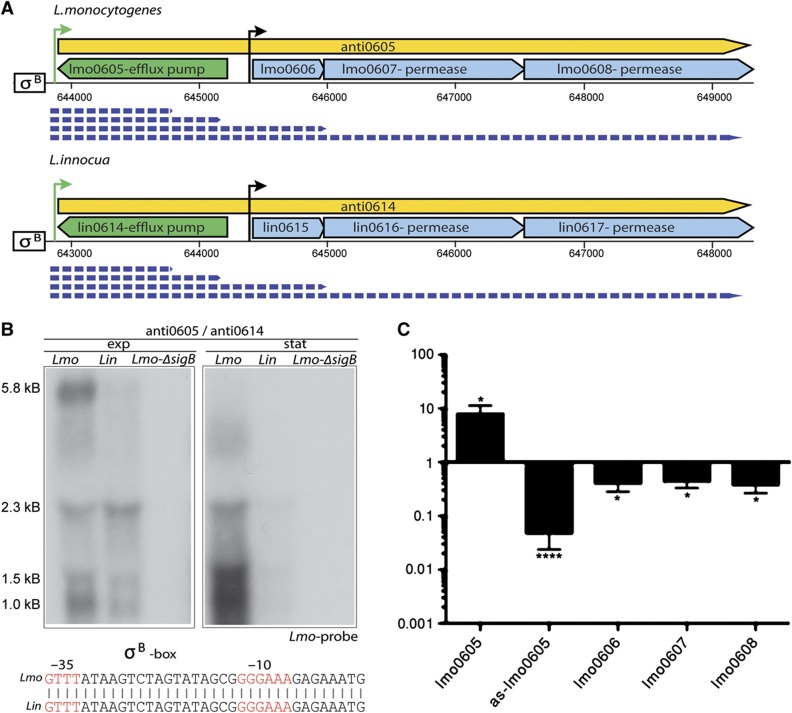
Excludon lasRNA in L. monocytogenes and L. innocua. Long antisense RNA (yellow box-arrows) encoded in the opposite strand of L. monocytogenes (Lmo) and L. innocua (Lin) genes (blue box-arrows) are defined by green arrows corresponding to TSSs. The size of the transcripts (dashed blue arrow) was estimated from northern blots. (A) anti0605 is transcribed opposite to lmo0605 in L. monocytogenes and lin0614 in L. innocua. (B) Northern blots were carried out with probes specific to L. monocytogenes (shown) or L. innocua (not shown). As probes cross-hybridized with RNA from both species, and showed identical hybridization patterns, only the blot with L. monocytogenes-specific probe is shown. Transcription appeared to be dependent on SigB in L. monocytogenes as indicated by the failure to detect bands in the ΔsigB mutant. This SigB-dependent regulation is likely conserved in L. innocua due to the conservation of the SigB box (σ-B) within the promoter region (as shown). Several transcripts were detected with the longest being ∼5.8 kb and likely corresponding to the antisense RNA plus the polycistronic mRNA of lmo0606-07-08 operon. lmo0605 codes for MatE (NorM), an Na+-driven multidrug efflux pump whereas the downstream operon codes for a predicted transcriptional regulator (lmo0606) and an ABC-type multidrug transport system (lmo0607, lmo0608). (C) Fold-change expression of the antisense transcript, lmo0605 and lmo0606-07-08 operon in the WT and ΔsigB mutant was measured by strand-specific qRT–PCR analysis. In the ΔsigB mutant where the lasRNA is not expressed, expression of lmo0605 is upregulated whereas lmo0606-07-08 is downregulated as shown.
Figure 6.
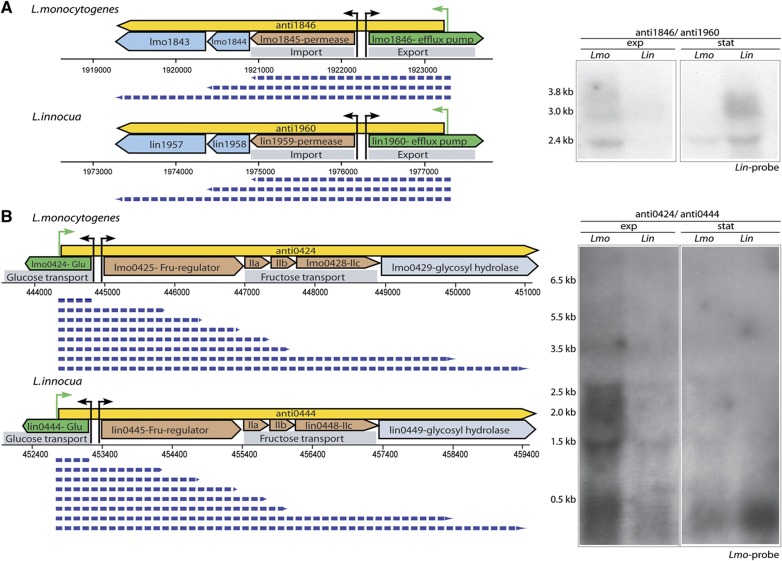
Excludon lasRNAs are a common phenomenon in the L. monocytogenes and L. innocua genomes. LasRNAs (yellow box-arrows) encoded opposite to L. monocytogenes (Lmo) and L. innocua (Lin) genes (blue box-arrows) are defined by green arrows corresponding to TSSs. Transcript sizes (dashed blue arrow) were estimated from the northern blots. Northern blots were carried out with probes specific to L. monocytogenes or L. innocua as indicated, however where probes cross-hybridized with RNA from both species, and showed identical patterns of bands, only a blot with a single probe is shown. (A) anti1846 is a lasRNA transcribed as an antisense to lmo1846, conserved and expressed in both species. Northern blot shows that anti1846 is transcribed as 3800, nt long and two shorter, 3000 and 2400, nt transcripts. Its transcription originates on the opposite strand to the coding sequence of lmo1846 encoding efflux pump from the MatE family and the distal part of the transcript contains the coding sequence of the downstream lmo1845 xanthine-uracil permease and 1843-44 encoding lipoprotein signal peptidase and ribosomal subunit synthase. (B) anti0424 is present in eight different lengths ranging from 500 to 6500 nt. Its transcription starts opposite to the coding sequence of lmo0424, a glucose permease, and continues to the adjacent operon encoding genes involved in fructose metabolism. The first gene of the operon, Imo0425 codes for a fructose-sensor/transcriptional antiterminator that regulates genes involved in fructose metabolism. The following four genes lmo0426-27-28, constitute the PTS lla, llb, llc components of a fructose permease and lmo0429, glycosyl hydrolase. Interestingly, both transcripts were upregulated during exponential phase growth in L. monocytogenes whereas they showed upregulation during stationary phase growth in L. innocua.
We also showed that six of the detected asRNAs are regulated by SigB, since their expression was eliminated in the ΔsigB strain. Direct regulation by SigB was supported by presence of a SigB box in the promoters of all these six asRNAs, as well as by the northern blots (Figures 4 and 5; Supplementary Table S5).
Comparison between L. monocytogenes and L. innocua revealed 36 asRNAs (51%) that are conserved and expressed in both species. Northern blot analysis on two of the 34 non-conserved asRNAs, anti2092 (anti-betL) and anti667 confirmed their specificity to L. monocytogenes (Figure 4D; Supplementary Figure S6A). Analysis of their promoters showed substantial sequence divergence in L. innocua when compared with L. monocytogenes, which as for the sRNAs, might explain the lack of expression in L. innocua.
Detailed examination of the studied asRNAs has surprisingly revealed two major asRNA species, which could be divided according to the nature of antisense overlap. asRNAs belonging to the first group are relatively short, overlap (partly or fully) a single ORF, and generally terminate near the TSS of the cis-encoded gene (Figure 4; Supplementary Figure S6). Six of the nine northern-blot verified asRNAs belong to this group, and their sizes range between 250 and 1400, nt. The second group consists of lasRNAs, which overlap more than one ORF, and can extend to 6.5 kb (Figures 5 and 6). Three of the nine northern-validated transcripts belong to this group.
Interestingly, lasRNAs demonstrated a typical, recurring pattern, where the lasRNA overlapping one ORF serves as the 5′ UTR of a neighboring, divergently encoded, ORF. Moreover, in all studied cases the two divergent genes had closely related or similar functions, yet mutually exclusive expression patterns. We further analyzed the expression of three lasRNAs that were expressed in both L. monocytogenes and L. innocua: anti0605, anti1846, and anti0424.
anti0605 originates from a SigB-dependent promoter located in the intergenic region between lmo0604 and lmo0605, generating a long 5.8 kb transcript and three shorter transcripts. SigB-dependent expression of all four transcripts in L. monocytogenes is supported by the northern blot analysis, confirming that they originate from the same TSS. The proximal part of anti0605 overlaps the gene lmo0605, a multidrug efflux pump from the MatE family, whereas the distal part of the 5.8-kb lasRNA transcript continues as the polycistronic mRNA of the lmo0606-07-08 operon, which also encodes efflux pumps (of the ABC-type, lmo0607-08) as well as a transcriptional regulator (lmo0606) (Figure 5A and B). Based on the genomic organization and the functional relatedness of the genes in this locus, we hypothesized that anti0605 has a dual function, acting both as an asRNA negatively regulating the expression of lmo0605, and at the same time as an mRNA simultaneously expressing lmo0606-07-08.
To test whether anti0605 indeed negatively regulates lmo0605, we quantified the transcript levels of lmo0605 and the lmo0606-07-08 operon by strand-specific qRT–PCR in WT bacteria and bacteria lacking the sigmaB transcription factor, where the anti0605 lasRNA is not expressed (Figure 5C). In the ΔsigB mutant, anti0605 shows a 50-fold downregulation (no expression) when compared with the expression in the WT, whereas lmo0605 transcript shows 10-fold upregulation, confirming the negative effect of anti0605 on lmo0605 transcript levels. In contrast, the downstream operon lmo0606-07-08 has the same expression profile as the anti0605 transcript (Figure 5C). It is downregulated in the ΔsigB mutant showing that the SigB-dependent lasRNA expression significantly contributes to the lmo0606-07-08 transcript levels, in addition to its separate SigB-independent promoter.
Similarly to anti0605, the 3.8-kb anti1846 transcript (Figure 6A) originates on the opposite strand to the coding sequence of lmo1846 and extends into the coding sequence of lmo1843-45, thus forming the 5′ UTR of the lmo1843-45 operon. While lmo1846 codes for an efflux pump of the MatE family, the opposing operon encodes for a xanthine-uracil permease (lmo1845), suggesting a regulatory mechanism by which an activation of the permease results in a simultaneous silencing of the associated efflux pump.
The 6.5-kb anti0424 is another example of a lasRNA linking two functionally related operons (Figure 6B). Its transcription starts opposite to the coding sequence of lmo0424, a glucose permease, and continues to the adjacent operon encoding genes involved in fructose metabolism. The first gene of the operon, lmo0425 codes for a fructose sensor/transcriptional antiterminator that regulates genes involved in fructose metabolism. The following four genes constitute the PTS components of a fructose permease (lmo0426-27-28) and a glycosyl hydrolase (lmo0429). Altogether, these examples suggest that lasRNA transcription is a general regulatory mechanism in Listeria where lasRNAs may have dual functions, acting both as asRNAs that negatively regulate expression of one gene, and simultaneously as mRNAs expressing another group of genes that are functionally related to the repressed gene. We had discovered the first example of such a lasRNA acting as an mRNA during the tiling-array analysis (Toledo-Arana et al, 2009; see Discussion below).
Discussion
Our results represent the first comparative transcriptomic analysis of two related bacteria, the pathogenic species L. monocytogenes and its non-pathogenic relative L. innocua. A publicly available genome browser was generated allowing direct visualization of transcriptome data from this study together with previously generated tiling-array data, in both species simultaneously, in different growth conditions and in different genetic backgrounds. In this study, we identified in the L. monocytogenes genome 113 sRNAs (among which 33 are novel) and 70 cis-encoded asRNAs (53 novel) that are expressed under various growth conditions. Comparison of the two species revealed both species specific and conserved regulatory RNA elements. Interestingly, several sRNAs and asRNAs are conserved at the genomic level, but expressed in only one of the Listeria species, revealing a hidden layer of divergence between the pathogenic and non-pathogenic species. Finally, our study proposes a new mechanism of regulation of divergent genes by long antisense transcripts.
Divergence of the L. monocytogenes and L. innocua non-coding genomes
We found in this study that 10 known L. monocytogenes virulence genes carry long 5′ UTRs (>100 nt), previously predicted to be the site of post-transcriptional regulation (Loh et al, 2006). These observations are in line with another study of a pathogenic bacterium demonstrating that larger 5′ UTRs are frequently associated with genes involved in pathogenesis (Sharma et al, 2010). Interestingly, 80 (9.5%) of the genes shared and expressed by both L. monocytogenes and L. innocua display different 5′ UTR lengths. As a consequence of the length and sequence changes, their 5′ UTRs have different predicted conformations suggesting that their expression might be regulated differently in the two species. One prominent example is that of lmo1654 encoding a secreted metalloprotease whose homolog in L. innocua has 5′ UTR shorter by 72 nucleotides. One could hypothesize that the shortening of the 5′ UTRs in L. innocua might contribute to the transition from pathogenic to non-pathogenic species.
Transcriptomic comparisons between the two species also revealed a number of species-specific differences between non-coding RNAs. Notably, a subset of sRNAs and asRNAs showed conservation at the genomic level but species-specific expression. Two sRNAs, Rli87 and Rli146, which are present in the genomes of both L. innocua and L. monocytogenes, are expressed only in L. monocytogenes in the tested growth conditions. The target prediction algorithm (TargetRNA; Tjaden, 2008) indicated that a putative target of Rli87 might be the L. monocytogenes surface protein InlA, which is a well-characterized virulence factor absent in L. innocua (Gaillard et al, 1991). It is thus tempting to speculate that Rli87 functions by base pairing with InlA and that the loss of its expression in L. innocua resulted from the loss of its target gene, or occurred simultaneously which remains to be established. Species-specific expression was also observed for two asRNAs, anti2092 and anti0667. Interestingly, anti2092 is encoded opposite to the betL gene which is one of the principal osmolyte uptake systems that contribute to Listeria growth and survival during the gastrointestinal phase of infection (Watson et al, 2009). Observed differences in the expression of the sRNA and asRNA regulatory elements could contribute to the differences in the pathogenicity of the two species.
We demonstrated that the differences in expression can frequently be explained by changes within the promoter sequence of the respective transcript in the L. innocua genome. This implies that, compared with protein regulators, RNA regulators might more easily originate or disappear as a consequence of only a small number of point mutations, suggesting that regulation by non-coding RNAs might provide an evolutionary strategy to adapt rapidly to changed regulatory needs in distinct environments.
lasRNAs and a new mechanism of bacterial gene regulation
In L. monocytogenes, only a few asRNAs had previously been annotated, mainly due to technical limitations. However, our strand-specific TSS mapping protocol allowed for the detection of 70 asRNAs, among which 53 were novel. Interestingly, these ranged in size from less than a hundred nucleotides to several kilobases. We classified these asRNAs as either small asRNAs or long lasRNAs. Small asRNAs overlap one ORF, often covering the Shine-Dalgarno sequence, and might function similarly to trans-acting sRNAs. Indeed, a number of small asRNAs have previously been found to affect transcription, RNA stability, and/or protein translation in other bacteria (Storz et al, 2011).
In contrast, lasRNA transcripts typically cover more than one ORF. In L. monocytogenes, only three lasRNAs have been previously reported (Toledo-Arana et al, 2009), but our study shows they are more abundant than previously thought. We studied three novel examples of loci exhibiting a particular gene topology, where an extremely long lasRNA could serve as the 5′ UTR of the downstream gene. In one of these cases (anti0605), we demonstrated that transcription of the lasRNA inhibits expression of the gene encoded on the opposite strand and also ensures expression of the downstream operon. Therefore, a single lasRNA transcript holds a dual function. We termed such a structure as an ‘excludon’ to describe this unique regulatory element as an ‘expression-excluding operon’. Such a dual function for a lasRNA transcript was first described in L. monocytogenes for the lasRNA regulating flagellum biosynthesis (Toledo-Arana et al, 2009). This lasRNA is transcribed opposite to the genes encoding the flagellum export apparatus whereas its distal part encompasses the coding sequence for a repressor of flagellum synthesis genes, MogR. When the lasRNA is transcribed, it negatively regulates the expression of the flagellum export apparatus genes and concurrently drives the expression of the MogR repressor, ensuring by several regulatory layers that flagella are not produced.
As illustrated by the example of MogR, genes whose regulation is linked by lasRNAs often have opposite or related functions. For instance, anti0605 transcription leads to expression of an ATP-driven multidrug efflux pump and inhibits expression of a MatE-family multidrug efflux pump. Gene expression studies had previously revealed naturally occurring conditions where these two efflux systems have opposite expression patterns (Toledo-Arana et al, 2009), including in human blood where the MatE pump is expressed and the ATP-driven pump is repressed, and in the intestine of germ-free mice where the expression pattern is reversed. Similarly, anti1846 is transcribed opposite to a MatE-family efflux pump, while its distal region codes for a permease, suggesting that the anti1846 lasRNA might lead to expression of the permease while expression of the efflux pump is repressed. Although the substrates of these two transport systems are not known, the excludon structure we detected might facilitate a controlled balancing of export (efflux pump) and import (permease) of specific molecules in Listeria. Our study also describes anti0424, a lasRNA that encodes genes involved in importing and metabolizing fructose (lmo0425-lmo0428) and whose 5′ UTR overlaps, in the antisense orientation, to a glucose-specific permease (lmo0424). Expression of the lasRNA could facilitate a switch between the utilization of the two carbon sources. Furthermore, conservation of the same genetic organization in L. grayi and the more distant organism Clostridium difficile suggests this mechanism may be more widespread (Supplementary Figure S7).
Until recently, asRNAs in bacteria were considered rare (Brantl, 2007), owing largely to a lack of methods to detect their presence (Sorek and Cossart, 2010). Following application of strand-specific high-throughput techniques for bacterial transcriptomics, it is now clear that asRNAs are much more abundant in bacteria than previously thought, as they were recently reported in several organisms such as Helicobacter pylori (Sharma et al, 2010), Synechocystis (Georg et al, 2009), Salmonella enterica (Lee and Groisman, 2010), Prochlorococcus sp. MED4 (Stazic et al, 2011), Staphylococcus aureus (Lasa et al, 2011), and Sulfolobus solfataricus (Wurtzel et al, 2010b). However, a general role for long asRNAs has remained so far obscure. Our results might assign a function for at least a subset of these asRNAs, as it is possible that the ‘excludon’ paradigm of lasRNA regulation extends to other organisms, and may represent a common method of linking regulation of physically adjacent genes that have opposing functions.
Materials and methods
Growth of Listeria
Strains used in this study were Listeria monocytogenes EGD-e (BUG1600), its isogenic mutants ΔprfA (BUG2214) and ΔsigB (BUG2215) (Mandin et al, 2007) and Listeria innocua (CIP107775 CLIP11262, BUG499) (Glaser et al, 2001). Bacteria were grown overnight in BHI medium at 37°C with shaking at 200 r.p.m. Cultures were subsequently diluted 1/500 into 100 ml BHI and grown at either 37°C or 30°C until mid-exponential phase (OD600=1.0) or stationary phase (OD600=3.0 for L. monocytogenes and OD600=2.5 for L. innocua). For conditions of low oxygen (hypoxia), cultures were jar-sealed with CampyGen sachet (Oxoid). At the appropriate OD600, bacteria were pelleted by centrifugation at 10 000 g for 5 min, flash frozen in liquid nitrogen and stored at −80°C until RNA was to be extracted.
RNA isolation and preparation of cDNA libraries
Bacterial pellets were resuspended in 400 μl solution A (½ vol Glucose 20%+½ vol Tris 25 mM pH 7.6+EDTA 10 mM) to which an additional 60 μl of 0.5 M EDTA was added. RNA was subsequently extracted using TRIzol reagent (Invitrogen) as described previously (Toledo-Arana et al, 2009). RNA integrity was verified via analysis of intact ribosomal RNA bands via the Experion Automated Electrophoresis system (Bio-Rad). Removal of 16 and 23S rRNA from total RNA was performed using MICROBExpress™ Bacterial mRNA Purification Kit (Ambion) according to manufacturer’s protocol (He et al, 2010). RNA concentrations and quality were determined by using the 2100 Bioanalzyer (Agilent).
Each RNA sample was divided into two subsamples (hereby TAP+ and TAP−) containing 2.5 μg of rRNA-depleted RNA. The RNA of TAP+ sub-sample was incubated with 2 U of TAP (Epicenter) for 1.5 h at 37°C to generate 5′ monophosphate RNAs. The other subsample (TAP−) was not treated with TAP. cDNA sequencing libraries were prepared as previously described (Wurtzel et al, 2010b), with an addition of a unique 4-base index sequence to the 5′ RNA adapter used for each subsample (Supplementary Figure S1A). cDNA libraries of every two subsamples were mixed in equal amounts, and sent to Illumina sequencing. The resulting sequences were sorted to the correct subsample by their index.
Read mapping
Sequencing reads were mapped as described previously (Wurtzel et al, 2010a, 2010b) to the corresponding reference genome of each subsample (TAP+ and TAP−) separately (Genbank: NC_003210 and NC_003212 for L. monocytogenes and L. innocua, respectively). Briefly, sequencing reads of either 34 or 36 bases were mapped using BLASTN with e-value of 0.0001 and the ‘−F F’ flag, requiring minimal alignment length of 28 nt, while allowing up to four mismatches. The alignment with the best score was considered as the correct mapping, and reads that were mapped at equal scoring to more than one position were discarded from further analysis. In cases where the read alignment did not begin in the first base of the read, read position was corrected to represent the genuine read beginning. The reads mappings were validated using Novoalign 2.0.5.22 as previously described (Avrani et al, 2011).
Determination of TSSs and comparison of 5′ UTRs length
To differentiate between reads initiating from TSSs versus RNA processing sites, a set of 5′ ends that most likely represented real TSSs (positive set) was defined and compared with a group of 5′ ends most likely represented processed sites (negative set). The positive set included genomic positions located upstream to previously defined transcriptional units (Toledo-Arana et al, 2009) (up to 50 nt), in which at least 10 reads were mapped (n=1140). If multiple genomic positions were located in a distance of <10 bases, the position with the highest number of supporting reads was selected as the TSS. The negative set for TAP+:TAP− enrichment analysis was defined as genomic positions located at least 50 nt downstream to ORF beginnings, in which at least 10 reads were mapped (n=33 826). The quality of the positive and negative sets was further validated by manual inspection of 200 randomly selected candidates from each set in published tiling-array expression data of L. monocytogenes grown in identical conditions (Toledo-Arana et al, 2009) using the integrated Listeria browser. Positions with genuine TSSs were characterized by sharp increase in the measured expression of their overlapping tiling-array probes, while processed sites, generally, did not show such pattern. For each site, the ratio between the number of 5′ end reads in the TAP+ versus TAP− samples was calculated (Supplementary Figure S1B). A significant enrichment was detected in the TAP+ samples for real TSSs (mean ratios of 6.5 and 0.5 for the positive and negative sets, respectively; P=3.2 × 10−117) (Supplementary Figure S1C). On the basis of these distributions, the minimal TAP+ enrichment threshold for a TSS was determined as 1.97. This threshold resulted in 98% specificity (e.g., <2% false positives) and 71% sensitivity (i.e., identified 71% of the positive set). This threshold was applied over the entire data of 5′ ends derived from Illumina sequencing for defining real TSSs (n=2018 TSSs across the L. monocytogenes transcriptome).
Since operon definition was not available for L. innocua, conservation of sites in L. monocytogenes was taken as a measure for the definition of the positive TSSs set in L. innocua. Conservation of TSSs was determined as follows. First, the distance of each L. monocytogenes 5′ end in the positive set (n=1140) from the beginning of the closest downstream ORF was recorded. Next, orthologous 5′ ends between the organisms were determined as sites located in the same distance (±5 nt) upstream to their homolog genes (homolog gene list was downloaded from OMAbrowser; Altenhoff et al, 2011). Positions supported by <10 mapped sequences were discarded from subsequent analysis. A total of 618 5′ ends were found in orthologous positions and formed the positive set. The negative set was constructed as described for L. monocytogenes, and a TAP+ to TAP− ratio that excluded 98% of the negative background was determined as 6.55, while preserving 73% of the sites in positive background, similarly to the 71% preserved sites in L. monocytogenes.
The TAP+ to TAP− ratio of every 5′ end was calculated, and 5′ ends with ratios larger than the determined enrichment factors, based on the positive and negative sets (1.97 and 6.55 to L. monocytogenes and L. innocua, respectively) were determined as TSSs. In addition, 5′ ends that were determined as TSSs in one of the organisms but not in the other, and were conserved (±5 nt) in position and represented the most highly represented 5′ end upstream to the gene, were annotated as TSSs in both organisms (213 and 398 such TSSs for L. monocytogenes and L. innocua, respectively; the higher number of 5′ ends of this kind in L. innocua is the result of less sequencing reads than in the L. monocytogenes samples). For each TSS, the distance of the TSS from the start codon (length of 5′ UTR) was recorded. Differences in 5′ UTR lengths between organisms were calculated as the absolute difference between the 5′ UTR lengths of both organisms.
To assess the reproducibility of the TSS mapping, the positions of TSSs in WT and ΔsigB L. monocytogenes grown to mid-log phase at 37°C were compared. The TSS mapping of genes known to be independent of SigB (Oliver et al, 2009) reproduced to a single-nucleotide resolution in 97% of the TSSs supported by over 10 reads in the WT bacteria. Additional comparison of WT and the ΔprfA TSSs showed a reproducibility of over 98%.
Detection of ncRNAs and divergence of the start sites
A list of the detected 5′ ends from L. monocytogenes, which were (i) supported by over 10 reads, (ii) and located 100 nt or more from the gene downstream to them (in case they were on the same strand), or 50 nt or more (in case they were on opposite strands), was compiled. In case several 5′ ends were detected within 30 nt from one another, they were clustered together for further analysis. The list was used for manual inspection of the transcriptome using the unified Listeria browser in its comparative L. monocytogenes/L. innocua mode. Each 5′ end in the list was evaluated as an sRNA or a long 5′ UTR by examining the expression of the tiling-array data (Toledo-Arana et al, 2009), derived from the same growth conditions, as well as transcriptome sequencing data (Oliver et al, 2009). Where the expression downstream to the 5′ ends appeared continuous with the downstream gene, it was determined as a 5′ UTR, and in cases the expression was separated by at least three tiling-array probes or was markedly different from the downstream gene, it was determined as an sRNA. Sizes of sRNAs were estimated by number of tiling-array probes downstream to the detected 5′ end that had similar expression. Homologs of the sRNAs were searched L. innocua were detected using BLASTN with flags (−e 0.001 −W 4 −F F) demanding query coverage greater or equal to 50%. If no sequence similarity was found then the sRNA was determined as non-conserved in sequence. If a similar sequence was found in L. innocua but no 5′ end was detected (±30 nt) then the sRNA was determined as conserved in sequence but not in expression. Otherwise, when a 5′ end was on the same strand of the sequence, it was assigned as the TSS of the sRNA homolog. Finally, the difference between the positions of 5′ ends relative to the beginnings of the sRNAs in both organisms was recorded as the divergence of the TSSs.
asRNAs were determined as 5′ ends, which overlap ORFs, supported by at least 10 sequencing reads, or which were detected in more than one sequencing condition. The L. innocua homologs of genes that were overlapped by asRNAs in L. monocytogenes were extracted from OMAbrowser (Altenhoff et al, 2011). Each homolog was searched for antisense transcription. If no asRNA was detected in the L. innocua homolog then the asRNA was determined as non-conserved. When antisense transcription was present in both organisms, the distance from beginning of the ORF was determined and the divergence of the transcription was determined as the absolute difference between the positions of the transcripts.
Relative expression of sRNA
The tiling arrays used in this study have been previously described (Toledo-Arana et al, 2009). Briefly, arrays were normalized with DNA reference normalization method (Huber et al, 2006) using the wild-type strain grown in BHI at 37°C generating log-transformed expression values. Median expression values of tiling data were adjusted to zero. Relative expression values (Supplementary Figure S4) were calculated by subtracting the probe expression values for a given sRNA in the wild-type condition from the probe expression values in a particular biological condition. As expression values are already log transformed this subtraction is equivalent to the log2 (fold change) between a particular biological condition and the wild-type condition.
Northern blot analysis
For all samples, 10–20 μg of total RNA was mixed with two volumes of Formaldehyde Loading Buffer (Ambion) followed by denaturation at 65°C for 15 min. For smaller transcripts, samples were electrophoresed on 5% TBE-Urea polyacrylamide gels (Criterion-Bio-Rad) at 100 V for 2 h in 1 × TBE running buffer followed by overnight transfer at 4°C/100 mA to Nytran membranes (Sigma). For larger transcripts, samples were electrophoresed on 1% formaldehyde-denaturing agarose gels at 50 V for 2 h in MOPS running buffer followed by downward passive transfer to Nytran membranes (Sigma). Membranes were subsequently UV crosslinked and probed with RNA probes. Briefly, RNA probes were synthesized and [α32P]UTP labeled using the Maxiscript T7 RNA polymerase kit (Ambion) with PCR-generated templates according to manufacturer’s instructions. Membranes were prehybridized for 30 min in Ultrahyb buffer (Ambion) as instructed for, and hybridizations were performed overnight at 60°C. Following hybridization, membranes were washed twice for 5 min with 2 × SSC, 0.1% SDS and twice for 15 min in 1 × SSC, 0.1% SDS at 60°C.
Strand-specific qRT–PCR
For quantitative PCR with reverse transcription, 1 μg of bacterial RNA was reverse transcribed using SuperScript III Reverse Transcriptase (Invitrogen) for gene-specific cDNA synthesis. The cDNAs were used as templates for PCR in the presence of SsoFast EvaGreen supermix (Bio-Rad) in the MyiQ single-color RT iCycler PCR detection system (Bio-Rad) and data were analyzed with the MyiQ optical system software (Bio-Rad). Expression of individual mRNAs was normalized to expression of the gyrA gene. Experiment was repeated several times on three independent RNA extracts and data statistically evaluated by two-tailed Student’s t-test.
Data availability
The sequencing data from this publication have been submitted to the Gene Expression Omnibus (URL: http://www.ncbi.nlm.nih.gov/geo/) and assigned the series identifier accession GSE36060, and samples accession GSM880615 to GSM880630. The Listeria Transcriptome Browser is available at: http://www.weizmann.ac.il/molgen/Sorek/listeria_browser/.
Supplementary Material
TSSs of genes detected in this study.
Comparison of known 5' UTRs lengths with 5' UTRs determined in this study.
All sRNAs identified in this study.
All asRNAs identified in this study.
Acknowledgments
We thank Adi Stern and Shany Doron for helpful discussions, and Shirley Horn-Saban and Daniella Amann-Zalcenstein for Illumina sequencing. This work was supported by a grant from the Pasteur-Weizmann program (to PC and RS), an ERC (Starting grant 260432) (RS), ERC (Advanced grant 233348) (PC), ANR (Bacregrna 09-BLAN-0024-02) (PC), Fondation Le Roch Les Mousquetaires (PC) and the Fondation Jeantet (PC). OW was supported by an Azrieli fellowship. NS was supported by INRA fellowship (Institute National de la recherché Agronomique).
Author contributions: OW, NS, PC, and RS designed the study. NS, JRM, and CA performed experiments. IK and SE constructed the sequencing libraries. OW and RS analyzed the deep sequencing data and performed bioinformatic analyses with contributions from CB. OW constructed the online browser. OW, NS, JRM, PC, and RS wrote the manuscript and incorporated comments from all authors.
Footnotes
The authors declare that they have no conflict of interest.
References
- Altenhoff AM, Schneider A, Gonnet GH, Dessimoz C (2011) OMA 2011: orthology inference among 1000 complete genomes. Nucleic Acids Res 39: D289–D294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avrani S, Wurtzel O, Sharon I, Sorek R, Lindell D (2011) Genomic island variability facilitates Prochlorococcus-virus coexistence. Nature 474: 604–608 [DOI] [PubMed] [Google Scholar]
- Brantl S (2007) Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr Opin Microbiol 10: 102–109 [DOI] [PubMed] [Google Scholar]
- Cossart P (2011) Illuminating landscape of host pathogen interactions with the bacterium Listeria monocytogenes. Proc Natl Acad Sci USA 108: 19484–19491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossart P, Toledo-Arana A (2008) Listeria monocytogenes, a unique model in infection biology: an overview. Microbes Infect 10: 1041–1050 [DOI] [PubMed] [Google Scholar]
- Desvaux M, Dumas E, Chafsey I, Chambon C, Hebraud M (2010) Comprehensive appraisal of the extracellular proteins from a monoderm bacterium: theoretical and empirical exoproteomes of Listeria monocytogenes EGD-e by secretomics. J Proteome Res 9: 5076–5092 [DOI] [PubMed] [Google Scholar]
- Doumith M, Cazalet C, Simoes N, Frangeul L, Jacquet C, Kunst F, Martin P, Cossart P, Glaser P, Buchrieser C (2004) New aspects regarding evolution and virulence of Listeria monocytogenes revealed by comparative genomics and DNA arrays. Infect Immun 72: 1072–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussurget O (2008) New insights into determinants of Listeria monocytogenes virulence. Int Rev Cell Mol Biol 270: 1–38 [DOI] [PubMed] [Google Scholar]
- Felden B, Vandenesch F, Bouloc P, Romby P (2011) The Staphylococcus aureus RNome and its commitment to virulence. PLoS Pathog 7: e1002006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard JL, Berche P, Frehel C, Gouin E, Cossart P (1991) Entry of L. monocytogenes into cells is mediated by internalin, a repeat protein reminiscent of surface antigens from gram-positive cocci. Cell 65: 1127–1141 [DOI] [PubMed] [Google Scholar]
- Gardner PP, Daub J, Tate J, Moore BL, Osuch IH, Griffiths-Jones S, Finn RD, Nawrocki EP, Kolbe DL, Eddy SR, Bateman A (2011) Rfam: Wikipedia, clans and the "decimal" release. Nucleic Acids Res 39: D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georg J, Voss B, Scholz I, Mitschke J, Wilde A, Hess WR (2009) Evidence for a major role of antisense RNAs in cyanobacterial gene regulation. Mol Syst Biol 5: 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser P, Frangeul L, Buchrieser C, Rusniok C, Amend A, Baquero F, Berche P, Bloecker H, Brandt P, Chakraborty T, Charbit A, Chetouani F, Couve E, de Daruvar A, Dehoux P, Domann E, Dominguez-Bernal G, Duchaud E, Durant L, Dussurget O et al. (2001) Comparative genomics of Listeria species. Science 294: 849–852 [DOI] [PubMed] [Google Scholar]
- Gottesman S (2005) Micros for microbes: non-coding regulatory RNAs in bacteria. Trends Genet 21: 399–404 [DOI] [PubMed] [Google Scholar]
- Guell M, van Noort V, Yus E, Chen WH, Leigh-Bell J, Michalodimitrakis K, Yamada T, Arumugam M, Doerks T, Kuhner S, Rode M, Suyama M, Schmidt S, Gavin AC, Bork P, Serrano L (2009) Transcriptome complexity in a genome-reduced bacterium. Science 326: 1268–1271 [DOI] [PubMed] [Google Scholar]
- Hain T, Steinweg C, Chakraborty T (2006a) Comparative and functional genomics of Listeria spp. J Biotechnol 126: 37–51 [DOI] [PubMed] [Google Scholar]
- Hain T, Steinweg C, Kuenne CT, Billion A, Ghai R, Chatterjee SS, Domann E, Karst U, Goesmann A, Bekel T, Bartels D, Kaiser O, Meyer F, Puhler A, Weisshaar B, Wehland J, Liang C, Dandekar T, Lampidis R, Kreft J et al. (2006b) Whole-genome sequence of Listeria welshimeri reveals common steps in genome reduction with Listeria innocua as compared to Listeria monocytogenes. J Bacteriol 188: 7405–7415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamon M, Bierne H, Cossart P (2006) Listeria monocytogenes: a multifaceted model. Nat Rev Microbiol 4: 423–434 [DOI] [PubMed] [Google Scholar]
- He S, Wurtzel O, Singh K, Froula JL, Yilmaz S, Tringe SG, Wang Z, Chen F, Lindquist EA, Sorek R, Hugenholtz P (2010) Validation of two ribosomal RNA removal methods for microbial metatranscriptomics. Nat Methods 7: 807–812 [DOI] [PubMed] [Google Scholar]
- Heroven AK, Bohme K, Rohde M, Dersch P (2008) A Csr-type regulatory system, including small non-coding RNAs, regulates the global virulence regulator RovA of Yersinia pseudotuberculosis through RovM. Mol Microbiol 68: 1179–1195 [DOI] [PubMed] [Google Scholar]
- Huber W, Toedling J, Steinmetz LM (2006) Transcript mapping with high-density oligonucleotide tiling arrays. Bioinformatics 22: 1963–1970 [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS (2009) Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irnov I, Sharma CM, Vogel J, Winkler WC (2010) Identification of regulatory RNAs in Bacillus subtilis. Nucleic Acids Res 38: 6637–6651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmierczak MJ, Mithoe SC, Boor KJ, Wiedmann M (2003) Listeria monocytogenes {sigma} B regulates stress response and virulence functions. J Bacteriol 185: 5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsford CL, Ayanbule K, Salzberg SL (2007) Rapid, accurate, computational discovery of Rho-independent transcription terminators illuminates their relationship to DNA uptake. Genome Biol 8: R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni RV, Kulkarni PR (2007) Computational approaches for the discovery of bacterial small RNAs. Methods 43: 131–139 [DOI] [PubMed] [Google Scholar]
- Lasa I, Toledo-Arana A, Dobin A, Villanueva M, de Los Mozos IR, Vergara-Irigaray M, Segura V, Fagegaltier D, Penades JR, Valle J, Solano C, Gingeras TR (2011) Genome-wide antisense transcription drives mRNA processing in bacteria. Proc Natl Acad Sci USA 108: 20172–20177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EJ, Groisman EA (2010) An antisense RNA that governs the expression kinetics of a multifunctional virulence gene. Mol Microbiol 76: 1020–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livny J, Waldor MK (2007) Identification of small RNAs in diverse bacterial species. Curr Opin Microbiol 10: 96–101 [DOI] [PubMed] [Google Scholar]
- Loh E, Gripenland J, Johansson J (2006) Control of Listeria monocytogenes virulence by 5′-untranslated RNA. Trends Microbiol 14: 294–298 [DOI] [PubMed] [Google Scholar]
- Mandin P, Repoila F, Vergassola M, Geissmann T, Cossart P (2007) Identification of new noncoding RNAs in Listeria monocytogenes and prediction of mRNA targets. Nucleic Acids Res 35: 962–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellin JR, Cossart P (2012) The non-coding RNA world of the bacterial pathogen Listeria monocytogenes. RNA Biol 9: 4 ( http://www.ncbi.nlm.nih.gov/pubmed/22336762) [DOI] [PubMed] [Google Scholar]
- Mitschke J, Georg J, Scholz I, Sharma CM, Dienst D, Bantscheff J, Voss B, Steglich C, Wilde A, Vogel J, Hess WR (2011) An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc Natl Acad Sci USA 108: 2124–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mraheil MA, Billion A, Mohamed W, Mukherjee K, Kuenne C, Pischimarov J, Krawitz C, Retey J, Hartsch T, Chakraborty T, Hain T (2011) The intracellular sRNA transcriptome of Listeria monocytogenes during growth in macrophages. Nucleic Acids Res 39: 4235–4248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen JS, Olsen AS, Bonde M, Valentin-Hansen P, Kallipolitis BH (2008) Identification of a sigma B-dependent small noncoding RNA in Listeria monocytogenes. J Bacteriol 190: 6264–6270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver HF, Orsi RH, Ponnala L, Keich U, Wang W, Sun Q, Cartinhour SW, Filiatrault MJ, Wiedmann M, Boor KJ (2009) Deep RNA sequencing of L. monocytogenes reveals overlapping and extensive stationary phase and sigma B-dependent transcriptomes, including multiple highly transcribed noncoding RNAs. BMC Genomics 10: 641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, Vogel J (2010) Regulatory RNA in bacterial pathogens. Cell Host Microbe 8: 116–127 [DOI] [PubMed] [Google Scholar]
- Pichon C, Felden B (2008) Small RNA gene identification and mRNA target predictions in bacteria. Bioinformatics 24: 2807–2813 [DOI] [PubMed] [Google Scholar]
- Pizarro-Cerda J, Cossart P (2006) Subversion of cellular functions by Listeria monocytogenes. J Pathol 208: 215–223 [DOI] [PubMed] [Google Scholar]
- Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermuller J, Reinhardt R, Stadler PF, Vogel J (2010) The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464: 250–255 [DOI] [PubMed] [Google Scholar]
- Sorek R, Cossart P (2010) Prokaryotic transcriptomics: a new view on regulation, physiology and pathogenicity. Nat Rev Genet 11: 9–16 [DOI] [PubMed] [Google Scholar]
- Stazic D, Lindell D, Steglich C (2011) Antisense RNA protects mRNA from RNase E degradation by RNA-RNA duplex formation during phage infection. Nucleic Acids Res 39: 4890–4899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz G, Vogel J, Wassarman KM (2011) Regulation by Small RNAs in Bacteria: Expanding Frontiers. Mol Cell 43: 880–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjaden B (2008) TargetRNA: a tool for predicting targets of small RNA action in bacteria. Nucleic Acids Res 36: W109–W113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K, Barthelemy M, Vergassola M, Nahori MA, Soubigou G, Regnault B, Coppee JY, Lecuit M, Johansson J, Cossart P (2009) The Listeria transcriptional landscape from saprophytism to virulence. Nature 459: 950–956 [DOI] [PubMed] [Google Scholar]
- Watson D, Sleator RD, Casey PG, Hill C, Gahan CG (2009) Specific osmolyte transporters mediate bile tolerance in Listeria monocytogenes. Infect Immun 77: 4895–4904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzel O, Dori-Bachash M, Pietrokovski S, Jurkevitch E, Sorek R (2010a) Mutation detection with next-generation resequencing through a mediator genome. PLoS One 5: e15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzel O, Sapra R, Chen F, Zhu Y, Simmons BA, Sorek R (2010b) A single-base resolution map of an archaeal transcriptome. Genome Res 20: 133–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Chen L, Sun L, Yu J, Jin Q (2008) VFDB 2008 release: an enhanced web-based resource for comparative pathogenomics. Nucleic Acids Res 36: D539–D542 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
TSSs of genes detected in this study.
Comparison of known 5' UTRs lengths with 5' UTRs determined in this study.
All sRNAs identified in this study.
All asRNAs identified in this study.
Data Availability Statement
The sequencing data from this publication have been submitted to the Gene Expression Omnibus (URL: http://www.ncbi.nlm.nih.gov/geo/) and assigned the series identifier accession GSE36060, and samples accession GSM880615 to GSM880630. The Listeria Transcriptome Browser is available at: http://www.weizmann.ac.il/molgen/Sorek/listeria_browser/.