Abstract
A high throughput method based on flow injection analysis was developed and validated for the quantification of the peptide Bβ15-42 in cellular samples comparing different labeling strategies and detection methods. The used labels were 1,4,7,10-tetraazacyclododecane-N, N′, N′′, N′′′-tetraaceticacid (In-DOTA) and 2-(4-isothiocyanatobenzyl) - 1,4,7,10-tetraazacyclododecane-N, N′, N′′, N′′′-tetraacetic acid (In-DOTA-Bn) for elemental labeling. 6-Hydroxy-9-(2-carboxyphenyl)- (3H)-xanthen-3-on (fluorescein) was employed as fluorescence label. The explored peptide (mass = 3 kD) is a novel candidate drug, which shows an anti-inflammatory effect after an event of myocardial infarction. The analysed samples were fractioned cell compartments of human umbilical cord vein endothelial cells (HUVEC) maintained via lysis with Triton X buffer. In order to enhance sensitivity and selectivity of peptide quantification via flow injection the peptide was labeled prior to incubation using elemental and fluorescence labels. Quantification of the elemental and fluorescence labeled peptide was performed via flow injection analysis combined with inductive coupled plasma sector field mass spectrometry (FIA-ICP-SFMS) or fluorescence detection (FIA-FLD), respectively. The employed quantification strategies were external calibration in the case of fluorescence detection and external calibration with and without internal standardization and on-line IDMS in the case of ICP-MS detection
The limit of detection (LOD) for FIA-ICP-MS was 9 pM In-DOTA-Bβ15-42 (0.05 fmol absolute) whereas FIA-FLD showed a LOD of 100 pM (3 fmol absolute) for the fluorescein labeled peptide. Short term precision of FIA-ICP-MS was superior for all ICP-MS based quantification strategies compared to FIA-FLD (FIA-ICP-SFMS: 0.3-3.3%; FIA-FLD: 6.5%). Concerning long term precision FIA-ICP-SFMS with on-line IDMS and internal standardization showed the best results (3.1 and 4.6%, respectively) whereas the external calibration of both applied methodological approaches was only in the range of 10 %.
The concentrations in the Triton X soluble fraction relative to the applied amount of Indium in the cell culture were in the range of 0.75-1.8% for In-DOTA or 0.30-0.79% for the 2-(4-isothiocyanatobenzyl) - 1,4,7,10-tetraazacyclododecane-N, N′, N′′, N′′′-tetraacetic acid (In-DOTA-Bn) labeled peptide Bβ15-42. In the Triton X insoluble fraction the relative concentrations of Indium were 0.03-0.18% for the In-DOTA labeled peptide and 0.03-0.13% for Bβ15-42-In-DOTA-Bn.
Keywords: ICP-MS, quantification, peptide, labeling, fluorescence
1. Introduction
Quantitative analysis is one of the key issues in a wide range of biological studies. In this context, the labeling of bio-molecules represents an upcoming analytical research field with increasing interest 1, 2. Labeling of bio-molecules can be pursued via metabolic 1, 3, 4, enzymatic 1, 5 or chemical reactions 6, 7, 8, 9, 10. Principally, a biological system can be studied by introduction of labeled probes. Labeling can be conducted directly - that means that the labeled bio-molecule is readily detectable before its introduction into a biological system- or indirectly – meaning that first a substituent is covalently linked onto the bio-molecule, which alone is not detectable but requires additional development steps that are conducted ex-vivo at the end of the experiment. Examples for direct labels are the fluorescein, In-DOTA or In-DOTA-Bn coupled peptides used in this study. Examples for indirect labeling are e.g. an ionophor which is linked to the bio-molecule and incubated with lanthanides to form a complex after the experiments, or an antigen linked to the bio-molecule visualized using specific antibodies. For studying the molecular or elemental inventory of a native unaltered biological system a plenty of derivatization techniques are common especially in quantitative proteomics using e.g. lanthanide tags that allow the analysis of several proteomes labeled by different lanthanides in the same electrophoresis gel. Finally using ICP-MS techniques the native distribution of metals and heteroelements can be studied 11, 12, 13. Both, pre- and post-labeling approaches offer general advantages like an increased selectivity, sensitivity and robustness in complex matrices. Labeling of bio-molecules with selective labels even allows rapid quantification using flow injection analysis (e.g. for the determination of drug concentrations) accompanied by reduced sample preparation procedures. Moreover, the flow injection set-up is characterised by low sample consumption, reduced reagent and material costs and convincing analysis speed which offers the possibility for high throughput analysis 14, 15.
Elemental labels in combination with ICP-MS detection were first introduced by Baranov et al. 16. In this work labeling strategies for ICP-MS using conventional fluorphores containing lanthanide elements or antibodies labeled with gold clusters were described. In the element coded affinity tag (ECAT) approach which was published by Whetstone et al.8, 1,4,7,10- tetraazacyclododecane-N, N′, N′′, N′′′-tetraaceticacid (DOTA; which forms a stable complex with lanthanides) is used to tag a protein 8. In metal element chelate tag (MECT) Liu et al. described a comparable attempt using Diethylenetriaminepentaacetic dianhydride (DTPA) as a metal chelating reagent for tagging peptides 9, 10. DOTA and DTPA based reagents are often used for labeling with lanthanides because of their high complex formation constancy 8, 9, 10. The option of isotope dilution analysis (IDA) and the capability for multiplexed analysis with wide ranging potential 1, 17, 18, 19 ,20, 21, 22 are unique features of elemental labels 15, 23, 24.
In this work labeling was implemented in a pre-clinical study to quantify the total Indium concentrations in cell extracts after exposure to the candidate drug peptide Bβ15-42 in cell models. The analysed peptide Bβ15-42 consists of 28 amino acids corresponding to the N-terminal sequence of the β-chain of fibrin and originates after cleavage of fibrin following plasmin digestion. The peptide is a competitor to the fibrin fragment N-terminal disulfide knot-II which binds to vascular endothelial cadherin (VE-cadherin) and therefore is a potential anticoagulant selective for intravascular coagulation. Especially it is being developed to prevent reperfusion injury after myocardial infarction. The peptide Bβ15-42 has already shown protective effects in models of myocardial reperfusion injury and was capable of impairing reducing infarct size, leukocyte infiltration and scar formation in vivo. In addition the peptide is able to improve survival under septic conditions and reduces heart and lung injury in models for hemorrhagic shock25.
Studies dealing with the quantification of candidate drug contents in different cell extracts are an important step during the drug development. Such endeavors are hampered by the complexity of the matrix demanding for high selectivity, small sample volumes and low concentrations 1, 26, 27, 28. Previous studies in our group have shown the analytical challenges arising in quantitative analysis of peptides in cell models and the drawbacks of LC-MS (liquid chromatography mass spectrometry) based techniques regarding matrix effects24. As a novelty in this work two different labels (elemental and fluorescence) were applied and compared in terms of analytical figures of merit. The detection of the fluorescein labeled peptide Bβ15-42 was performed via flow injection analysis combined with fluorescence detection (FIA-FLD). We have chosen the FIA-FLD approach because it is fully automatable and does not require time consuming sample preparation steps, which is the case for fluorescence readers with microtiter plates.
The determination of the In-DOTA labeled peptide Bβ15-42 was carried out with flow injection analysis combined with inductive coupled plasma sector field mass spectrometry (FIA-ICP-SFMS) comparing three different quantification strategies.
2. Experimental
2.1. Synthesis of the labeled peptides
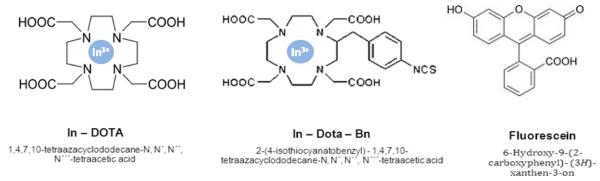
Peptide synthesis and labeling was performed on solid support as described in a previous publication24. The employed labels are depicted in Figure 1. The elemental labels In-DOTA (1,4,7,10-tetraazacyclododecane-N, N′, N′′, N′′′-tetraaceticacid) and In-DOTA-Bn (2-(4-isothiocyanatobenzyl) - 1,4,7,10-tetraazacyclododecane-N, N′, N′′, N′′′-tetraacetic acid) consist of DOTA as chelating moiety and In as elemental component. In-DOTA-Bn carries an additional SCN-Bn (p-isothiocyanatobenzyl) group leading to four free carboxyl groups after binding to the peptide. The connection between the In-DOTA and In-DOTA-Bn labels and the peptide Bβ15-42 was established via amid binding. The attachment of the fluorescence label fluorescein (6-Hydroxy-9-(2-carboxyphenyl)-(3H)-xanthen-3-on) to the peptide Bβ15-42 was performed via an additional carboxylate group. All employed labels were synthesized by PICHEM G.m.b.H. (Graz, Austria).
Figure 1.
Chemical structure of the employed elemental and fluorescence labels.
2.2. Reagents and standards
The ammonia formate buffer, which was employed as an eluent for FIA-FLD analysis was prepared using suprapure ammonia (Merck, Dammstadt, Germany) and suprapure formic acid (Merck). A stock solution of approximately 0.5 mg of the fluorescein labeled peptide was freshly prepared in 1 mL water in an amber glass vial. The standards with the fluorescein labeled peptide for FIA-FLD measurements were diluted with water prior to analysis. For the preparation of the eluents for FIA-ICP-SFMS analysis subboiled nitric acid (Merck) and suprapure methanol LC-MS grade (Honeywell, New Jersey, USA) were used. Labeled and unlabeled peptides were purchased at Pichem (Graz, Austria) and were especially synthesized for our purposes. The applied In (Merck) and Rhodium (Fluka, Buchs, Switzerland) standards were 1000 mg L−1 single element ICP-MS standards. For IDMS measurements an 113In spike was purchased at Isotec (Merseyside, England) with a purity of 93.1 atom%. The In metal was dissolved in HNO3 and diluted with ultra pure water (final acidity 5% HNO3 w/w). After dilution to approximately 2 μg L−1 the spike was characterized via reverse isotope dilution mass spectrometry (IDMS). The quantification with reverse IDMS revealed a final In concentration of 2.10 μg L−1 In which corresponds to 18.6 nM. Water was pre-cleaned by reverse osmosis and afterwards purified using a purification system (SG Ultra Clear Basic, SG Wasseraufbereitung und Regeneration GmbH, Barsbüttel, Germany).
2.3. Cell culture experiment
Primary human umbilical cord vein endothelial cells human umbilical cord vein endothelial cells (HUVEC) were raised in 6-well cell culture plates (approximately 500.000 cells per mL). After reaching confluence the cultured cells were incubated in parallel experiments with 500 μL cell culture medium (IMDM with phenol red as an indicator) containing 15 μg of the either In-DOTA- or In-DOTA-Bn or fluorescein labeled peptide Bβ15-42 for different exposure times. The experiment was aborted at six different points of time (0, 2, 4, 16, 64,128 min) by removal of the supernatants and by washing the cells with phosphate buffer solution (PBS, pH 7.4). The harvest of the cells was executed by lysis with Triton X buffer followed by centrifugation to generate different fractions of the cell lysate. The analysed cell lysate fractions were Triton X soluble fraction and Triton X insoluble fraction. In addition the supernatant stimulation medium and the wash solution were pooled and investigated.
2.5. Sample preparation for FIA-ICP-SFMS analysis
The Triton X soluble and Triton X insoluble fraction containing the In-DOTA and In-DOTA-Bn labeled peptide Bβ15-42 were centrifuged and diluted 1:10 with 2% nitric acid before analysis. All investigated samples were prepared as clear solutions and showed a good solubility in 2% nitric acid. Additionally 10 μL Rhodium (1 μg L−1) were added to each sample as an internal standard for external calibration. For on-line IDMS 10 μL min−1 113In spike (2.10 μg L−1) were added after the injection valve utilizing a syringe pump and a low void volume T-piece (IDEX, Illinois, USA).
2.6. Instrumental
2.6.1. FIA-FLD
For quantification of the fluorescein labeled peptide an inert titanium HPLC gradient system (Ultimate 3000 Standard HPLC System, Dionex, Sunnyvale, California) with an implemented high precision autosampler and a fluorescence detector (Dionex Fluorescence Detector 3000) was used. For all connections PEEK capillaries (IDEX) with an internal diameter of 0.254 mm. were used. The operation parameters for FIA-FLD are listed in Table 1. In order to prevent damage of the fluorescence flow cell a pressure limit of 20 bar was set within the software of the method and the pressure curve was monitored over the whole runtime.
Table 1.
Operation parameters for FIA-FLD.
| Carrier solvent | Ammonia formate (pH 9.25) |
| Flow rate | 0.5 mL min−1 |
| Injection volume | 25 μL |
| Injection loop volume | 50 μL |
| Tray temperature | 6 °C |
|
| |
| Absorbance wavelength | 485 nm |
| Emission wavelength | 514 nm |
| Data point s-1 | 10 |
| Cycle time | 1.5 min |
2.6.2. FIA-ICP-SFMS
For the determination of In-DOTA-Bβ15-42 and In-DOTA-Bn-Bβ15-42 an inert titanium HPLC gradient system (Rheos 2000, Flux Instruments AG, Basel, Switzerland) in combination with a metal-free autosampler (HTC PAL Autosampler, Thermo Fisher Scientific Inc., Waltham, USA) was combined with a high resolution ICP-SFMS (Element 2, Thermo Scientific Inc., Bremen, Germany). For direct connection of the HPLC effluent to the PFA-nebulizer (PFA-ST, Elemental Scientific Inc., Omaha, Nebrasca, USA) a teflon capillary (IDEX) with an internal diameter of 0.2 mm was used. The PFA-nebulizer was operated in a cooled (5°C) cyclonic silica glass spray chamber (PC3, ISA Elemental Scientific). The FIA-ICP-SFMS operating parameters are specified in Table 2.
Table 2.
Operation parameters for FIA-ICP-SFMS.
| Carrier solvent | 2% nitric acid |
| Flow rate | 0.4 mL min−1 |
| Injection volume | 5 μL |
| Injection loop volume | 5 μL |
| Tray - temperature | 6 °C |
|
| |
| Nebulizer | PFA-ST microconcentric |
| Spray chamber | Cyclonic, 5 °C |
| Nebulizer gas flow | 1.10 L min−1 |
| Auxillary gas flow | 1.00 L min−1 |
| Plasma gas | 18.00 L min−1 |
| ICP RF power | 1230 W |
| Scan mode | EScan |
| m/z measured | 102.91, 112.90, 114.90 |
| Dwell time per isotope | 0.1 s |
| Data points s-1 | 3 |
| Cycle time | 1.5 min |
2.7. Data Evaluation
Chromlink software (Version 2.1, Perkin Elmer SCIEX, Massachusetts, USA) in combination with Totalchrom software (Version 6.2, Perkin Elmer SCIEX) were used for the generation and export of FIA-ICP-SFMS chromatograms. The generation of FIA-FLD results and the integration of all chromatographic data from FIA-ICP-SFMS and FIA-FLD analysis were carried out using Chromeleon software (Version 6.7, Dionex, Sunnyvale, California, USA).
2.8. Quantification strategies
For quantification of the fluorescence labeled peptide external calibration with fluorescein- B β15-42 was performed. In-DOTA-Bβ15-42 and In-DOTA-Bn- Bβ15-42 were quantified via external calibration with and without internal standardization (Rhodium) and species unspecific on-line IDMS. For on-line IDMS the 113In spike was continuously added via an external pump with a flow rate of 10 μL min−1 (CMA 140 microdialysis pump, CMA/Microdialysis AB, Solna, Sweden). The concentration of Indium labeled peptides was quantified according to equation 1:
| 1 |
The resulting analyte mass flow Mxj was calculated at any point transient profile using equation 1 and an analyte mass flow Mx versus time diagram was established. Rx stands for the isotope ratio in the sample and Ry stands for the isotope ratio in the spike. Rb stands for the blend ratio which is calculated at any point of the chromatogram and h represents the isotopic abundance of the spike isotope in the spike (y) and in the sample (x). The integration of the peaks in the obtained chromatogram resulted in the absolute amount of the labeled peptide Bβ15-42 injected on the column and the labeled peptide Bβ15-42 concentration was calculated by dividing the absolute amount through the injection volume (5 μL). Blank correction of the integrated peaks was not necessary due to the low carryover of the autosampler. The contribution of peak integration to the total combined uncertainty of the calculated isotope ratios is depending on the number of data points available per chromatographic peak and was evaluated according to Koellensperger et al.29. A contribution of < 5 % to the total combined uncertainty of the isotope ratio was obtained.
3. Results and discussion
Both applied strategies, FIA-FLD and FIA-ICP-SFMS, used a pre-labeling approach where the investigated peptide Bβ15-42 was labeled with a fluorescence or an element marker prior to analysis of the candidate drug in samples resulting from a pre-clinical cell experiment.
3.1. Comparison of FIA-FLD and FIA-ICP-SFMS
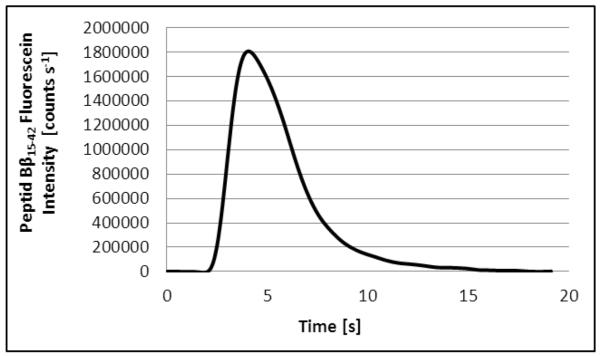
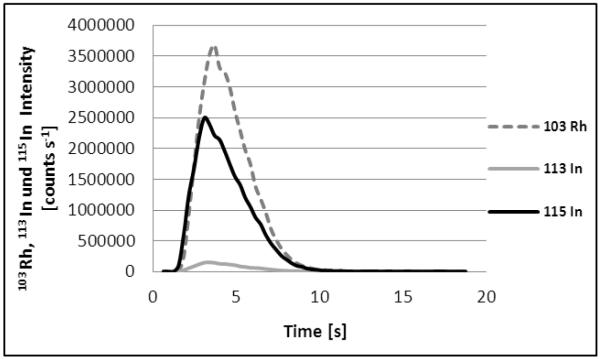
The signals obtained with the two methods, i.e. FIA-FLD and FIA-ICP-SFMS are depicted in Figure 2 (All measured Indium blanks were consistent and no blank correction was necessary). The two methods revealed a peak width (@ 50% peak height) of 5.9 ± 0.2 sec and 6.2 ± 0.2 seconds, respectively. The narrow peaks demanded for a high data collection rate of 3 and 10 datapoints s−1. The overall cycle time of the two methods was very short (1.5 min) allowing high throughput analysis of the examined cellular samples (40 samples per hour). It is noteworthy that sample throughput could be further enhanced for both methods with regard to the acquisition speed of the detection systems. However, this enhancement is currently limited by the cycle times of the utilized autosamplers. Nevertheless, the presented set-up is advantageous in comparison to conventional ICP-MS analysis as it allows much lower cycle times and minimum sample consumption in the low μL-range. As an additional benefit sample preparation for FIA-ICP-MS can be reduced to simple dilution of liquid biological samples.
Figure 2.
a: FIA-FLD signal of 10 nM fluorescein labeled peptide Bβ15-42 (injection volume 25 μL, absolute amount 0.25 pmol).
b: FIA-ICP-SFMS signal of 10 nM In (injection volume 5 μL, absolute amount 0.05 pmol). 1.05 ng/mL Rhodium were added to all measured samples and standards.
A comparison of the two examined detection methods revealed that detection via ICP-SFMS was superior regarding sensitivity and LODs and LOQs compared to the FLD set-up. It is noteworthy that the five-fold injection volume (25 μL) was used in the case of the FIA-FLD system. The LODs were 9 pM (0.05 fmol absolute) for FIA-ICP-SFMS respectively 100 pM (3 fmol absolute) for FIA-FLD (Tab. 3). The correlation coefficient of FIA-FLD gathered with external calibration over a working range of 0.4-100 nM using the fluorescein labeled peptide Bβ15-42 was determined as 0.9948 (with a slope of 13200) whereas the correlation coefficient of the FIA-ICP-SFMS set-up was 0.9998 (Slope: 17000) for external calibration with In standard solution and a working range of 0.03-100 nM.
In the next step long and short term repeatability precision have been determined by measurement of cell lysate samples (Triton X soluble fraction) from cells incubated with the fluorescein and the In labeled peptide Bβ15-42. The measurements (n=5) revealed a short term precision of 1.7 % for the elemental labeling approach and FIA-ICP-SFMS detection (external calibration), and a short term precision of 6.5% for the fluorescence labeling approach. The superior precision of the FIA-ICP-SFMS may be related to the different injection modes i.e. full loop versus partial loop injection. In order to evaluate both methods regarding long term precision 6 repetitive injections were performed within a measurement sequence of 4 hours. The long term precision obtained was approximately in the same range (FIA-ICP-SFMS: 9.5%; FIA-FLD: 9.3%) and can be related to instrument drift which was not compensated via internal standardization at this point.
As a general result most of the analytical figures of merit of the two different approaches were comparable - however, elemental labeling with FIA-ICP-SFMS detection is favorable because of significantly lower absolute LODs (factor of 60) and the availability of different quantification and internal standardization strategies. Moreover it has to be stressed that the selectivity of elemental detection in combination with flow injection is superior compared to fluorescence detection, as treatment media or buffers often contain chemicals, which lead to unspecific fluorescence signals and wrong-positive results.
3.2. Comparison of different FIA-ICP-SFMS quantification strategies
Due to the general lack of adequate matrix reference materials and reference substances in bio-analysis it is beneficial, if alternative quantification strategies are on hand. ICP-MS offers different quantification and standardization strategies e.g. internal standardization with reference elements, species specific IDA and species unspecific IDA. In the present work two different standardization strategies - i.e. internal standardization with Rhodium and species unspecific on-line IDMS via addition of 113In - were compared with external calibration for quantification of the In labeled peptide with FIA-ICP-SFMS.
For an assessment of the three quantification strategies an in-house reference sample was prepared by pooling the cellular fraction (Triton X soluble fraction) of 3 cell lysates obtained after incubation of HUVEC cells with DOTA-In- Bβ15-42. The reference concentration of the In labeled peptide Bβ15-42 in the sample was determined via conventional ICP-SFMS analysis after microwave digestion of two aliquots of the pooled sample. Accurate quantification was carried out via IDMS (113In spike added before digestion) as this strategy is regarded as a primary method of measurement30. The reference concentration of the peptide in the digested samples (n=2 digestions and n=3 measurements per digested sample) was 82.3 nM with a relative standard deviation of 0.3%.
Subsequently several aliquots of the identical, pooled sample were diluted by a factor of 10 and subjected to FIA-ICP-SFMS without further treatment. The values obtained with the three quantification strategies are listed in Table 3. The results compare good, but it is evident that the two approaches without internal standardization of the sample (i.e. external calibration and on-line addition of the indium spike) are significantly lower as the reference value. This can be explained by a bias originating from the injection process due to viscosity effects, which is not corrected for by these approaches. As a consequence all investigated cellular samples were quantified after internal standardization with rhodium.
Table 3.
Analytical figures of merit of FIA – FLD and FIA – ICP – SFMS comparing different quantification strategies.
| FIA-FLD | FIA-ICP- SFMS |
|
|---|---|---|
| LOD | 100 pM | 9 pM |
| LOQ | 400 pM | 30 pM |
| LOD absolute | 3 fmol | 0.05 fmol |
| LOQ absolute | 10 fmol | 0.15 fmol |
| Working range | 0.4 - 100 nM | 0.03 - 100 nM |
| FIA-FLD | FIA-ICP-SFMS | ICP-SFMS | |||
|---|---|---|---|---|---|
| External calibration |
External calibration |
External calibration with internal Standard |
online-IDMS | Reference * | |
|
Concentration of
Indium in cellular sample [nM] |
n.a. | 70.3 ± 1.2 | 87.4 ± 0.8 | 71.9 ± 2.4 | 82.3 ± 0.3 |
|
Repeatability [%]
(short term) (n=5) |
6.5 | 1.7 | 0.9 | 3.3 | |
|
Repeatability [%]
(long term) (n=5) |
9.3 | 9.5 | 4.6 | 3.1 | |
|
Repeatability [%]
of biological replicates (n=3) |
15 | 13 | |||
determined via ICP-SFMS after microwave digestion utilizing isotope dilution analysis as quantification strategy.
In the next step the three quantification strategies were evaluated regarding short and long term precision. As can be seen in Table 3 all strategies revealed comparable short term repeatability (n=5 repetitive injections) in the low percentage range. The approach combining external calibration with internal standardization resulted in the lowest value of 0.9%, since it corrects for any variance associated with the injection process of the autosampler. Regarding long term repeatability quantification of the elemental labeled peptide Bβ15-42 with external calibration yielded 9.5% whereas the use of an internal standard resulted in 4.6% because in this case signal drift is compensated. This is also valid for on-line IDMS, which resulted in a long term precision of 3.3%, accordingly.
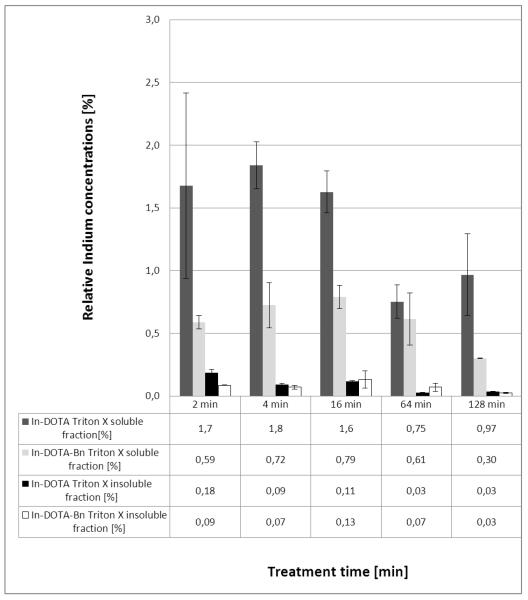
3.3. Relative Indium concentrations in cell extracts after exposure to the labeled peptide Bβ15-42
In the present study the total Indium concentrations in cell extracts (Triton X soluble and insoluble fraction) after exposure to the differently labeled peptide Bβ15-42 by HUVEC cells were measured (Fig. 4). Due to unspecific fluorescence emission of the treatment medium in FIA-FLD, it was not possible to calculate mass balances in this case. Hence, only results for the two elemental labeled peptides are depicted. As can be readily observed in Figure 4 the repeatability of biological experiments was excellent. It can be clearly seen that the relative Indium concentrations in the investigated cell extracts were higher in the first minutes after stimulation. These findings agree with the previous published results obtained via cell binding assay31. The total Indium concentrations in the Triton X soluble fraction were in the range of 30-220 nM (In-DOTA labeled peptide) and 31-82 nM (In-DOTA-Bn). In the Triton X insoluble fraction the total Indium concentrations ranged from 5-62 nM (In-DOTA) and 8-41 nM (In-DOTA-Bn). The relative Indium concentrations (depicted as percentage of total Indium in all fractions) in the Triton X soluble fraction (cell associated fraction of peptide after exposure, washing, cell lysis and extraction with Triton X buffer) was 0.75-1.8% for the In-DOTA labeled peptide Bβ15-42 and 0.30-0.79% for the In-DOTA-Bn labeled peptide Bβ15-42. The relative Indium concentrations in the Triton X insoluble fraction were determined as 0.03-0.18% for the In-DOTA and 0.03-0.13% for the In-DOTA-Bn label. The lower cellular Indium concentrations at 64 and 128 min can be interpreted as a result of excretion of either intact peptide or Indium containing peptide fragments after metabolization. Our results clearly show higher Indium concentrations in the Triton X soluble fraction of the cell lysate suggesting only a loose binding of the Indium labeled peptide Bβ15-42 to cell structures. The higher total Indium concentrations after exposure to the DOTA labeled peptide - compared to exposure to the DOTA-Bn labeled peptide - may be due to the higher negative charge of the DOTA labeled peptide at neutral pH which may result in a tighter attachment to positively charged structures of the cell. This effect seems to overweight covalent cross-linking via the SCN group and this pure electrostatic binding may also explain the higher Indium concentrations in the Triton X insoluble fraction after exposure to the In-DOTA labeled peptide compared to exposure to the In-DOTA-Bn peptide. The gained results show that the relative Indium concentrations are higher in the Triton X soluble fraction in contrast to the Triton X insoluble fraction.
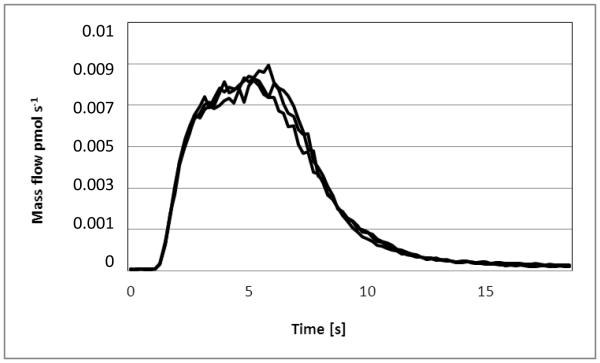
Figure 3.
Mass flow (overlay of 3 signals of the same Trition X soluble fraction) obtained via processing the signals of 115In and 113In (on-line addition of the 113In spike was accomplished by an external syringe pump). The depicted curve represents the mass flow in pmol s−1.
4. Conclusion
Peptide quantification strategies based on fluorescence and elemental labeling in combination with FLD or ICP-SFMS detection show high potential for future research and for different applications in bio-analysis. As a benefit the used flow injection set-up is characterised by low sample consumption and less sample preparation procedures in combination with a lower cycle time, which facilitates high throughput analysis. Additionally the flow injection set-up is directly applicable for speciation analytics.
In this study we have demonstrated that elemental labeling with ICP-MS detection in comparison with fluorescence detection combined with fluorescence labeling is an accurate and precise tool for quantification of drug concentrations in biological samples. But it is also noteworthy that the labeling approach is only reasonable if the functional capabilities of the labeled peptide are not altered. At the moment the potential influence of the different labels on the functionality of the peptide Bβ15-42 is under investigation and we are looking forward to get more information about this condition.
The superior results of FIA-ICP-MS can be explained by the fact that the detection technique is less prone to matrix effects allowing species unspecific calibration and accurate quantification independent of the sample matrix. Additionally, the very low sample intake of the FIA set-up further reduces unwanted signal drift or suppression. Another advantage of elemental labeling and ICP-MS detection is the possibility to implement different quantification strategies e.g. species specific or unspecific IDMS and on-line IDMS, which indicate favorable effects on repeatability. However, our data show that internal standardization is comparable to the IDMS approaches for quantification of labeled compounds in biological systems, both in terms of precision and accuracy. This is due to the fact, that the quantification strategy itself has only a small impact on the total combined uncertainty of measurement, in the case of probing and quantifying biological replicates of cell experiments.
Compared to fluorescence based labeling and detection ICP-MS offers more possibilities for multiplexing because of the variety of elements suitable for labeling, while FLD is limited by the number of available fluorescence dyes. Additionally it should be mentioned that ICP-MS detection combined with elemental labeling endorses a less tedious sample handling because the elemental label is not labile against light radiation.
Our next experiments will address speciation analysis of the samples regarding label stability targeting at free ionic In, DOTA bound Indium and the intact In-DOTA-peptide Bβ15-42.
Supplementary Material
Figure 4.
Relative Indium concentrations in the Triton X soluble fraction and Triton X insoluble fraction of cell extracts after exposure to the In-DOTA and In-DOTA-Bn labeled peptide Bβ15-42. The given uncertainties represent the standard deviation of n=3 biological replicates.
Acknowledgements
We gratefully acknowledge the Austrian Science Found (Project P21739-B11) for providing financial support and Thermo Scientific for providing the HTC PAL Autosampler.
6. References
- 1.Prange A, Pröfrock D. Chemical labels and natural element tags for quantitative analysis of bio-molecules. Journal of Analytical Atomic Spectrometry. 2008;23:432–459. [Google Scholar]
- 2.Hempen C, Karst U. Labeling strategies for bioassays. Analytical and Bioanalytical Chemistry. 2006;384:572–583. doi: 10.1007/s00216-005-3392-0. [DOI] [PubMed] [Google Scholar]
- 3.Ong S, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Molecular and Cellular Proteomics. 2002;1.5:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 4.Mann M, et al. Functional and quantitative proteomics using SILAC. Nature Publishing Group. 2006;7:952–958. doi: 10.1038/nrm2067. [DOI] [PubMed] [Google Scholar]
- 5.Wang M, et al. ICP-MS-based strategies for protein quantification. Mass Spectrometry Reviews. 2010;29:326–348. doi: 10.1002/mas.20241. [DOI] [PubMed] [Google Scholar]
- 6.Gygi S, et al. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nature Biotechnology. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 7.Wiese S, et al. Protein labeling by iTRAQ: A new tool for quantitative mass spectrometry in proteome research. Proteomics. 2007;7:340–350. doi: 10.1002/pmic.200600422. [DOI] [PubMed] [Google Scholar]
- 8.Whetstone P, et al. Element-coded affinity tags for peptides and proteins. Bioconjugate Chemistry. 2004;15:3–6. doi: 10.1021/bc034150l. [DOI] [PubMed] [Google Scholar]
- 9.Liu H, et al. Method for quantitative proteomics research by using metal element chelated tags coupled with mass spectrometry. Analytical Chemistry. 2006;78:6614–6621. doi: 10.1021/ac060895j. [DOI] [PubMed] [Google Scholar]
- 10.Terenghi M, et al. Multiplexed determination of protein biomarkers using metal-tagged antibodies and size exclusion chromatography-inductive coupled plasma mass spectrometry. Analytical Chemistry. 2009;81:9440–9448. doi: 10.1021/ac901853g. [DOI] [PubMed] [Google Scholar]
- 11.Hutchinson R, et al. Imaging and spatial distribution of β-amyloid peptide and metal ions in Alzheimer’s plaques by laser ablation-inductively coupled plasma-mass spectrometry. Analytical Biochemistry. 2005;346:225–233. doi: 10.1016/j.ab.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 12.Becker JS, et al. Bioimaging of metals in brain tissue by laser ablation inductively coupled plasma mass spectrometry (LA-ICP-MS) and metallomics. Metallomics. 2010;2:104–111. doi: 10.1039/b916722f. [DOI] [PubMed] [Google Scholar]
- 13.Becker JS, et al. Bioimaging of metals by laser ablation inductively coupled plasma mass spectrometry (LA-ICP-MS) Mass Spectrometry Reviews. 2010;29:156–175. doi: 10.1002/mas.20239. [DOI] [PubMed] [Google Scholar]
- 14.Tanner M, Günther D. Short transient signals, a challenge for inductive coupled mass spectrometry, a review. Analytica Chimica Acta. 2009;633:19–28. doi: 10.1016/j.aca.2008.11.041. [DOI] [PubMed] [Google Scholar]
- 15.Benkhedda K, et al. Inductive coupled plasma mass spectrometry using flow injection online preconcentration and time of flight mass analyser. Trends in Analytical Chemistry. 2002;21:332–342. [Google Scholar]
- 16.Baranov V, et al. The potential of elemental analysis in biotechnology. Journal of Analytical Atomic Spectrometry. 2002;17:1148–1152. [Google Scholar]
- 17.Tholey A, Schaumlöffel D. Metal labeling for quantitative protein and proteome analysis using inductively-coupled plasma mass spectrometry. Trends in Analytical Chemistry. 2010;29:399–408. [Google Scholar]
- 18.Matar K, El Hashim A, Edafiogho I. Liquid chromatography-tandem mass spectrometric method for determination of E121 in mouse plasma and its application to pharmacokinetics. Journal of Separational Sciences. 2010;33:1888–1896. doi: 10.1002/jssc.200900800. [DOI] [PubMed] [Google Scholar]
- 19.Schaumlöffel D, Lobinski R. Isotope dilution technique for quantitative analysis of endogenous element species in biological systems. International Journal of Mass Spectrometry. 2005;242:217–233. [Google Scholar]
- 20.Wang X, Wang Y. LC-MS/MS coupled with stable isotope dilution method for the quantification of 6-Thioguanine and S6-Methylthioguanine in genomic DNA of human cancer cells treated with 6-Thioguanine. Analytical Chemistry. 2010;82:5797–5803. doi: 10.1021/ac1008628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bettmer J. Application of isotope dilution ICP-MS techniques to quantitative proteomics. Analytical and Bioanalytical Chemistry. 2010;397:3495–3502. doi: 10.1007/s00216-010-3861-y. [DOI] [PubMed] [Google Scholar]
- 22.Mounicou S, Szpunar J, Lobinski R. Inductive-coupled plasma mass spectrometry in proteomics, metabollomics and metallomics studies. European Journal of Mass Spectrometry. 2010;16:243–253. doi: 10.1255/ejms.1059. [DOI] [PubMed] [Google Scholar]
- 23.Lou X, et al. Polymer based elemental tags for sensitive bioassays. Angewandte Chemie International Edition. 2007;46:6111–6114. doi: 10.1002/anie.200700796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quinn Z, et al. Simultaneous determination of proteins using element-tagged immunoassay coupled with ICP-MS detection. Journal of Analytical Atomic Spectrometry. 2002;17:892–896. [Google Scholar]
- 25.Petzelbauer P, et al. The fibrin-derived peptide Bβ15-42 protects the myocardium against ischemia-reperfusion injury. Nature Medicine. 2005;11:298–304. doi: 10.1038/nm1198. [DOI] [PubMed] [Google Scholar]
- 26.Koellensperger G, et al. Quantification of elemental labeled peptides in cellular uptake studies. Journal of Analytical Atomic Spectrometry. 2009;24:97–102. [Google Scholar]
- 27.Ornatsky O, et al. Development of analytical methods for multiplex bio-assay with inductive coupled plasma mass spectrometry. Journal of Analytical Atomic Spectrometry. 2008;23:463–469. doi: 10.1039/b710510j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szökö E, Tabi T. Analysis of biological samples by capillary electrophoresis with laser induced fluorescence detection. Journal of Pharmaceutical and Biomedical Analysis. 2010;53:1180–1192. doi: 10.1016/j.jpba.2010.07.045. [DOI] [PubMed] [Google Scholar]
- 29.Koellensperger G, et al. Uncertainty of species unspecific quantification strategies in hyphenated ICP-MS analysis. Journal of Analytical Atomic Spectrometry. 2003;18:1047–1055. [Google Scholar]
- 30.Comité Consultatif pour la Quantité de Matiere . Rapport de la 1ersession, BIPM, Sèvre. France: 1995. [Google Scholar]
- 31.Bach T, et al. Endothelial cell VE-cadherin functions as a receptor for the β15-42 sequence of fibrin. Journal of Biological Chemistry. 1998;273:30719–30728. doi: 10.1074/jbc.273.46.30719. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.