Abstract
RNA interference (RNAi) is a collection of small RNA directed mechanisms that result in sequence specific inhibition of gene expression. The notion that RNAi could lead to a new class of therapeutics caught the attention of many investigators soon after its discovery. The field of applied RNAi therapeutics has moved very quickly from lab to bedside. The RNAi approach has been widely used for drug development and several phase I and II clinical trials are under way. However, there are still some concerns and challenges to overcome for therapeutic applications. These include the potential for off-target effects, triggering innate immune responses and most importantly obtaining specific delivery into the cytoplasm of target cells. This review focuses on the current status of RNAi-based therapeutics, the challenges it faces and how to overcome them.
Keywords: RNAi, delivery, siRNA, therapeutics, shRNA
Introduction
Prior to 1980, RNA was viewed as an inert nucleic acid intermediate for protein production. A dramatic change in our perception came about in the early 1980s with Altman's and Cech's Nobel Prize winning discoveries of catalytic RNAs (Cech et al, 1981; Guerrier-Takada et al, 1983). This development inspired many researchers to shift their attention towards this nucleic acid. The recognition of RNA as a regulator of gene expression culminated with the discovery of RNAi (Fire et al, 1998) for which Fire and Mello were awarded the Nobel Prize in 2006. This discovery paved the way for many additional findings, including microRNA (miRNA) regulation of translation, chromatin remodelling directed by small RNAs and seminal observations in 2001 that small interfering RNA duplexes of 19–23 base pairs (siRNAs) could trigger sequence specific gene inhibition in mammalian cells (Caplen et al, 2001; Elbashir et al, 2001).
This latter property of siRNAs carries an immense therapeutic potential. Cellular genes involved in human diseases can be targeted and silenced by exogenous introduction of siRNAs or by introduction of gene constructs expressing short hairpin RNAs (shRNA) that are converted into siRNAs by the RNAi machinery.
Currently, the RNAi-based drugs under investigation (see Table 1 for summary) are for the most part synthetic small interfering RNAs, although expressed short hairpins and at least one anti-miRNA antisense are in trials. The siRNAs are double stranded molecules, consisting of a guide strand that is perfectly complementary to a target mRNA and a passenger strand. They range in size from approximately 20–30 nucleotides (nt) and suppress target-specific gene expression by promoting mRNA degradation (Elbashir et al, 2001; Hannon & Rossi, 2004). Core components of this siRNA-mediated post-transcriptional silencing (PTGS) include the RNAse III enzyme Dicer and its co-factor transactivating response RNA-binding protein (TRBP) along with the Argonaute family of proteins, in particular Argonaute 2 (Ago 2), which is the catalytic engine of the RNA induced silencing complex (RISC). Dicer converts dsRNAs into 21–25 nucleotide duplexes with 3′ 2nt overhangs. The siRNA is then incorporated into one or more of the Argonaute proteins in RISC where the RNA serves as a sequence specific guide for complementary base pairing with the target and guides RISC for sequence specific target degradation or translational inhibition (Hammond et al, 2000; Tuschl et al, 1999). In the laboratory and in current trials the siRNAs are most often chemically synthesized, bypassing the Dicer cleavage step for entry into RISC.
Table 1.
RNAi-based therapeutics
 |
The key challenge for RNAi application: delivery
As with the older oligonucleotide technologies using antisense DNA or catalytic RNAs, delivery to the target cells and tissues is a major challenge for RNAi based drugs. Thus, delivery of siRNAs to specific cells needs to be addressed for safe and effective in vivo RNAi drug applications. The negative charge of siRNAs as well as their size makes it difficult for them to cross the cell membrane. Various delivery strategies include, but are not limited to, nanoparticles, cationic lipids, antibodies, cholesterol, aptamers and viral vectors for short hairpin RNAs (shRNAs) (Fig. 2). These are discussed below and summarized in Table 2.
Figure 2. Delivery strategies for RNAi.
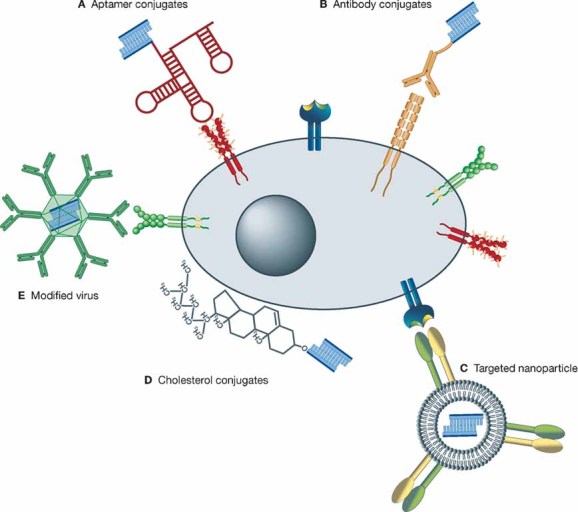
The cell (grey ellipse) contains a nucleus (dark circle) and a cell membrane (dark ellipse). Cell surface molecules such as receptors are present on the cell surface (shown in colour). RNAi therapeutics (mainly siRNA (blue)) can be targeted to the cell surface molecules via different delivery vehicles. They can be conjugated to aptamers (A), which can bind specifically to cell surface molecules and be internalized. siRNAs can also be conjugated to cell specific antibodies (B) and be delivered to the target cells via recognition of cell surface molecules by the specific antibody followed by internalization through endocytosis. Targeted nanoparticles (C) transport RNAi therapeutics to specific cells. The modifications of the nanoparticles (targeting ligand) can interact with receptors on the cell surface and the nanoparticle with its load can be internalized. Cholesterol conjugated siRNAs (D) can be delivered to cells and be internalized by the interaction of the cholesterol with the membrane through hydrophobic interactions, triggering Clathrin-dependent endocytosis. Modified viruses (E) can also be used for cell specific delivery of RNAi therapeutics by cell specific cell surface interactions triggering endocytosis.
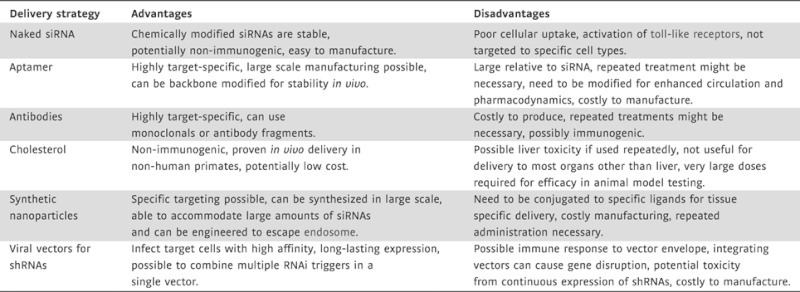
Table 2.
Pros and cons of delivery approaches
 |
Tissue delivery
Some of the delivery strategies that have been successfully utilized in animal models during pre-clinical studies have not been tried yet in a patient setting. Various strategies of targeted and non-targeted delivery are presented in Figure 2. The notion of simply attaching a ligand to an siRNA for targeted delivery to specific tissues or cells is very enticing. For instance, the use of cholesterol-conjugated siRNAs was shown to effectively deliver anti-Apo B siRNAs to the liver and other organs in a rodent model (Soutschek et al, 2004; Zimmermann et al, 2006). Cholesterol has also been successfully applied to deliver siRNAs topically in murine vaginal mucosal tissue for prevention as well as inhibition of a potentially lethal herpes simplex type 2 infection (Wu et al, 2009) and it has been shown to be an effective means for delivering anti-microRNA oligomers or antagomirs to a variety of tissues in mouse models (Krutzfeldt et al, 2007; Krutzfeldt et al, 2005). ApoB siRNAs conjugated to cholesterol and other fatty acids such as stearoyl or docosanyl conjugates mediated cellular siRNA uptake and gene silencing in vivo (Wolfrum et al, 2007). The expression of Sid-1 homologues on the cell surface also enhances systemic cellular uptake of siRNA conjugates and gene silencing efficacy (Duxbury et al, 2005; Wolfrum et al, 2007). Sid-1 is a transmembrane protein responsible for systemic uptake of dsRNA in Caenorhabditis elegans (Winston et al, 2002). The activity of Sid-1 as an siRNA uptake system in mammals could prove to be beneficial for the delivery of siRNAs conjugated to cell membrane interaction compounds such as cholesterol.
Glossary
- Adeno-associated virus
Small single stranded DNA virus used as a vector for gene therapy.
- Adenoviruses
Non-enveloped icosahedral viruses containing double stranded DNA genome that are commonly used as vectors for gene therapy.
- Antagomirs
Backbone modified antisense oligomers complementary to microRNAs, blocking their function.
- Antisense DNA
Short DNA strands complementary to target mRNAs.
- ApoB
Apolipoprotein B is the main apolipoprotein of low density lipoproteins that transport cholesterol and triglycerides through the blood stream.
- Blood–brain barrier
A selectively permeable barrier between the capillaries and the brain that acts as a filter preventing many substances from entering the central nervous system.
- Catalytic RNAs
Ribonucleic acids with enzymatic activity.
- Endosome
A membrane-bound organelle that sorts endocytosed material and recycles it back to the cell surface or delivers it to other compartments as the lysosome where such cargo can be degraded.
- Lentiviral vectors
Gene therapy vectors engineered from HIV or other members of the lentivirus family (a class of retrovirus).
- Locked nucleic acid technology
Backbone modification of nucleic acids that locks the sugars into a single constrained conformation, making the oligomers they are associated with nuclease-resistant and forming thermodynamically stable hybrids with ribonucleic acids.
- microRNA
Short RNAs processed from primary transcripts that guide RISC to complementary sequences in 3′-UTR of target messages.
- Off-target effects
Undesired down regulation of non-targeted transcripts by miRNAs or siRNAs.
- Polycation polymers
Positively charged polymers that interact with negatively charged nucleic acids and can be used as delivery vehicle for gene therapy.
- RISC
Proteins associated with the RNA in the RNA interference pathway are termed the RNA induced silencing complex or RISC.
- RNA-aptamer
In vitro evolved, 2' Fl backbone modified RNA molecules that take on three-dimensional shapes allowing them to bind with high affinities to targeted proteins.
- RNA interference
A cellular process in which small double stranded RNAs are used as precursors for selection of a guide strand that directs sequence specific base pairing to complementary sequences.
- short hairpin RNA
Precursors for miRNAs and small interfering RNAs (siRNAs) both of which serve as guides for RISC.
- SNALPs
Stable nucleic acid lipid particles that can be modified with targeting ligands and are used as delivery vehicle for gene therapy.
- Toll-like receptors
Proteins that recognize pathogen molecules and activate immune cell responses.
- Vector
Carrier and delivery vehicle for gene therapy.
Another strategy that has been used to lower ApoB levels and hopefully decrease the cardiovascular complications associated with high LDL-cholesterol levels is to deliver α-tocopherol (vitamin E) directly conjugated to a backbone modified ApoB targeted siRNA to the liver with resultant lowering of cholesterol in a rodent model (Nishina et al, 2008). Given the simplicity of conjugating cholesterol or a vitamin to an appropriately backbone modified siRNA and the very encouraging results in animal model testing of these approaches, it is unclear why this approach has not been used in any of the human clinical trials to date.
A second highly effective approach for the delivery of siRNAs to the liver is the use of lipid carriers. Particularly encouraging are results obtained from the delivery of anti-ApoB siRNAs using stable nucleic acid lipid particles or SNALPs (Zimmermann et al, 2006). SNALPs are lipid nanoparticles that fully encapsulate and systemically deliver a variety of nucleic acid molecules such as siRNAs, shRNAs or aptamers. They are stable and have a long circulation half-life in the blood stream. Intravenous delivery of SNALP encapsulated siRNAs targeting ApoB was shown to provide potent knockdown of the ApoB mRNA for over 48 h post delivery, and a reduction in circulating cholesterol for almost two weeks in a non-human primate test (Zimmermann et al, 2006).
Targeted cellular delivery
Targeted delivery is another important goal for siRNA therapeutics. Positively charged protamine peptides have been conjugated to single chain antibodies targeting various cellular receptors, including ERB2, HIV gp120 and the lymphocyte specific LFA1 receptor (Peer et al, 2007; Song et al, 2005). The positively charged peptide also binds the therapeutic siRNAs, which are functionally delivered into target cells both in culture and in animals (Peer et al, 2007; Song et al, 2005). In another example, a CD7 specific antibody conjugated to a nine arginine peptide that bound and delivered to T-lymphocytes siRNAs targeting CD4, CCR5 or viral transcripts was used for their systemic delivery in a humanized mouse model and was effective in blocking CD4+ T cell HIV infection (Kumar et al, 2008; Kumar et al, 2007).
The blood–brain barrier constitutes a real challenge for delivering exogenous molecules to the brain. To transmit siRNAs across the blood–brain barrier, Kumar et al (2007) fused a rabies glycoprotein peptide to nine arginines. While the nine arginines bound siRNAs targeting the Japanese encephalitis virus, the rabies peptide selectively binds to and is internalized by the acetylcholine receptor, resulting in retrograde transport of the siRNAs in neuronal axons and protection from the lethal viral infection (Kumar et al, 2007). Another approach breaching the blood–brain barrier for siRNA delivery, was reported by Bonoiu et al. (2009). They used gold nanoparticles (GNP) complexed to siRNA molecules, called nanoplexes, for modulation of the dopaminergic signaling pathway in an in vitro model as potential treatment for drug addiction therapy (Bonoiu et al, 2009).
Another means by which therapeutic siRNAs can be delivered to target cells via receptor mediated binding and internalization is to use receptor binding aptamers (Chu et al, 2006; McNamara et al, 2006; Zhou et al, 2008). Aptamers are single stranded, three-dimensional oligonucleotide structures that bind with high affinity to a wide variety of target molecules (Gold et al, 1995). The systematic evolution of ligands by exponential enrichment (SELEX) process generates aptamers from libraries of approximately 1015 nucleic acid molecules by in vitro selection (McNamara et al, 2006; Tuerk & Gold, 1990). For example, McNamara et al (2006) have shown that an aptamer which binds and is internalized by prostate cancer cells via the prostate specific membrane antigen (PSMA) receptor achieves intratumoural delivery of siRNAs and inhibition of tumour growth in a mouse xenograft. In all the cases published so far where aptamers have been used to deliver RNA molecules, the aptamers provided highly selective delivery only to cells expressing the receptor at the cell surface.
Polycation polymers such as cyclodextrin or dynamic polyconjugates are another promising delivery method for siRNAs. Delivery of siRNA to hepatocytes with siRNA dynamic polyconjugates, a membrane-active polymer, was non-toxic and efficient in vitro and in vivo (Rozema et al, 2007). Cyclodextrin nanoparticles coupled to transferin were also used successfully for systemic delivery of siRNAs to tumours in mice (Bartlett et al, 2007; Hu-Lieskovan et al, 2005). The transferin can bind to the transferin receptor which is highly expressed on tumour cells due to their high demand of iron. In fact, Calando Pharmaceuticals is currently employing transferin complexed cyclodextrin particles to deliver therapeutic siRNAs to solid tumours in phase I clinical trials.
Expression of shRNAs that get processed into siRNAs is yet another alternative strategy for RNAi based treatment of disease. For this to be an effective strategy, the shRNA expression units generally need to be incorporated into a viral vector. Systemic delivery of adeno-associated virus (AAV) and lentiviral vectors into animals have been carried out for pre-clinical studies of RNAi treatment of HBV, neurological and other organ-based diseases.
Viral vectors such as those derived from murine retroviruses or lentiviruses stably integrate therapeutic transgenes into the host genome (Boris-Lawrie & Temin, 1994; Samulski et al, 1989). The integration allows long lasting transgene expression, which may have a therapeutic advantage. However, random integration may also cause problems via gene disruption or trans-activation of oncogenes. Other viral vectors based upon AAV and adenovirus do not stably integrate, but persist as episomes until diluted out by cell division. A limitation with these viral vectors is the re-targeting to specific tissues via genetic modifications of the coat proteins so as not to perturb a virus particle assembly and function. Furthermore, a major proportion of the human population has antibodies against AAVs and adenovirus from previous infections, thus limiting the number of viral vector applications that can be made in a patient setting (Chirmule et al, 1999). To date, there is one known ongoing clinical trial with viral vector introduced shRNAs to treat HIV infection in AIDS/lymphoma patients for HIV being conducted at the City of Hope National Medical Center in California and supported in part by Benitec, Inc. In this trial, patient autologous haematopoietic stem cells are genetically modified with a lentiviral vector harbouring an anti-HIV shRNA along with two other anti-HIV RNAs (Anderson et al, 2007; Li et al, 2005). As virally delivered shRNAs will be expressed for long periods of time, it is critical that the disease being treated will benefit from persistent shRNA expression. There are numerous disease models for which gene expressed shRNAs or microRNA mimics are being tested in animal models. It is reasonable to expect more clinical trials in the near future.
Everything concerning delivery discussed so far requires some form of injection into animals or patients. Ideally, an oral delivery would make development of siRNAs into drugs more widely acceptable from a patient perspective and indeed some groups are exploring oral delivery strategies. Aerosol-mediated delivery is also being tested in animals and in human clinical trials. Aerosol delivery of a minimally modified anti-Akt1 siRNA was performed successfully with polyester amine polymers in mice to treat lung cancer (Xu et al, 2008). In 2006, Alnylam Inc. initiated the first phase 1 clinical study using minimally modified, unencapsulated siRNAs to treat RSV (respiratory syncytial virus) infection using intranasal delivery of their drug ALN-RSV01 (http://www.alnylam.com). In summary, there are a variety of already proven delivery options for siRNAs, some of which provide cell and tissue specific delivery. Only a couple of these options are currently being tested in clinical trials. The important considerations for use of a delivery modality are the safety, efficacy and cost of materials. Each of these has to be addressed separately before a vehicle will be commercially viable. Minimal backbone modifications for stabilizing siRNAs and shielding them from activation of toll-like receptors (TLR) do not substantially increase the cost, whereas packaging or complexing the siRNAs into/with delivery vehicles requires separate manufacturing of the siRNAs and the carriers, thereby substantially increasing the price. The important issues of safety are best addressed with the 2′-OMe backbone modifications described above. When selectively placed within the siRNA backbone, this modification can prevent off-target miRNA-like functions and prevent the activation of some of the TLRs. Viral delivery strategies for shRNAs or microRNA mimics have the inherent cost of vector manufacturing under FDA approved conditions. This can be costly, and if repeat administrations are required, the expense is increased significantly for patient applications. Thus, the best strategy for delivery has to take into account the disease being treated and the need for long-term expression (viral vectors) versus transient target knockdowns (siRNAs).
The endosome
Once delivered to the right tissue or the right cell, another obstacle that has to be overcome with siRNA delivery systems is the endosome. The siRNAs need to be delivered to the cytoplasm to be effective and most of the delivery systems which use receptor mediated endocytosis or even pinocytosis result in endosomal internalization and eventually elimination via the Golgi network. Escape from the endosome is therefore a critical factor for efficient therapeutic applications. A few approaches have been made trying to tackle this issue. Some polymers are designed to escape the endosome by the so-called ‘proton-sponge effect’ and direct membrane interaction of polycation and sulphoglucanes which enhances the escape (Russ et al, 2008). Receptor ligands, that could also be used as delivery vehicle, can be expressed with His-tags that enables the protein–siRNA complex to escape the endosome (Tarwadi et al, 2008). Viruses such as adenoviruses escape the endosome through their ability to lyse lipid bilayer membranes at a certain pH, which is present in the sorting endosome (Blumenthal et al, 1986; Seth et al, 1984; Wickham et al, 1994). SNALPs are designed to escape the endosome by membrane fusion and releasing their siRNA load directly into the cytoplasm. They undergo an interaction with the endosomal membrane. The lipids of the SNALP bilayer interact with the endosomal membrane and the two combine (http://www.protivabio.com/snalp/). Nevertheless, the endosome escape remains a critical factor for RNAi therapeutics and needs to be carefully addressed.
RNAi in the clinic
There are currently only two clinical trials using ex vivo delivery, whereas most of the trials employ systemic delivery including injections directly into the target tissues such as the eyes for treatment of age-related macular degeneration (AMD) or directly into tumours, inhalation or infusion with targeted delivery vehicles with incorporated siRNAs (Fig 1).
Figure 1. Delivery of therapeutics to patients.
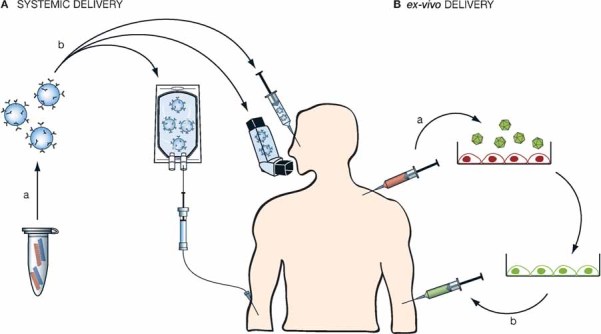
- Systemic delivery: (a) double stranded siRNA is packaged into delivery vehicle (targeted nanoparticles, polymers, liposomes, etc.). (b) It is then given intravenously, via inhalator or direct injection into the target tissue (the eye, tumour, etc.).
- ex-vivo delivery: (a) cells (dendritic cells, haematopoietic stem cells, etc.) are extracted from the patient and transduced with a virus containing shRNAs. (b) The genetically modified cells are then re-infused into the patient.
AMD is the leading cause of legal blindness in people over 55 years in the US. More than 1.75 million are affected and more than 200,000 new cases occur every year. Exudative AMD, the ‘wet’ form of AMD, causes loss of vision due to abnormal blood vessel growth (choroidal neovascularization) behind the retina and macula. Bleeding or leaking of fluids from these newly formed blood vessels causes the macula to bulge or lift, resulting in irreversible damage and loss of vision if left untreated. Anti-vascular endothelial growth factor (VEGF) agents cause regression of abnormal blood vessel growth and improvement of vision. In 2004 the FDA approved Pegaptanib (Macugen®, OSI pharmaceuticals, Pfizer), a VEGF targeting RNA-aptamer. This drug was the first intravitreal injection to be marketed. It opened the door for patients, their doctors, and scientists to further improve the treatment for AMD. Bevasiranib (Acuity Pharmaceuticals), a VEGF siRNA, has shown promising results in mice (Shen et al, 2006) and also stabilization of patients' condition and improved vision. It is currently being tested in a Phase III clinical trial. In contrast to targeting VEGF mRNA, AGN211745 (Sirna-027) developed by Allergen and Merck's Sirna Therapeutics targets the VEGF receptor (VEGFR1). This siRNA drug is in a Phase II clinical trial. Also, Quark Pharmaceuticals in collaboration with Silence Therapeutics developed an siRNA (RTP801i-14) for the treatment of AMD targeting the hypoxia-inducible RTP801, also known as DNA-damage inducible transcript 4 (DDIT4) that is involved in AMD disease progression. RTP801i-14 is presently being evaluated in a phase I clinical trial.
Another siRNA drug from Quark pharmaceuticals is I5NP (QPI-1002). This drug was developed to protect patients from acute kidney injury after cardiac bypass surgery and to prevent delayed graft function in patients undergoing deceased donor kidney transplantation by inhibiting p53 expression. The renal failure that occurs in 2 percent of patients undergoing heart surgery is caused by a reduction in the blood flow to the kidneys during surgery and hence its tissue can be damaged. Expression of p53 causes the removal of the damaged tissue and can therefore cause kidney failure after surgery. Presently, Quark Pharmaceuticals has enrolled patients for phase I/II dose escalation and safety studies.
Respiratory syncytial virus (RSV) is a single-stranded RNA virus of the family paramyxoviridae, which causes respiratory tract infections in patients of all ages. It affects approximately 300, 000 people in the USA and 75,000-120,000 children are hospitalized every year (Falsey, 2005; Shay et al, 1999, 2001). Alnylam Pharmaceuticals' front-running siRNA-based drug ALN-RSV01 targets the nucleocapsid encoding gene of the virus and therefore inhibits viral replication in the lung. In their phase I clinical trial, intranasal siRNA delivery was employed and ALN-RSV01 was well tolerated by adults (http://www.alnylam.com). In early 2008, they initiated a Phase II trial in lung transplant patients naturally infected with RSV and they plan to advance ALN-RSV01 into a pediatric patient population.
Pachyonychia congenital disorder (PC) is a rare but very painful disorder that primarily affects the skin, nails and mouth. Symptoms include blistering on the hands and feet, mouth sores and cysts of various types (http://www.pachyonychia.org/). It is caused by mutations in one of four keratin (K) genes (K6a, K6b, K16 or K17). The most common mutation in keratin causing PC is K6a. Trans Derm, Inc. has developed an siRNA that selectively inhibits a mutant allele of K6a (TD101) (Leachman et al, 2008). By eliminating the mutant form of keratin, they hope to treat PC in patients harbouring that allelic mutation. Trans Derm K6a is in a phase I clinical trial as a collaborative effort between Trans Derm and Charity PC Project.
Worldwide, an estimated 150–200 million people are infected with Hepatitis C virus (HCV). HCV accounts for approximately 15 percent of acute viral hepatitis, between 60 and 70 percent of chronic hepatitis, and up to 50 percent of cirrhosis, end-stage liver disease and liver cancer. Adult liver cancer is the third leading cause of cancer deaths worldwide. Patients harbouring chronic HCV are treated with a combination of ribovarin, an oral antiviral agent and interferon α, which can have considerable toxicity. A novel approach to treating HCV infection has been developed by Santaris Pharmaceuticals using their locked nucleic acid (LNA) technology for antisense oligomers. They have developed an LNA antisense oligomer (SPC3649), which targets miR-122, an miRNA that enhances HCV replication and translation (Chang et al, 2008; Henke et al, 2008). Such anti-microRNA antisense agents are termed antagomirs and can be potent inhibitors of microRNA function (Krutzfeldt et al, 2005). Incorporated into oligonucleotides, LNA dramatically enhances the binding affinity to complementary RNA sequences and their stability (Elmen et al, 2005; Grunweller et al, 2003; Wahlestedt et al, 2000). The greater potency of LNA in binding RNA means that LNA oligonucleotide drugs can be significantly shorter than conventional antisense phosphorothioate oligomers. These shorter RNA antagomirs are taken up efficiently by cells and tissues, thereby overcoming many of the delivery problems of RNAi to date and can be administered ‘naked’. SPC3649, the miR-122 antagomir is the first anti-microRNA being tested for therapeutic application and is currently being investigated in a phase I clinical trial.
In all of the above trials, ‘naked RNAs’ are being delivered directly to the tissue of interest. Currently, there is only one clinical trial employing systemic delivery via infusion using targeted nanoparticles for siRNA delivery to tumours. Calando pharmaceuticals is currently treating patients with relapsed or refractory solid tumours (such as breast cancer, prostate cancer, lung cancer, sarcomas and lymphomas) in a phase I clinical trial (http://www.clinicaltrials.gov). They developed CALAA-01, an siRNA that targets the ribonucleotide reductase subunit M2 (RRM2). This reductase catalyses the formation of deoxyribonucleotides from ribonucleotides and inhibition of this pathway results in loss of cell proliferation (Heidel et al, 2007). Over-expression of RRM2 significantly enhances the invasive and metastatic potential of solid tumours and is also implicated in angiogenesis (Zhang et al, 2009). CALAA-01 siRNA is delivered in cyclodextrin nanoparticles coated with transferrin, which directs and enhances uptake into tumour cells that express high levels of the transferrin receptor. These particles have been previously successfully tested for siRNA delivery to tumours in mice (Bartlett et al, 2007). Results from the phase I trial are currently being evaluated.
Ex vivo delivery is addressed by two clinical trials, one treating HIV-1 infection in AIDS lymphoma patients and the other for treatment of metastatic melanoma. The City of Hope National Medical center in collaboration with Benitec, Inc. started a pilot feasibility study for the treatment of HIV-1 infection in AIDS/lymphoma patients. The tat and rev shared exon of HIV-1 is targeted by an RNA Polymerase III expressed short hairpin RNA which is used in combination with a Pol III expressed TAR decoy and a chimeric VA1-ribozyme targeting CCR5 (Li et al, 2005). Patient derived haematopoietic (blood) stem cells (HSCs) are transduced with a lentiviral vector that incorporates the shRNA, VA1-ribozyme and the TAR decoy into the genomes of the HSCs. These genetically modified cells are then infused into patients whose bone marrow has been ablated for treatment of their AIDS-related lymphoma. To date, four patients have been treated and three of the four patients have persistent expression of the anti-tat/rev shRNA.
Duke University is conducting a phase I trial using siRNA for metastatic melanoma, a form of cancer that originates in melanocytes. Patients are being treated with autologous dendritic cells transfected with siRNAs plus a tumour antigen encoding mRNA. The goal of the siRNA therapy is the induction of an anti-melanoma immune response by alterating the proteasome-mediated antigen processing (Abdel-Wahab et al, 2005; Dannull et al, 2007). Monocytes of patients will be harvested and tranfected ex vivo with siRNAs that mediate the downregulation of the inducible immunoproteasome subunits LMP2, LMP7 and MECL1. In vitro, these cells are differentiated into dendritic cells (DC). After the induction of maturation, the DCs will be transfected with mRNAs encoding melanoma antigens MART, MAGE-3, tyrosinase and gp100. Since dendritic cells are powerful antigen presenting cells, the siRNA knockdown of the proteasome will enhance the melanoma antigen presentation with the goal of provoking a strong immune response against the melanoma cells in these patients.
Unexpected problems and possible solutions for RNAi based therapies
Given that RNAi was first described slightly over a decade ago and the mechanisms of this phenomenon are still being unraveled, it is quite amazing that there is such a collection of ongoing clinical trials. Although there is no FDA approved siRNA drug so far, this may change within the next couple of years, and should open the doors for more approved therapeutic siRNAs. As with any fast-moving technology, there are often unforeseen problems and the RNAi field is not recalcitrant to such problems. The first indication or cautionary note came from studies conducted by Merck-Rosetta using gene arrays to look for off-target effects of siRNAs. Their results showed that ectopically applied siRNAs can alter the expression levels of dozens of non-targeted transcripts, and suggested that even short complementary stretches of siRNAs with non-targeted transcripts can affect their expression levels (Jackson et al, 2003). Once it was understood about how microRNAs affect down-regulation of target proteins via binding to the 3′-UTR, which in turn can inhibit protein translation and in some instances trigger non-specific degradation, it was quickly realized that ectopically applied siRNAs were affecting non-targeted gene expression via microRNA-like functions (Jackson et al, 2006b). This off-targeting by siRNAs can be easily controlled by a 2′-OMe modification at the second ribose from the 5′-end of the siRNA (Jackson et al, 2006a). This simple solution should be used for all in vivo siRNA applications, but surprizingly this is not the case. This may in part be due to the long time it takes to develop a compound for clinical trial, and many of the siRNAs currently in the clinic were developed prior to the publication of the 2′-OMe findings.
Bridge the Gap
The Gap
RNAi-based therapeutics are currently being tested in various phase I and II clinical trials, however there are still several problems to overcome before their clinical application becomes widespread. Obtaining specific and effective delivery into the cytoplasm of target cells is one of the critical challenges. The enthusiasm with which the RNAi field has explored its clinical applications has led to the identification of several deleterious responses often triggered by these molecules such as potential for off-target effects and triggering host innate immune responses, both of which can lead to damaging secondary side-effects in the patient.
The Bridge
This review describes the current status of RNAi-based therapeutics and summarizes the many approaches the field is taking to tackle the issue of the RNA delivery. Lipid carriers, viruses and ingenious means to promote selective internalization of the RNA molecules by cellular receptors are some examples. The latter must be further explored in order to allow simple systemic delivery of si, sh and miRNAs. Targeted delivery is a major issue in many other more developed drug delivery fields and considering the approaches developed in other research areas may prove beneficial for RNA delivery.
The incredible leap we have faced in the understanding of the basic mechanisms of RNAi has brought a solution for most of the harmful effects identified previously, e.g., a 2′-OMe backbone modifications to prevent off-target effects. Strategies that capitalize upon the endogenous mechanism without disrupting the natural pathway should be used to achieve maximal benefit from RNAi-based therapeutics and fundamental research in this area is bound to continue to bring novel translational ideas and solutions.
The next alarming finding with respect to RNAi therapeutic applications came from studies using Pol III expressed shRNAs delivered in an AAV delivered by tail vein injection into mice, which resulted in lethality due to acute liver failure in many of the animals (Grimm et al, 2006). The lesson learnt from this study was that the levels of ectopic expression of therapeutic shRNAs has to be carefully controlled since these are processed into siRNAs which can elicit off target effects and effectively compete with the endogenous microRNAs for the RNAi machinery.
If the above concerns were not enough, two reports in the literature showed that certain sequence motifs in siRNAs can trigger type I interferon production via activation of toll-like receptors (TLRs) 7 and 8, thereby compromising the sequence specific knockdown effects of the RNAi pathway (Hornung et al, 2005; Judge et al, 2005; Robbins et al, 2008). Again, the solution to preventing activation of these two TLRs seems to be simple enough, the inclusion of at least one 2′-OMe in either the sense or anti-sense strand of the siRNA (Judge & MacLachlan, 2008; Robbins et al, 2007).
Abrogating the interferon stimulation mediated by a different TLR, TLR3 which recognizes double stranded RNAs is not so simple though, albeit still feasible. A somewhat surprising finding came from a group studying a murine model for siRNA treatment of AMD, presumably mediated by knocking down the expression of VEGF or the VEGF receptor as described above (Kleinman et al, 2008). This study showed that the inhibition of neo-vascularization was not due to specific knockdown of these targets (almost any siRNA gave the same phenotypic results), but was caused by double stranded RNA activation of TRL3, triggering interferon-γ and interleukin 12-production, with subsequent anti-neo-vascularization effects. Discouragingly, backbone modifications of the siRNAs did not ameliorate this response (Kleinman et al, 2008). The take home lesson from this study is that the use of ‘naked’ siRNAs in vivo is potentially problematic and the use of delivery vehicles, which shields the siRNAs from the cell surface TLR3, needs to be explored and tested. When siRNAs are delivered via some carrier mechanism, including tethering to cholesterol or encapsulation in bilayer lipids, no interferon types of responses to in vivo siRNA delivery have been observed (Soutschek et al, 2004; Zimmermann et al, 2006).
As an interesting caveat to the concerns about siRNAs activating interferons, it has recently been shown that activating the immune system can also be exploited for therapeutic purposes. A Bcl-2 siRNA with 5′-triphosphate ends provoked massive apoptosis of melanoma tumour cells in lung metastasis in vivo (Poeck et al, 2008). The 5′-triphosphates are recognized by the retinoic acid induced protein I (Rig-I) which activates innate immune cells such as dendritic cells and therefore an interferon response. This interferon response in combination with the Bcl-2 gene knockdown results in the apoptosis of tumour cells (Poeck et al, 2008).
Looking to the future
The promise of RNAi as a powerful new approach for therapeutic treatment of diseases has propelled early stage clinical testing of siRNAs for a variety of diseases. It is still too soon to evaluate whether or not RNAi based therapeutics will live up to their expectations. The most important aspect of developing a therapeutic strategy such as RNAi is to have a good understanding of the basic mechanisms of RNAi. Since this is a highly evolved endogenous mechanism for regulating gene expression, it is important to fully understand how the various RNAi based mechanisms function. By using strategies that capitalize upon the endogenous mechanism without disrupting the natural pathway, we can expect to achieve maximal benefit from RNAi based therapeutics. In the future, we should see stand alone RNAi therapeutics, but more likely we will see RNAi being used in combination with therapies already in place. The power of sequence specific inhibition of gene expression is of course a goal worth achieving for the treatment of all diseases. Overcoming the obstacles for achieving this is still a formidable task, but great strides are being made.
The authors declare that they have no conflict of interest.
For more Information
Clinical trials: http://www.clinicaltrials.gov
Pachyonchia charity/patients help: http://www.pachyonychia.org/
Various cancer info: http://www.cancer.gov
Center for disease control: http://www.cdc.gov/
US Food and Drug Administration: http://www.fda.gov/
World Health Organization: http://www.who.int/
References
- Abdel-Wahab Z, Cisco R, Dannull J, Ueno T, Abdel-Wahab O, Kalady MF, Onaitis MW, Tyler DS, Pruitt SK. Cotransfection of DC with TLR4 and MART-1 RNA induces MART-1-specific responses. J Surg Res. 2005;124:264–273. doi: 10.1016/j.jss.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Anderson J, Li MJ, Palmer B, Remling L, Li S, Yam P, Yee JK, Rossi J, Zaia J, Akkina R. Safety and efficacy of a lentiviral vector containing three anti-HIV genes–CCR5 ribozyme, tat-rev siRNA, and TAR decoy–in SCID-hu mouse-derived T cells. Mol Ther. 2007;15:1182–1188. doi: 10.1038/sj.mt.6300157. [DOI] [PubMed] [Google Scholar]
- Bartlett DW, Su H, Hildebrandt IJ, Weber WA, Davis ME. Impact of tumor-specific targeting on the biodistribution and efficacy of siRNA nanoparticles measured by multimodality in vivo imaging. Proc Natl Acad Sci USA. 2007;104:15549–15554. doi: 10.1073/pnas.0707461104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal R, Seth P, Willingham MC, Pastan I. pH-dependent lysis of liposomes by adenovirus. Biochemistry. 1986;25:2231–2237. doi: 10.1021/bi00356a057. [DOI] [PubMed] [Google Scholar]
- Bonoiu AC, Mahajan SD, Ding H, Roy I, Yong KT, Kumar R, Hu R, Bergey EJ, Schwartz SA, Prasad PN. Nanotechnology approach for drug addiction therapy: gene silencing using delivery of gold nanorod-siRNA nanoplex in dopaminergic neurons. Proc Natl Acad Sci USA. 2009;106:5546–5550. doi: 10.1073/pnas.0901715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boris-Lawrie K, Temin HM. The retroviral vector. Replication cycle and safety considerations for retrovirus-mediated gene therapy. Ann N Y Acad Sci. 1994;716:59–70. doi: 10.1111/j.1749-6632.1994.tb21703.x. discussion 71. [DOI] [PubMed] [Google Scholar]
- Caplen NJ, Parrish S, Imani F, Fire A, Morgan RA. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc Natl Acad Sci USA. 2001;98:9742–9747. doi: 10.1073/pnas.171251798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech TR, Zaug AJ, Grabowski PJ. In vitro splicing of the ribosomal RNA precursor of Tetrahymena: involvement of a guanosine nucleotide in the excision of the intervening sequence. Cell. 1981;27:487–496. doi: 10.1016/0092-8674(81)90390-1. [DOI] [PubMed] [Google Scholar]
- Chang J, Guo JT, Jiang D, Guo H, Taylor JM, Block TM. Liver-specific microRNA miR-122 enhances the replication of hepatitis C virus in nonhepatic cells. J Virol. 2008;82:8215–8223. doi: 10.1128/JVI.02575-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirmule N, Propert K, Magosin S, Qian Y, Qian R, Wilson J. Immune responses to adenovirus and adeno-associated virus in humans. Gene Ther. 1999;6:1574–1583. doi: 10.1038/sj.gt.3300994. [DOI] [PubMed] [Google Scholar]
- Chu TC, Twu KY, Ellington AD, Levy M. Aptamer mediated siRNA delivery. Nucleic Acids Res. 2006;34:e73. doi: 10.1093/nar/gkl388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannull J, Lesher DT, Holzknecht R, Qi W, Hanna G, Seigler H, Tyler DS, Pruitt SK. Immunoproteasome down-modulation enhances the ability of dendritic cells to stimulate antitumor immunity. Blood. 2007;110:4341–4350. doi: 10.1182/blood-2007-04-083188. [DOI] [PubMed] [Google Scholar]
- Duxbury MS, Ashley SW, Whang EE. RNA interference: a mammalian SID-1 homologue enhances siRNA uptake and gene silencing efficacy in human cells. Biochem Biophys Res Commun. 2005;331:459–463. doi: 10.1016/j.bbrc.2005.03.199. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Elmen J, Thonberg H, Ljungberg K, Frieden M, Westergaard M, Xu Y, Wahren B, Liang Z, Orum H, Koch T, et al. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. 2005;33:439–447. doi: 10.1093/nar/gki193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsey AR. Respiratory syncytial virus infection in elderly and high-risk adults. Exp Lung Res. 2005;31((Suppl 1)):77. [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Gold L, Polisky B, Uhlenbeck O, Yarus M. Diversity of oligonucleotide functions. Annu Rev Biochem. 1995;64:763–797. doi: 10.1146/annurev.bi.64.070195.003555. [DOI] [PubMed] [Google Scholar]
- Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- Grunweller A, Wyszko E, Bieber B, Jahnel R, Erdmann VA, Kurreck J. Comparison of different antisense strategies in mammalian cells using locked nucleic acids, 2′-O-methyl RNA, phosphorothioates and small interfering RNA. Nucleic Acids Res. 2003;31:3185–3193. doi: 10.1093/nar/gkg409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Hannon GJ, Rossi JJ. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431:371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- Heidel JD, Liu JY, Yen Y, Zhou B, Heale BS, Rossi JJ, Bartlett DW, Davis ME. Potent siRNA inhibitors of ribonucleotide reductase subunit RRM2 reduce cell proliferation in vitro and in vivo. Clin Cancer Res. 2007;13:2207–2215. doi: 10.1158/1078-0432.CCR-06-2218. [DOI] [PubMed] [Google Scholar]
- Henke JI, Goergen D, Zheng J, Song Y, Schuttler CG, Fehr C, Junemann C, Niepmann M. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 2008;27:3300–3310. doi: 10.1038/emboj.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S, Noronha A, Manoharan M, Akira S, de Fougerolles A, Endres S, Hartmann G. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat Med. 2005;11:263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- Hu-Lieskovan S, Heidel JD, Bartlett DW, Davis ME, Triche TJ. Sequence-specific knockdown of EWS-FLI1 by targeted, nonviral delivery of small interfering RNA inhibits tumor growth in a murine model of metastatic Ewing's sarcoma. Cancer Res. 2005;65:8984–8992. doi: 10.1158/0008-5472.CAN-05-0565. [DOI] [PubMed] [Google Scholar]
- Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- Jackson AL, Burchard J, Leake D, Reynolds A, Schelter J, Guo J, Johnson JM, Lim L, Karpilow J, Nichols K, Marshall W, Khvorova A, Linsley PS. Position-specific chemical modification of siRNAs reduces ‘off-target’ transcript silencing. RNA. 2006a;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, Linsley PS. Widespread siRNA ‘off-target’ transcript silencing mediated by seed region sequence complementarity. RNA. 2006b;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judge A, MacLachlan I. Overcoming the innate immune response to small interfering RNA. Human gene therapy. 2008;19:111–124. doi: 10.1089/hum.2007.179. [DOI] [PubMed] [Google Scholar]
- Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- Kleinman ME, Yamada K, Takeda A, Chandrasekaran V, Nozaki M, Baffi JZ, Albuquerque RJ, Yamasaki S, Itaya M, Pan Y, Appukuttan B, Gibbs D, Yang Z, Kariko K, Ambati BK, Wilgus TA, DiPietro LA, Sakurai E, Zhang K, Smith JR, Taylor EW, Ambati J. Sequence- and target-independent angiogenesis suppression by siRNA via TLR3. Nature. 2008;452:591–597. doi: 10.1038/nature06765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzfeldt J, Kuwajima S, Braich R, Rajeev KG, Pena J, Tuschl T, Manoharan M, Stoffel M. Specificity, duplex degradation and subcellular localization of antagomirs. Nucleic Acids Res. 2007;35:2885–2892. doi: 10.1093/nar/gkm024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Kumar P, Ban HS, Kim SS, Wu H, Pearson T, Greiner DL, Laouar A, Yao J, Haridas V, Habiro K, Yang YG, Jeong JH, Lee KY, Kim YH, Kim SW, Peipp M, Fey GH, Manjunath N, Shultz LD, Lee SK, Shankar P. T cell-specific siRNA delivery suppresses HIV-1 infection in humanized mice. Cell. 2008;134:577–586. doi: 10.1016/j.cell.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Wu H, McBride JL, Jung KE, Kim MH, Davidson BL, Lee SK, Shankar P, Manjunath N. Transvascular delivery of small interfering RNA to the central nervous system. Nature. 2007;448:39–43. doi: 10.1038/nature05901. [DOI] [PubMed] [Google Scholar]
- Leachman SA, Hickerson RP, Hull PR, Smith FJ, Milstone LM, Lane EB, Bale SJ, Roop DR, McLean WH, Kaspar RL. Therapeutic siRNAs for dominant genetic skin disorders including pachyonychia congenita. J Dermatol Sci. 2008;51:151–157. doi: 10.1016/j.jdermsci.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MJ, Kim J, Li S, Zaia J, Yee JK, Anderson J, Akkina R, Rossi JJ. Long-term inhibition of HIV-1 infection in primary hematopoietic cells by lentiviral vector delivery of a triple combination of anti-HIV shRNA, anti-CCR5 ribozyme, and a nucleolar-localizing TAR decoy. Mol Ther. 2005;12:900–909. doi: 10.1016/j.ymthe.2005.07.524. [DOI] [PubMed] [Google Scholar]
- McNamara JO, 2nd, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E, Sullenger BA, Giangrande PH. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- Nishina K, Unno T, Uno Y, Kubodera T, Kanouchi T, Mizusawa H, Yokota T. Efficient in vivo delivery of siRNA to the liver by conjugation of alpha-tocopherol. Mol Ther. 2008;16:734–740. doi: 10.1038/mt.2008.14. [DOI] [PubMed] [Google Scholar]
- Peer D, Zhu P, Carman CV, Lieberman J, Shimaoka M. Selective gene silencing in activated leukocytes by targeting siRNAs to the integrin lymphocyte function-associated antigen-1. Proc Natl Acad Sci USA. 2007;104:4095–4100. doi: 10.1073/pnas.0608491104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poeck H, Besch R, Maihoefer C, Renn M, Tormo D, Morskaya SS, Kirschnek S, Gaffal E, Landsberg J, Hellmuth J, et al. 5′-Triphosphate-siRNA: turning gene silencing and Rig-I activation against melanoma. Nat Med. 2008;14:1256–1263. doi: 10.1038/nm.1887. [DOI] [PubMed] [Google Scholar]
- Robbins M, Judge A, Ambegia E, Choi C, Yaworski E, Palmer L, McClintock K, Maclachlan I. Misinterpreting the therapeutic effects of siRNA caused by immune stimulation. Human gene therapy. 2008 doi: 10.1089/hum.2008.131. [DOI] [PubMed] [Google Scholar]
- Robbins M, Judge A, Liang L, McClintock K, Yaworski E, MacLachlan I. 2′-O-methyl-modified RNAs act as TLR7 antagonists. Mol Ther. 2007;15:1663–1669. doi: 10.1038/sj.mt.6300240. [DOI] [PubMed] [Google Scholar]
- Rozema DB, Lewis DL, Wakefield DH, Wong SC, Klein JJ, Roesch PL, Bertin SL, Reppen TW, Chu Q, Blokhin AV, et al. Dynamic PolyConjugates for targeted in vivo delivery of siRNA to hepatocytes. Proc Natl Acad Sci USA. 2007;104:12982–12987. doi: 10.1073/pnas.0703778104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ V, Elfberg H, Thoma C, Kloeckner J, Ogris M, Wagner E. Novel degradable oligoethylenimine acrylate ester-based pseudodendrimers for in vitro and in vivo gene transfer. Gene Ther. 2008;15:18–29. doi: 10.1038/sj.gt.3303046. [DOI] [PubMed] [Google Scholar]
- Samulski RJ, Chang LS, Shenk T. Helper-free stocks of recombinant adeno-associated viruses: normal integration does not require viral gene expression. J Virol. 1989;63:3822–3828. doi: 10.1128/jvi.63.9.3822-3828.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth P, Willingham MC, Pastan I. Adenovirus-dependent release of 51Cr from KB cells at an acidic pH. J Biol Chem. 1984;259:14350–14353. [PubMed] [Google Scholar]
- Shay DK, Holman RC, Newman RD, Liu LL, Stout JW, Anderson LJ. Bronchiolitis-associated hospitalizations among US children, 1980–1996. JAMA. 1999;282:1440–1446. doi: 10.1001/jama.282.15.1440. [DOI] [PubMed] [Google Scholar]
- Shay DK, Holman RC, Roosevelt GE, Clarke MJ, Anderson LJ. Bronchiolitis-associated mortality and estimates of respiratory syncytial virus-associated deaths among US children, 1979–1997. J Infect Dis. 2001;183:16–22. doi: 10.1086/317655. [DOI] [PubMed] [Google Scholar]
- Shen J, Samul R, Silva RL, Akiyama H, Liu H, Saishin Y, Hackett SF, Zinnen S, Kossen K, Fosnaugh K, et al. Suppression of ocular neovascularization with siRNA targeting VEGF receptor 1. Gene Ther. 2006;13:225–234. doi: 10.1038/sj.gt.3302641. [DOI] [PubMed] [Google Scholar]
- Song E, Zhu P, Lee SK, Chowdhury D, Kussman S, Dykxhoorn DM, Feng Y, Palliser D, Weiner DB, Shankar P, et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat Biotechnol. 2005;23:709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harborth J, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- Tarwadi Jazayeri JA, Prankerd RJ, Pouton CW. Preparation and in vitro evaluation of novel lipopeptide transfection agents for efficient gene delivery. Bioconjug Chem. 2008;19:940–950. doi: 10.1021/bc700463q. [DOI] [PubMed] [Google Scholar]
- Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Tuschl T, Zamore PD, Lehmann R, Bartel DP, Sharp PA. Targeted mRNA degradation by double-stranded RNA in vitro. Genes Dev. 1999;13:3191–3197. doi: 10.1101/gad.13.24.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlestedt C, Salmi P, Good L, Kela J, Johnsson T, Hokfelt T, Broberger C, Porreca F, Lai J, Ren K, et al. Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc Natl Acad Sci USA. 2000;97:5633–5638. doi: 10.1073/pnas.97.10.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham TJ, Filardo EJ, Cheresh DA, Nemerow GR. Integrin alpha v beta 5 selectively promotes adenovirus mediated cell membrane permeabilization. J Cell Biol. 1994;127:257–264. doi: 10.1083/jcb.127.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston WM, Molodowitch C, Hunter CP. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science. 2002;295:2456–2459. doi: 10.1126/science.1068836. [DOI] [PubMed] [Google Scholar]
- Wolfrum C, Shi S, Jayaprakash KN, Jayaraman M, Wang G, Pandey RK, Rajeev KG, Nakayama T, Charrise K, Ndungo EM, et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat Biotechnol. 2007;25:1149–1157. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
- Wu Y, Navarro F, Lal A, Basar E, Pandey RK, Manoharan M, Feng Y, Lee SJ, Lieberman J, Palliser D. Durable protection from Herpes Simplex Virus-2 transmission following intravaginal application of siRNAs targeting both a viral and host gene. Cell Host Microbe. 2009;5:84–94. doi: 10.1016/j.chom.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu CX, Jere D, Jin H, Chang SH, Chung YS, Shin JY, Kim JE, Park SJ, Lee YH, Chae CH, et al. Poly(ester amine)-mediated, aerosol-delivered Akt1 small interfering RNA suppresses lung tumorigenesis. Am J Respir Crit Care Med. 2008;178:60–73. doi: 10.1164/rccm.200707-1022OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Hu S, Wu J, Chen L, Lu J, Wang X, Liu X, Zhou B, Yen Y. Overexpression of RRM2 decreases thrombspondin-1 and increases VEGF production in human cancer cells: implication of RRM2 in angiogenesis. Mol Cancer. 2009;8:11. doi: 10.1186/1476-4598-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Li H, Li S, Zaia J, Rossi JJ. Novel dual inhibitory function aptamer-siRNA delivery system for HIV-1 therapy. Mol Ther. 2008;16:1481–1489. doi: 10.1038/mt.2008.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, Harborth J, Heyes JA, Jeffs LB, John M, et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]


