Abstract
The intraglomerular renin-angiotensin system (RAS) is linked to the pathogenesis of progressive glomerular diseases. Glomerular podocytes and mesangial cells play distinct roles in the metabolism of angiotensin (ANG) peptides. However, our understanding of the RAS enzymatic capacity of glomerular endothelial cells (GEnCs) remains incomplete. We explored the mechanisms of endogenous cleavage of ANG substrates in cultured human GEnCs (hGEnCs) using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and isotope-labeled peptide quantification. Overall, hGEnCs metabolized ANG II at a significantly slower rate compared with podocytes, whereas the ANG I processing rate was comparable between glomerular cell types. ANG II was the most abundant fragment of ANG I, with lesser amount of ANG-(1–7) detected. Formation of ANG II from ANG I was largely abolished by an ANG-converting enzyme (ACE) inhibitor, whereas ANG-(1–7) formation was decreased by a prolylendopeptidase (PEP) inhibitor, but not by a neprilysin inhibitor. Cleavage of ANG II resulted in partial conversion to ANG-(1–7), a process that was attenuated by an ACE2 inhibitor, as well as by an inhibitor of PEP and prolylcarboxypeptidase. Further fragmentation of ANG-(1–7) to ANG-(1–5) was mediated by ACE. In addition, evidence of aminopeptidase N activity (APN) was demonstrated by detecting amelioration of conversion of ANG III to ANG IV by an APN inhibitor. While we failed to find expression or activity of aminopeptidase A, a modest activity attributable to aspartyl aminopeptidase was detected. Messenger RNA and gene expression of the implicated enzymes were confirmed. These results indicate that hGEnCs possess prominent ACE activity, but modest ANG II-metabolizing activity compared with that of podocytes. PEP, ACE2, prolylcarboxypeptidase, APN, and aspartyl aminopeptidase are also enzymes contained in hGEnCs that participate in membrane-bound ANG peptide cleavage. Injury to specific cell types within the glomeruli may alter the intrarenal RAS balance.
Keywords: angiotensin-converting enzyme, prolylendopeptidase, endothelium
the intrarenal renin-angiotensin system (RAS) is a critical modulator of blood pressure regulation and salt balance under normal physiological conditions. It is also linked to the pathogenesis of various disease states, including renovascular hypertension and progressive glomerular diseases (57). Angiotensin (ANG) II is the main effector of the system, with known pressor (48), anti-natriuretic (48), proliferative (42), and profibrotic properties (24). ANG II [ANG-(1–8)] is an octapeptide that is generated from the cleavage of its decapeptide precursor ANG I by a carboxypeptidase, primarily ANG-converting enzyme (ACE), or by other peptidases, such as chymase or cathepsin G. However, ANG I can be converted into other fragments by the action of various carboxy-, endo-, or aminopeptidases. An outline of the major RAS enzymes and corresponding cleaving sites are shown in Table 1. Importantly, some of those alternative ANG fragments are bioactive. For instance, ANG-(1–7) exerts vasodilatory (39), aquaretic (30), and anti-proliferative actions (52). Therefore, the process of enzymatic fragmentation of ANG peptides may impact the net production of bioactive molecules, thereby modulating RAS-related cellular events and local homeostasis. Thus the relative contribution of the peptidases involved in this process is of significance.
Table 1.
Outline of the major renin-angiotensin system enzymes indicating their corresponding cleaving sites within the angiotensin peptide sequence
| APA ↓ | TOP PEP NEP ↓ | Cathepsin G Tonin Chymase ACE ↓ | Cathepsin A CPA ACE2 ↓ | Mol. Wt. | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ANG I | Asp | — | Arg | — | Val | — | Tyr | — | Ile | — | His | — | Pro | — | Phe | — | His | — | Leu | 1,296 |
| ANG-(2-10) | Arg | — | Val | — | Tyr | — | Ile | — | His | — | Pro | — | Phe | — | His | — | Leu | 1,181 | ||
| ANG-(1-9) | Asp | — | Arg | — | Val | — | Tyr | — | Ile | — | His | — | Pro | — | Phe | — | His | 1,183 | ||
| ANG-II | Asp | — | Arg | — | Val | — | Tyr | — | Ile | — | His | — | Pro | — | Phe | 1,046 | ||||
| ANG-(1-7) | Asp | — | Arg | — | Val | — | Tyr | — | Ile | — | His | — | Pro | 899 |
| DAPAPA↓ | PCPPEPACE2 ↓ | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ANG II | Asp | — | Arg | — | Val | — | Tyr | — | Ile | — | His | — | Pro | — | Phe | 1,046 | ||||
| ANG III | Arg | — | Val | — | Tyr | — | Ile | — | His | — | Pro | — | Phe | 931 | ||||||
| ANG-(1-7) | Asp | — | Arg | — | Val | — | Tyr | — | Ile | — | His | — | Pro | 899 |
| APBAPN ↓ | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ANG III | Arg | — | Val | — | Tyr | — | Ile | — | His | — | Pro | — | Phe | 931 | ||||||
| ANG IV | Val | — | Tyr | — | Ile | — | His | — | Pro | — | Phe | 665 | ||||||||
| Amino acid position | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
ANG, angiotensin; APA, aminopeptidase A; TOP, thimet oligopeptidase; PEP, prolylendopeptidase; NEP, neprilysin; ACE, angiotensin-converting enzyme; CPA, carboxypeptidase A; ACE2, angiotensin-converting enzyme 2; DAP, aspartyl aminopeptidase; PCP, prolylcarboxypeptidase, APB, aminopeptidase B; APN, aminopeptidase N.
Aside from the tubular compartment, the kidney glomerulus has the capacity to metabolize ANG peptides. Among the different cell types that reside inside the glomerular tuft, we and others have shown that podocytes possess enzymatic capability to metabolize ANG I and ANG II (12, 28, 59). In particular, podocytes are equipped with membrane-bound enzymatic machinery that favors formation of ANG-(2–10) and ANG-(1–7) from ANG I and promotes rapid degradation of ANG II (58). Similarly, mesangial cells are known to contain active ANG-cleaving ectoenzymes (47). However, little is known about the intrinsic ability of glomerular endothelial cells (GEnCs) to process ANG peptides.
The vascular endothelium represents a major reservoir of ACE (8). Although GEnCs have some unique structural features that distinguish them from other types of endothelial cells, such as fenestrations that preserve basal lamina but lack diaphragm (41), they are also known to express ACE. Indeed, early workers employed an ACE activity assay to characterize human GEnCs (hGEnCs) in vitro (53). Nevertheless, studies focused on intrinsic RAS enzyme profiling in hGEnCs solely explored the expression of ACE, but did not determine whether other functional RAS-related enzymatic pathways are contained in them (18, 33). The purpose of our investigation was to characterize the enzymatic cleavage of ANG peptides by hGEnCs using a mass spectrometry (MS)-based technique as analytical tool. We hypothesize that ACE activity represents only a fraction of the total RAS-related enzymatic machinery of hGEnCs, and that that machinery differs from that contained in podocytes.
METHODS
Cell culture.
Primary hGEnCs were obtained from ScienCell Research Laboratories (Carlsbad, CA). Passages 2–5 were used for the experiments. Cells were grown at 37°C with 5% CO2 on plates coated with fibronectin (2 μg/cm2) using a 5% fetal bovine serum-containing media supplied by the manufacturer. Identity of hGEnCs was confirmed by verifying the expression of platelet/endothelial cell adhesion molecule-1 by immunofluorescence (data not shown).
For the experiments, cells were grown in 12-well plates to subconfluence. After a 1-h starvation period, cells were maintained in serum-free medium and spiked with 1 μM ANG I [ANG-(1–10)], ANG II, ANG III [ANG-(2–8)], or ANG-(1–7), for 15 min to 12 h. Human mesangial cells (hMesCs) generously provided by Dr. Hannah E. Abboud (University of Texas, San Antonio, TX), and conditionally immortalized human podocytes (hPODs) generously provided by Dr. Moim Saleem (University of Bristol, Bristol, UK), were cultured under their standard conditions (38, 45) and used for comparison in selected experiments. Cells were incubated in the presence or absence of RAS enzyme inhibitors. Inhibitors included the following: 100 μM captopril (ACE inhibitor), 100 μM chymostatin (chymase inhibitor), 10 μM benzylsuccinate [carboxypeptidase A (CPA) inhibitor], 1 μM DX600 (ACE2 inhibitor), 1–10 μM thiorphan [neprilysin (NEP) inhibitor], 1–10 μM SCH39370 (NEP inhibitor), 100 nM Z-Pro-prolinal (ZPP) [prolylendopeptidase (PEP) inhibitor], 100 μM amastatin [aminopeptidase A (APA) inhibitor], 100 μM to 1 mM bestatin [aminopeptidase N (APN) inhibitor], 10–100 μM arphamenine B [aminopeptidase B (APB) and puromycin-sensitive aminopeptidase inhibitor], 1 mM dithiothreitol (DTT) [used as aspartyl aminopeptidase (DAP) inhibitor], and 100 μM phosphoramidon, 100 nM pepstatin, 100 μM leupeptin, 1 mM EDTA, and 1 mM 1,10-phenathroline (all broad metalloprotease inhibitors). All of the inhibitors were purchased from Sigma-Aldrich (St. Louis, MO), except DX600 (Phoenix Pharmaceuticals) and SCH39370 (generous gift from Dr. Mark Chappell, Wake Forest University, Winston-Salem, NC). When appropriate, cells were preincubated for 20 min with inhibitors before the addition of the ANG substrate. Conditioned media were collected at various time points and stored at −20°C until assayed. In addition, to determine whether the observed peptide disappearance from the cell media could be partially attributable to receptor binding and internalization or direct endocytosis, selected experiments were conducted in the presence of 10 μM losartan (gift from Merck, Whitehouse Station, NJ) or 400 mM sucrose.
ANG metabolite analysis and quantification.
Samples obtained at various time points were assayed by matrix-assisted laser/desorption/ionization time-of-flight (MALDI-TOF) MS, as previously described (58). Briefly, peptides were purified by C18-Zip-Tip columns (Millipore, Billerica, MA). Columns were equilibrated with 100% acetonitrile, washed with 0.1% trifluoroacetic acid, loaded with 20 μl of sample, washed with 10 μl of 0.1% trifluoroacetic acid, and eluted with a low-pH MALDI matrix compound (10 g/l α-cyano-4-hydroxycinnamic acid in a 1:1 mixture of 100% acetonitrile and 0.1% trifluoroacetic acid). Eluted matrix (1.5 μl) was applied to the surface of a target plate in triplicate and air-dried. Spectra were collected in reflectron mode using a M@LDI MALDI-TOF mass spectrometer (Waters, Milford, MA). Results were analyzed using MassLynx 2.0 software (Waters). Peptide identities were confirmed by MS/MS de novo sequencing from ion series generated by MALDI-TOF-TOF (ABI 4700 Series) and linear trap quadrupole MS (Thermo Scientific, West Palm Beach, FL). Quantifications of observed peaks were performed using customized isotopic absolute quantification (AQUA) peptides (Sigma-Aldrich) as internal standards. AQUA peptides are 6 Da larger than the native peptide as a result of [13C.15N]valine incorporation into the amino acid sequence. A cocktail of AQUA peptides was mixed with each sample of conditioned buffer before MALDI-TOF MS analysis. The final concentration of each AQUA peptide in the mix was set between 10 and 250 nM, depending on the qualitative appearance of the peptide peaks on the mass spectra. For determination of the abundance of each native peptide, the intensity of the major monoisotopic peak was divided by the intensity of the major peak of the corresponding AQUA peptide. The ratio was multiplied by the known concentration of the AQUA peptide to estimate the native peptide concentration. This method has been validated previously by others (17). Peak intensities that did not exceed a signal-to-noise ratio < 9.0 were deemed below the limit of quantification and were not used for quantification. Peak abundances were normalized to total cell protein.
RAS-related enzyme expression.
In parallel to the analytic studies, we searched for gene expression of a panel of candidate RAS-related enzymes in hGEnCs. For messenger RNA (mRNA) detection, total RNA was extracted using an RNeasy mini kit (Qiagen, Austin, TX) and reverse transcribed with an Advantage RT-for-PCR kit (Clontech, Palo Alto, CA), starting with 500 ng of total RNA. RT-PCR was performed using Taq 2× Master Mix (New England BioLaboratories, Ipswich, MA). PCR primers for the amplification of ACE, ACE2, NEP, PEP, prolylcarboxypeptidase (PCP), APA, and APN were custom designed and purchased from Sigma-Genosys (St. Louis, MO). We also searched for intracellular RAS components upstream of ANG I cleavage, namely angiotensinogen (AGT) and cathepsin D. Primer sequences included: ACE, (F) 5′ ACTACGGGGCCCAGCACATC 3′, (R) 5′ AGGGAAGGGCACCACCAAGTC 3′; ACE2, (F) 5′ ATGTCCCGGAGCCGTATCAAT 3′, (R) 5′ GCCAACCACTATCACTCCCATCAC 3′; PCP, (F) 5′ ATGGGCCGCCGAGCCCTCCTG 3′, (R) 5′ GGTTGGTTGGCAAGTGTAGG 3′; PEP, (F) 5′ GAGCCAAGAGTTTTCCGAGAGGTGA 3′, (R) 5′ CCAGGATACCACCCATGTGTCTCAC 3′; NEP, (F) 5′ AGCAGCCTCAGCCGAACCTACA 3, (R) 5′ CCACATAAAGCCTCCCCACAGC 3′; APA, (F) 5′ TTCCTCCGTGTTAGGGTTTGCG 3′, (R) 5′ ACCGATACACCAGAAGCCTGAG 3′; APN, (F) 5′ GTGAACAACCGGTCCATCCAACT 3′; (R) 5′ GAATTCTGAGGGGCGCGTAACA 3′. Each 25 μl reaction consisted of 10 μl of cDNA, 1 μl of forward primer, 1 μl of reverse primer, and 12.5 μl of Master Mix. Final primer concentrations were 400 nM. Cycling conditions were as follows: 95°C, 5 min; 95°C, 30 s, 70°C, 30 s, 68°C, 1 min ×34; 68°C, 5 min; 4°C hold. The reaction was modified for ACE2 by changing the annealing temperature to 65°C and extending the PCR reaction to 36 cycles. PCR products were run on a 2% agarose gel with ethidium bromide (0.5 μl/75 ml gel) at 200 V for 30 min. Sample loading dye was 2 g sucrose in 5 ml Tris-acetate-EDTA buffer. In addition, for confirmation of presence of protein in hGEnCs, Western blotting was performed as previously published (58) with minor modifications. Each well was loaded with 25 μg of cell protein. Primary antibodies used included the following: goat anti-ACE (N-20, dilution 1:200), goat anti-ACE2 (C-18, dilution 1:500), goat anti-DAP (DNPEP, dilution 1:500), and goat anti-cathepsin D (C-20, dilution 1:1,000) from Santa Cruz Biotechnology (Santa Cruz, CA); rabbit anti-PEP (dilution 1:1,000), mouse anti-PCP (dilution 1:500), and rabbit anti-APA (BP1, dilution 1:500) from Abcam (Cambridge, MA); rabbit anti-NEP (CD10, dilution 1:500) from Millipore; rabbit anti-APN (CD13, dilution 1:1,000) from Origene (Rockville, MD), and goat anti-AGT (dilution 1:1,000) from R&D Systems (Minneapolis, MN). Whole normal human kidney lysate was obtained from Novus Biologicals (Littleton, CO).
Search for additional RAS-related enzymes.
To search for other RAS-related enzymes contained in hGEnCs that were not initially included in our set of candidate peptidases, we utilized tandem MS. Human glomerular endothelial cells obtained from two different vendors [ScienCell and Cell Systems (Kirkland, WA)] were used for these experiments. Cell pellets were lysed in 500 μl of a buffer containing 10 mM HEPES, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, and protease inhibitor cocktail, following a 5-min incubation on ice to allow the cells to swell. The detergent NP-40 was added to a final concentration of 0.1%. Lysate was dounced and centrifuged twice at 3,000 g for 10 min at 4°C. Pooled supernatant was then centrifuged at 17,000 g for 30 min, also at 4°C. Proteins were then precipitated in acetone and resuspended in 100 mM ammonium bicarbonate buffer with 0.1% SDS. Protein concentration was determined using the Bradford method. Twenty micrograms of protein were digested overnight following reduction and alkylation. Tryptic peptides were isolated using solid phase extraction Strata-X cartridges (Phenomenex, Torrance, CA). Digests were acidified with 0.1% formic acid and adsorbed to solid-phase extraction columns, washed three times with 0.1% formic acid, and eluted sequentially in 20, 30, 40, 50, 60, and 70% acetonitrile:0.1% formic acid. Sequentially eluted peptides were dried in a vacuum concentrator and resuspended in mobile phase A (2% acetonitrile:0.2% formic acid). Peptide concentration was estimated using absorbance at 280 nm and ∼2 μg of peptide/fraction was injected for reverse phase chromatography using an Eksigent nano ultra 2D (AB/SCIEX, Framingham, MA). Peptides were trapped on a Nano Trap Column 100-μm inner diameter × 1 cm c18 PepMap100 (Dionex, Bannockburn, IL) and separated over 55 min on an Acclaim PepMap100 75 μm inner diameter × 15 cm c18, 3 μm, 100A column (Dionex) from 5 to 60% mobile phase B (95% ACN:0.2% formic acid) and then 10 min 60 to 85% mobile phase B. Spectra were acquired on a Triple TOF mass spectrometer with nanospray source (AB/SCIEX), and the top 20 parent ions between 300 and 1,250 mass-to-charge ratio (m/z) were fragmented using information-dependent acquisition mode. Product ion accumulation times were set to 50 ms (cycle time 1.3 s), and parent ions were dynamically excluded for 10 s. Data files were converted to mascot generic format and searched against the Homo sapiens proteome (Swissprot release 2,011–6), including common contaminants using MASCOT (version 2.2.07). Search parameters included the following: enzyme = trypsin, allow up to two missed cleavages, fixed modification = carbamidomethyl (C), variable modifications = acetyl (N-term), oxidation (M), deamidation (NQ), pyro-glu (N term EQ), peptide tolerance = 20 ppm, MS/MS tolerance = 0.5 Da, decoy database search selected. MASCOT search results were uploaded into SCAFFOLD 3 (Proteome Science, Portland, OR) and searched with X! Tandem CYCLONE (2010.12.01.1) using parameters assigned with MASCOT. Both searches were combined, and protein identification probabilities were computed by peptide prophet and protein prophet.
Statistical analyses.
Data were analyzed by Student's t-test with the assumption of equal variance or ANOVA with post hoc analysis by Bonferroni's correction. Difference in peptide disappearance was analyzed using two-factor (inhibitor or cell type, and time) ANOVA for repeated measures and Scheffé test. Values are means ± SE. P < 0.05 was considered statistically significant.
RESULTS
ANG I metabolism.
Incubation of hGEnCs, hPODs, and hMesCs in the presence of ANG I revealed differences in the pattern of ANG I fragmentation among cell types. ANG II and ANG-(1–7) were the predominant metabolites of ANG I in hGEnCs and hMesCs, whereas hPODs primarily converted ANG I into ANG-(2–10) and ANG-(1–7), with significantly less ANG II formation detected (Fig. 1). The pattern observed in hPODs mimicked that of mouse podocytes (58). The overall rate of ANG I disappearance was not different among glomerular cell types (data not shown).
Fig. 1.
Comparison of angiotensin (ANG) I metabolites detected in conditioned media from human glomerular endothelial cells (hGEnCs; A), human mesangial cells (hMesCs; B), or human podocytes (hPODs; C), exposed to 1 μM ANG I. Measurements done by matrix-assisted laser/desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (MS) were assisted with absolute quantification (AQUA) peptides. Values are means ± SE.
ANG II metabolism.
Incubation of hGEnCs, hPODs, and hMesCs in the presence of ANG II revealed differences in the pattern of ANG II fragmentation among cell types. ANG-(1–7) was the predominant metabolite of ANG II in hGEnCs and hMesCs, whereas hPODs primarily converted ANG II into ANG III (Fig. 2). Small amounts of ANG IV [ANG-(3–8)] were also detected in samples obtained from all three cell types. Furthermore, the rate of disappearance of ANG II during exposure to hGEnCs was significantly slower compared with the rate observed when ANG II was exposed to hPODs (Fig. 3). Addition of losartan did not modify the mass spectra (data not shown), suggesting that the disappearance of the peptide was not explained by binding to its receptor and subsequent internalization. Similarly, since sucrose had no effect on the observed spectra (data not shown), peptide trapping by endocytosis did not seem to play a role in ANG peptide disappearance.
Fig. 2.
Comparison of ANG II metabolites detected in conditioned culture media from hGEnCs (A), hMesCs (B), or hPODs (C), exposed to 1 μM ANG II. Measurements were done by MALDI-TOF MS assisted with AQUA peptides. Values are means ± SE.
Fig. 3.
ANG II disappearance from conditioned media in hGEnCs and hPODs exposed to 1 μM ANG II. Measurements were done by MALDI-TOF MS assisted with AQUA peptides. Values are means ± SE. *P < 0.05 vs. hPODs.
Identification of enzymes activity in hGEnCs.
The most prominent peaks observed under incubation of hGEnCs with ANG I were 1,046 and 899 m/z, corresponding to ANG II and ANG-(1–7), respectively (Fig. 4A). Small peaks of 1,181 and 1,183 m/z, corresponding to ANG-(2–10) and ANG-(1–9), were also detected. Coincubation of ANG I with an ACE inhibitor (captopril) led to a substantial reduction in the ANG II abundance (Fig. 4), whereas no effect on the spectra was provoked by a chymase inhibitor (data not shown). These findings indicate that ACE is largely responsible for the conversion of ANG I into ANG II by hGEnCs. Because NEP (EC 3.4.24.11) is known to be an important enzyme responsible of cleaving ANG I into ANG-(1–7) in kidney cells (6, 58), a search for NEP activity in hGEnCs was conducted. Thus cells were exposed to ANG I and coincubated with either thiorphan or SCH39370, both NEP inhibitors. No effect on ANG I or ANG-(1–7) abundance was detected with either inhibitor (data not shown). On the other hand, the formation of ANG-(1–7) was significantly decreased in the presence of ZPP, a PEP inhibitor (Fig. 5), suggesting that PEP converts ANG I to ANG-(1–7) in hGEnCs. However, the disappearance rate of ANG I was not altered by ZPP (Fig. 5C). Exposure to broad metalloprotease inhibitors, 1 mM EDTA and 1 mM 1,10-phenanthroline, significantly reduced the formation of ANG II and ANG-(1–7) (data not shown), confirming the enzymatic origin of the detected metabolites. Inhibition of APA or APN with amastatin or bestatin did not significantly affect the spectra, neither did DX600 or benzylsuccinate, inhibitors of ACE2 and CPA, respectively (data not shown). These findings suggest that the contribution of APA, APN, ACE2, and CPA to ANG I cleavage in hGEnCs is not as robust as that of ACE and PEP.
Fig. 4.
A: MALDI-TOF mass spectrum generated from conditioned media collected after incubation of human glomerular endothelial cells with 1 μM ANG I in the absence (top) or presence (bottom) of 100 μM captopril [ANG-converting enzyme (ACE) inhibitor] for 4 h. Inset: quantitative inspection of ANG II [1,046 mass-to-charge ratio (m/z)] peaks using 100 nM AQUA-ANG II (1,052 m/z) as internal standard. B: quantification of observed differences at 4 h (average from 5 separate batches of cells). C: disappearance curve of ANG I, with or without ACE inhibition. Values are means ± SE. *P < 0.0001 vs. control. ¥P < 0.01 vs. control.
Fig. 5.
A: MALDI-TOF mass spectrum generated from conditioned media collected after incubation of hGEnCs with 1 μM ANG I in the absence (top) or presence (bottom) of 100 nM Z-prolyl prolinal [prolylendopeptidase (PEP) inhibitor] for 4 h. Inset: quantitative inspection of ANG 1–7 (899 m/z) peaks using 100 nM AQUA-ANG 1–7 (905 m/z) as internal standard. B: quantification of observed differences at 4 h (average from 3 separate batches of cells), including the effect of 10 μM SCH39370 [neprilysin (NEP)] inhibitor. C: disappearance curve of ANG I, with or without PEP inhibition. Values are means ± SE. *P < 0.01 vs. control.
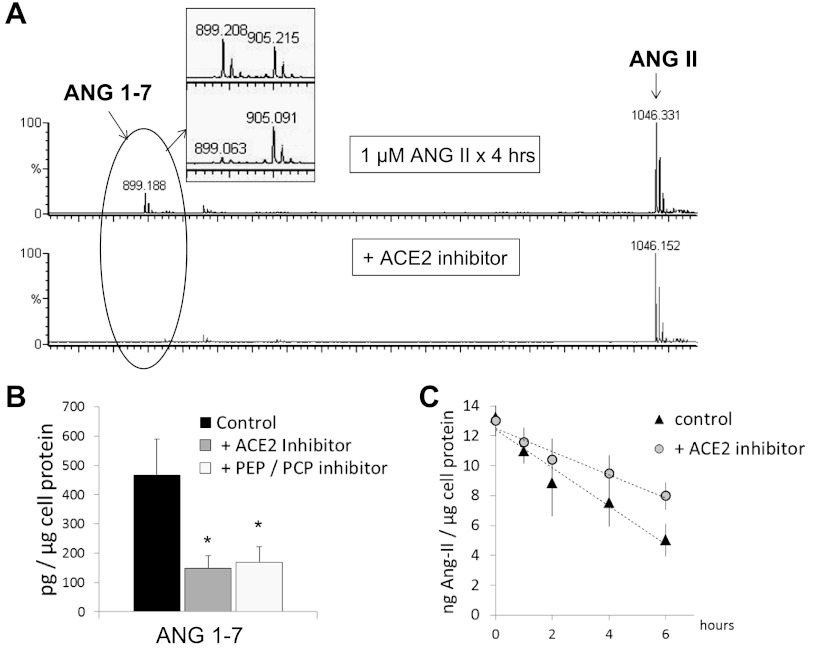
As mentioned above, 899 m/z was the most abundant peak detected upon exposure of ANG II to hGEnCs, reflecting ANG-(1–7) formation. Coincubation of ANG II with either an ACE2 inhibitor (DX600) or a PEP inhibitor (ZPP) led to significant reduction in ANG-(1–7) abundance. Besides, ACE2 inhibition modified the slope of ANG II disappearance, although the rate was not significantly different than control (Fig. 6). ZPP can also blunt PCP activity (44); therefore, PCP (EC 3.4.16.2) could also be a contributor to the fragmentation of ANG II in hGEnCs. Significant reduction in the conversion of ANG II to ANG-(1–7) was also observed during coincubation of ANG II with 1,10-phenanthroline, confirming the proteolytic origin of the detected peptide fragment. Furthermore, addition of 1,10-phenanthroline allowed for a better detection of the 931 m/z peak that corresponds to ANG III. The abundance of ANG III was partially decreased when a nonspecific DAP inhibitor (DTT) was added to 1,10-phenanthroline (Fig. 7), whereas it remained unaltered when amastatin or bestatin, APA and APN inhibitors, respectively, were added (data not shown).
Fig. 6.
A: MALDI-TOF mass spectrum generated from conditioned media collected after incubation of hGEnCs with 1 μM ANG II in the absence (top) or presence (bottom) of 1 μM DX600 (ACE2 inhibitor) for 4 h. Inset: quantitative inspection of ANG 1–7 (899 m/z) peaks using 50 nM AQUA-ANG 1–7 (905 m/z) as internal standard. B: quantification of observed differences at 4 h (average from 3 separate batches of cells), including the effect of 100 nM Z-prolyl prolinal [PEP and prolylcarboxypeptidase (PCP) inhibitor]. C: disappearance curve of ANG II, with or without ACE2 inhibition. Values are means ± SE. *P < 0.005 vs. control.
Fig. 7.
A: MALDI-TOF mass spectrum generated from conditioned media collected after incubation of hGEnCs with 1 μM ANG II in the presence of 1 mM 1,10-phenanthroline alone (top) or the combination of 1 mM 1,10-phenanthroline and 1 mM DTT (bottom) for 4 h. Dashed box: quantitative inspection of ANG III (931 m/z) peaks using 10 nM AQUA-ANG III (937 m/z) as internal standard. B: quantification of observed differences at 4 h (average from 3 separate batches of cells). Values are means ± SE. *P < 0.01.
Similarly, we analyzed the fragmentation of relevant ANG-derived heptapeptides. It was observed that ANG-(1–7) was primarily converted into ANG-(1–5) by ACE, since captopril significantly inhibited the reaction (Fig. 8). In addition, cleavage of ANG III into ANG IV was mediated by APN, since the conversion was diminished by bestatin (Fig. 9). Arphamenine B, inhibitor of APB and puromycin-sensitive aminopeptidase, had no effect on the cleavage or ANG III, nor did it on ANG I, ANG II, or ANG-(1–7) cleavage (data not shown).
Fig. 8.
A: MALDI-TOF mass spectrum generated from conditioned media collected after incubation of hGEnCs with 1 μM ANG 1–7 in the absence (top) or presence (bottom) of 100 μM captopril (ACE inhibitor) for 4 h. Inset: quantitative inspection of ANG 1–5 (664 m/z) peaks using 10 nM AQUA-ANG 1–5 (670 m/z) as internal standard. B: quantification of observed differences at 4 h (average from 3 separate batches of cells). C: disappearance curve of ANG I, with or without ACE inhibition. Values are means ± SE. *P < 0.001. ¥P < 0.01 vs. control.
Fig. 9.
A: MALDI-TOF mass spectrum generated from conditioned media collected after incubation of hGEnCs with 1 μM ANG III in the absence (top) or presence (bottom) of 1 mM bestatin [aminopeptidase N (APN) inhibitor] for 4 h. Inset: quantitative inspection of ANG IV (775 m/z) peaks using 50 nM AQUA-ANG IV (781 m/z) as internal standard. B: quantification of observed differences at 4 h (average from 3 separate batches of cells). C: disappearance curve of ANG III, with or without APN inhibition. Values are means ± SE. *P < 0.001. ¥P < 0.01 vs. control.
Gene expression of RAS enzymes.
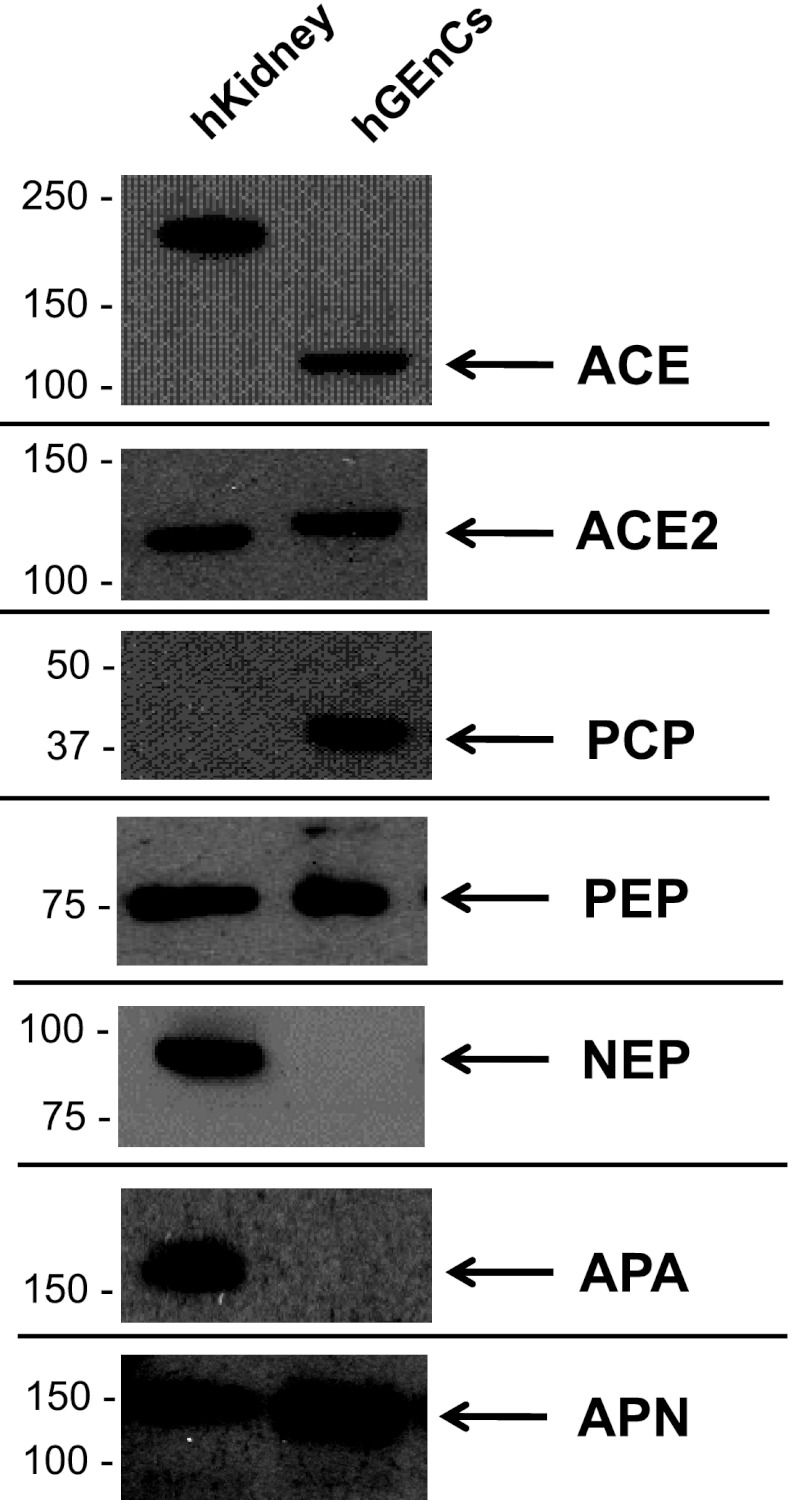
The presence of candidate RAS enzymes in hGEnCs that were implicated by the MALDI-TOF MS experiments was confirmed by RT-PCR (Fig. 10) and Western blotting (Fig. 11). Expression of AGT and cathepsin D in hGEnCs was also demonstrated by Western blotting (Fig. 12, top).
Fig. 10.

mRNA encoded for renin-ANG system (RAS) enzymes detected in hGEnCs extracts as examined by RT-PCR. APA, aminopeptidase A.
Fig. 11.
Gene expression of RAS enzymes in hGEnCs extracts examined by Western blotting. Each well was loaded with 25 μg of cell protein. hKidney, human kidney extract lysate.
Fig. 12.
Top: gene expression of RAS elements upstream ANG I metabolism, angiotensinogen (AGT) and cathepsin D, in hGEnCs verified by Western blotting. Bottom: expression of the RAS enzyme aspartyl aminopeptidase (DAP; which was identified by tandem MS) in hGEnCs verified by Western blotting. Each lane was loaded with 25 μg of cell protein, except hKidney lane of blot probed for cathepsin D (5 μg).
Peptidase discovery by tandem MS.
Crude fractions of hGEnCs cell lysate from two different cell lines revealed few peptidases identified at high confidence (>95%) traditionally associated with ANG cleavage (Table 2). Based on number of unique peptides, APN, cathepsin D, and cathepsin B are likely to be the most abundant RAS-related peptidases in hGEnCs. Among the remainder of the detected peptidases, we searched for an enzyme responsible for the modest aspartyl aminopeptidase activity observed in our experiments, activity that was not mediated by APA. DAP was detected at low protein confidence levels from a single peptide, which scored at >95% confidence. Its presence was further confirmed by Western blotting (Fig. 12, bottom). Although other aminopeptidases were identified, they did not seem to account for any ANG cleavage detected by MALDI-TOF MS, based on their known substrate/inhibitor properties (BRENDA, www.brenda-enzymes.org). Accordingly, DAP was selected as candidate enzyme for the conversion of ANG I to ANG-(2–10) and ANG II to ANG III in hGEnCs. However, those conversions were not found to be nearly as dominant as those mediated by ACE, PEP, or ACE2 in our studies.
Table 2.
Peptidases identified from human glomerular endothelial cells by tandem mass spectrometry showing the number of unique peptides and the corresponding protein identification probability
| Protein and Gene Symbol | Accession No. | No. Unique Peptides | PIP, % |
|---|---|---|---|
| Cytosol aminopeptidase LAP3 | P28838 | 11 | 100 |
| Cathepsin D CTSD | P07339 | 8 | 100 |
| Cathepsin B CTSB | P07858 | 8 | 100 |
| Puromycin-sensitive aminopeptidase NPEPPS | P55786 | 9 | 100 |
| Aminopeptidase N ANPEP | P15144 | 11 | 100 |
| Methionine aminopeptidase 2 METAP2 | P50579 | 2 | 99.8 |
| Xaa-Pro aminopeptidase 1 XPNPEP1 | Q9NQW7 | 2 | 98.6 |
| Aspartyl aminopeptidase DNPEP | Q9ULA0 | 1 | 67 |
| Methionine aminopeptidase 1 METAP1 | P53582 | 1 | 67 |
| Aminopeptidase B RNPEP | Q9H4A4 | 1 | 67 |
| Platelet endothelial cell adhesion molecule (PECAM-1) | P16284 | 7 | 100 |
| von Willebrand factor (vWF) | P04275 | 8 | 100 |
Low confidence protein identifications (< 95%) were also provided if peptide confidence exceeded 50%. Endothelial cell phenotype markers are listed at the bottom (PECAM-1 and vWF). PIP, protein identification probability.
DISCUSSION
Our study is a comprehensive examination of the enzymatic pathways responsible for the processing of ANG peptides by hGEnCs. Herein, we described the expression and activity of various RAS-related enzymes in an in vitro system. These studies highlight, for the first time, the differences in the pattern of ANG peptide metabolism among resident glomerular cells, specifically, between GEnCs and podocytes. In addition, they expand the traditional understanding of ANG metabolizing pathways beyond ACE and demonstrate an important contribution of PEP in the glomerular metabolism of ANG I and ANG II, supporting our hypothesis that ACE activity represents only a fraction of the total RAS enzymatic capacity of hGEnCs. A schematic that summarizes our observations is presented in Fig. 13. Nevertheless, it should be recognized that cultured cells may not reflect the cellular events that occur in the in vivo setting.
Fig. 13.
Proposed schematic of ANG-related enzymatic machinery contained in hGEnCs (A) and hPODs (B). Thicknesses of arrows and boxes are proportional to the magnitude of the corresponding enzymatic conversion based on our observations.
The role of the vascular endothelium in the regulation of the RAS has been widely recognized. ACE (EC 3.4.15.1), an ectoenzyme responsible for the conversion of ANG I into ANG II, is a key component of the vascular RAS. By virtue of its localization in endothelial cells distributed across all vascular beds, ACE regulates the concentration of circulating ANG II, as well as the amount of ANG II delivered to tissue compartments. As described in endothelial cells from other sites, such as pulmonary endothelium (21) and umbilical vein (22, 26), we found that ACE is expressed in hGEnCs and is largely responsible for the generation of ANG II by these cells. No evidence for other ANG II-forming enzyme was found in our study. Although chymase has been found to be an important ANG I-converting enzyme in normal and injured blood vessels, its localization seems to be circumscribed to mast cells (5, 19). Furthermore, we also found that ACE is partly responsible for the metabolism of ANG-(1–7) in hGEnCs, converting ANG-(1–7) into ANG-(1–5), as previously described in a lung preparation (10) and rat urine (13). Using kidney brush border membranes, Allred et al. (1) also showed ACE-mediated ANG-(1–7) cleavage, but they did not explore the occurrence of that reaction in a glomerular endothelial preparation. Overall, the robust ACE activity displayed by hGEnCs is in contrast to that contained in hPODs. Therefore, the balance of the intraglomerular RAS may be affected by disruption of the ability of either cell type to cleave ANG peptides.
PEP (EC 3.4.21.26), formerly known as postproline cleaving enzyme activity, is one of several endopeptidases capable of metabolizing ANG I and ANG II (3). Previous work revealed that NEP is largely responsible for conversion of ANG I to ANG-(1–7) in isolated rat glomerular suspensions (59). However, PEP activity was not explored in that study. An early publication reported postproline cleaving enzyme activity in human kidney (23). Utilizing enzyme purified from porcine kidney, it was later recognized that ANG II is a substrate of PEP (60). Later, Casarini et al. (9) described PEP-mediated hydrolysis along the rat nephron. Similarly, PEP activity was detected in human urine (36). More recently, expression and activity of PEP was localized in mouse kidney medulla and certain cortical regions (32). Specifically, the authors reported histological evidence of PEP in podocytes, but not in other glomerular cell types. Our data indicate that PEP is contained in hGEnCs and participates in the cleavage of ANG I and ANG II, converting both peptides to ANG-(1–7). Although previous reports suggested a role of PEP in the fragmentation of vasoactive peptides in the vascular endothelium (4, 35, 40), ours is the first demonstration of PEP expression and activity in endothelial cells originated from human glomeruli. Despite finding evidence of NEP mRNA in hGEnCs, no enzymatic activity attributable to NEP was identified in this study. Thus PEP may account for an important fraction of the fragmentation of ANG peptides within the glomerular compartment.
ACE2 (3.4.17.23) has been increasingly recognized as a key component of the intrarenal RAS (14, 25, 49). By converting ANG II into ANG-(1–7), ACE2 activity is determinant in opposing ANG II-mediated effects. Our findings revealed that ACE2 is contained in hGEnCs, and that, in addition to PEP, it contributes to the degradation of ANG II. Expression and activity of ACE2 has been previously demonstrated in renal cortex and vasculature (31, 50, 62). In terms of renal endothelial cell expression, Ye et al. (63) did not find histological evidence of ACE2 expression in mouse GEnCs. On the other hand, although Lely et al. (27) found positive staining for ACE2 restricted to visceral and parietal epithelia in normal human glomeruli, they reported neo-expression of ACE2 in glomerular endothelia in kidneys from primary and secondary glomerulopathies. Furthermore, expression of ACE2 was reported in human aortic endothelial cells in culture (20). Therefore, GEnCs could be an additional source of glomerular ACE2, along with podocytes (58, 63) and mesangial cells (2).
Involvement of PCP, also called angiotensinase C, as an ANG II-degrading enzyme cannot be discarded, since the inhibitor of PEP can also inhibit PCP, and PCP mRNA and protein were indeed detected in the cells. PCP has been purified from human kidney. Moreover, others have reported PCP in endothelial cells from other vascular beds (34, 44, 65). Interestingly, a polymorphism in the PCP gene has been associated with blood pressure response to an ACE inhibitor in hypertensive subjects (64).
While ANG-(1–7) was the predominant fragment resulting from ANG II metabolism in our studies, significantly smaller peaks corresponding to ANG III and ANG IV were also detected. Such pattern of ANG II fragmentation was previously observed in bovine adrenal endothelial cells (11). Our failure to detect APA expression (EC 3.4.11.7) or activity suggested that other enzymes must be responsible for the modest aspartyl aminopeptidase activity observed in hGEnCs. Our data indicate that DAP (EC 3.4.11.21) may be partly responsible for such activity. In agreement with our findings, one study found that rat kidney must contain a DAP distinct from APA (37), whereas others purified DAP from mammalian kidney (61). DAP activity was also reported in human renal cell carcinoma cells (56) and membrane fractions of pancreatic islet cells (7). Downstream of ANG III, we found that its conversion to ANG IV is mediated by APN (EC 3.4.11.2). Importantly, APN was found in the cell surface of human umbilical vein (15, 16) and aortic endothelial cells (15). In the kidney, APN is highly expressed in glomerular epithelial (51, 55) and tubular epithelial cells (43), but it has not been previously reported to be contained in GEnCs. Although other aminopeptidases, i.e., APB (EC 3.4.11.6), puromycin-sensitive aminopeptidase (EC 3.4.11.14), etc., were detected in hGEnCs by tandem MS, activity of those enzymes was not observed by our methods. However, those enzymes likely participate in intracellular RAS-related enzymatic processes that were not examined herein, but that appear to be relevant in certain disease states (46). Upstream of ANG I conversion, we also found evidence of AGT and cathepsin D expression in hGEnCs, suggesting that these cells possess the necessary components for intracellular de novo generation of ANG I and its metabolites. In agreement with our findings, AGT was reported to be expressed in human glomerular endothelium (54), whereas cathepsin D activity was present in rat preparations containing glomerular endothelial cells (29).
During our experiments of ANG II incubation, we observed that the detection of the substrate and its metabolites was not affected either by blockade of ANG II receptor binding and internalization, or by interruption of peptide trapping via endocytosis, suggesting that, even if a fraction of the peptides traveled across the cell membrane, intracellular enzymatic activity had little to no impact on the detected mass spectra. Therefore, the enzymatic mechanisms described here seem to mostly reflect reactions mediated by peptidases known to be membrane bound.
In summary, our studies demonstrate that hGEnCs possess a subset of peptidases that are capable of fragmenting ANG substrates in a relatively distinct pattern. Among those peptidases, the contribution of ACE to the overall ANG peptide cleavage seems to be of utmost importance, although other ectoenzymes were also found to play an important role in the degradation process. Notably, the enzymatic artillery of GEnCs differs from that contained in podocytes in that GEnCs favor ANG II formation, whereas PODs are mainly equipped to degrade ANG II or shunt ANG I conversion away from ANG II generation (Fig. 13). We speculate that injury to specific cell types within the glomerulus may lead to distinct effects on the homeostasis of the intraglomerular RAS or on the net delivery of filtered ANG peptides to the tubular compartment. Further work is needed to explore this contention.
GRANTS
J.C.V. is supported by the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health, Grant DK080944–04, Dialysis Clinics, Inc., and the Ralph H. Johnson Veterans Affairs Med. Ctr. Research Enhancement Award Program. M.G.J. was supported by a Veterans Affairs CDA-2 Career Development Award and the Nephcure Foundation Young Investigator Award.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
J.C.Q.V., T.A.M., J.M.A., J.R.R., and M.G.J. conception and design of research; J.C.Q.V., J.L.I., and A.M.B. performed experiments; J.C.Q.V. and M.G.J. analyzed data; J.C.Q.V. interpreted results of experiments; J.C.Q.V. and M.G.J. prepared figures; J.C.Q.V. drafted manuscript; J.C.Q.V., T.A.M., J.M.A., J.R.R., and M.G.J. edited and revised manuscript; J.C.Q.V., J.L.I., A.M.B., T.A.M., J.M.A., J.R.R., and M.G.J. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Shane Wing for technical assistance. We also thank Dr. John Schwacke for insightful discussion about the angiotensin peptide network.
REFERENCES
- 1. Allred AJ, Diz DI, Ferrario CM, Chappell MC. Pathways for angiotensin-(1—7) metabolism in pulmonary and renal tissues. Am J Physiol Renal Physiol 279: F841–F850, 2000 [DOI] [PubMed] [Google Scholar]
- 2. Aragao DS, Cunha TS, Arita DY, Andrade MC, Fernandes AB, Watanabe IK, Mortara RA, Casarini DE. Purification and characterization of angiotensin converting enzyme 2 (ACE2) from murine model of mesangial cell in culture. Int J Biol Macromol 49: 79–84, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ardaillou R. Active fragments of angiotensin II: enzymatic pathways of synthesis and biological effects. Curr Opin Nephrol Hypertens 6: 28–34, 1997 [DOI] [PubMed] [Google Scholar]
- 4. Bausback HH, Ward PE. Vascular, post proline cleaving enzyme: metabolism of vasoactive peptides. Adv Exp Med Biol 198: 397–404, 1986 [DOI] [PubMed] [Google Scholar]
- 5. Borland JA, Kelsall C, Yacoub MH, Chester AH. Expression, localisation and function of ACE and chymase in normal and atherosclerotic human coronary arteries. Vascul Pharmacol 42: 99–108, 2005 [DOI] [PubMed] [Google Scholar]
- 6. Burns KD. The emerging role of angiotensin-converting enzyme-2 in the kidney. Curr Opin Nephrol Hypertens 16: 116–121, 2007 [DOI] [PubMed] [Google Scholar]
- 7. Cai WW, Wang L, Chen Y. Aspartyl aminopeptidase, encoded by an evolutionarily conserved syntenic gene, is colocalized with its cluster in secretory granules of pancreatic islet cells. Biosci Biotechnol Biochem 74: 2050–2055, 2010 [DOI] [PubMed] [Google Scholar]
- 8. Caldwell PR, Seegal BC, Hsu KC, Das M, Soffer RL. Angiotensin-converting enzyme: vascular endothelial localization. Science 191: 1050–1051, 1976 [DOI] [PubMed] [Google Scholar]
- 9. Casarini DE, Boim MA, Stella RC, Schor N. Endopeptidases (kininases) are able to hydrolyze kinins in tubular fluid along the rat nephron. Am J Physiol Renal Physiol 277: F66–F74, 1999 [DOI] [PubMed] [Google Scholar]
- 10. Chappell MC, Pirro NT, Sykes A, Ferrario CM. Metabolism of angiotensin-(1–7) by angiotensin-converting enzyme. Hypertension 31: 362–367, 1998 [DOI] [PubMed] [Google Scholar]
- 11. Cui L, Nithipatikom K, Campbell WB. Simultaneous analysis of angiotensin peptides by LC-MS and LC-MS/MS: metabolism by bovine adrenal endothelial cells. Anal Biochem 369: 27–33, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Durvasula RV, Shankland SJ. Activation of a local renin angiotensin system in podocytes by glucose. Am J Physiol Renal Physiol 294: F830–F839, 2008 [DOI] [PubMed] [Google Scholar]
- 13. Ferrario CM, Averill DB, Brosnihan KB, Chappell MC, Iskandar SS, Dean RH, Diz DI. Vasopeptidase inhibition and ANG-(1–7) in the spontaneously hypertensive rat. Kidney Int 62: 1349–1357, 2002 [DOI] [PubMed] [Google Scholar]
- 14. Ferrario CM, Varagic J. The ANG-(1–7)/ACE2/mas axis in the regulation of nephron function. Am J Physiol Renal Physiol 298: F1297–F1305, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fukasawa K, Fujii H, Saitoh Y, Koizumi K, Aozuka Y, Sekine K, Yamada M, Saiki I, Nishikawa K. Aminopeptidase N (APN/CD13) is selectively expressed in vascular endothelial cells and plays multiple roles in angiogenesis. Cancer Lett 243: 135–143, 2006 [DOI] [PubMed] [Google Scholar]
- 16. Gao JJ, Xue X, Gao ZH, Cui SX, Cheng YN, Xu WF, Tang W, Qu XJ. LYP, a bestatin dimethylaminoethyl ester, inhibited cancer angiogenesis both in vitro and in vivo. Microvasc Res 82: 122–130, 2011 [DOI] [PubMed] [Google Scholar]
- 17. Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol 17: 994–999, 1999 [DOI] [PubMed] [Google Scholar]
- 18. Ide N, Hirase T, Nishimoto-Hazuku A, Ikeda Y, Node K. Angiotensin II increases expression of IP-10 and the renin-angiotensin system in endothelial cells. Hypertens Res 31: 1257–1267, 2008 [DOI] [PubMed] [Google Scholar]
- 19. Ihara M, Urata H, Kinoshita A, Suzumiya J, Sasaguri M, Kikuchi M, Ideishi M, Arakawa K. Increased chymase-dependent angiotensin II formation in human atherosclerotic aorta. Hypertension 33: 1399–1405, 1999 [DOI] [PubMed] [Google Scholar]
- 20. Iizuka K, Kusunoki A, Machida T, Hirafuji M. Angiotensin II reduces membranous angiotensin-converting enzyme 2 in pressurized human aortic endothelial cells. J Renin Angiotensin Aldosterone Syst 10: 210–215, 2009 [DOI] [PubMed] [Google Scholar]
- 21. Johnson AR. Human pulmonary endothelial cells in culture. Activities of cells from arteries and cells from veins. J Clin Invest 65: 841–850, 1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Johnson AR, Erdos EG. Metabolism of vasoactive peptides by human endothelial cells in culture. Angiotensin I converting enzyme (kininase II) and angiotensinase. J Clin Invest 59: 684–695, 1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kato T, Okada M, Nagatsu T. Distribution of post-proline cleaving enzyme in human brain and the peripheral tissues. Mol Cell Biochem 32: 117–121, 1980 [DOI] [PubMed] [Google Scholar]
- 24. Ketteler M, Noble NA, Border WA. Transforming growth factor-beta and angiotensin II: the missing link from glomerular hyperfiltration to glomerulosclerosis? Annu Rev Physiol 57: 279–295, 1995 [DOI] [PubMed] [Google Scholar]
- 25. Koitka A, Cooper ME, Thomas MC, Tikellis C. Angiotensin converting enzyme 2 in the kidney. Clin Exp Pharmacol Physiol 35: 420–425, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Koumbadinga GA, Bawolak MT, Marceau E, Adam A, Gera L, Marceau F. A ligand-based approach to investigate the expression and function of angiotensin converting enzyme in intact human umbilical vein endothelial cells. Peptides 31: 1546–1554, 2010 [DOI] [PubMed] [Google Scholar]
- 27. Lely AT, Hamming I, van Goor H, Navis GJ. Renal ACE2 expression in human kidney disease. J Pathol 204: 587–593, 2004 [DOI] [PubMed] [Google Scholar]
- 28. Liebau MC, Lang D, Bohm J, Endlich N, Bek MJ, Witherden I, Mathieson PW, Saleem MA, Pavenstadt H, Fischer KG. Functional expression of the renin-angiotensin system in human podocytes. Am J Physiol Renal Physiol 290: F710–F719, 2006 [DOI] [PubMed] [Google Scholar]
- 29. Lovett DH, Ryan JL, Kashgarian M, Sterzel RB. Lysosomal enzymes in glomerular cells of the rat. Am J Pathol 107: 161–166, 1982 [PMC free article] [PubMed] [Google Scholar]
- 30. Magaldi AJ, Cesar KR, de Araujo M, Simoes e Silva AC, Santos RA. Angiotensin-(1–7) stimulates water transport in rat inner medullary collecting duct: evidence for involvement of vasopressin V2 receptors. Pflügers Arch 447: 223–230, 2003 [DOI] [PubMed] [Google Scholar]
- 31. Mizuiri S, Hemmi H, Arita M, Aoki T, Ohashi Y, Miyagi M, Sakai K, Shibuya K, Hase H, Aikawa A. Increased ACE and decreased ACE2 expression in kidneys from patients with IgA nephropathy. Nephron Clin Pract 117: c57–c66, 2011 [DOI] [PubMed] [Google Scholar]
- 32. Myohanen TT, Venalainen JI, Garcia-Horsman JA, Piltonen M, Mannisto PT. Distribution of prolyl oligopeptidase in the mouse whole-body sections and peripheral tissues. Histochem Cell Biol 130: 993–1003, 2008 [DOI] [PubMed] [Google Scholar]
- 33. Nestoridi E, Kushak RI, Tsukurov O, Grabowski EF, Ingelfinger JR. Role of the renin angiotensin system in TNF-alpha and Shiga-toxin-induced tissue factor expression. Pediatr Nephrol 23: 221–231, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ngo ML, Mahdi F, Kolte D, Shariat-Madar Z. Upregulation of prolylcarboxypeptidase (PRCP) in lipopolysaccharide (LPS) treated endothelium promotes inflammation. J Inflamm (Lond) 6: 3, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Palmieri FE, Ward PE. Dipeptidyl(amino)peptidase IV and post proline cleaving enzyme in cultured endothelial and smooth muscle cells. Adv Exp Med Biol 247A: 305–311, 1989 [DOI] [PubMed] [Google Scholar]
- 36. Quinto BM, Juliano MA, Hirata I, Carmona AK, Juliano L, Casarini DE. Characterization of a prolyl endopeptidase (kininase) from human urine using fluorogenic quenched substrates. Int J Biochem Cell Biol 32: 1161–1172, 2000 [DOI] [PubMed] [Google Scholar]
- 37. Ramirez M, Prieto I, Martinez JM, Vargas F, Alba F. Renal aminopeptidase activities in animal models of hypertension. Regul Pept 72: 155–159, 1997 [DOI] [PubMed] [Google Scholar]
- 38. Saleem MA, O'Hare MJ, Reiser J, Coward RJ, Inward CD, Farren T, Xing CY, Ni L, Mathieson PW, Mundel P. A conditionally immortalized human podocyte cell line demonstrating nephrin and podocin expression. J Am Soc Nephrol 13: 630–638, 2002 [DOI] [PubMed] [Google Scholar]
- 39. Sampaio WO, Nascimento AA, Santos RA. Systemic and regional hemodynamic effects of angiotensin-(1–7) in rats. Am J Physiol Heart Circ Physiol 284: H1985–H1994, 2003 [DOI] [PubMed] [Google Scholar]
- 40. Santos RA, Brosnihan KB, Jacobsen DW, DiCorleto PE, Ferrario CM. Production of angiotensin-(1–7) by human vascular endothelium. Hypertension 19: II56–II61, 1992 [DOI] [PubMed] [Google Scholar]
- 41. Satchell SC, Braet F. Glomerular endothelial cell fenestrations: an integral component of the glomerular filtration barrier. Am J Physiol Renal Physiol 296: F947–F956, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schelling P, Fischer H, Ganten D. Angiotensin and cell growth: a link to cardiovascular hypertrophy? J Hypertens 9: 3–15, 1991 [PubMed] [Google Scholar]
- 43. Scherberich JE, Wiemer J, Herzig C, Fischer P, Schoeppe W. Isolation and partial characterization of angiotensinase A and aminopeptidase M from urine and human kidney by lectin affinity chromatography and high-performance liquid chromatography. J Chromatogr 521: 279–289, 1990 [DOI] [PubMed] [Google Scholar]
- 44. Shariat-Madar Z, Mahdi F, Schmaier AH. Recombinant prolylcarboxypeptidase activates plasma prekallikrein. Blood 103: 4554–4561, 2004 [DOI] [PubMed] [Google Scholar]
- 45. Shultz PJ, DiCorleto PE, Silver BJ, Abboud HE. Mesangial cells express PDGF mRNAs and proliferate in response to PDGF. Am J Physiol Renal Fluid Electrolyte Physiol 255: F674–F684, 1988 [DOI] [PubMed] [Google Scholar]
- 46. Singh R, Choubey D, Chen J, Leehey DJ. Inhibition of intracellular angiotensin II formation blocks high glucose effect on mesangial matrix. Regul Pept 158: 103–109, 2009 [DOI] [PubMed] [Google Scholar]
- 47. Singh R, Singh AK, Alavi N, Leehey DJ. Mechanism of increased angiotensin II levels in glomerular mesangial cells cultured in high glucose. J Am Soc Nephrol 14: 873–880, 2003 [DOI] [PubMed] [Google Scholar]
- 48. Siragy HM. Angiotensin II compartmentalization within the kidney: effects of salt diet and blood pressure alterations. Curr Opin Nephrol Hypertens 15: 50–53, 2006 [DOI] [PubMed] [Google Scholar]
- 49. Soler MJ, Wysocki J, Batlle D. Angiotensin-converting enzyme 2 and the kidney. Exp Physiol 93: 549–556, 2008 [DOI] [PubMed] [Google Scholar]
- 50. Soler MJ, Ye M, Wysocki J, William J, Lloveras J, Batlle D. Localization of ACE2 in the renal vasculature: amplification by angiotensin II type 1 receptor blockade using telmisartan. Am J Physiol Renal Physiol 296: F398–F405, 2009 [DOI] [PubMed] [Google Scholar]
- 51. Stefanovic V, Vlahovic P, Ardaillou N, Ronco P, Ardaillou R. Cell surface aminopeptidase A and N activities in human glomerular epithelial cells. Kidney Int 41: 1571–1580, 1992 [DOI] [PubMed] [Google Scholar]
- 52. Strawn WB, Ferrario CM, Tallant EA. Angiotensin-(1–7) reduces smooth muscle growth after vascular injury. Hypertension 33: 207–211, 1999 [DOI] [PubMed] [Google Scholar]
- 53. Striker GE, Soderland C, Bowen-Pope DF, Gown AM, Schmer G, Johnson A, Luchtel D, Ross R, Striker LJ. Isolation, characterization, and propagation in vitro of human glomerular endothelial cells. J Exp Med 160: 323–328, 1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Takamatsu M, Urushihara M, Kondo S, Shimizu M, Morioka T, Oite T, Kobori H, Kagami S. Glomerular angiotensinogen protein is enhanced in pediatric IgA nephropathy. Pediatr Nephrol 23: 1257–1267, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tauc M, Chatelet F, Verroust P, Vandewalle A, Poujeol P, Ronco P. Characterization of monoclonal antibodies specific for rabbit renal brush-border hydrolases: application to immunohistological localization. J Histochem Cytochem 36: 523–532, 1988 [DOI] [PubMed] [Google Scholar]
- 56. Varona A, Blanco L, Lopez JI, Gil J, Agirregoitia E, Irazusta J, Larrinaga G. Altered levels of acid, basic, and neutral peptidase activity and expression in human clear cell renal cell carcinoma. Am J Physiol Renal Physiol 292: F780–F788, 2007 [DOI] [PubMed] [Google Scholar]
- 57. Velez JC. The importance of the intrarenal renin-angiotensin system. Nat Clin Pract Nephrol 5: 89–100, 2009 [DOI] [PubMed] [Google Scholar]
- 58. Velez JC, Bland AM, Arthur JM, Raymond JR, Janech MG. Characterization of renin-angiotensin system enzyme activities in cultured mouse podocytes. Am J Physiol Renal Physiol 293: F398–F407, 2007 [DOI] [PubMed] [Google Scholar]
- 59. Velez JC, Ryan KJ, Harbeson CE, Bland AM, Budisavljevic MN, Arthur JM, Fitzgibbon WR, Raymond JR, Janech MG. Angiotensin I is largely converted to angiotensin (1–7) and angiotensin (2–10) by isolated rat glomeruli. Hypertension 53: 790–797, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ward PE, Bausback HH, Odya CE. Kinin and angiotensin metabolism by purified renal post-proline cleaving enzyme. Biochem Pharmacol 36: 3187–3193, 1987 [DOI] [PubMed] [Google Scholar]
- 61. Wilk S, Wilk E, Magnusson RP. Purification, characterization, and cloning of a cytosolic aspartyl aminopeptidase. J Biol Chem 273: 15961–15970, 1998 [DOI] [PubMed] [Google Scholar]
- 62. Wysocki J, Ye M, Soler MJ, Gurley SB, Xiao HD, Bernstein KE, Coffman TM, Chen S, Batlle D. ACE and ACE2 activity in diabetic mice. Diabetes 55: 2132–2139, 2006 [DOI] [PubMed] [Google Scholar]
- 63. Ye M, Wysocki J, William J, Soler MJ, Cokic I, Batlle D. Glomerular localization and expression of Angiotensin-converting enzyme 2 and Angiotensin-converting enzyme: implications for albuminuria in diabetes. J Am Soc Nephrol 17: 3067–3075, 2006 [DOI] [PubMed] [Google Scholar]
- 64. Zhang Y, Hong XM, Xing HX, Li JP, Huo Y, Xu XP. E112D polymorphism in the prolylcarboxypeptidase gene is associated with blood pressure response to benazepril in Chinese hypertensive patients. Chin Med J (Engl) 122: 2461–2465, 2009 [PubMed] [Google Scholar]
- 65. Zhu L, Carretero OA, Liao TD, Harding P, Li H, Sumners C, Yang XP. Role of prolylcarboxypeptidase in angiotensin II type 2 receptor-mediated bradykinin release in mouse coronary artery endothelial cells. Hypertension 56: 384–390, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]