Abstract
The mechanisms leading to neuronal death in neurodegenerative disease are poorly understood. Many of these disorders, including Alzheimer’s (AD), Parkinson’s (PD) and prion diseases, are associated with the accumulation of misfolded disease-specific proteins. The unfolded protein response (UPR) is a protective cellular mechanism triggered by rising levels of misfolded proteins. One arm of this pathway results in the transient shutdown of protein translation, through phosphorylation of the alpha subunit of eukaryotic translation initiation factor, eIF2α. UPR activation and/or increased eIF2α–P levels are seen in patients with AD, PD and prion disease 1-4, but how this links to neurodegeneration is unknown. Here we show that accumulation of prion protein (PrP) during prion replication causes persistent translational repression of global protein synthesis by eIF2α–P, associated with synaptic failure and neuronal loss in prion-diseased mice. Further, we show that promoting translational recovery in hippocampi of prion-infected mice is neuroprotective. Over-expression of GADD34, a specific eIF2α–P phosphatase, as well as reduction of PrP levels by lentivirally-mediated RNAi, reduced eIF2α–P levels. As a result, both approaches restored vital translation rates during prion disease, rescuing synaptic deficits and neuronal loss, and thereby significantly increasing survival. In contrast, salubrinal, an inhibitor of eIF2α-P dephosphorylation5 increased eIF2α-P levels, exacerbating neurotoxicity and significantly reducing survival in prion diseased mice. Given the prevalence of protein misfolding and UPR activation in several neurodegenerative diseases, our results suggest that manipulation of common pathways such as translational control, rather than disease-specific approaches, may lead to new therapies preventing synaptic failure and neuronal loss across the spectrum of these disorders.
Neurodegenerative diseases pose an ever-increasing challenge for society and health care systems worldwide, but their molecular pathogenesis is still largely unknown and no curative treatments exist. Alzheimer’s (AD), Parkinson’s (PD) and prion diseases are separate clinical and pathological conditions, but it is likely they share common mechanisms leading to neuronal death. Mice with prion disease show misfolded prion protein (PrP) accumulation and develop extensive neurodegeneration (with profound neurological deficits), in contrast to mouse models of AD or PD, in which neuronal loss is rare. Uniquely therefore, prion-infected mice allow access to mechanisms linking protein misfolding with neuronal death. Prion replication involves the conversion of cellular PrP, PrPC, to its misfolded, aggregating conformer, PrPSc, a process leading ultimately to neurodegeneration6. We have previously shown rescue of neuronal loss and reversal of early cognitive and morphological changes in prion-infected mice by depleting PrP in neurons, preventing prion replication and abrogating neurotoxicity7-9. However, the molecular mechanisms underlying both the progression of disease, and those underlying recovery in PrP-depleted animals, were unknown.
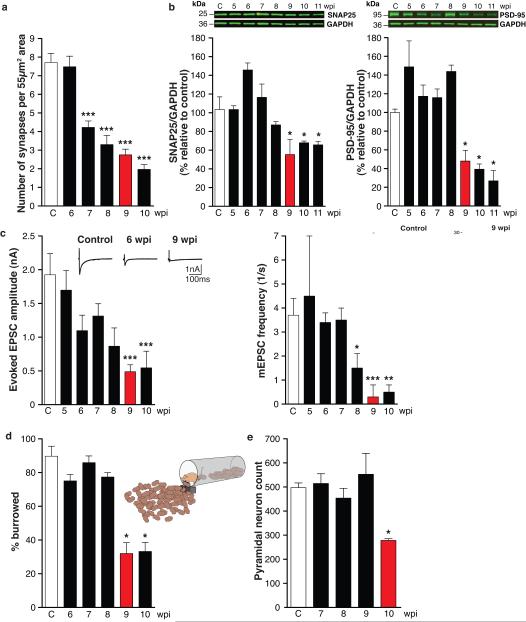
In order to understand these processes better, we now analysed the evolution of neurodegeneration in prion diseased mice. We examined hippocampi from prion-infected tg37 mice used in our previous experiments7-10, in which the time course of impairment and recovery are clearly defined. Hemizygous tg37 mice express mouse PrP at ~3x wild type levels and succumb to RML (Rocky Mountain Laboratory) prion infection within 12 weeks post infection (wpi)10. They first develop behavioural signs with decreased burrowing activity at ~9wpi, following reduction in hippocampal synaptic transmission and first neuropathological changes7,8. This is the window of reversibility when diseased neurons can still be rescued: PrP depletion up to 9wpi, but not later, rescues neurotoxicity, as by 10wpi neuronal loss is established7-9. We measured PrP levels, synapse number, levels of synaptic proteins and synaptic transmission in prion-infected mice weekly from 5wpi, and burrowing behaviour from 6wpi. We examined brains histologically and counted CA1 neurons. (Cohorts of at least 30 animals were used per group; biochemical and histological analyses were done on 3 mice per time point, burrowing behaviour on 12, n for other analyses is indicated in figure legends). We found an early decline in synapse number in asymptomatic animals at 7wpi to ~55% of control levels (Fig. 1a), despite unchanged levels of several pre- and post-synaptic marker proteins (Fig. 1b). Reduced synapse number with normal synaptic protein levels is likely to reflect impaired structural plasticity of synapses at this early stage of disease. At 9wpi, however, there was a sudden decline in synaptic protein levels to ~50% of control levels for several pre- (SNAP-25 and VAMP-2) and post-synaptic (PSD-95 and NMDAR1) proteins (Fig. 1b and Supplementary Fig. 1b). This was associated with further decline in synapse number, and the critical reduction in synaptic transmission, both in amplitude of evoked excitatory post-synaptic currents (EPSCs) and in the number of spontaneous EPSCs (mEPSCs) in CA1 neurons (Fig.1c and Supplementary Fig. 1e). This was coincident with behavioural change (Fig. 1d) and first spongiform pathology (Supplementary Fig. 1d), and was rapidly followed by the onset of neurodegeneration, resulting in 50% reduction in hippocampal pyramidal neurons at 10wpi (Fig. 1e). All animals developed overt motor signs and were terminally sick by 12wpi.
Fig. 1. Sudden decline of synaptic proteins is the key event leading to synaptic transmission failure and neuronal death in prion-diseased mice.
a, Synapse number in the stratum radiatum of the hippocampal CA1 region (imaged by electron microscopy; Supplementary Fig. 1a), declined from 7wpi (n=2 mice; 32 sections per timepoint; p < 0.0001***). b, Levels of pre-synaptic (SNAP-25) and post-synaptic (PSD-95) proteins measured relative to GAPDH declined abruptly at 9wpi. Representative western blots are shown. (n=3 mice per timepoint, p = 0.05*, Student’s t test; 2 tails). c, Whole-cell recordings from CA1 neurons showed significant reduction at 9wpi in amplitude of both evoked excitatory post synaptic currents (EPSC) and frequency of spontaneous miniature mEPSCs. Representative raw traces of evoked EPSCs are shown (left panel, inset). Right panel inset shows amplitude (top) and decay histograms (bottom) showing fewer events per recorded neuron from prion-infected mice, with unchanged mean amplitudes and decay kinetics. (n = 2 mice, 4-8 cells per timepoint. p <0.05* ; p <0.005**, p <0.0001***). d, Burrowing behaviour declines abruptly at 9wpi (n=12 mice; p < 0.001**, Student’s t test; 2 tails) and e, number of CA1 pyramidal neurons is reduced by ~50% at 10wpi (n = 3 mice, 3 sections per mouse; p = 0.04*). All data show mean ± s.e.m. One-way ANOVA with Tukey’s post-test was used unless otherwise stated. Control mice were injected with normal brain homogenate (NBH) and examined at each time point. Data from controls at all time points was averaged, due to lack variability over the time course, to simplify figures. For electrophysiological recordings and EM a single control time point at 10wpi was used.
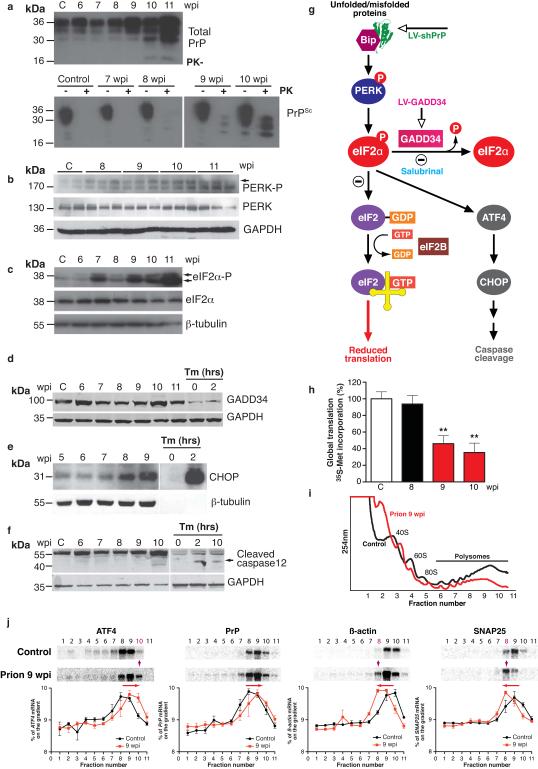
The abrupt loss of synaptic proteins at 9wpi appeared to be a critical factor in the evolution of disease, occurring when synapse number and transmission were already declining. This could result from increased degradation, or decreased synthesis. Prion infection in mice is known to impair the ubiquitin proteasome system, causing reduction - not increase - in protein degradation11. We therefore asked if protein synthesis was reduced through translational control mechanisms. Given that total PrP levels rise during disease (Fig. 2a), and that PrP is synthesized in the endoplasmic reticulum (ER), we examined the translational repression pathway of the UPR. Rising levels of unfolded proteins detected by BiP/Grp78 (BiP) in the ER membrane cause auto-phosphorylation of protein kinase-like ER kinase (PERK). PERK-P phosphorylates eIF2α, which blocks the initiation step of translation, reducing new protein synthesis12. eIF2α–P then induces ATF4 and CHOP expression, ultimately leading to caspase 12 cleavage, and expression of GADD34, the stress-induced eIF2α-P specific phosphatase and key effector of a negative feedback loop that terminates UPR signaling, allowing translational recovery (Fig. 2g).
Fig. 2. Prion replication induces the unfolded protein response (UPR) and results in eIF2α-P-mediated translational repression.
a, Total PrP and PrPSc levels (detected by addition of proteinase K (PK)) increase during prion infection; b, PERK-P and c, eIF2α-P levels rise during disease, d, GADD34 levels do not change but, e, CHOP levels increase throughout disease and f, caspase-12 is cleaved at 10wpi. g, Scheme depicting the translational repression pathway of the UPR showing points for intervention by LV-shPrP, LV-GADD34 and salubrinal. Activation of the pathway results in reduction of global translation at 9wpi, h, determined by 35S-methionine incorporation into hippocampal slices (n = 3 mice, 6 slices; p =0.003**). i, Polysomal profiles from hippocampi show a reduction in active polysomes in fractions 6-11 from prion-infected mice at 9wpi. j, Northern blots on polysomal fractions show increased translation of ATF4 with shift into fraction 10, and decreased translation of SNAP-25 and β-actin (shift into fraction 8) in prion-infected mice at 9wpi. PrP translation is essentially unchanged. Quantitative line plots of Northern blots show percentage of mRNA in each fraction found on the gradients, Tunicamycin (Tm) treated HeLa cells were analysed at 0, 2 and 10 hours as a control for UPR activation. All data show mean ± s.e.m. One-way ANOVA with Tukey’s post-test was used unless otherwise stated. Control mice at 11wpi are shown on western blots, and at 9wpi for Northern blots and 35S met labeling; n = 3 mice for all experiments. For quantification of western blots see Supplementary Fig. 2.
Upregulation of various steps in the pathway are seen in human prion cases13,14 and in prion-infected mice13; and increased phosphorylation of eIF2α occurs in AD and PD1-4. We characterized this pathway in prion diseased mice. We found that PERK-P and eIF2α-P increased throughout the course of disease (Fig. 2b,c and Supplementary Fig. 2b-d), in parallel with rising levels of total PrP, and the presence of detectable protease-resistant PrPSc (Fig. 2a). GADD34 levels did not change, despite rising eIF2α–P levels, suggesting insufficient GADD34 for dephosphorylation of increased eIF2α–P (Fig. 2d, Supplementary Fig. 2e). Caspase 12 cleavage occurred at 10wpi, following rising CHOP expression (Fig. 2e,f), coincident with onset of neuronal loss (Fig. 1e; see also Hetz et al13). However, the exact effector mechanism of neuronal death is unclear: we found neither apoptosis, nor autophagy, nor necrosis on examination of hippocampal slices (Supplementary Fig. 3); and neither Bax deletion, nor Bcl-2 overexpression15, nor caspase 12 deficiency16 are neuroprotective in prion disease.
We asked what effects the marked rise in eIF2α–P levels at 9wpi had on overall protein synthesis in hippocampi. We found abrupt, significant reduction in global translation rates, with a 50% decline in 35S-methionine incorporation in hippocampal slices from prion-infected mice at 9wpi compared to mice at 8wpi and uninfected controls (Fig. 2h), confirming sudden onset of reduced protein synthesis. We also looked at translation of specific mRNAs. We extracted polysomes from hippocampi of prion-infected mice. In successive polysomal fractions, mRNAs are associated with increased numbers of actively translating ribosomes. The change from a single fraction to the next reflects a large change in translation rate for any specific mRNA. Polysomal profiles at 9wpi showed a reduction in the overall number of actively translating ribosomes, represented by the smaller area under the curve between fractions 6-11 in prion-infected mice (Fig. 2i). Northern blots for specific mRNAs in individual polysomal fractions confirmed changes in actively translated messages consistent with eIF2α–P induction. Thus SNAP-25 and β-actin mRNAs showed a left shift to a lower polysomal fraction (Fig. 2j), representing reduced active translation (Fig. 1b). In contrast, ATF4 mRNA (which escapes eIF2α–P-mediated inhibition of translation, due to the presence of upstream open reading frames (uORFs) in its 5′ UTR17,18), showed increased active translation, represented by a right shift to a higher polysomal fraction (Fig. 2j, Supplementary Fig. 4 and 5). PrP mRNA did not show reduced translation, possibly due to the existence of similar translational control elements within the PrP gene. Indeed, human PrP mRNA has multiple upstream AUG uORFs in its 5′ UTR, which could allow it to escape eIF2α-P translational inhibition in the same way as ATF4 does17,18 (Supplementary Fig. 6).
Overall, these findings confirm that reduction in protein synthesis in prion disease is controlled at the translational, not the transcriptional, level, as it is rates of translation not levels of total mRNA that change (Supplementary Figs. 1c and 2g,h).
We propose that the key trigger to prion neurodegeneration is the continued unchecked activation of the UPR due to rising levels of PrP during disease, with fatal repression of translation rates. Importantly, prion neurotoxicity relates in a dose-dependent manner to PrP expression19-21. We therefore asked if levels of eIF2α-P and onset of neurodegeneration were related to levels of PrP in different strains of mice. We found that in homozygous tg37 mice, which over-express PrP ~6-fold, eIF2α-P was induced at 6wpi, and mice succumbed to prion infection at ~8wpi. In wild type C57/Bl6 mice, which express 1x levels of PrP, eIF2α-P was induced at 16wpi and animals succumbed at ~22wpi. Thus, as for hemizygous tg37 mice where PrP was expressed at 3x wild type levels and eIF2α-P was induced at 9wpi followed by death at 12wpi (Fig 2c), in each case there was a corresponding critical decline in synaptic proteins and synapse number after eIF2α-P induction (Supplementary Fig. 7; Fig 1a,b).
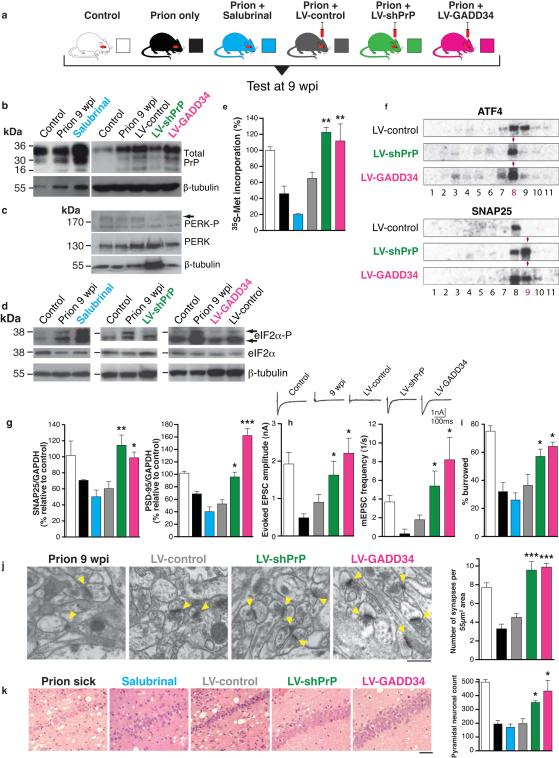
Transient eIF2α phosphorylation is beneficial to cells overloaded with misfolded proteins - reducing protein synthesis and increasing availability of chaperones, promoting refolding22,23. However, persistently high levels of eIF2α-P are detrimental in vitro24. To directly test the role of eIF2α-P in prion neurodegeneration in vivo, we first asked if reduction of eIF2α-P levels in prion disease would be neuroprotective. We used two approaches. We overexpressed GADD34, the eIF2α-P specific phosphatase to directly reduce eIF2α-P levels. In a separate experiment, we used targeted RNAi of PrP to abrogate UPR activation and prevent eIF2α-P formation. We then asked if increased levels of eIF2α-P exacerbate prion neurotoxicity by using salubrinal, a specific small molecule inhibitor of eIF2α-P dephosphorylation5, in infected mice. Salubrinal penetrates the blood brain barrier (Supplementary Fig. 8) and has been used for modulation of eIF2α-P dependent effects in ER stress-mediated processes in the central nervous system in vivo after peripheral administration25-27. Mice were inoculated with prions and received hippocampal injections of lentiviruses expressing GADD34 (LV-GADD34), anti-PrP shRNA (LV-shPrP), or YFP only (LV-control) at 5wpi, allowing 4 weeks for lentiviral expression to occur, before testing the effects of treatment on eIF2α-P levels and neurotoxicity at 9wpi. (All virally expressed constructs were driven by the CAMKII promoter for neuron-specific expression, Supplementary Fig. 9a-c). Another group of prion-infected mice received daily intra-peritoneal injections of salubrinal (1mg/kg), for 1 week, from 8wpi, with controls receiving vehicle alone. Two further control groups received normal brain homogenate, or RML prion inoculation alone (Fig. 3a).
Fig. 3. Preventing eIF2α-P formation or promoting its dephosphorylation in prion-diseased mice rescues synaptic failure and neuronal loss, while increased eIF2α-P levels exacerbate neurotoxicity.
a, Mice were infected with RML prions and treated with salubrinal (blue) or stereotaxically injected with lentiviruses expressing anti-PrP shRNA (LV-shPrP; green) or GADD34 (LV-GADD34; pink) or no insert (LV-control; grey) into both hippocampi. Control groups received no virus (prion only; black) or normal brain homogenate (NBH; control; white). Mice were tested at 9wpi. b, LV-shPrP reduced total PrP and prevented UPR induction, reducing levels of PERK-P c and eIF2α-P, d. LV-GADD34 reduced eIF2α-P despite PERK-P induction, and salubrinal increased eIF2α-P. e, Both LV-GADD34 and LV-shPrP prevented reduction in global translation at 9wpi, but salubrinal reduced translation rates even further. f, LV-GADD34 and LV-shPrP reversed prion-induced eIF2α-P-mediated translational changes of specific mRNAs in polysomal fractions shown on Northern blots. g, Synaptic protein levels; h, synaptic transmission, i, burrowing behaviour, j, and synapse number were protected by GADD34 treatment and PrP knockdown. Salubrinal exacerbated protein loss. Representative EM images (arrowheads denote individual synapses) and quantification are shown (n = 2 mice, 32 slices per mouse for each analysis). k, LV-GADD34 and LV-shPrP resulted in extensive neuroprotection of hippocampal CA1 pyramidal neurons and spongiosis (left hand panels, haematoxylin and eosin stained sections) and chart, right, when the animals were dying of scrapie at nearly 14wpi. Salubrinal accelerated neurodegeneration with extensive neuronal loss seen at 9wpi, earlier than in prion sick animals at 12wpi. Scale bar: 50μm and 2μm for EM images. All data in bar charts show mean ± s.e.m. One-way ANOVA with Tukey’s post test was used for multiple comparisons; p <0.05*; p <0.005**, p <0.0001***. For all experiments n=3 mice, unless otherwise stated. All controls are at 9wpi. For quantification of Northern and western blots see Supplementary Figs. 4 and 9.
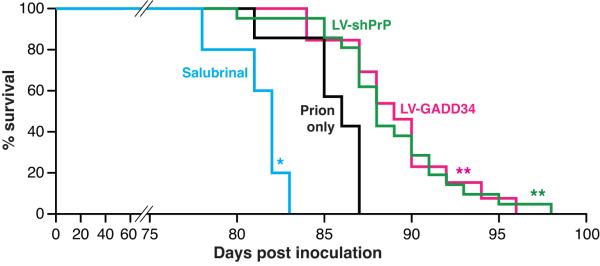
We examined mice from each group at 9wpi when eIF2α–P-mediated translational repression occurs (Fig. 2c,h). LV-GADD34 treatment did not reduce PrP levels (Fig. 3b) and PERK-P levels were equivalent to those in prion only or LV-control treated animals (Fig. 3c and Supplementary Fig. 9d), confirming UPR activation in these mice. Critically, however, eIF2α–P levels were reduced (Fig. 3d and Supplementary Fig. 9e), strongly supporting its dephosphorylation by lentivirally-mediated GADD34 expression. LV-shPrP treatment reduced PrP levels (Fig. 3b) and prevented the PrP-induced rise in PERK-P and eIF2α–P seen in untreated animals (Fig. 3c,d), confirming prevention of UPR activation. Both GADD34 over-expression and PrP knockdown prevented prion-induced eIF2α–P-mediated translational repression, with restoration of global rates at 9wpi (Fig. 3e) and prevention of eIF2α–P-induced changes in translation of specific mRNAs (Fig. 3f and Supplementary Fig. 4). As a result, synaptic protein levels, synaptic transmission and synapse number in prion-diseased mice treated with GADD34 or PrP knockdown were protected and equivalent to levels in uninfected control mice (Fig. 3g,h,j). Burrowing deficits were prevented (Fig. 3i) and there was extensive neuronal protection in the hippocampus, with no neuronal loss and markedly reduced spongiform change (Fig. 3k). Further, targeted expression of LV-GADD34 and focal PrP knockdown had a modest, but highly significant, effect on survival, increasing this to 90±3 days and 92±5 days respectively, compared to 83±2 days for prion only mice (Fig. 4), and to 82±2 days for LV-control injected mice (Supplementary Fig. 10). For both LV-GADD34 and LV-shPrP, treatment was localised to the dorsal hippocampus, a very small area of the brain, so prion infection in the rest of the brain was fatal, but neuroprotection in GADD34 treated animals was seen even when the animals were terminally sick (Fig. 3k). More extensive brain-wide delivery of GADD34, or targeting of this pathway, would be predicted to further increase survival and give more widespread neuroprotection. Critically, treatment with salubrinal had the opposite effect, by preventing dephosphorylation of eIF2α-P. Thus, eIF2α-P levels were markedly higher at 9wpi than in prion-only controls (Fig. 3d and Supplementary Fig. 9e), causing further repression of global translation (Fig. 3e) and reduction of synaptic proteins (Fig. 3g). Salubrinal treatment resulted in earlier severe neuronal loss (Fig. 3k), and significantly accelerated disease, compared to untreated prion-infected mice (Fig. 4).
Fig. 4. Reducing eIF2α-P levels in prion-diseased mice significantly increases survival.
Kaplan-Meier survival plots for prion-infected mice treated with salubrinal (n=10), prions alone (n=7), hippocampal injections of LV-GADD34 (n=13) or LV-shPrP (n=14). Focal treatment with LV-GADD34 and LV-shPrP resulted in significantly increased survival compared to prion-infected mice with no treatment (p = 0.007**; Student’s t test); salubrinal treatment reduced survival (p =0.03*). For plots for LV-control and DMSO controls see Supplementary Fig. 10.
In conclusion, we have shown that PrP replication causes sustained UPR induction with persistent, deleterious expression of eIF2α-P in prion disease. The resulting chronic blockade of protein synthesis leads to synaptic failure, spongiosis and neuronal loss. Promoting eIF2α-P dephosphorylation rescues vital translation rates and is thereby neuroprotective, while preventing this further reduces translation and enhances neurotoxicity. The data support the development of generic proteostatic approaches22,28 to therapy - fine-tuning protein synthesis - in prion, and perhaps other neurodegenerative disorders involving protein misfolding.
Methods summary
Prion infection of mice
All animal work conformed to UK regulations and institutional guidelines, performed under Home Office guidelines. tg3710 and C57/Bl6N mice (Harlan) were inoculated with 1% brain homogenate of Chandler/RML (Rocky Mountain Laboratories) prions aged 3-4 weeks, as described7. Animals were culled when they developed clinical signs of scrapie. Control mice received 1% normal brain homogenate. Hippocampi were processed for protein, RNA or histological analysis7-10. For all analyses n=3 mice unless otherwise stated.
Lentiviruses and stereotaxic surgery
Lentiviral plasmids were generated using the Invitrogen Gateway cloning system29. The neuron-specific promoter CAMKII was used to drive shPrP expression, C-terminal GADD34 expression, or YFP alone (control virus). Viruses were injected stereotaxically into the CA1 region of the hippocampus as described9.
Salubrinal treatment
Mice received daily intraperitoneal injections of 1mg/kg of salubrinal (Calbiochem), or vehicle (diluted DMSO in saline (Sigma))25, for 7 days from 8wpi.
Experimental analyses
Synapse numbers were counted in EM images of the stratum radiatum of the hippocampal CA1 region30. Synaptic marker proteins, UPR pathway proteins and PrP were analysed by immunoblotting of brain homogenates. PrPSc was detected after PK digestion7. Whole-cell recordings were done in acute hippocampal slices to measure synaptic transmission31. Global translation levels were detected using 35S−Methionine incorporation in acute hippocampal slices and translation of specific transcripts in polysomal fractions from hippocampi were analysed by Northern blotting32. Neuronal counts were determined by quantifying NeuN positive pyramidal CA1 neurons9. All analyses were performed using hippocampi from 3 mice in triplicate unless otherwise stated. Burrowing behaviour was performed as described on groups of 10 or more mice8. Statistical analyses were performed using using Prism v5 software, using Student’s t test for data sets with normal distribution and a single intervention. ANOVA testing was performed using one-way analysis with Tukey’s post-hoc test for multiple comparisons.
Extended methods
Generation of lentiviral plasmids
Lentiviral plasmids were generated by using the Invitrogen Gateway cloning system as described29. att-flanked cassettes were constructed by PCR amplification using att recombination site sequences and 20-25 template specific sequences. The C-terminus GADD34 cassette was amplified from FLAG-tagged C-term GADD34 using attB5 5′-ggggacaactttgtatacaaaagttggcaccatgcgttcaggagaggcgtccga-3′; and attB4 5′-ggggacaactttgtatagaaaagttgggtgttggtctcagccacgcctcccac3′ primers; WPRE and YFP cassettes were amplified from pLL3.7-shPrP plasmid using attB3 5′- ggggacaactttgtataataaagttgtcaacctctggattacaaaatttgt-3′ and attB2 5′- ggggaccactttgtacaagaaagctgggtatgcggggaggcggcccaaagggaga-3′ primers for WPRE and attB4r 5′ accatggtgagcaagggcga-3′ and attB3r 5′ ttacttgtacagctcgtccatgccg-3′ primers for YFP . The CAMKII promoter was amplified from using attB1 5′- ggggacaagtttgtacaaaaaagcaggctacttgtggactaagtttgttcgc-3′ and attB5r primer 5′- ggggacaacttttgtatacaaagttgtctgcccccagaactaggg-3′primers. Cassettes were amplified and recombined into appropriate pDONR entry vectors. Once the entry plasmids were generated they were recombined with the pLenti6/BLOCK-iT/DEST vector (Invitrogen) to construct lentiviral plasmids. We used lentiviral plasmids containing shPrP and empty vector constructs as described9. Lentiviruses were generated using DNA/Ca2+ phosphate transfection of HEK293 cells33. Additional stocks of virus were generated by GenTarget, Inc. (San Diego, CA) and titre determined using FACS (BD FACS Calibur). Viruses were used with a final titre of 0.6-1.5 × 108 TU.
Stereotaxic injection
Mice were anesthetized using isofluorane and injected into CA1 region of the hippocampus as described9.
Electron microscopy
Semi-thin sections for electron microscopy were obtained by terminal perfusion of mice as described30. Ultrathin sections (70 nm) were examined in a JEOL 100-CXII electron microscope (JEOL (UK) Ltd) equipped with a ‘Megaview III’ digital camera (Olympus Soft Imaging Solutions GmbH, Munster, Germany). Synapses were scored using criteria of structures showing a postsynaptic density, containing synaptic vesicles and a synaptic junction. 32 images from two mice were used for scoring.
Electrophysiology
Whole-cell recordings were made from identified CA1 neurons and recording performed as described30. In brief, neurons were voltage-clamped using a Multiclamp 700B amplifier and pClamp 10.3 software (Molecular Devices) and EPSCs were evoked by stimulation with bipolar platinum electrode at 37°C. Pipettes (2.5-3.5MΩ) were filled with a solution containing (mM): KCl 110, HEPES 40, EGTA 0.2, MgCl2 1, CaCl2 0.1, pH was adjusted to 7.2 with KOH. Neurons were visualized with 60x objectives on a Nikon FS600 microscope fitted with differential interference contrast (DIC) optics. 4-8 cells were measured per mouse in at least 2 animals per experiment.
Immunoblotting
Protein samples were isolated from hippocampi using protein lysis buffer (50mM Tris, 150mM NaCl, 2mM EDTA, 1mM MgCl2, 100mM NaF, 10% glycerol, 1% Triton X-100, 1% Na deoxycholate, 0.1% SDS and 125mM sucrose) supplemented with Phos-STOP and protease inhibitors (Roche). Synaptic protein levels were determined by resolving 20mg of protein on SDS-PAGE gels, transferred onto nitrocellulose membrane and incubated with primary antibodies, SNAP25, (1:10000; Abcam), VAMP2 (1:5000; Synaptic Systems), NMDA-R1 (1:1000; Sigma) and PSD95 (1:1000; Millipore). Odyssey IRDye800 seconday antibodies (1:5000; LI-COR) were applied, visualized and quantitated using Odyssey infrared imager (LI-COR; software v3.0). Protein for PrP levels and UPR pathway activation were determined using the primary antibodies, 8H4 for total PrP (1:1000; Abcam), ICSM35 for PrPSc (1:10,000; D-GEN), PERK-P, total PERK, P-eIF2a, eIF2a (1:1000; Cell Signaling), CHOP (1:1000; ThermoScientific), Caspase12 (1:1000; Exalpha), BiP/Grp78 (1:1000; Stressgen) and GADD34 (1:1000; ProteinTech). Horseradish peroxidase (HRP) conjugated secondary antibodies (1:5000; DAKO) were applied and protein visualized using enhanced chemiluminescence (GE Healthcare) and quantitated using ImageJ. Antibodies against GAPDH (1:5000; Santa Cruz) or β-tubulin (1:5000; Millipore) were used to determine loading.
Hippocampal slice preparation and 35S-methionine labeling
Slices were dissected in an oxygenated cold (2–5°C) sucrose artificial cerebrospinal fluid (ACSF) containing (mm): 26mM NaHCO3, 2.5mM KCl, 4 mM MgCl2, 0.1 mM CaCl2, and 250mM sucrose. Hippocampal slices were prepared using a tissue chopper (McIlwain). Slices were allowed to recover in normal ACSF buffer while being oxygenated at 37°C for 1hr, then incubated with [35S]-Methionine label for one hour, then homogenized. Proteins were TCA precipitated and incorporation of radiolabel was measured by scintillation counting (Winspectal, Wallac Inc.).
Polysomal fraction preparation and Northern blots
Sucrose density gradient centrifugation was used to separate hippocampal homogenates into polysomal and subpolysomal fractions. Polysomal fractions were isolated as described32. Briefly, hippocampi were dissected in ice-cold gradient buffer (0.3 M NaCl, 15 mM MgCl2, 15 mM Tris-HCL (pH7.4), 0.1 mg/ml cyclohexamide, and 1 mg/ml heparin). The hippocampal tissue was homogenised in gradient buffer containing RNase inhibitors and 1.2% TritonX-100 added. Samples were centrifuged and the supernatants layered onto 10-60 % sucrose gradients. The gradients were sedimented at 38,000 rpm for 120 minutes at 4°C. 1 ml fractions were collected from the gradients into 3 ml of 7.7 M guanidine-HCL using a Foxy R1 gradient fractionator (Teledyne ISCO; ISCO peak Trak v1.10 software) with continuous measurement of the absorbance at 254 nm. RNA was then isolated and equal volumes of each fraction were analysed by Northern blot analysis32.
Neuropathological examination of brain sections
Paraffin embedded brains were either stained with NeuN antibody (1:200; Millipore) for neuronal counts. CA1 pyramidal neuron counts were determined using three serial sections from three separate miceAll images were taken on using Axiovision 4.8 software (Zeiss) and counted using volocity imaging system.
Burrowing
Burrowing was performed as described, at least 10 mice were burrowed for each individual experimental group10,34.
Statistical analysis
Student’s t tests were applied to all data sets with two tails (two samples; unequal variance). ANOVA testing was performed using one-way analysis with Tukey’s post-hoc test for group effects. Statistical tests were performed using Prism v5. All data in bar charts show mean ± s.e.m.
Supplementary Material
Supplementary information: Supplementary figures 1-10 and legends, Supplementary methods and references.
Acknowledgements
We thank David Read for imaging analysis, and Jenny Edwards, Tim Smith, Judy McWilliam, Paul Glynn and Colin Molloy for technical assistance; Professor John Collinge (MRC Prion Unit) for the original RML prion inoculum and Professor Ken Liddle for critical reading of the manuscript. This work was funded by the Medical Research Council, UK.
Footnotes
Author contributions: JAM did most of the experimental work and analysis. NV and MGM performed stereotaxic surgery and prion inoculations. HR, DP, MH and JM performed various experiments, DD performed EM analyses, JRS performed electrophysiological analysis, CAO and DAB performed mass spectrometry analysis, PT and AB worked with JAM in Cambridge, AEW and MB contributed expertise and direction on translational control mechanisms, GRM directed and supervised the project. JAM and GRM wrote the paper. All authors contributed to discussion and analysis of data and final draft of paper.
The authors declare no conflict of interest.
References
- 1.Hoozemans JJ, et al. Activation of the unfolded protein response in Parkinson’s disease. Biochem Biophys Res Commun. 2007;354:707–711. doi: 10.1016/j.bbrc.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 2.Hoozemans JJ, et al. The unfolded protein response is activated in pretangle neurons in Alzheimer′s disease hippocampus. 2009. [DOI] [PMC free article] [PubMed]
- 3.Hoozemans JJ, et al. The unfolded protein response is activated in Alzheimer′s disease. Acta Neuropathol. 2005;110:165–172. doi: 10.1007/s00401-005-1038-0. [DOI] [PubMed] [Google Scholar]
- 4.Unterberger U, et al. Endoplasmic reticulum stress features are prominent in Alzheimer disease but not in prion diseases in vivo. J Neuropathol Exp Neurol. 2006;65:348–357. doi: 10.1097/01.jnen.0000218445.30535.6f. [DOI] [PubMed] [Google Scholar]
- 5.Boyce M, et al. A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science. 2005;307:935–939. doi: 10.1126/science.1101902. [DOI] [PubMed] [Google Scholar]
- 6.Prusiner SB. Molecular biology of prion diseases. Science. 1991;252:1515–1522. doi: 10.1126/science.1675487. [DOI] [PubMed] [Google Scholar]
- 7.Mallucci G, et al. Depleting neuronal PrP in prion infection prevents disease and reverses spongiosis. Science. 2003;302:871–874. doi: 10.1126/science.1090187. [DOI] [PubMed] [Google Scholar]
- 8.Mallucci GR, et al. Targeting cellular prion protein reverses early cognitive deficits and neurophysiological dysfunction in prion-infected mice. Neuron. 2007;53:325–335. doi: 10.1016/j.neuron.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 9.White MD, et al. Single treatment with RNAi against prion protein rescues early neuronal dysfunction and prolongs survival in mice with prion disease. Proc Natl Acad Sci U S A. 2008;105 doi: 10.1073/pnas.0802759105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mallucci GR, et al. Post-natal knockout of prion protein alters hippocampal CA1 properties, but does not result in neurodegeneration. EMBO J. 2002;21:202–210. doi: 10.1093/emboj/21.3.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kristiansen M, et al. Disease-associated prion protein oligomers inhibit the 26S proteasome. Mol Cell. 2007;26:175–188. doi: 10.1016/j.molcel.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 12.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 13.Hetz C, Russelakis-Carneiro M, Maundrell K, Castilla J, Soto C. Caspase-12 and endoplasmic reticulum stress mediate neurotoxicity of pathological prion protein. EMBO J. 2003;22:5435–5445. doi: 10.1093/emboj/cdg537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoo BC, et al. Overexpressed protein disulfide isomerase in brains of patients with sporadic Creutzfeldt-Jakob disease. Neurosci Lett. 2002;334:196–200. doi: 10.1016/s0304-3940(02)01071-6. [DOI] [PubMed] [Google Scholar]
- 15.Steele AD, et al. Diminishing apoptosis by deletion of Bax or overexpression of Bcl-2 does not protect against infectious prion toxicity in vivo. J Neurosci. 2007;27:13022–13027. doi: 10.1523/JNEUROSCI.3290-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steele AD, et al. Prion pathogenesis is independent of caspase-12. Prion. 2007;1:243–247. doi: 10.4161/pri.1.4.5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harding HP, et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 18.Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Mol Cell. 2010;40:228–237. doi: 10.1016/j.molcel.2010.09.028. [DOI] [PubMed] [Google Scholar]
- 19.Bueler H, et al. Mice devoid of PrP are resistant to scrapie. Cell. 1993;73:1339–1347. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- 20.Manson JC, Clarke AR, McBride PA, McConnell I, Hope J. PrP gene dosage determines the timing but not the final intensity or distribution of lesions in scrapie pathology. Neurodegeneration. 1994;3:331–340. [PubMed] [Google Scholar]
- 21.Sandberg MK, Al-Doujaily H, Sharps B, Clarke AR, Collinge J. Prion propagation and toxicity in vivo occur in two distinct mechanistic phases. Nature. 2011;470:540–542. doi: 10.1038/nature09768. [DOI] [PubMed] [Google Scholar]
- 22.Tsaytler P, Harding HP, Ron D, Bertolotti A. Selective inhibition of a regulatory subunit of protein phosphatase 1 restores proteostasis. Science. 2011;332:91–94. doi: 10.1126/science.1201396. [DOI] [PubMed] [Google Scholar]
- 23.Chafekar SM, Hoozemans JJ, Zwart R, Baas F, Scheper W. Abeta 1-42 induces mild endoplasmic reticulum stress in an aggregation state-dependent manner. Antioxid Redox Signal. 2007;9:2245–2254. doi: 10.1089/ars.2007.1797. [DOI] [PubMed] [Google Scholar]
- 24.Wiseman RL, et al. Flavonol activation defines an unanticipated ligand-binding site in the kinase-RNase domain of IRE1. Mol Cell. 2010;38:291–304. doi: 10.1016/j.molcel.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sokka AL, et al. Endoplasmic reticulum stress inhibition protects against excitotoxic neuronal injury in the rat brain. J Neurosci. 2007;27:901–908. doi: 10.1523/JNEUROSCI.4289-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saxena S, Cabuy E, Caroni P. A role for motoneuron subtype-selective ER stress in disease manifestations of FALS mice. Nat Neurosci. 2009;12:627–636. doi: 10.1038/nn.2297. [DOI] [PubMed] [Google Scholar]
- 27.Zhu Y, et al. Eif-2a protects brainstem motoneurons in a murine model of sleep apnea. J Neurosci. 2008;28:2168–2178. doi: 10.1523/JNEUROSCI.5232-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 29.White MD, Milne RV, Nolan MF. A Molecular Toolbox for Rapid Generation of Viral Vectors to Up- or Down-Regulate Neuronal Gene Expression in vivo. Front Mol Neurosci. 2011;4:8. doi: 10.3389/fnmol.2011.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haustein MD, et al. Acute hyperbilirubinaemia induces presynaptic neurodegeneration at a central glutamatergic synapse. J Physiol. 2011;588:4683–4693. doi: 10.1113/jphysiol.2010.199778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steinert JR, et al. Nitric oxide is an activity-dependent regulator of target neuron intrinsic excitability. Neuron. 2011;71:291–305. doi: 10.1016/j.neuron.2011.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johannes G, Sarnow P. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA. 1998;4:1500–1513. doi: 10.1017/s1355838298981080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang M, Deng L, Chen G. High Ca(2+)-phosphate transfection efficiency enables single neuron gene analysis. Gene Ther. 2004;11:1303–1311. doi: 10.1038/sj.gt.3302305. [DOI] [PubMed] [Google Scholar]
- 34.Cunningham C, et al. Synaptic changes characterize early behavioural signs in the ME7 model of murine prion disease. Eur J Neurosci. 2003;17:2147–2155. doi: 10.1046/j.1460-9568.2003.02662.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.