Abstract
In recent years, our understanding of the pathogenesis of inflammatory bowel diseases (IBD) has greatly increased. Hallmarks of IBD include loss of intestinal barrier function, increased cytokine production and failed resolution of tissue damage. Lasting treatments are still lacking and therefore, a better understanding of the underlying molecular mechanisms is necessary to design novel therapeutic approaches. Apart from transcriptional and post-transcriptional regulation of relevant genes, mammals have evolved a complex and efficient series of mechanisms to rapidly modify newly made proteins for the purposes of signaling and adaptation. These post-translational protein modifications include, amongst others, phosphorylation, hydroxylation, neddylation and cytokine cleavage by the inflammasome. This review focuses on our current understanding of post-translational protein modifications with a particular focus on their relevance to IBD pathogenesis.
Introduction
The inflammatory bowel diseases (IBD), comprised of ulcerative colitis (UC) and Crohn’s disease (CD) are chronic mucosal inflammatory disorders which result from a dysregulated immune response in a genetically predisposed host(1). Although the etiology of CD and UC remains unclear, accumulating evidence suggests that dysfunction of the mucosal immune system contributes fundamentally to the pathogenesis of IBD. Significant evidence implicates impaired innate immunity (granulocytes, macrophages and dendritic cells) in IBD, particularly CD(2). Such defects have the potential to allow abnormal microbial invasion and pathological T-cell mediated chronic inflammation. In this context, signaling defects and the protein processing associated with such signaling has become an area of increasing interest and importance. In this context, the modification of proteins at the post-translational level significantly extends the regulatory possibilities of a given pathway. Protein processing through a variety of means allows for tight control and rapid responses to inflammation beyond that of direct transcriptional control and de novo protein synthesis. This review will focus on protein phosphorylation, neddylation and hydroxylation as a means to control the inflammatory response (Overview in Figure 1). We will also summarize some of the recent findings with regard to cytokine cleavage by the inflammasome.
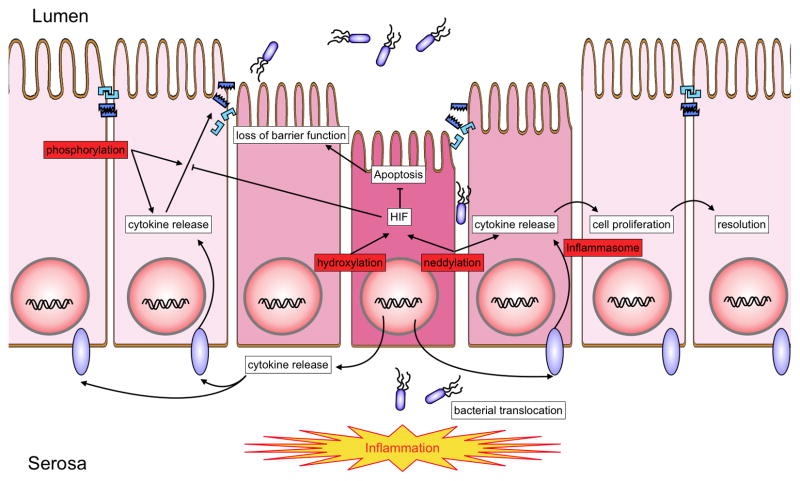
Figure 1. Overview of posttranslational protein modifications relevant to IBD.
A number of posttranslational protein modifications relevant to IBD (highlighted in red). These pathways include phosphorylation, neddylation hydroxylation and cleavage of cytokine precursor forms by the inflammasome. The interplay of these rapid response mechanisms enables rapid adaptation to incoming inflammatory signals. Cytokine induced barrier breakdown allows for bacterial translocation to the basal aspect of intestinal epithelial cells. Bacterial antigens and endogenous danger signals are recognized by the adaptive and innate immune system, triggering a variety of reactions including apoptosis, increased cytokine release, loss of tight junctional proteins and barrier breakdown.
Phosphorylation
Inflammatory pathways are regulated, in part, through a relatively small number of transcription factors, of which the best understood is NF-κB (3). Upon cellular stimulation by a variety of mediators including cytokines, bacterial toxins, or oxidative stress, a signaling transduction cascade is activated, leading to the phosphorylation of IκBα on Ser 32 and 36 by the multimeric IKK (IκB kinase) complex (4). Phosphorylation of IκBα is followed by ubiquitination via the E3 ligase SCFβTRCP and IκBα is targeted for proteosomal degradation by the 26S proteasome (5). Once IκBα is degraded, NF-κB translocates to the nucleus and binds to the promoter regions of several pro-inflammatory genes, inducing their expression and thus amplifying the inflammatory response (5–7) (Figure 2). Mouse models of intestinal inflammation have been quite revealing, and even surprising, with regard to NF-κB signaling. While most studies have suggested that the activation of NF-κB is a strong pro-inflammatory biomarker, studies with mice bearing intestinal epithelial defects in either the phosphorylation of IκBα or the activation of NF-κB have been somewhat surprising. For example, it is notable that conditional deletion of the NF-κB pathway in intestinal epithelial cells in mice leads to an increased susceptibility to colitis(8). This observation strongly implicates epithelial NF-κB in a prominently protective role in colitis, likely through the expression of anti-apoptotic genes in intestinal epithelial cells and through enhanced epithelial barrier function.
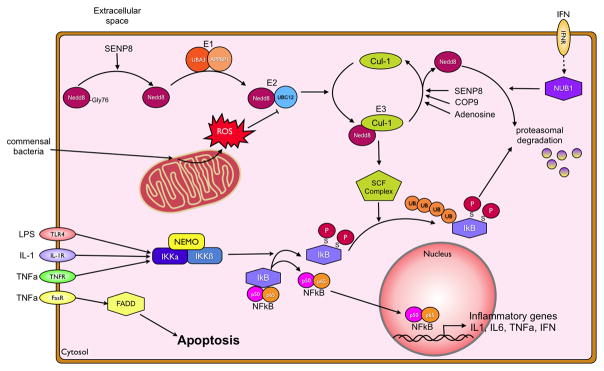
Figure 2. Phosphorylation and neddylation pathways.
Recognition of pro-inflammatory stimuli, presence of microbial cell wall components, secreted TNFα or various other cytokines, through their respective receptors triggers a pro-inflammatory first response of the IEC. The NFκB pathway is activated through the phosphorylation of IκB by the Iκ-kinases α and β. This phosphorylation allows for its recognition by the neddylated Skp-Cullin-F-Box (SCF) complex, polyubiquitination and subsequent proteasomal degradation. Neddylation of Cullins is achieved through a multienzyme process, conjugating a NEDD8 moiety to the target protein. NEDD8, in order to be conjugated has to be processed from its pro-form to the mature form by the isopeptidase SENP8. The same protein, in addition to the COP9 signalosome removes NEDD8 from Cullins. Neddylated Cullins are integrated into SCF complex and activate it to enable SCF complex mediated ubiquitination. The NFκB heterodimer can then translocate to the nucleus and bind to the promoter regions of various pro- and anti-inflammatory cytokines, including TNFα, IL-1β and interferon-γ. This can further promote inflammation, e.g. by TNFα induced apoptosis through the FADD pathway, and barrier breakdown, but also abrogate the incoming stimulus by induction of NUB1 through the interferon receptor. Anti-inflammatory mechanisms include the induction of deneddylation of Cullin-proteins through adenosine and ROS mediated inhibition of the E2 ligase NEDD8/UBC12.
The tight regulation of phosphorylation of IκBα and NF-κB activity are also important for gut homeostasis, particularly through gut microflora interactions with the host. For example, Neish et al. demonstrated that enteric microorganisms (e.g. nonvirulent Salmonella strains) interactions with epithelial cells attenuate synthesis of inflammatory effector molecules elicited by diverse proinflammatory stimuli (9). These studies revealed that although phosphorylation of IκBα occurs, subsequent polyubiquitination necessary for regulated IκBα degradation was completely abrogated by non-virulent enteric organisms. Likewise, work by Kelly and colleagues showed that non-pathogenic bacteria in the gut, and their regulation of the NF-κB pathway, are important for maintaining intestinal homeostasis. They were able to show, that commensal Bacteroides thetaiotaomicron species dampen inflammatory processes by the translocation of the NF-κB subunit RelA from the nucleus to the cytoplasm in a PPARγ dependent manner (10).
Another important example of posttranslational modification by phosphorylation in IBD is the phosphorylation of myosin light chain kinases (MLCK) through atypical protein kinases C (aPKC)(11). The breakdown of intestinal barrier integrity through loss of tight junctions (TJs) is one of the hallmarks of IBD (12–14) and is in part cytokine-mediated (15–17). Several groups have by now provided evidence, that MLCKs are important for this downregulation of tight junction function by mediating incomplete maturation and distribution of TJs (18–20) (Figure 3). aPKCs have been shown to be necessary for the proper alignment of tight junctions in epithelia since lack of aPKCs through dominant negative expression results in atypical localization of tight junction proteins (21). aPKC proteins are posttranslationally modified through PDK-1 mediated phosphorylation and dephosphorylation. Dephosphorylated PKCs are subsequently degraded in a ubiquitin dependent matter (22). Part of the dephosphorylated PKCs can be salvaged and rephosphorylated by heat shock protein-70 (Hsp70) (23). This chaperoning activity of Hsp’s is necessary to prevent ubiquitination and degradation as has been shown by Mashukova and colleagues (24). Recently the same group provided evidence that TNFα actively downregulates aPKC activity in a human colon cancer cell line (CaCo2) and in murine dextran sodium sulfate (DSS)-induced colitis. Furthermore, they showed that TNFα signaling inhibits the Hsp70 activity required for the rephosphorylation of aPKC subsequently disrupting epithelial barrier function as indicated by a loss of transepithelial resistance (25). This provides evidence, that inflammation induced intestinal barrier breakdown, at least in part, relies on a posttranslational mechanism (Figure 3).
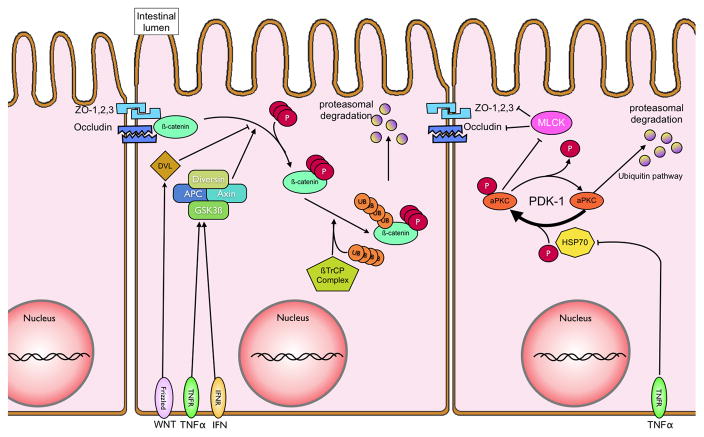
Figure 3. Intestinal barrier regulation in iflammation.
Following NFκB activation, various cytokines, including TNFα, are secreted in a paracrine fashion. Recognition of TNFα by its surface receptor TNFR1, inhibits heat shock protein 70 (HSP70). HSP70 mediates the re/-phosphorylation of atypical Protein Kinase C (aPKC) by serving as a chaperoning agent for the unphosphorylated aPKC. Phosphorylated aPKC inhibits Myosin light chain kinases, which prevent full maturation of tight junctional proteins (zonula occludens 1,2,3 and occludins) leading to decreased transepithelial resistance and loss of barrier function. Tight junctional integrity also relies on the interaction between β-catenin and the cytoskeleton. TNF-α and interferon-gamma, secreted during inflammation, can activate a multienzyme complex comprised of diversin, adenomatous polyposis coli (APC), Axin and glycogen synthase 3b (GSK3β). The GSK3β subunit phosphorylates β-catenin, allowing for its recognition and ubiquitination by the β-TrCP/E3 ligase. Ubiquinated β-catenin is subsequently proteasomally degraded, and thus unavailable for enhancing barrier function.
Finally, there is much recent interest in beta-catenin signaling in both intestinal inflammation and intestinal cancer (26, 27). Like NF-κB, beta-catenin is tightly controlled by phosphorylation. In the absence of a Wnt signal, the expression of beta-catenin is controlled to low levels through proteasomal degradation. Beta-catenin is targeted for ubiquitination by the beta-transducin repeat containing protein (βTrCP) and is then degraded by the proteosome. Beta-catenin is phosphorylated by the serine/threonine kinases casein kinase 1 (CK1) and glycogen synthase 3β (GSK3β) in a multi-protein complex (the destruction complex) comprised of axin, adenomatous polyposis coli (APC) and diversin. CK1 phosphorylates β-catenin on Ser45, which primes β-catenin for subsequent phosphorylation by GSK-3 (28). GSK-3β destabilizes β-catenin by phosphorylation at Ser33, Ser37, and Thr41 (29) (Figure 3). Mutations at these sites result in the stabilization of β-catenin protein levels and have been implicated in tumor growth and upregulation of proliferative genes (30, 31). Following activation by a Wnt signal, the protein dishevelled prevents degradation of beta-catenin that in turn displaces GSK3β from the destruction complex. Thus, inhibition of GSK3β might serve as a potential therapeutic target in different disease models, as recently reviewed by Klamer et al. (32). Recent studies have suggested that in addition to its role in cancer progression, beta-catenin significantly contributes to epithelial homeostasis during inflammation. For instance, Nava et al, recently showed that interferon-gamma (IFNγ) and TNFα synergistically inhibit epithelial proliferation during modeled inflammation in vitro and during DSS through a mechanism involving beta-catenin phosphorylation (33).
Taken together, these studies strongly implicate protein phosphorylation events as central control points for both pro- and anti-inflammatory signaling in intestinal inflammatory models.
Neddylation
A mechanism of post-translational modification of much recent interest is neddylation, i.e. the reversible conjugation of a NEDD8 (Neural precursor cell expressed, developmentally down-regulated 8) (34) moiety to proteins (35). Neddylation and deneddylation responses are highly conserved exist in a wide variety of cell types (36) and species (37–40). Activating the inactive Nedd8-precursor through cleavage a carboxy-terminal glycine residue by UCH-L3 or SENP8 enables it to be conjugated to the E1 UBA3-APPBP1 heterodimer (41–44). Subsequently NEDD8 is conjugated to its specific E2 Ubc12 (ubiquitin conjugating enzyme) (45) and afterwards linked to the E3 complex (46, 47) (Figure 2). Neddylation plays a large role in the post-translational modification of Cullin-RING-ligases (48) involved in the ubiquitin pathway. Cullins act as scaffolding proteins and are essential for the assembly of the ubiquitin E3 ligase complex conjugating ubiquitin to target proteins and thus marking them for proteasomal degradation (49). New insights into potential roles for Cullin-deneddylation in inflammation have come of interest in recent years. The work by Neish et al. alluded to above have demonstrated that commensal bacteria-associated attenuation of NFκB is Cullin-de-neddylation-dependent (9). Furthermore, Kumar et al. were able to demonstrate that commensal bacteria can influence the neddylation status of Cullin-1 (Cul1) through generation of reactive oxygen species (ROS). Initially the demonstrated, that epithelial cells react with increases in ROS when co-cultured with commensal bacteria. This resulted in a transient and reversible deneddylation of Cul1 and subsequent decrease of NFκB pathway end products. Interestingly, they were able to show, that different commensal bacterial strains differ in the amount of ROS they generate. Since there is an altered microbiota in patients with IBD compared to healthy subjects, and commensal bacterial strains also differ in their primary location in the gut, there might be different amounts of ROS in different parts of the intestine altering the inflammatory response in IBD (50).
Adenosine receptor signaling has also been linked to neddylation. While signal transduction through the various adenosine receptors is well characterized, less is known about post-receptor events (51). One particularly intriguing mechanism suggests that adenosine inhibits NF-κB through actions on proteasomal degradation of IkB proteins (52). These findings were based on studies addressing adenosine signaling mechanisms which revealed that adenosine and adenosine analogs display a dose-dependent deneddylation of Cul-1 with rank order of receptor potencies A2BAR >A1AR≫A2AAR = A3AR (52). Our current understanding is that deneddylation reactions on Cullin targets via CSN-associated proteolysis is increasingly implicated as a central point for Cullin-mediated E3 ubiquitylation (48). Notably, other pathways for deneddylation have been reported. For example, the identification of the Nedd8-specific proteases NEDP1 and DEN1 have provided new insight into this emerging field. NEDP1/DEN1 appear to contain isopeptidase activity capable of directly deneddylating Cullin targets (53, 54). How adenosine influences NEDP1/DEN1 activity is not currently known.
Neddylation of other Cullin proteins (i.e. Cul-2) have also been implicated in mucosal inflammation, particularly related to hypoxia-inducible factor (HIF). HIF is a member of the Per-ARNT-Sim (PAS) family of basic helix-loop-helix (bHLH) transcription factors. HIF activation is dependent upon stabilization of an O2-dependent degradation domain of the α subunit and subsequent nuclear translocation to form a functional complex with HIF-1β and cofactors such as CBP and its ortholog p300 (55). Under conditions of adequate oxygen supply, iron and oxygen dependent hydroxylation of two prolines (Pro564 and Pro402) within the oxygen-dependent degradation domain (ODD) of HIF-1α initiates the association with the von Hippel-Lindau tumor suppressor protein (pVHL) and rapid degradation via ubiquitin-E3 ligase proteasomal targeting (56, 57). A second hypoxic switch operates in the carboxy terminal transactivation domain of HIF-1α Here, hypoxia blocks the hydroxylation of asparagine (Asn80), thereby facilitating the recruitment of CBP/p300 (58). The proteasomal degradation of α-subunit of HIF provides a particularly intriguing example of post-translational modification (Figure 4). The E3 SCF ubiquitin ligase specific to HIFα-family members are comprised of Elongin B/C, RBX, CUL2, and the F-box domain of pVHL, and are responsible for the polyubiquitination of HIFα (59). Regulation of the E3 SCF is maintained by the covalent modification of NEDD8. The functional E3-SCF requires the COP9 signalosome (CSN) to bind Nedd8 to Cul2, which can be de-neddylated by Den1/SenP8 (60, 61). In recent work by MacManus et al (62), it was shown that the HIF target gene adrenomedullin (ADM) functions as an endogenously generated vascular mediator that serves as a mucosal protective factor through fine-tuning of HIF. The underlying mechanism involved ADM-mediated deneddylation of Cul2, resulting in less pVHL activity and subsequent fine-tuning of HIF expression. Exogenous administration of ADM in a DSS colitis model resulted in decreased tissue and serum levels of pro-inflammatory cytokines, identifying the Cul2 pathway as another potential therapeutic target for IBD (62).
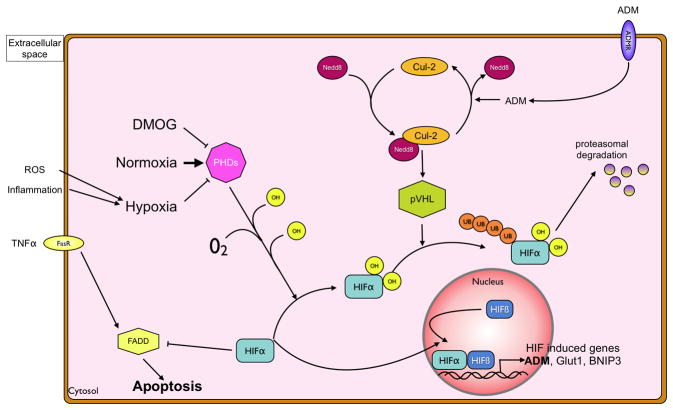
Figure 4. Regulation of hypoxia-inducible factor by hydroxylation.
Under normoxic conditions, the HIFα subunit is hydroxylated through prolylhydroxylases 1–3 (PHDs) or the Factor-inhibiting-HIF (FIH). Hydroxylated HIF is recognized by the von Hippel-Lindau (pVHL) protein, which in its activated state contains a neddylated Culllin-2 subunit. This leads to ubiquitination of the HIF1α subunit and its degradation by the proteasome. During inflammation, local oxygen is limited due to increased metabolism and also due to neutrophil derived reactive oxygen species. This hypoxic environment renders PHDs impotent, as does pharmacological inhibition with DMOG, preventing ubiquitination of HIFα by pVHL. Non-ubiquitinated HIFα translocates to the nucleus, binds to the beta-subunit and the heterodimer functions as a transcription factor of a variety of genes, including adrenomedullin (ADM). Furthermore, HIFα inhibits Fas-associated-death-domain (FADD) induced apoptosis, thereby increasing intestinal barrier function. The autocrine release of ADM can, through the ADM receptor-mediated deneddylation of Cul2, serve as a negative feedback mechanism.
Hydroxylation
It is recently appreciated that studies of the oxygenation profile of the gut may provide important insight into the pathogenesis of IBD. While the intestinal tract is highly vascularized, blood flow is tightly regulated and fluctuates multiple times during the day (e.g. postprandial increases in blood flow). As a result, the amount of oxygen available to the intestinal tissue changes in fundamental ways as part of our normal physiology. It is thought that the steep gradient between the highly metabolic serosa and the anaerobic lumen of the gut primes the intestinal epithelium for rapid responses to changes in tissue oxygenation (63). In particular, inflammatory processes can rapidly increase the demand for oxygen in inflamed tissue, thereby leading to profound hypoxia (64), so called “inflammatory hypoxia” (65). Adaptation to hypoxia is, at least in part, mediated through the HIF-1 and HIF-2 (66, 67). Many cell types, including intestinal epithelial cells (IEC) (68), express both HIF1α and HIF2α and murine genetic studies suggest that these proteins have non-redundant roles (69). Some have suggested that distinct transcriptional responses mediated by HIF1α and HIF2α may be integrated in ways that support particular adaptations to hypoxia. For example, the transcriptional responses that coordinate the glycolytic pathways include more than 11 target genes and seem to be more selective for the HIF-1α than for the HIF-2α isoform (69). Likewise, studies addressing the selectivity of the two isoforms of HIFα suggest greater selectivity of HIF-2 for both erythropoietin production (69) and for intestinal iron transport (70).
The stability of the HIFα subunit is post-translationally regulated by four prolylhydroxylases (PHD1–3 and Factor Inhibiting HIF, FIH), all of which are present in intestinal epithelial cells (71–74). Under normoxic conditions, these enzymes hydroxylate HIF1-α at specific prolines (PHDs) and/or at a specific asparaginyl (FIH) residue (66, 71). This hydroxylation leads to interaction with the von Hippel-Lindau protein, poly-ubiquitination of HIF-α subunit and subsequent proteasomal degradation (75) (Figure 4). Several studies have shown that HIF triggers the transcription of a number of genes that enable IEC to function as an effective barrier. Guided initially by microarray analysis of hypoxic IEC (76), these studies have been validated in animal models of intestinal inflammation (72, 73, 77–80) and in human intestinal inflammation tissues (81–83). Interestingly, the functional proteins encoded by a number of uniquely hypoxia-inducible genes in intestinal epithelia localize primarily to the most luminal aspect of polarized epithelia, providing significant support for the hypothesis that hypoxia supports a barrier-protective phenotype. Molecular studies of these hypoxia-elicited pathway(s) have shown a dependence on HIF-mediated transcriptional responses. Notably, epithelial barrier protective pathways driven by HIF tend not to be the classical regulators of barrier function, such as the tight junction proteins occludin or claudins. Rather, the HIF-regulated molecules include molecules which support overall tissue integrity and include increased mucin production, (84) molecules that modify mucin (e.g. intestinal trefoil factor)(85), promote xenobiotic clearance via P-glycoprotein (86), enhance nucleotide metabolism (by ecto-5′-nucleotidase and CD73)(76, 87) and drive nucleotide signaling (e.g. adenosine A2B receptor) (87).
As an extension of the original studies identifying HIF stabilization within the intestinal mucosa, transgenic mice expressing either mutant Hif1α (causing constitutive repression of Hif1α) or mutant von Hippel-Lindau (causing constitutive overexpression of HIF) were targeted to the IEC (78). Loss of epithelial HIF-1α resulted in a more severe colitic phenotype than wild-type animals, including increased epithelial permeability, enhanced loss of bodyweight, and decreased colon length. Constitutively active intestinal epithelial HIF (mutant Vhl) was protective for each of these individual parameters. These findings may be somewhat model-dependent, since epithelial HIF-based signaling has also been shown to promote inflammation in another study (80). Nonetheless, these findings have revealed that IEC can adapt to even severe hypoxia and that HIF contributes in fundamental ways to this adaptation.
The identification of HIF-selective PHDs has provided unique opportunities for the development of PHD-based therapies (88, 89). While there is wide interest in developing HIF inhibitors as potential cancer therapies, opportunities also exist to selectively stabilize HIF in an attempt to promote inflammatory resolution (90). For example, 2-OG analogues stabilize HIF-α (88) and effectively promote the resolution of colitis in mouse models (72). Interestlingly, the protection afforded by PHD inhibitors (e.g. decreased tissue inflammatory cytokines, increased barrier function, decreased epithelial apoptosis) may involve both HIF and NF-κB activities. For example, in a genetic screen of PHD isoform deficient animals, Tambuwala et al. revealed that Phd1−/− mice were less susceptible to the development of DSS colitis, likely through decreased epithelial cell apoptosis (91), which was originally shown to be NF-κB-dependent (72). Likewise, it was shown that hydroxylase inhibiton inhibits TNF-α induced barrier breakdown. Hindryckx and colleagues demonstrated, that DMOG repressed FADD (Fas-associated death domain protein), a linkage protein for the TNFα-receptor-1. This inhibition reduced TNF-α induced apoptosis and restored, or prevented loss of, epithelial barrier function. This response was HIF1-α mediated, and not dependent on abrogation of the NFκB pathway, since siRNA inhibition of HIF1-α diminished the protective function of DMOG despite a fully functional NFκB pathway (92). To date, selective inhibitors for particular PHD isoforms have not become available.
The inflammasome
Recently, the inflammasome has generated some interest in its role in IBD. The inflammasome consists of several receptors of the NLR family (nucleotide-binding domain leucin rich repeat), a class of more than 20 receptors, who differ in their N-terminal domains (93–95). One subset of the NLR family, containing NLRP1, NLRP3, and NLRC4 are termed the inflammasome, defined by its ability to trigger the activation of capase-1 (96–98). Activated caspase-1 post-translationally cleaves pro-IL-1β and pro-IL-18, resulting in active forms of IL-1 and IL-18. Similar to TNF-α, IL-1β has been shown to be capable of increased epithelial barrier breakdown through the NFκB-pathway (99). From this perspective, the inflammasome provides an interesting post-translational modification of importance to disease pathogenesis.
Numerous exogenous and endogenous activators of the NLRP3 inflammasome have been identified, including bacteria (e.g. commensal E. coli (100)), different bacterial toxins (e.g. LPS), bacterial RNA and poly(I:C)(101), reactive oxygen species (102) and ATP(103) (Figure 5). Recently, Bauer and colleagues demonstrated a strong role for the NRLP3 inflammasome for murine experimental colitis. Macrophages lacking different components of the inflammasome (e.g. NRLP3, ASC or caspase-1) were unable to secrete the mature form of IL-1β following DSS stimulation. Using mice deficient for NRLP3, they were able to show that DSS-induced colitis was significantly less severe compared to wild-type mice, revealed by less severe weight loss and reduced pro-inflammatory cytokine levels in colonic tissue. Similar results were achieved by utilizing pralnacasan to pharmacologically inhibit caspase-1 (104). This study stands in contrast to other studies showing a hyper-susceptible phenotype of NLRP3 deficient mice for DSS-induced colitis, potentially due to the lack of pro-resolution effects of IL-1β and IL-18 (105–107). Especially the contribution of IL-18 to the etiology of IBD remains unclear. Initially it was believed, that high levels of IL-18 favored increased disease activity since inhibition of IL-18 protected mice from experimental colitis (108–110). However, more recent data by Dupaul-Chicoine and colleagues, hints at a more protective role of IL-18 and the inflammasome. They observed, that mice lacking caspase-1 developed more severe colitis following DSS-treatment and that administration of IL-18 attenuated this effect (106). Lack of the inflammasome (Nlrp3-knockout mice) and thus the inability to activate IL-18 increases bacterial translocation, i.e. decreases barrier function of IECs, underlining a more protective role of IL-18 (111).
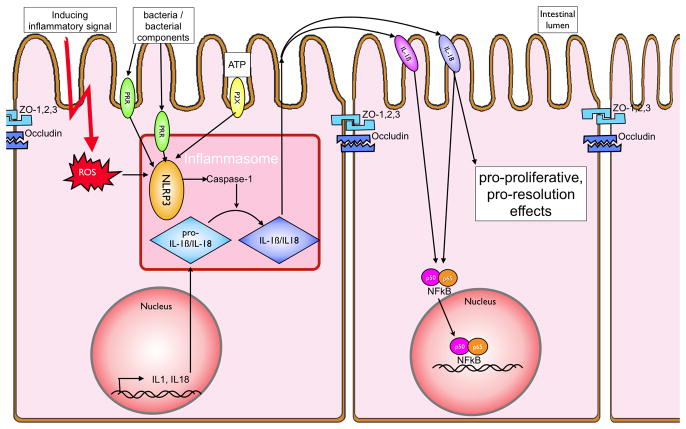
Figure 5. The inflammasome and IBD.
Activation of the NLRP3 inflammasome by either cell surface (e.g. TLR4 via the TRIF pathway) or intracellular (e.g. TLR9 or NOD-receptors) pattern recognition receptors leads to the cleavage of the IL-1β and IL-18 precursors to their mature form. This activation is achieved through pathogen associated molecular patterns (PAMPs) and also through endogenous danger associated molecular patterns (DAMPs) such as extracellular ATP (via the P2X7 receptor) or intracellular reactive oxygen species (ROS). IL-1β and IL-18, when secreted, bind to their respective receptors and activate the NFκB pathway. In addition, there are possible anti-inflammatory influences attributed to IL-18 and enhanced cell proliferation, thereby leading to more rapid resolution of intestinal barrier breakdown.
Genetic variants and their implications for post-translational protein modifications in IBD
The identification of genetic variants is an area of intense investigation in IBD. To date, genome wide association studies have identified nearly 100 susceptibility loci for IBD (112, 113). Of particular interest for protein post-translational modifications in IBD is the caspase-recruitment containing domain 9 (CARD9) gene product (114). CARD9 has been strongly implicated in apoptosis and NFκB signaling as a convergence signal for TLR and NOD2 pathways(115), the latter of which has been prominently associated as a genetic variant in IBD (112, 113). Numerous studies have indicated that CARD9 signaling is strongly associated with dectin-1 receptor signaling, which relays incoming β-glucan signals to the inflammasome and leads to increased IL-1β synthesis (116). Furthermore, Bi et al. recently demonstrated, that CARD9 is able to relay Dectin-2 signals and associated increases in IκBα kinase ubiquitination (117). As outlined above, increased ubiquitination leads to degradation of IκB and subsequently increased NFκB levels. This observation nicely compliments the previous finding by Glocker and colleagues that the homozygous mutation of CARD9 results in increased susceptibility to fungal infections (118). CARD9 has now been identified as one of the many susceptibility loci for CD (119). This might be due to the impaired NFκB signaling and subsequent loss of barrier function. As Nenci and colleagues have shown, the IEC specific loss of NFκB signaling prevents appropriate responses to inflammatory stimuli and results in a pro-inflammatory phenotype with increased apoptosis and loss of barrier integrity (120). Thus, a loss of CARD9 signaling via genetic mutation could result in a loss of NFκB signaling and the associated loss of barrier. Additionally, CARD9 has been linked to pVHL function. Yang et al. showed that pVHL is able to downregulate NFκB activity by increasing phosphorylation of CARD9. This leads to decreased CARD9 activity and thus decreased NFκB levels (121). Using CARD9 as an example, genetic variations can significantly alter post-translational modifications such as ubiquitination and phosphorylation, leading to altered NFκB levels, which have been shown to be critical for the development of IBD.
Conclusion
The gastrointestinal mucosa provides an interesting example for which to study post-translation protein modifications in IBD. In this review, we have outlined the evidence for post-translation modifications as a priming signal within the intestinal mucosa and their associated changes in models of IBD and in tissue from both CD and UC patients. These studies have documented several important examples of protein post-translation modifications as significant components of the inflammatory microenvironment, both directly and indirectly associated with protein post-translational modifications. This work has resulted in the identification and validation of a number of potentially important therapeutic targets along the length of the gastrointestinal tract. Ongoing studies to define the differences and similarities between innate and adaptive immune responses between and within IBD patients will continue to teach us important lessons about the complexity of this organ system. Such information will provide new insight into the pathogenesis of disease and importantly, will refine these targets as templates for the development of therapies for human disease.
Acknowledgments
This work was supported by a German Research Foundation (DFG) grant (EH 371/1-1) to S.E. and by National Institutes of Health Grants DK50189, HL60569 and by funding from the Crohn’s and Colitis Foundation of America.
Footnotes
Disclosure statement: The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as influencing the objectivity of this review.
References
- 1.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 2.Marks DJ, Rahman FZ, Sewell GW, et al. Crohn’s disease: an immune deficiency state. Clin Rev Allergy Immunol. 2010;38:20–31. doi: 10.1007/s12016-009-8133-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnes PJ, Karin M. Nuclear factor-κB - A pivitol transcription factor in chronic inflammatory disease. N Engl J Med. 1997;336:1066–1071. doi: 10.1056/NEJM199704103361506. [DOI] [PubMed] [Google Scholar]
- 4.Wojcik C, Di Napoli M. Ubiquitin-proteasome system and proteasome inhibition: new strategies in stroke therapy. Stroke. 2004;35:1506–1518. doi: 10.1161/01.STR.0000126891.93919.4e. Epub 2004 Apr 1529. [DOI] [PubMed] [Google Scholar]
- 5.Chen Z, Hagler J, Palombella VJ, et al. Signal-induced site-specific phosphorylation targets I kappa B alpha to the ubiquitin-proteasome pathway. Genes Dev. 1995;9:1586–1597. doi: 10.1101/gad.9.13.1586. [DOI] [PubMed] [Google Scholar]
- 6.Brown K, Gerstberger S, Carlson L, et al. Control of I kappa B-alpha proteolysis by site-specific, signal-induced phosphorylation. Science. 1995;267:1485–1488. doi: 10.1126/science.7878466. [DOI] [PubMed] [Google Scholar]
- 7.DiDonato JA, Hayakawa M, Rothwarf DM, et al. A cytokine-responsive IkappaB kinase that activates the transcription factor NF-kappaB. Nature. 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 8.Zaph C, Troy AE, Taylor BC, et al. Epithelial-cell-intrinsic IKK-beta expression regulates intestinal immune homeostasis. Nature. 2007;446:552–556. doi: 10.1038/nature05590. [DOI] [PubMed] [Google Scholar]
- 9.Neish AS, Gewirtz AT, Zeng H, et al. Prokaryotic regulation of epithelial responses by inhibition of IkappaB- alpha ubiquitination. Science. 2000;289:1560–1563. doi: 10.1126/science.289.5484.1560. [DOI] [PubMed] [Google Scholar]
- 10.Kelly D, Campbell JI, King TP, et al. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-gamma and RelA. Nat Immunol. 2004;5:104–112. doi: 10.1038/ni1018. [DOI] [PubMed] [Google Scholar]
- 11.Marchiando AM, Graham WV, Turner JR. Epithelial barriers in homeostasis and disease. Annu Rev Pathol. 2010;5:119–144. doi: 10.1146/annurev.pathol.4.110807.092135. [DOI] [PubMed] [Google Scholar]
- 12.Bruewer M, Samarin S, Nusrat A. Inflammatory bowel disease and the apical junctional complex. Ann N Y Acad Sci. 2006;1072:242–252. doi: 10.1196/annals.1326.017. [DOI] [PubMed] [Google Scholar]
- 13.Laukoetter MG, Bruewer M, Nusrat A. Regulation of the intestinal epithelial barrier by the apical junctional complex. Curr Opin Gastroenterol. 2006;22:85–89. doi: 10.1097/01.mog.0000203864.48255.4f. [DOI] [PubMed] [Google Scholar]
- 14.Schmitz H, Barmeyer C, Fromm M, et al. Altered tight junction structure contributes to the impaired epithelial barrier function in ulcerative colitis. Gastroenterology. 1999;116:301–309. doi: 10.1016/s0016-5085(99)70126-5. [DOI] [PubMed] [Google Scholar]
- 15.Schmitz H, Fromm M, Bentzel CJ, et al. Tumor necrosis factor-alpha (TNFalpha) regulates the epithelial barrier in the human intestinal cell line HT-29/B6. J Cell Sci. 1999;112 ( Pt 1):137–146. doi: 10.1242/jcs.112.1.137. [DOI] [PubMed] [Google Scholar]
- 16.Bruewer M, Luegering A, Kucharzik T, et al. Proinflammatory cytokines disrupt epithelial barrier function by apoptosis-independent mechanisms. J Immunol. 2003;171:6164–6172. doi: 10.4049/jimmunol.171.11.6164. [DOI] [PubMed] [Google Scholar]
- 17.Wang F, Schwarz BT, Graham WV, et al. IFN-gamma-induced TNFR2 expression is required for TNF-dependent intestinal epithelial barrier dysfunction. Gastroenterology. 2006;131:1153–1163. doi: 10.1053/j.gastro.2006.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turner JR. Molecular basis of epithelial barrier regulation: from basic mechanisms to clinical application. Am J Pathol. 2006;169:1901–1909. doi: 10.2353/ajpath.2006.060681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye D, Ma I, Ma TY. Molecular mechanism of tumor necrosis factor-alpha modulation of intestinal epithelial tight junction barrier. Am J Physiol Gastrointest Liver Physiol. 2006;290:G496–504. doi: 10.1152/ajpgi.00318.2005. [DOI] [PubMed] [Google Scholar]
- 20.Ma TY, Boivin MA, Ye D, et al. Mechanism of TNF-{alpha} modulation of Caco-2 intestinal epithelial tight junction barrier: role of myosin light-chain kinase protein expression. Am J Physiol Gastrointest Liver Physiol. 2005;288:G422–430. doi: 10.1152/ajpgi.00412.2004. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki A, Ishiyama C, Hashiba K, et al. aPKC kinase activity is required for the asymmetric differentiation of the premature junctional complex during epithelial cell polarization. J Cell Sci. 2002;115:3565–3573. doi: 10.1242/jcs.00032. [DOI] [PubMed] [Google Scholar]
- 22.Newton AC. Regulation of the ABC kinases by phosphorylation: protein kinase C as a paradigm. Biochem J. 2003;370:361–371. doi: 10.1042/BJ20021626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao T, Newton AC. Invariant Leu preceding turn motif phosphorylation site controls the interaction of protein kinase C with Hsp70. J Biol Chem. 2006;281:32461–32468. doi: 10.1074/jbc.M604076200. [DOI] [PubMed] [Google Scholar]
- 24.Mashukova A, Oriolo AS, Wald FA, et al. Rescue of atypical protein kinase C in epithelia by the cytoskeleton and Hsp70 family chaperones. J Cell Sci. 2009;122:2491–2503. doi: 10.1242/jcs.046979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mashukova A, Wald FA, Salas PJ. Tumor necrosis factor alpha and inflammation disrupt the polarity complex in intestinal epithelial cells by a posttranslational mechanism. Mol Cell Biol. 2011;31:756–765. doi: 10.1128/MCB.00811-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bossard C, Souaze F, Jarry A, et al. Over-expression of neurotensin high-affinity receptor 1 (NTS1) in relation with its ligand neurotensin (NT) and nuclear beta-catenin in inflammatory bowel disease-related oncogenesis. Peptides. 2007;28:2030–2035. doi: 10.1016/j.peptides.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 27.Tsuchiya K, Nakamura T, Okamoto R, et al. Reciprocal targeting of Hath1 and beta-catenin by Wnt glycogen synthase kinase 3beta in human colon cancer. Gastroenterology. 2007;132:208–220. doi: 10.1053/j.gastro.2006.10.031. [DOI] [PubMed] [Google Scholar]
- 28.Xu C, Kim NG, Gumbiner BM. Regulation of protein stability by GSK3 mediated phosphorylation. Cell Cycle. 2009;8:4032–4039. doi: 10.4161/cc.8.24.10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu C, Li Y, Semenov M, et al. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 2002;108:837–847. doi: 10.1016/s0092-8674(02)00685-2. [DOI] [PubMed] [Google Scholar]
- 30.Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]
- 31.He TC, Sparks AB, Rago C, et al. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 32.Klamer G, Song E, Ko KH, et al. Using small molecule GSK3beta inhibitors to treat inflammation. Curr Med Chem. 2010;17:2873–2881. doi: 10.2174/092986710792065090. [DOI] [PubMed] [Google Scholar]
- 33.Nava P, Koch S, Laukoetter MG, et al. Interferon-gamma regulates intestinal epithelial homeostasis through converging beta-catenin signaling pathways. Immunity. 2010;32:392–402. doi: 10.1016/j.immuni.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar S, Tomooka Y, Noda M. Identification of a set of genes with developmentally down-regulated expression in the mouse brain. Biochem Biophys Res Commun. 1992;185:1155–1161. doi: 10.1016/0006-291x(92)91747-e. [DOI] [PubMed] [Google Scholar]
- 35.Kamitani T, Kito K, Nguyen HP, et al. Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J Biol Chem. 1997;272:28557–28562. doi: 10.1074/jbc.272.45.28557. [DOI] [PubMed] [Google Scholar]
- 36.Mikus P, Zundel W. COPing with hypoxia. Semin Cell Dev Biol. 2005;16:462–473. doi: 10.1016/j.semcdb.2005.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones D, Crowe E, Stevens TA, et al. Functional and phylogenetic analysis of the ubiquitylation system in Caenorhabditis elegans: ubiquitin-conjugating enzymes, ubiquitin-activating enzymes, and ubiquitin-like proteins. Genome Biol. 2002;3:RESEARCH0002. doi: 10.1186/gb-2001-3-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osaka F, Saeki M, Katayama S, et al. Covalent modifier NEDD8 is essential for SCF ubiquitin-ligase in fission yeast. EMBO J. 2000;19:3475–3484. doi: 10.1093/emboj/19.13.3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tateishi K, Omata M, Tanaka K, et al. The NEDD8 system is essential for cell cycle progression and morphogenetic pathway in mice. J Cell Biol. 2001;155:571–579. doi: 10.1083/jcb.200104035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ou CY, Lin YF, Chen YJ, et al. Distinct protein degradation mechanisms mediated by Cul1 and Cul3 controlling Ci stability in Drosophila eye development. Genes Dev. 2002;16:2403–2414. doi: 10.1101/gad.1011402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wada H, Kito K, Caskey LS, et al. Cleavage of the C-terminus of NEDD8 by UCH-L3. Biochem Biophys Res Commun. 1998;251:688–692. doi: 10.1006/bbrc.1998.9532. [DOI] [PubMed] [Google Scholar]
- 42.Huang DT, Miller DW, Mathew R, et al. A unique E1-E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8. Nat Struct Mol Biol. 2004;11:927–935. doi: 10.1038/nsmb826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mendoza HM, Shen LN, Botting C, et al. NEDP1, a highly conserved cysteine protease that deNEDDylates Cullins. J Biol Chem. 2003;278:25637–25643. doi: 10.1074/jbc.M212948200. [DOI] [PubMed] [Google Scholar]
- 44.Wu K, Yamoah K, Dolios G, et al. DEN1 is a dual function protease capable of processing the C terminus of Nedd8 and deconjugating hyper-neddylated CUL1. J Biol Chem. 2003;278:28882–28891. doi: 10.1074/jbc.M302888200. [DOI] [PubMed] [Google Scholar]
- 45.Liakopoulos D, Doenges G, Matuschewski K, et al. A novel protein modification pathway related to the ubiquitin system. EMBO J. 1998;17:2208–2214. doi: 10.1093/emboj/17.8.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hori T, Osaka F, Chiba T, et al. Covalent modification of all members of human cullin family proteins by NEDD8. Oncogene. 1999;18:6829–6834. doi: 10.1038/sj.onc.1203093. [DOI] [PubMed] [Google Scholar]
- 47.Jones J, Wu K, Yang Y, et al. A targeted proteomic analysis of the ubiquitin-like modifier nedd8 and associated proteins. J Proteome Res. 2008;7:1274–1287. doi: 10.1021/pr700749v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parry G, Estelle M. Regulation of cullin-based ubiquitin ligases by the Nedd8/RUB ubiquitin-like proteins. Semin Cell Dev Biol. 2004;15:221–229. doi: 10.1016/j.semcdb.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 49.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 50.Kumar A, Wu H, Collier-Hyams LS, et al. Commensal bacteria modulate cullin-dependent signaling via generation of reactive oxygen species. EMBO J. 2007;26:4457–4466. doi: 10.1038/sj.emboj.7601867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eltzschig HK, Rivera-Nieves J, Colgan SP. Targeting the A2B adenosine receptor during gastrointestinal ischemia and inflammation. Expert Opin Ther Targets. 2009;13:1267–1277. doi: 10.1517/14728220903241666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khoury J, Ibla JC, Neish AS, et al. Antiinflammatory adaptation to hypoxia through adenosine-mediated cullin-1 deneddylation. J Clin Invest. 2007;117:703–711. doi: 10.1172/JCI30049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mendoza HM, Shen LN, Botting C, et al. NEDP1, a highly conserved cysteine protease that deNEDDylates Cullins. J Biol Chem. 2003;278:25637–25643. doi: 10.1074/jbc.M212948200. Epub 22003 May 25631. [DOI] [PubMed] [Google Scholar]
- 54.Wu K, Yamoah K, Dolios G, et al. DEN1 is a dual function protease capable of processing the C terminus of Nedd8 and deconjugating hyper-neddylated CUL1. J Biol Chem. 2003;278:28882–28891. doi: 10.1074/jbc.M302888200. Epub 22003 May 28819. [DOI] [PubMed] [Google Scholar]
- 55.Semenza GL. HIF-1, O(2), and the 3 PHDs: how animal cells signal hypoxia to the nucleus. Cell. 2001;107:1–3. doi: 10.1016/s0092-8674(01)00518-9. [DOI] [PubMed] [Google Scholar]
- 56.Maxwell PH, Wiesener MS, Chang GW, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 57.Tanimoto K, Makino Y, Pereira T, et al. Mechanism of regulation of the hypoxia-inducible factor-1 alpha by the von Hippel-Lindau tumor suppressor protein. Embo J. 2000;19:4298–4309. doi: 10.1093/emboj/19.16.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lando D, Peet DJ, Whelan DA, et al. Asparagine hydroxylation of the HIF transactivation domain: a hypoxic switch. Science. 2002;295:858–861. doi: 10.1126/science.1068592. [DOI] [PubMed] [Google Scholar]
- 59.Mikus P, Zundel W. COPing with hypoxia. Semin Cell Dev Biol. 2005;16:462–473. doi: 10.1016/j.semcdb.2005.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chiba T, Tanaka K. Cullin-based ubiquitin ligase and its control by NEDD8-conjugating system. Curr Protein Pept Sci. 2004;5:177–184. doi: 10.2174/1389203043379783. [DOI] [PubMed] [Google Scholar]
- 61.Wei N, Deng XW. The COP9 signalosome. Annu Rev Cell Dev Biol. 2003;19:261–286. doi: 10.1146/annurev.cellbio.19.111301.112449. [DOI] [PubMed] [Google Scholar]
- 62.MacManus CF, Campbell EL, Keely S, et al. Anti-inflammatory actions of adrenomedullin through fine tuning of HIF stabilization. FASEB J. 2011;25:1856–1864. doi: 10.1096/fj.10-170316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Taylor CT, Colgan SP. Hypoxia and gastrointestinal disease. J Mol Med (Berl) 2007;85:1295–1300. doi: 10.1007/s00109-007-0277-z. [DOI] [PubMed] [Google Scholar]
- 64.Hatoum OA, Binion DG, Gutterman DD. Paradox of simultaneous intestinal ischaemia and hyperaemia in inflammatory bowel disease. Eur J Clin Invest. 2005;35:599–609. doi: 10.1111/j.1365-2362.2005.01567.x. [DOI] [PubMed] [Google Scholar]
- 65.Eltzschig HK, Carmeliet P. Hypoxia and inflammation. N Engl J Med. 2011;364:656–665. doi: 10.1056/NEJMra0910283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Semenza GL. Oxygen homeostasis. Wiley Interdiscip Rev Syst Biol Med. 2010;2:336–361. doi: 10.1002/wsbm.69. [DOI] [PubMed] [Google Scholar]
- 67.Semenza GL, Wang GL. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol Cell Biol. 1992;12:5447–5454. doi: 10.1128/mcb.12.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mastrogiannaki M, Matak P, Keith B, et al. HIF-2alpha, but not HIF-1alpha, promotes iron absorption in mice. J Clin Invest. 2009 doi: 10.1172/JCI38499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ratcliffe PJ. HIF-1 and HIF-2: working alone or together in hypoxia? J Clin Invest. 2007;117:862–865. doi: 10.1172/JCI31750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mastrogiannaki M, Matak P, Keith B, et al. HIF-2alpha, but not HIF-1alpha, promotes iron absorption in mice. J Clin Invest. 2009;119:1159–1166. doi: 10.1172/JCI38499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 72.Cummins EP, Seeballuck F, Keely SJ, et al. The hydroxylase inhibitor dimethyloxalylglycine is protective in a murine model of colitis. Gastroenterology. 2008;134:156–165. doi: 10.1053/j.gastro.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 73.Robinson A, Keely S, Karhausen J, et al. Mucosal protection by hypoxia-inducible factor prolyl hydroxylase inhibition. Gastroenterology. 2008;134:145–155. doi: 10.1053/j.gastro.2007.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cummins EP, Berra E, Comerford KM, et al. Prolyl hydroxylase-1 negatively regulates IkappaB kinase-beta, giving insight into hypoxia-induced NFkappaB activity. Proc Natl Acad Sci U S A. 2006;103:18154–18159. doi: 10.1073/pnas.0602235103. Epub 12006 Nov 18117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Semenza GL. Oxygen sensing, homeostasis, and disease. N Engl J Med. 2011;365:537–547. doi: 10.1056/NEJMra1011165. [DOI] [PubMed] [Google Scholar]
- 76.Synnestvedt K, Furuta GT, Comerford KM, et al. Ecto-5′-nucleotidase (CD73) regulation by hypoxia-inducible factor-1 (HIF-1) mediates permeability changes in intestinal epithelia. J Clin Invest. 2002;110:993–1002. doi: 10.1172/JCI15337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Han IO, Kim HS, Kim HC, et al. Synergistic expression of inducible nitric oxide synthase by phorbol ester and interferon-gamma is mediated through NF-kappaB and ERK in microglial cells. J Neurosci Res. 2003;73:659–669. doi: 10.1002/jnr.10706. [DOI] [PubMed] [Google Scholar]
- 78.Karhausen JO, Furuta GT, Tomaszewski JE, et al. Epithelial hypoxia-inducible factor-1 is protective in murine experimental colitis. J Clin Invest. 2004;114:1098–1106. doi: 10.1172/JCI21086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Morote-Garcia JC, Rosenberger P, Nivillac NM, et al. Hypoxia-inducible factor-dependent repression of equilibrative nucleoside transporter 2 attenuates mucosal inflammation during intestinal hypoxia. Gastroenterology. 2009;136:607–618. doi: 10.1053/j.gastro.2008.10.037. [DOI] [PubMed] [Google Scholar]
- 80.Shah YM, Ito S, Morimura K, et al. Hypoxia-inducible factor augments experimental colitis through an MIF-dependent inflammatory signaling cascade. Gastroenterology. 2008;134:2036–2048. 2048 e2031–2033. doi: 10.1053/j.gastro.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Giatromanolaki A, Sivridis E, Maltezos E, et al. Hypoxia inducible factor 1alpha and 2alpha overexpression in inflammatory bowel disease. J Clin Pathol. 2003;56:209–213. doi: 10.1136/jcp.56.3.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mariani F, Sena P, Marzona L, et al. Cyclooxygenase-2 and Hypoxia-Inducible Factor-1alpha protein expression is related to inflammation, and up-regulated since the early steps of colorectal carcinogenesis. Cancer Lett. 2009;279:221–229. doi: 10.1016/j.canlet.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 83.Matthijsen RA, Derikx JP, Kuipers D, et al. Enterocyte shedding and epithelial lining repair following ischemia of the human small intestine attenuate inflammation. PLoS One. 2009;4:e7045. doi: 10.1371/journal.pone.0007045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Louis NA, Hamilton KE, Canny G, et al. Selective induction of mucin-3 by hypoxia in intestinal epithelia. J Cell Biochem. 2006;99:1616–1627. doi: 10.1002/jcb.20947. [DOI] [PubMed] [Google Scholar]
- 85.Furuta GT, Turner JR, Taylor CT, et al. Hypoxia-inducible factor 1-dependent induction of intestinal trefoil factor protects barrier function during hypoxia. J Ex Med. 2001;193:1027–1034. doi: 10.1084/jem.193.9.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Comerford KM, Wallace TJ, Karhausen J, et al. Hypoxia-inducible factor-1-dependent regulation of the multidrug resistance (MDR1) gene. Cancer research. 2002;62:3387–3394. [PubMed] [Google Scholar]
- 87.Eltzschig HK, Ibla JC, Furuta GT, et al. Coordinated adenine nucleotide phosphohydrolysis and nucleoside signaling in posthypoxic endothelium: role of ectonucleotidases and adenosine A2B receptors. J Ex Med. 2003;198:783–796. doi: 10.1084/jem.20030891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mole DR, Schlemminger I, McNeill LA, et al. 2-oxoglutarate analogue inhibitors of HIF prolyl hydroxylase. Bioorganic & medicinal chemistry letters. 2003;13:2677–2680. doi: 10.1016/s0960-894x(03)00539-0. [DOI] [PubMed] [Google Scholar]
- 89.Masson N, Ratcliffe PJ. HIF prolyl and asparaginyl hydroxylases in the biological response to intracellular O(2) levels. Journal of cell science. 2003;116:3041–3049. doi: 10.1242/jcs.00655. [DOI] [PubMed] [Google Scholar]
- 90.Colgan SP, Taylor CT. Hypoxia: an alarm signal during intestinal inflammation. Nat Rev Gastroenterol Hepatol. 2010;7:281–287. doi: 10.1038/nrgastro.2010.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tambuwala MM, Cummins EP, Lenihan CR, et al. Loss of prolyl hydroxylase-1 protects against colitis through reduced epithelial cell apoptosis and increased barrier function. Gastroenterology. 2010;139:2093–2101. doi: 10.1053/j.gastro.2010.06.068. [DOI] [PubMed] [Google Scholar]
- 92.Hindryckx P, De Vos M, Jacques P, et al. Hydroxylase inhibition abrogates TNF-alpha-induced intestinal epithelial damage by hypoxia-inducible factor-1-dependent repression of FADD. J Immunol. 2010;185:6306–6316. doi: 10.4049/jimmunol.1002541. [DOI] [PubMed] [Google Scholar]
- 93.Ting JP, Lovering RC, Alnemri ES, et al. The NLR gene family: a standard nomenclature. Immunity. 2008;28:285–287. doi: 10.1016/j.immuni.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ye Z, Ting JP. NLR, the nucleotide-binding domain leucine-rich repeat containing gene family. Curr Opin Immunol. 2008;20:3–9. doi: 10.1016/j.coi.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 95.Stutz A, Golenbock DT, Latz E. Inflammasomes: too big to miss. J Clin Invest. 2009;119:3502–3511. doi: 10.1172/JCI40599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Agostini L, Martinon F, Burns K, et al. NALP3 forms an IL-1beta-processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity. 2004;20:319–325. doi: 10.1016/s1074-7613(04)00046-9. [DOI] [PubMed] [Google Scholar]
- 97.Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 98.Martinon F, Mayor A, Tschopp J. The inflammasomes: guardians of the body. Annu Rev Immunol. 2009;27:229–265. doi: 10.1146/annurev.immunol.021908.132715. [DOI] [PubMed] [Google Scholar]
- 99.Al-Sadi RM, Ma TY. IL-1beta causes an increase in intestinal epithelial tight junction permeability. J Immunol. 2007;178:4641–4649. doi: 10.4049/jimmunol.178.7.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Petrilli V, Papin S, Dostert C, et al. Activation of the NALP3 inflammasome is triggered by low intracellular potassium concentration. Cell Death Differ. 2007;14:1583–1589. doi: 10.1038/sj.cdd.4402195. [DOI] [PubMed] [Google Scholar]
- 101.Kanneganti TD, Ozoren N, Body-Malapel M, et al. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature. 2006;440:233–236. doi: 10.1038/nature04517. [DOI] [PubMed] [Google Scholar]
- 102.Cruz CM, Rinna A, Forman HJ, et al. ATP activates a reactive oxygen species-dependent oxidative stress response and secretion of proinflammatory cytokines in macrophages. J Biol Chem. 2007;282:2871–2879. doi: 10.1074/jbc.M608083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Perregaux D, Gabel CA. Interleukin-1 beta maturation and release in response to ATP and nigericin. Evidence that potassium depletion mediated by these agents is a necessary and common feature of their activity. J Biol Chem. 1994;269:15195–15203. [PubMed] [Google Scholar]
- 104.Bauer C, Duewell P, Mayer C, et al. Colitis induced in mice with dextran sulfate sodium (DSS) is mediated by the NLRP3 inflammasome. Gut. 2010;59:1192–1199. doi: 10.1136/gut.2009.197822. [DOI] [PubMed] [Google Scholar]
- 105.Hirota SA, Ng J, Lueng A, et al. NLRP3 inflammasome plays a key role in the regulation of intestinal homeostasis. Inflamm Bowel Dis. 2011;17:1359–1372. doi: 10.1002/ibd.21478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dupaul-Chicoine J, Yeretssian G, Doiron K, et al. Control of intestinal homeostasis, colitis, and colitis-associated colorectal cancer by the inflammatory caspases. Immunity. 2010;32:367–378. doi: 10.1016/j.immuni.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 107.Zaki MH, Vogel P, Body-Malapel M, et al. IL-18 production downstream of the Nlrp3 inflammasome confers protection against colorectal tumor formation. J Immunol. 2010;185:4912–4920. doi: 10.4049/jimmunol.1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kanai T, Watanabe M, Okazawa A, et al. Macrophage-derived IL-18-mediated intestinal inflammation in the murine model of Crohn’s disease. Gastroenterology. 2001;121:875–888. doi: 10.1053/gast.2001.28021. [DOI] [PubMed] [Google Scholar]
- 109.Siegmund B, Lehr HA, Fantuzzi G, et al. IL-1 beta -converting enzyme (caspase-1) in intestinal inflammation. Proc Natl Acad Sci U S A. 2001;98:13249–13254. doi: 10.1073/pnas.231473998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Siegmund B, Fantuzzi G, Rieder F, et al. Neutralization of interleukin-18 reduces severity in murine colitis and intestinal IFN-gamma and TNF-alpha production. Am J Physiol Regul Integr Comp Physiol. 2001;281:R1264–1273. doi: 10.1152/ajpregu.2001.281.4.R1264. [DOI] [PubMed] [Google Scholar]
- 111.Zaki MH, Boyd KL, Vogel P, et al. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity. 2010;32:379–391. doi: 10.1016/j.immuni.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Franke A, McGovern DP, Barrett JC, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cho JH, Brant SR. Recent insights into the genetics of inflammatory bowel disease. Gastroenterology. 2011;140:1704–1712. doi: 10.1053/j.gastro.2011.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bertin J, Guo Y, Wang L, et al. CARD9 is a novel caspase recruitment domain-containing protein that interacts with BCL10/CLAP and activates NF-kappa B. J Biol Chem. 2000;275:41082–41086. doi: 10.1074/jbc.C000726200. [DOI] [PubMed] [Google Scholar]
- 115.Ruland J. CARD9 signaling in the innate immune response. Ann N Y Acad Sci. 2008;1143:35–44. doi: 10.1196/annals.1443.024. [DOI] [PubMed] [Google Scholar]
- 116.Gross O, Poeck H, Bscheider M, et al. Syk kinase signalling couples to the Nlrp3 inflammasome for anti-fungal host defence. Nature. 2009;459:433–436. doi: 10.1038/nature07965. [DOI] [PubMed] [Google Scholar]
- 117.Bi L, Gojestani S, Wu W, et al. CARD9 mediates dectin-2-induced IkappaBalpha kinase ubiquitination leading to activation of NF-kappaB in response to stimulation by the hyphal form of Candida albicans. J Biol Chem. 2010;285:25969–25977. doi: 10.1074/jbc.M110.131300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Glocker EO, Hennigs A, Nabavi M, et al. A homozygous CARD9 mutation in a family with susceptibility to fungal infections. N Engl J Med. 2009;361:1727–1735. doi: 10.1056/NEJMoa0810719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhernakova A, Festen EM, Franke L, et al. Genetic analysis of innate immunity in Crohn’s disease and ulcerative colitis identifies two susceptibility loci harboring CARD9 and IL18RAP. Am J Hum Genet. 2008;82:1202–1210. doi: 10.1016/j.ajhg.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nenci A, Becker C, Wullaert A, et al. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature. 2007;446:557–561. doi: 10.1038/nature05698. [DOI] [PubMed] [Google Scholar]
- 121.Yang H, Minamishima YA, Yan Q, et al. pVHL acts as an adaptor to promote the inhibitory phosphorylation of the NF-kappaB agonist Card9 by CK2. Mol Cell. 2007;28:15–27. doi: 10.1016/j.molcel.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]