Abstract
A common variant on chromosomal region 15q24–25.1, marked by rs1051730, was found to be associated with lung cancer risk. Here, we attempted to confirm the second variant on 15q24–25.1 in several large sporadic lung cancer populations and determined what percentage of additional risk for lung cancer is due to the genetic effect of the second variant. SNPs rs1051730 and rs481134 were genotyped in 2,818 lung cancer cases and 2,766 controls from four populations. Joint analysis of these two variants (rs1051730 and rs481134) on 15q24–25.1 identified three major haplotypes (G_T, A_C, and G_C) and provided stronger evidence for association of 15q24–25.1 with lung cancer (P = 9.72 × 10−9). These two variants represent three levels of risk associated with lung cancer. The most common haplotype G_T is neutral; the haplotype A_C is associated with increased risk for lung cancer with 5.0% higher frequency in cases than in controls [P = 1.68 × 10−7; odds ratio (OR), 1.24; 95% confidence interval (95% CI), 1.14–1.35]; whereas the haplotype G_C is associated with reduced risk for lung cancer with 4.4% lower frequency in cases than in controls (P = 7.39 × 10−7; OR, 0.80; 95% CI, 0.73–0.87). We further showed that these two genetic variants on 15q24–25.1 independently influence lung cancer risk (rs1051730: P = 4.42 × 10−11; OR, 1.60; 95% CI, 1.46–1.74; rs481134: P = 7.01 × 10−4; OR, 0.81; 95% CI, 0.72–0.92). The second variant on 15q24–25.1, marked by rs481134, explains an additional 13.2% of population attributable risk for lung cancer.
Introduction
Lung cancer is the leading cause of cancer-related death in the United States. In 2009, there will be an estimated 219,000 cases of lung cancer diagnosed, and only 15% of those patients are expected to survive for more than 5 years (1). Genetic factors play important roles in lung cancer susceptibility (2). Recent genome-wide association studies have identified an association of a common variant on chromosomal region 15q24–25.1 with lung cancer susceptibility (3–7). This locus spans ~200 kb with high linkage disequilibrium (LD) and contains IREB2, LOC123688, PSMA4, and the neuronal nicotinic acetylcholine receptor (nAChR) genes CHRNA5, CHRNA3, and CHRNB4. Two SNPs, rs1051730 and rs8034191, on the 15q24–25.1 locus were reported to be associated with both familial and sporadic lung cancer (5). These two SNPs are in high LD (r2 = 0.89), thereby representing the same association signal on the 15q24–25.1 locus. In addition to this association signal, a second variant (rs588765) on the 15q24–25.1 locus was recently implicated in familial lung cancer (8). A haplotype that contains the major allele of rs588765 was identified to be associated with reduced risk of lung cancer in the analysis of 194 familial lung cancer patients and 217 disease-free controls collected by the Genetic Epidemiology of Lung Cancer Consortium (GELCC; ref. 8).
Here, we validated the second variant in 2,818 lung cancer cases and 2,766 controls. These cases were composed of three groups: cases with family history of lung cancer in Caucasians and sporadic lung cancer in Caucasians and African Americans. These two genetic variants on the 15q24–25.1 locus have independent genetic effects on lung cancer risk.
Materials and Methods
Study samples
Four study populations were used in this study. Detailed information on these study subjects is presented in Supplementary Table S1. All the case patients in this study are histologically confirmed non–small cell lung cancer. Briefly, 194 familial lung cancer cases and 217 disease-free controls were recruited from the GELCC. Each case patient with familial lung cancer was chosen from one high-risk lung cancer family with three or more members with lung cancer (5). All the subjects from the GELCC are Caucasians. Eight hundred ninety sporadic lung cancer cases and 865 controls were from the Mayo Clinic (Rochester, MN). These samples are part of the Mayo Clinic Lung Cancer Cohort collected from an ongoing case-control study (9). All the samples from the Mayo Clinic are Caucasians (MCC). The Texas cohorts include 1,466 sporadic cases and 1,389 controls in Caucasians (TXC) and 286 sporadic cases and 293 controls in African Americans (TXA). These samples are part of an ongoing case-control study at the University of Texas M.D. Anderson Cancer Center since 1991 (Houston, TX; ref. 3). Informed consent was obtained from all participants in the studies. SNPs rs481134 and rs588765 are in high LD (r2 = 0.98), and thus we chose rs481134 representing the second variants in our study. SNPs rs1051730 and rs481134 were genotyped in the four populations. Detailed methods for SNP genotyping were described previously (3, 8).
Statistical analyses
To rule out confounding effects on lung cancer risk, the association analysis was adjusted by sex, age, and smoking status with the use of logistic regression analysis. If P is the probability of being a case, then our linear logistic model has the following form:
where α is the intercept, g represents SNP genotype and is coded as the number of copies of the disease allele, and β0 is additive genetic effects at that SNP, xi is covariate (such as sex, age, and smoking), and βi is the coefficient associated with xi. Allelic odds ratio (OR) associated with each SNP and 95% confidence intervals (95% CI) were estimated. In the combined association analysis, the association was further adjusted for race and study sites. Population stratification was not detected in our previous studies (3, 5). To confirm the results from our logistic models, multiple case-control groups were combined using a Mantel-Hazenszel model in which the groups were allowed to have different population frequencies for alleles and genotypes but were assumed to have common relative risks (10). Because SNPs rs1051730 and rs481134 are in moderate LD, we also performed subgroup association analysis to eliminate confounding effects due to their LD. In the subgroup analysis, marginal ORs and their 95% CIs were estimated with adjustment for the same covariates.
Both omnibus/global association statistic and haplotype-specific statistic were computed. The haplotype-specific analysis tests each haplotype at one time (each versus all others) using χ2 tests with 1 degree of freedom (df). Totally, H-1 haplotype-specific tests were done at each location where H is the number of haplotypes at that location. The omnibus association analysis jointly estimates all haplotype effects at a location with a single χ2 with H-1 df. Similarly, the association was adjusted by the same covariates as the single-marker analysis in the logistic regression model. Haplotype-specific ORs and their 95% CIs were estimated. In addition to the haplotype analysis, we performed a logistic regression analysis modeling both main effects and their interaction of the two SNPs, rs1051730 and rs481134. The goodness-of-fit of logistic model was evaluated based on Akaike information criterion and χ2 test.
The population genetic attributable risk percent (PAR) was estimated for each variant, which defines what percentage of the total risk for lung cancer is due to genetic effect of that variant:
where pi is the prevalence of that i-th genotype associated with lung cancer among control subjects and ORi is OR associated with that genotype (11, 12). We used the lowest-risk genotype as the reference to estimate ORs in the above logistic regression model with adjustment of covariates. Similarly, we also jointly estimated PAR for the two loci (rs1051730 and rs481134) using haplotype-specific ORs.
Possible haplotype phases for a given genotype were inferred using the Expectation-Maximization algorithm (13). The LD statistic (r2) was calculated based on haplotype frequencies estimated via the Expectation-Maximization algorithm. Hardy-Weinberg equilibrium for each SNP was examined among control subjects with the use of Fisher exact test. SNPs rs1051730 and rs481134 genotyped in the four populations were in Hardy-Weinberg equilibrium. All statistical analyses were implemented in PLINK software (http://pngu.mgh.harvard.edu/~purcell/plink/; ref. 13).
Results
Associations between rs481134 and rs1051730 and lung cancer
The SNP rs481134 is in high LD with rs588765 (r2 = 0.98). The SNPs rs1051730 and rs481134 were genotyped in four populations: GELCC, MCC, TXC, and TXA (Supplementary Table S1). In single-marker association analysis, rs1051730 is consistently and strongly associated with lung cancer among the four study populations (P = 1.56 × 10−7;OR, 1.24; 95% CI, 1.15–1.35, from the combined analysis), whereas rs481134 shows no association (P = 0.514). The minor allele of rs105730 A is associated with increased risk of lung cancer and is, on average, 5% more frequent in cases than in controls (Table 1). To confirm the results from our logistic regression model where study site was incorporated as a covariate, the combined association was further examined by Mantel-Hazenszel tests (ref. 10; Supplementary Table S2). Mantel-Hazenszel tests yielded consistent results with those from the logistic model.
Table 1.
Single-marker association analysis of SNPs rs1051730 and rs481134 with lung cancer
| Samples/SNPs | Alleles* | Allele frequency |
P† | OR (95% CI) | |
|---|---|---|---|---|---|
| Cases | Controls | ||||
| GELCC (194 familial cases and 217 controls, Caucasians) | |||||
| rs1051730 | G/A | 0.4094 | 0.2938 | 2.16 × 10−3 | 1.88 (1.26–2.82) |
| rs481134 | T/C | 0.5632 | 0.5714 | 0.750 | 0.94 (0.59–1.29) |
| MCC (890 sporadic cases and 865 controls, Caucasians) | |||||
| rs1051730 | G/A | 0.3975 | 0.3651 | 0.040 | 1.16 (1.01–1.33) |
| rs481134 | T/C | 0.5863 | 0.5891 | 0.897 | 0.99 (0.85–1.23) |
| TXC (1,466 sporadic cases and 1,389 controls, Caucasians) | |||||
| rs1051730 | G/A | 0.3767 | 0.3315 | 2.79 × 10−4 | 1.23 (1.10–1.37) |
| rs481134 | T/C | 0.5806 | 0.5621 | 0.141 | 1.08 (0.98–1.19) |
| TXA (268 sporadic cases and 293 controls, African Americans) | |||||
| rs1051730 | G/A | 0.1456 | 0.0911 | 1.83 × 10−3 | 1.91 (1.27–2.86) |
| rs481134 | T/C | 0.6818 | 0.7000 | 0.466 | 0.90 (0.62–1.18) |
| Combined (2,818 cases and 2,794 controls)‡ | |||||
| rs1051730 | G/A | 0.3635 | 0.3136 | 1.56 × 10−7 | 1.24 (1.15–1.35) |
| rs481134 | T/C | 0.5909 | 0.5859 | 0.514 | 1.03 (0.95–1.10) |
Bold letter is the reference allele used for estimating OR.
Association was adjusted by sex, age, and smoke using logistic regression model.
Association was adjusted by sex, age, smoke, race, and study sites using logistic regression model.
Interestingly, haplotype analysis of SNPs rs1051730 and rs481134 identified three major haplotypes (G_T, A_C, and G_C) and provided stronger evidence for association with lung cancer by a global P value of 9.72 × 10−9. Individual haplotype tests by examining the distribution differences for each haplotype showed that distinct haplotypes differ significantly between lung cancer cases and controls. These observations result from different distributions of the haplotypes A_C and G_C. The haplotype A_C is associated with increased risk for lung cancer with 5.0% higher frequency in cases than in disease-free controls (P = 1.68 × 10−7; haplo-type-specific OR, 1.24; 95% CI, 1.14–1.35). This haplotype contains the risk allele of rs1051730 A (Tables 1 and 2). The haplotype G_C is associated with reduced risk for lung cancer with 4.4% lower frequency in cases than in controls (P = 7.39 × 10−7; OR, 0.80; 95% CI, 0.73–0.87; Table 2). As compared with the most common haplotype G_T, the protective haplotype G_C contains the allele of rs481134 C that associated with reduced risk of lung cancer. As a whole, these haplotypes containing SNPs rs481134 and rs1051730 account for a total of 31.6% of population attributable risk for lung cancer (Supplementary Table S3).
Table 2.
Haplotype-based association analysis of SNPs rs1051730 and rs481134 with lung cancer
| Samples | Haplotypes* | Frequency |
P† | OR (95% CI) | |
|---|---|---|---|---|---|
| Cases | Controls | ||||
| GELCC | G_T | 0.4303 | 0.4272 | 0.826 | 0.96 (0.66–1.40) |
| A_C | 0.4095 | 0.2936 | 2.87 × 10−3 | 1.85 (1.23–2.77) | |
| G_C | 0.1602 | 0.2792 | 1.59 × 10−3 | 0.47 (0.29–0.75) | |
| Omnibus‡ | — | — | 1.21 × 10−3 | — | |
| MCC | G_T | 0.4128 | 0.4101 | 0.894 | 1.01 (0.88–1.16) |
| A_C | 0.3975 | 0.3644 | 0.037 | 1.16 (1.01–1.33) | |
| G_C | 0.1897 | 0.2255 | 8.83 × 10−3 | 0.80 (0.68–0.95) | |
| Omnibus | — | — | 0.018 | — | |
| TXC | G_T | 0.4192 | 0.4384 | 0.131 | 0.92 (0.83–1.02) |
| A_C | 0.3752 | 0.3305 | 3.55 × 10−4 | 1.22 (1.10–1.37) | |
| G_C | 0.2055 | 0.2311 | 0.018 | 0.86 (0.75–0.93) | |
| Omnibus | — | — | 1.04 × 10−3 | — | |
| TXA | G_T | 0.3159 | 0.3010 | 0.534 | 1.09 (0.83–1.44) |
| A_C | 0.1453 | 0.0900 | 1.58 × 10−3 | 1.93 (1.28–2.91) | |
| G_C | 0.5388 | 0.6090 | 7.94 × 10−3 | 0.70 (0.54–0.91) | |
| Omnibus | — | — | 2.53 × 10−3 | — | |
| Combined | G_T | 0.4080 | 0.4139 | 0.446 | 0.97 (0.90–1.05) |
| A_C | 0.3627 | 0.3127 | 1.68 × 10−7 | 1.24 (1.14–1.35) | |
| G_C | 0.2292 | 0.2734 | 7.39 × 10−7 | 0.80 (0.73–0.87) | |
| Omnibus | — | — | 9.72 × 10−9 | — | |
Two-SNP (i.e., rs1051730-rs481134) haplotypes were analyzed.
Association was adjusted by the same covariates as the single-locus analysis.
Omnibus test (i.e., global test) jointly examines all haplotype effects at that locus.
Both genetic variants have independent effects on lung cancer risk
SNP rs1051730 shows moderate LD with rs481134 (r2 = 0.30), which may mask or change genetic effects on those loci in the association analysis. To rule out the confounding effect of rs1051730 on the association of rs481134, we performed the association analysis of rs481134 with lung cancer in subjects with genotype GG at rs1051730 (Table 3). This subgroup analysis eliminated the effect masked by rs1051730 due to its LD with rs481134 because all the subjects in the analysis carry the wild-type G alleles at rs1051730. This subgroup analysis essentially analyzed subjects with haplotypes G_T and/or G_C and allowed direct test on the allelic effects at rs481134. As a result, the analysis gave a significant association of rs481134 with lung cancer (P = 7.01 × 10−4 from the combined analysis). As expected in the haplotype analysis, the allele C of rs481134 is associated with reduced risk of lung cancer (marginal OR, 0.81; 95% CI, 0.72–0.92). In the TXC population, the second variant on 15q24–25.1, marked by rs481134, accounts for 13.2% of population attributable risk for lung cancer (Supplementary Table S3).
Table 3.
Subgroup association analysis of SNPs rs481134 and rs1051730 with lung cancer
| Samples/SNP | Alleles* | Frequency |
P† | Marginal OR (95% CI) | |
|---|---|---|---|---|---|
| Cases | Controls | ||||
| Subjects with genotypes GG at SNP rs1051730 | |||||
| GELCC (64 familial cases and 102 controls) | |||||
| rs481134 | T/C | 0.2937 | 0.4059 | 0.196 | 0.68 (0.38–1.22) |
| MCC (322 sporadic cases and 342 controls) | |||||
| rs481134 | T/C | 0.3224 | 0.3666 | 0.095 | 0.82 (0.66–1.03) |
| TXC (570 sporadic cases and 604 controls) | |||||
| rs481134 | T/C | 0.3224 | 0.3558 | 0.098 | 0.86 (0.72–1.03) |
| TXA (188 sporadic cases and 240 controls) | |||||
| rs481134 | T/C | 0.6183 | 0.6778 | 0.036 | 0.72 (0.52–0.98) |
| Combined (1,144 cases and 1,288 controls) | |||||
| rs481134 | T/C | 0.3694 | 0.4227 | 7.01 × 10−4 | 0.81 (0.72–0.92) |
| Subjects with genotypes CC at SNP rs481134 | |||||
| GELCC (62 familial cases and 69 controls) | |||||
| rs1051730 | G/A | 0.7411 | 0.4851 | 4.51 × 10−4 | 4.47 (3.64–5.31) |
| MCC (299 sporadic cases and 297 controls) | |||||
| rs1051730 | G/A | 0.6829 | 0.5997 | 1.82 × 10−3 | 1.46 (1.22–1.70) |
| TXC (488 sporadic cases and 422 controls) | |||||
| rs1051730 | G/A | 0.6695 | 0.5781 | 7.21 × 10−5 | 1.49 (1.29–1.68) |
| TXA (120 sporadic cases and 138 controls) | |||||
| rs1051730 | G/A | 0.2288 | 0.1168 | 2.20 × 10−4 | 2.83 (1.63–4.91) |
| Combined (969 cases and 926 controls) | |||||
| rs1051730 | G/A | 0.6232 | 0.5093 | 4.42 × 10−11 | 1.60 (1.46–1.74) |
Bold letter is the reference allele used for estimating OR.
Association was adjusted by the same covariates as the single-locus analysis.
Similarly, SNP rs481134 could affect the association of rs1051730 with lung cancer. In the subgroup analysis of subjects with genotype CC at rs481134, a consistent, stronger association of rs1057130 with lung cancer was observed in an even smaller sample size (P =4.42× 10−11). This subgroup analysis essentially analyzed subjects with haplotypes A_C and/or G_C and allowed direct test on the allelic effects at rs1051730. Interestingly, the allelic OR associated with the risk allele of rs1051730 A is increased to 1.60 (95% CI, 1.46–1.74) as compared with 1.24 (95% CI, 1.14–1.35) in the previous single-marker analysis (Tables 1 and 3). These results are consistent with those from the Mantel-Hazenszel tests (Supplementary Table S2). Based on the marginal OR estimate and genotype frequencies estimated from the TXC populations, the first variant on 15q24–25.1, marked by rs1051730, accounts for 25.8% of population attributable risk for lung cancer (Supplementary Table S3). These analyses showed that both variants independently influence risk for lung cancer.
We also performed a logistic regression analysis modeling the SNPs rs1051730 and rs481134 simultaneously (Supplementary Table S4). Generally, this logistic regression analysis yielded consistent results with the haplotype analysis (Table 2) and subgroup association analysis (Table 3). In addition to their effects independently influencing risk of lung cancer, the two variants may exert a synergistic effect (interaction) to modify the risk of lung cancer.
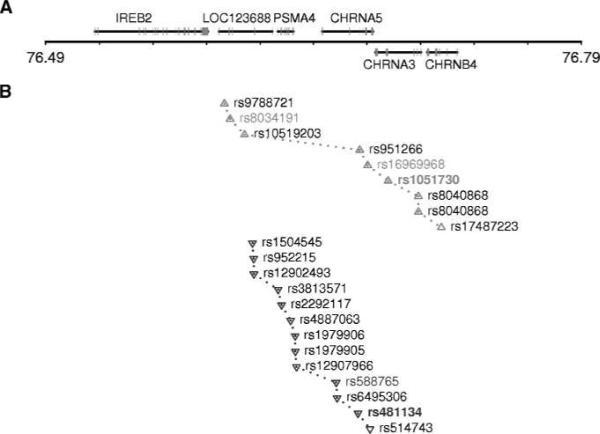
SNP rs1051730 is a synonymous SNP at codon 215 of CHRNA3, and rs481134 is located within the second intron of CHRNA5. Analysis of LD among the 15q24–25.1 locus revealed two independent sets of SNPs that are in high LD with these two variants (r2 > 0.7; Fig. 1). These two independent sets of SNP span across the chromosomal region 15q24–25.1 and illustrate the complexity of the 15q24–25.1 locus underlying lung cancer susceptibility.
Figure 1.
Chromosome 15q24–25.1 locus. A, genomic context of 15q24–25.1. The middle horizontal line is the physical position of chromosome (Mb; build 36.3). Genes shown on the upper side of the horizontal line are transcribed in the + orientation (from left to right), and those on the lower side in the − orientation (from right to left). B, SNP positions and LD among these SNPs. Up triangle, SNPs that are in high LD with the first variant, rs1051730 (r2 > 0.7). Down triangle, SNPs that are in high LD with the second variant, rs481134. Pairwise LD was estimated for 251 SNPs on 15q24–25.1 (from 76,016,651 to 76,992,181 bp) in 219 disease-free unrelated controls collected from the GELCC.
Association and OR of different haplotype configurations with lung cancer
Only three haplotypes (rs1051730-rs481134), G_T, A_C, and G_C, were observed in general populations (Table 2). As a result, six genotypes account for more than 99% subjects in the samples (Table 4). Using the genotype G_T/G_T as reference, we estimated the association and OR of different haplotype configurations with lung cancer. In general, subjects carrying two copies of either risk or protective haplotypes show significant association with lung cancer. For instance, subjects with genotypes A_C/A_C in TXC had a significantly higher risk (P = 2.71 × 10−3; OR, 1.24; 95% CI, 1.08–1.42) for developing lung cancer than subjects with genotypes G_T/G_T, whereas carriers of G_C/G_C had a reduced lung cancer risk by 18% (P = 0.049; OR, 0.82; 95% CI, 0.67–0.99).
Table 4.
Association and OR of different haplotype configurations with lung cancer
| Samples (cases/controls) | Genotypes* | Frequency | P† | OR (95% CI) | |
|---|---|---|---|---|---|
| Cases | Controls | ||||
| MCC | |||||
| 145/142 | G_T/G_T | — | — | — | 1.00 (reference) |
| 292/267 | G_T/A_C | 0.6682 | 0.6528 | 0.611 | 1.04 (0.90–1.18) |
| 140/110 | A_C/A_C | 0.4912 | 0.4365 | 0.168 | 1.13 (0.95–1.34) |
| 145/148 | G_C/G_T | 0.5000 | 0.5103 | 0.834 | 0.98 (0.82–1.15) |
| 127/135 | G_C/A_C | 0.4669 | 0.4874 | 0.586 | 0.95 (0.81–1.13) |
| 31/51 | G_C/G_C | 0.1761 | 0.2642 | 0.039 | 0.77 (0.60–0.99) |
| TXC | |||||
| 250/248 | G_T/G_T | — | — | — | 1.00 (reference) |
| 415/415 | G_T/A_C | 0.6241 | 0.6259 | 0.992 | 1.00 (0.89–1.11) |
| 212/140 | A_C/A_C | 0.4589 | 0.3608 | 2.71 × 10−3 | 1.24 (1.08–1.42) |
| 263/277 | G_C/G_T | 0.5127 | 0.5276 | 0.6895 | 0.98 (0.85–1.10) |
| 216/201 | G_C/A_C | 0.4635 | 0.4477 | 0.6201 | 1.03 (0.91–1.18) |
| 50/75 | G_C/G_C | 0.1667 | 0.2322 | 0.049 | 0.82 (0.67–0.99) |
h1/h2 indicates a genotype composed of two haplotypes, h1 and h2.
Association was adjusted by the same covariates as the single-locus analysis. GELCC and TXA populations were not analyzed because of their small sample sizes.
Discussion
After the recent discovery that common genetic variation in 15q24–25.1 influences inherited risk of lung cancer (3–7), we identified a second sequence variant at 15q24–25.1 associated with familial lung cancer (8) and further validated this new association in large sporadic lung cancer populations. We showed that these two genetic variants on 15q24–25.1 have independent genetic effects on lung cancer risk. The second variant on 15q24–25.1, marked by rs481134, explains an additional 13.2% of the population attributable risk for lung cancer. These results further confirm the complexity of the chromosomal region 15q24–25.1 underlying lung cancer susceptibility.
Interestingly, the second variant did not show association with lung cancer in single-marker analysis. However, haplo-type analysis of SNPs rs1051730 and rs481134 provided stronger evidence for association with lung cancer. SNPs rs1051730 and rs481134 are in moderate LD (r2 = 0.30), which can mask or change the genetic effects of those loci in the association analysis. This may explain why the association of rs481134 with lung cancer was not detected in the single-marker analysis. The neutral haplotype G_T contains a wild-type allele at each variant, the protective haplotype G_C contains a wild-type allele at rs1051730 and a protective allele C at rs481134, whereas the haplotype A_C contains both a protective allele C at SNP rs481134 and a risk allele A at rs1051730. This suggests that the rs1051730 risk allele outweighs the rs481134 protective allele, and the overall effect of this haplotype increases risk of lung cancer. As a result, the protective effect of SNP rs481134 at haplotype A_C was masked by rs1051730, and thus its association with lung cancer was not detected in the single-marker analysis. Comparison of haplotypes G_T (neutral) and G_C (protective), which have the same allele G at rs1051730 but have different alleles (C/T) at rs481134, clearly showed that the allele C of rs481134 is associated with reduced risk of lung cancer. Similarly, comparison of haplotypes A_C (risk) with G_C (protective), which have the same allele C at rs481134 but have different alleles at rs1051730, showed that the allele A of rs1051730 is associated with increased risk of lung cancer. Actually, these comparisons are the rationale behind subsequent subgroup analyses for estimating marginal OR associated with each of these two variants.
To eliminate the effect masked by rs1051730, we performed an association analysis in a subgroup of subjects with genotype GG at rs1051730 and identified a significant association of rs481134 with lung cancer. The allele C of rs481134 is associated with reduced risk of lung cancer (Table 3), which is consistent with the haplotype analysis. These results showed that the two variants independently influence risk for lung cancer. Interestingly, the allelic OR associated with the risk allele of rs1051730 A is much larger from subgroup analysis of subjects with genotype CC at rs481134 than that estimated from the whole samples (OR, 1.60 versus 1.24; Tables 1 and 3). This is because SNPs rs1051730 and rs481134 are in LD and they have opposite genetic effects on lung cancer risk. The effect of rs1051730 was offset by the effect of rs481134, and thus the effect size associated with rs1051730 was underestimated in previous studies (3–7). Therefore, the marginal ORs associated with these two variants were used in the calculation of PAR for lung cancer. However, it should be noted that the estimated PAR for individual SNPs may not represent PAR in the general population because the marginal OR was used. The effect size associated with rs481134 is generally less significant than that of rs1051730 and may need to be further validated in future studies. In addition to their effects independently influencing risk of lung cancer, the two variants may exert a synergistic effect to modify the risk of lung cancer (Supplementary Table S4). Our data emphasize the importance of the use of multilocus association models in association analyses.
Only three major haplotypes, G_T, A_C, and G_C, were observed at variants rs1051730 and rs481134 in all of the four populations. These three haplotypes accounted for more than 99% subjects in the samples. A very small number of individuals carry haplotype A_T. The risk for developing lung cancer is expected to be further increased among these individuals, although the number of individuals with this haplo-type was too small to formally estimate its effect size. In the subgroup analysis of subjects with genotype CC at rs481134, the allelic OR associated with the risk allele of rs1051730 A was estimated to be 1.60 (95% CI, 1.46–1.74). This allelic OR can be viewed as an indirect estimator of effect size of the haplotype A_T. Therefore, two risk haplotypes exist at 15q24–25.1: the common haplotype A_C confers moderate risk for increasing lung cancer and the rare haplotype A_T confers excessive risk for increasing lung cancer.
The 15q25.1 lung cancer susceptibility locus contains IREB2, LOC123688, PSMA4, CHRNA5, CHRNA3, and CHRNB4 (Fig. 1A). The nAChR genes encode for subunits of the nicotinic acetylcholine receptors, seem biologically relevant, and are attractive candidates. Recent candidate-gene studies of nicotine dependence also identified several variants in the CHRNA5-CHRNA3-CHRNB4 gene cluster on 15q24–25.1 that alter risk for nicotine dependence, including a missense SNP, rs16969968 (a change from aspartic acid to asparagine at codon 398 in CHRNA5; refs. 14, 15). SNP rs16969968 shows high LD with rs1051730 (r2 = 0.99) from the lung cancer association analyses. Nicotinic receptors containing the missense variant α4β2α5N398 of CHRNA5 exhibit reduced response to the nicotinic agonist epibatidine compared with receptors containing the more common variant α4β2α5D398 (16). Interestingly, SNP rs588765, in high LD with rs481134 (r2 = 0.98), was initially identified to be associated with mRNA levels of CHRNA5 in brain. The major allele of rs588765 is associated with higher expression of CHRNA5, conferring reduced risk of nicotine dependence (8). The same two genetic variants are also associated with lung cancer risk. These data point to the candidacy of CHRNA5 for the 15q24–25.1 locus underlying lung cancer susceptibility, although the other five candidate genes in this high LD region cannot be completely excluded (Fig. 1B).
Genetic variation in CHRNA5 can affect its functionality in two ways: (a) rendering the receptors differentially responsive to their ligands and thus affecting their downstream signal transduction (17), and (b) modifying expression and thus affecting the composition of nAChR subunits that govern the acute response to agonists, such as endogenous acetylcholine and exogenous nicotine (18). The mechanisms of regulation of CHRNA5 function or amount may underlie the genesis of lung cancer in smokers in two ways (Supplementary Fig. S1). First, polymorphisms in CHRNA5 may affect nicotine dependence and propensity to smoke and to develop lung cancer. Smokers who are addicted to nicotine usually use more tobacco than those who are not, and thus they have a higher likelihood of developing lung cancer resulting from increased exposure to carcinogens found in tobacco smoke. This mechanism suggests a possible indirect genetic factor (e.g., through smoking behavior) contributing to lung cancer. Second, nAChRs that are functionally present in human lung airway epithelial cells and in lung carcinomas may play a direct, functional role in lung carcinogenesis (19). In addition to nicotine, nitrosamines 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) and N-nitrosonornicotine (NNN) are two major carcinogens found in tobacco smoke. NNK and NNN are agonists for α-bungarotoxin α7 nAChR and the heteromeric, epibatidine-sensitive α-β nAChRs, respectively (20). The affinity of NNK for the α7 nAChR was found to be 1,300 times higher than that of nicotine, whereas the affinity of NNN for heteromeric α-β nAChRs was 5,000 times higher than that of nicotine (20, 21). Recent studies have shown that all these individual constituents of tobacco smoke can stimulate nAChR signaling in nonneuronal cells, including regulation of cell proliferation, angiogenesis, migration, invasion, and secretion (19, 22, 23). This mechanism suggests a possible direct gene-environment interaction factor (e.g., cell proliferation and apoptosis) contributing to lung cancer. That is, given the same amount of tobacco exposure, tobacco-induced nonneuronal nAChR signaling responds differentially in smokers with different variants in CHRNA5. This may account for individual differences in lung cancer susceptibility to the same environmental risk factors such as tobacco smoking. It is also possible that both of these two mechanisms exist during lung carcinogenesis. Defining the contribution of both mechanisms warrants further studies.
Supplementary Material
Acknowledgments
We thank the Fernald Medical Monitoring Program for sharing their biospecimens and data with us; the lung cancer families who participated in this research; Vanderbilt University Microarray Shared Resource, Washington University Genotyping Core, and Mayo Clinic Genotyping Shared Resource (supported in part by grant P30CA 15083) for their high-caliber service; Shaw Levy and Jennifer Baker for their assistance in various aspects of this work; and Jay Tichelaar and Michael James for their helpful criticisms and comments on the manuscript.
Grant Support NIH grants U01CA76293 (Genetic Epidemiology of Lung Cancer Consortium), R01CA058554, R01CA093643, R01CA099147, R01CA099187, R01ES012063, R01ES013340, R03CA77118, R01CA80127, P30ES06096, P50CA70907 (Specialized Program of Research Excellence), N01HG65404, N01-PC35145, P30CA22453, R01CA63700, and DE-FGB-95ER62060; Mayo Clinic intramural research funds; and Department of Defense VITAL grant. This study was supported in part by NIH, the Intramural Research Programs of the National Cancer Institute, and the National Human Genome Research Institute.
Footnotes
Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest No potential conflicts of interest were disclosed.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Matakidou A, Eisen T, Houlston RS. Systematic review of the relationship between family history and lung cancer risk. Br J Cancer. 2005;93:825–33. doi: 10.1038/sj.bjc.6602769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amos CI, Wu X, Broderick P, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616–22. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hung RJ, McKay JD, Gaborieau V, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–7. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 5.Liu P, Vikis HG, Wang D, et al. Familial aggregation of common sequence variants on 15q24–25.1 in lung cancer. J Natl Cancer Inst. 2008;100:1326–30. doi: 10.1093/jnci/djn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McKay JD, Hung RJ, Gaborieau V, et al. Lung cancer susceptibility locus at 5p15.33. Nat Genet. 2008;40:1404–6. doi: 10.1038/ng.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thorgeirsson TE, Geller F, Sulem P, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–42. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang JC, Cruchaga C, Saccone NL, et al. Risk for nicotine dependence and lung cancer is conferred by mRNA expression levels and amino acid change in CHRNA5. Hum Mol Genet. 2009;18:3125–35. doi: 10.1093/hmg/ddp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang P, Allen MS, Aubry MC, et al. Clinical features of 5,628 primary lung cancer patients: experience at Mayo Clinic from 1997 to 2003. Chest. 2005;128:452–62. doi: 10.1378/chest.128.1.452. [DOI] [PubMed] [Google Scholar]
- 10.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–48. [PubMed] [Google Scholar]
- 11.Hanley JA. A heuristic approach to the formulas for population attributable fraction. J Epidemiol Community Health. 2001;55:508–14. doi: 10.1136/jech.55.7.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirschfield GM, Liu X, Xu C, et al. Primary biliary cirrhosis associated with HLA, IL12A, IL12RB2 variants. N Engl J Med. 2009;360:2544–55. doi: 10.1056/NEJMoa0810440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saccone NL, Saccone SF, Hinrichs AL, et al. Multiple distinct risk loci for nicotine dependence identified by dense coverage of the complete family of nicotinic receptor subunit (CHRN) genes. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:453–66. doi: 10.1002/ajmg.b.30828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saccone SF, Hinrichs AL, Saccone NL, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bierut LJ, Stitzel JA, Wang JC, et al. Variants in nicotinic receptors and risk for nicotine dependence. Am J Psychiatry. 2008;165:1163–71. doi: 10.1176/appi.ajp.2008.07111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gotti C, Zoli M, Clementi F. Brain nicotinic acetylcholine receptors: native subtypes and their relevance. Trends Pharmacol Sci. 2006;27:482–91. doi: 10.1016/j.tips.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 18.Williams BM, Temburni MK, Levey MS, Bertrand S, Bertrand D, Jacob MH. The long internal loop of the α 3 subunit targets nAChRs to subdomains within individual synapses on neurons in vivo. Nat Neurosci. 1998;1:557–62. doi: 10.1038/2792. [DOI] [PubMed] [Google Scholar]
- 19.Catassi A, Servent D, Paleari L, Cesario A, Russo P. Multiple roles of nicotine on cell proliferation and inhibition of apoptosis: implications on lung carcinogenesis. Mutat Res. 2008;659:221–31. doi: 10.1016/j.mrrev.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 20.Schuller HM, Orloff M. Tobacco-specific carcinogenic nitrosamines. Ligands for nicotinic acetylcholine receptors in human lung cancer cells. Biochem Pharmacol. 1998;55:1377–84. doi: 10.1016/s0006-2952(97)00651-5. [DOI] [PubMed] [Google Scholar]
- 21.Arredondo J, Chernyavsky AI, Grando SA. Nicotinic receptors mediate tumorigenic action of tobacco-derived nitrosamines on immortalized oral epithelial cells. Cancer Biol Ther. 2006;5:511–7. doi: 10.4161/cbt.5.5.2601. [DOI] [PubMed] [Google Scholar]
- 22.Egleton RD, Brown KC, Dasgupta P. Nicotinic acetylcholine receptors in cancer: multiple roles in proliferation and inhibition of apoptosis. Trends Pharmacol Sci. 2008;29:151–8. doi: 10.1016/j.tips.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 23.Schuller HM. Is cancer triggered by altered signalling of nicotinic acetylcholine receptors? Nat Rev Cancer. 2009;9:195–205. doi: 10.1038/nrc2590. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.