Abstract
Background
During the multiyear progression to colorectal cancer, numerous genomic alterations arise in events ranging from single base mutations to gains or losses of entire chromosomes. A single genetic change might not stand out as an independent predictor of outcome. The goal of this study was to determine if more comprehensive measurements of genomic instability provide clinically relevant prognostic information.
Methods
Our study included 65 sporadic colorectal cancer patients diagnosed from 1987 to 1991 with last follow-up ascertained in 2006. We estimated an overall tally of alterations using the genome-wide sampling technique of inter-(simple sequence repeat [SSR]) polymerase chain reaction (PCR), and evaluated its relationship with all-cause survival. We also extended and sensitized the Bethesda criteria for microsatellite instability (MSI), by analyzing 348 microsatellite markers instead of the normal five. We expanded the MSI categories into four levels: MSI stable (MSS), very low-level MSI, moderately low-level MSI, and classical high-level MSI.
Results
Tumors with genomic instability above the median value of 2.6% as measured by inter-SSR PCR, were associated with far greater risk of death compared to tumors with lower levels of genomic instability. Adverse outcome was most pronounced for patients presenting with stage 3 disease. A gradient of increased survival was observed across increasing MSI levels but did not reach statistical significance.
Conclusion
Our findings suggest genomic instabilities quantified by inter-SSR PCR and increased precision in MSI values may be clinically useful tools for estimating prognosis in colorectal cancer.
DNA sequencing has confirmed earlier cytogenetic, microarray and genomic sampling approaches that had indicated carcinoma genomes are extensively damaged, with different damage profiles evident in each tumor.1,2 Ten years ago, by genomic sampling, we estimated about 11,000 genomic events had occurred per colorectal tumor.3 With approximately 1% of the genome representing coding sequences, these results are consistent with more recent DNA sequencing studies finding around one hundred mutated genes per tumor.4–6 This extensive overall alteration reflects the consequences of multiple genomic instabilities arising at various times in tumor progression, combined with unrepaired genomic damage resulting from environmental effects. One can anticipate that the nature and extent of genomic instability in a tumor could impact clinical outcomes, by altering the likelihood of deleterious genomic events occurring.
With high-level microsatellite instability (MSI-H), the tumor progression pathway is shifted and tumors with MSI-H are associated with superior outcomes.7,8 Because microsatellite instability (MSI) levels are generally determined by examination of only five markers, with two or more altered being taken as MSI-H according to current National Cancer Institute guidelines, lower level levels of MSI are difficult to precisely measure and possible outcome differences within this group have not been evaluated.9 For larger event instabilities such as copy number aberrations detected with microarray comparative genomic hybridization, more extensively altered tumors have more gene functions altered or lost. Although there is a greater likelihood of lethal events occurring in some members of the tumor cell population, there is also a greater likelihood that some members of the tumor cell population have evolved to more aggressive states. Studies of breast, colon, and prostate cancers have confirmed that higher levels of large event instabilities are associated with generally worse outcomes.10–12
Below we present the first report of the prognostic value of a tally of genomic instability as estimated by inter-(simple sequence repeat [SSR]) polymerase chain reaction (PCR) in colorectal tumors. We also evaluated the prognostic value of a more precise version of an established measure of genomic instability, MSI, defined by genome-wide allelotyping with 348 microsatellite markers.
METHODS
Tumors and DNA Extraction
Paired colorectal tumor and normal mucosa samples were analyzed in these experiments. The principal set of tissue samples were collected from 58 consecutive patients diagnosed with sporadic colorectal cancer (CRC) who underwent surgery at Roswell Park Cancer Institute, Buffalo, New York, between 1988 and 1995.13 These tissues were used for all of the MSI determinations. An additional seven tissues were randomly selected from a Yale study of patients diagnosed with primary CRC between January 1987 and April 1991.14 Both the Roswell Park and Yale specimens were used for the inter-SSR PCR measurements.15 Patients gave their written consent to obtain tissue specimens, as approved by the Roswell Park Cancer Institute institutional review board. Informed consent to obtain tissues from patients was waived by the Yale Human Investigations Committee and all participating hospital IRBs. Tissue samples were examined and tumor specific specimens were selected by pathologists. DNA was extracted from liquid nitrogen-frozen tumor samples and separately from adjacent normal tissue. Small pieces of the tissue (approximately 8 mm 3) were digested with 1 mg/ml proteinase K for 3 h at 65°C, followed by 0.5 mg/ml RNase treatment for 1 h. The DNA was then isolated via phenol:choroform:isoamyl alcohol extraction and ethanol precipitation.
Genomic Instability Measurements
Inter-SSR PCR Instability
Inter-SSR instability was determined for all sixty-five paired tumor and normal tissue DNAs as described.3,15 (CA)8RG and (CA)8RY (R = a 50:50 mixture of the purines adenine and guanine; Y = a 50:50 mixture of the pyrimidines cytosine and thymine) primers were end labeled with γ-32P-ATP via T4 polynucleotide kinase. Amplification of sequences between the repeat elements was carried out in a 20 μl reaction mixture containing 1 mM primer (1:5 labeled:unlabeled oligonucleotide), 50 ng genomic DNA, 0.3 units Taq polymerase (Invitrogen Corporation, Carlsbad, CA) in 1× PCR buffer (10 mM Tris-HCl, pH 9.0, 2% formamide, 50 mM KCl, 0.2 mM deoxynucleotide triphosphates (dCTP, dATP, dTTP, dGTP), 1.5 mM MgCl2, 0.01% gelatin, 0.01% Triton X-100). Amplification conditions were as follows: 2 min initial denaturation at 94°C, followed by 35 cycles of 30 s at 94°C, 45 s at 52°C, 2 min at 72°C, and a final extension of 5 min at 72°C. PCR products were mixed with 10% loading buffer (40% sucrose, 0.25% bromophenol blue) and 5 ml of each product was analyzed on a nondenaturing 8% polyacrylamide gel without urea, buffered with 1× TBE (0.89 mM Tris-borate, pH 8.3, 2 mM EDTA). Electrophoresis was performed for 22 min at 80 W, followed by 50 W for 3400 volt-h. The gels were dried for 1 h and autoradiographed with Kodak XAR film with an overnight exposure.
Low-Level MSI
Fractional allelic loss rate had been determined for the first 58 specimens by loss of heterozygosity electrophoretic gel analyses of PCR products, using 348 markers spaced an average 10 Mb apart, spanning the entire genome.13 The methodology and loss of heterozygosity raw data has been reported previously.13 By evaluation of banding patterns in the entire set of loss of heterozygosity gels for MSI at each of the 348 markers, precise MSI values were determined, including low-level MSI (MSI-L).
Statistical Analyses
Inter-SSR values were used to define two sets of patients according to genomic instability index (GII). One group consisted of GII above (GII high) and the second group GII below (GII low) the median level of 2.6% observed in the entire sample.
The distribution of MSI levels were divided into four sets: zero markers exhibiting MSI (MSI stable, or MSS), very low-level MSI (0.8–2.4% of informative markers), moderately low-level MSI (2.5–9.6%), and high-level MSI (>40%). High-level MSI is based on the Bethesda criteria.9 The MSI-low level was divided into two categories (above and below the median of MSI-low values) for our analyses. Patient age at diagnosis, American Joint Committee on Cancer stage at diagnosis, vital status, cause of death, sex, and race were ascertained from the medical chart. Survival time was calculated from date of diagnosis to date of last follow-up or of death. We last ascertained vital status in November 2006; median follow-up time was calculated among patients still living at that point to determine that adequate follow-up time had elapsed. Nonparametric tests were employed in descriptive analyses as a result of the relatively small sample size in each study group. Survival analyses employed the Cox proportional hazard test in multivariate analyses, and the method of Kaplan-Meier to create survival curves. Before conducting survival studies, two patients were excluded from analyses because they died less than 6 months after diagnosis to account for possible bias related to surgical failure. Characteristics of these patients are as follows: GII (1.3, 1.3), MSI (0.90, 3.14) stage (3, 4), survival in months (2.7 and 5.1), cause of death (both non-CRC) and age (48, 47). All-cause survival was employed as the outcome measure given the relatively small sample size. SPSS Ver. 18 was used in all analyses.
RESULTS
Inter-SSR PCR Instability Compared with Patient Outcomes
We first analyzed outcomes of patients previously assayed for genomic instability with inter-SSR PCR at the time of tumor resection, and followed clinically for approximately the next decade.15 Instability by this method is quantified by the percentage of inter-SSR PCR electrophoretic bands altered, defined as the GII. Clinicopathological characteristics according to GII levels of tumors are reported in Table 1. Patients whose tumors exhibited an increased GII level (GII high group) had a higher death rate from CRC compared to patients in the GII low group (51.6 vs. 35.3%, respectively) and a higher overall mortality rate (71.0 vs. 58.8%, respectively, data not shown) but these differences did not reach statistical significance. Patients in the two GII groups did not vary substantially by age, disease stage, sex, and race. Patients with GII high tumors tended to have reduced survival time compared to those with GII low tumors (64.1 vs. 94.5 months, respectively, P = 0.20) for all-cause mortality (Table 2). When examining CRC-specific death as the outcome of interest, the difference in survival time was slightly more pronounced (63.7 vs. 104.4 months, respectively, P = 0.11, data not shown.) Stratified analyses show reduced median survival time for patients with GII high tumors in each tumor, node, metastasis system (TNM) stage.
TABLE 1.
Clinicopathological characteristics of 65 patients with CRC according to GII status of tumor
| Characteristic | GII lowan = 34 | GII high n = 31 | P b,c |
|---|---|---|---|
| Vital status | |||
| Alive | 14 (41.2%) | 9 (29.0%) | 0.41 |
| Dead from CRC | 12 (35.3%) | 16 (51.6%) | |
| Dead from other causes | 8 (23.5%) | 6 (19.4%) | |
| Median follow-up (months)d | 149.9 | 133.3 | 0.61 |
| American Joint Committee on Cancer TNM stage at diagnosis | |||
| 1 | 10 (29.4%) | 9 (29.0%) | 0.83 |
| 2 | 7 (20.6%) | 6 (19.4%) | |
| 3 | 9 (26.5%) | 11 (35.5%) | |
| 4 | 8 (23.5%) | 5 (16.1%) | |
| Colon subsitee | |||
| Left | 22 (66.7%) | 19 (63.3%) | 0.78 |
| Right | 11 (33.3%) | 11 (36.7%) | |
| Age (years) (mean) | 62.3 | 60.3 | 0.41 |
| Sex | |||
| Female | 17 (50.0%) | 16 (51.6%) | 0.89 |
| Male | 17 (50.0%) | 15 (48.4%) | |
| Race | |||
| White | 32 (94.1%) | 27 (90.0%) | 0.54 |
| Black | 2 (5.9%) | 3 (10.0%) | |
GII as calculated from inter-SSR PCR assay. GII low and GII high categories are defined, respectively, as below and above median GII value for entire sample of patients
Fisher's exact test for proportions (two-sided)
Mann-Whitney U nonparametric test for continuous values
Among patients alive at end of study period
Left colon defined as rectum through splenic flexure, right colon as transverse colon through cecum
TABLE 2.
Inter-SSR PCR instability: multivariate RR of death according to stage among 63 patients with CRC
| Characteristic | n | Median survival (months) (SE) | Risk of death |
|
|---|---|---|---|---|
| Adjusted RR | 95% CIa | |||
| GII | ||||
| Low (0–2.6) | 32 | 94.5 (10.3) | 1.00 | 1.21–4.85 |
| High (>2.6) | 31 | 64.1 (9.3) | 2.42 | |
| Stage 1 | ||||
| GII low | 10 | 140.3 (15.1) | 1.00 | 0.37–12.31 |
| GII high | 9 | 106.3 (10.5) | 2.14 | |
| Stage 2 | ||||
| GII low | 7 | 132.1 (24.6) | 1.00 | 0.36–13.14 |
| GII high | 6 | 127.4 (22.2) | 2.17 | |
| Stage 3 | ||||
| GII low | 8 | 94.9 (17.2)b | 1.00 | 1.11–17.06 |
| GII high | 11 | 40.6 (5.2) | 4.35 | |
| Stage 4 | ||||
| GII low | 7 | 23.0 (10.7) | 1.00 | 0.21–1.44 |
| GII high | 5 | 11.8 (10.7) | 0.99 | |
Excluded were two patients who died less than 6 months after diagnosis (see “Methods” section)
Cox proportional hazard survival analyses controlling for age, race, and TNM stage. Stage excluded from the multivariate model in stratified analyses
Mann-Whitney U nonparametric test; P = 0.02 for stage 3 disease; all other tests, P > 0.20
Overall, patients with GII high tumors were more than twice as likely to die compared to patients with GII low tumors (relative risk [RR] 2.42, 95% confidence interval [CI] 1.21–4.85) after controlling for age, TNM stage and race (Table 2). For this analysis, we excluded two patients who died shortly after surgery, whose deaths were not attributed to CRC. Cox survival analyses stratified by TNM stage showed increased risk of death related to GII high tumors at stages 1 to 3, although findings reached statistical significance for stage 3 disease only (RR 4.35 95% CI 1.11–17.06). For patients with stage 1 disease, nearly all survived after tumor resection, regardless of GII level. For patients with stage 4 disease, distant metastases had already arisen before resection of the primary tumor, and the median survival time of all these patients was low in both GII groups. Differences in outcomes based on GII instability were most evident for stage 2 and 3 patients, presumably reflecting the likelihood that occult metastasis had already occurred at the time of primary tumor resection. Clinically, this information would be particularly useful for deciding which patients may most benefit from adjuvant therapy.
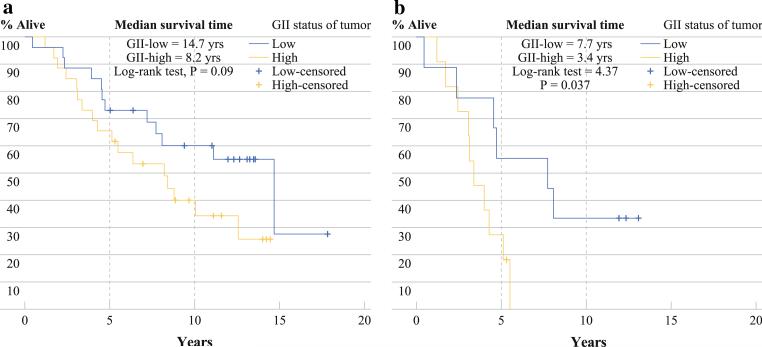
Figure 1a illustrates the comparatively adverse survival rate of CRC patients with GII high tumors (log rank test, P = 0.09) among those diagnosed with TNM stages 1, 2, and 3 (n = 52). The estimated 5- and 10-year survival rates for patients with GII high tumors was 65 and 35%, respectively, compared to 75 and 60%, respectively, for patients with GII low tumors. Overall, the first loss to follow-up occurred after the 5 years. Figure 1b presents the Kaplan-Meier survival curves among TNM stage 3 patients only (n = 20.) Notably, the 5-year survival rate for patients with GII high tumors was 25% compared to 60% for GII low tumors (log rank test, P = 0.037.) No stage 3 patients with GII high tumors survived past 6 years, whereas 35% of such patients with GII low tumors survived up to 10 years.
FIG. 1.
a Kaplan–Meier curves showing overall survival of GII high and GII low tumors among patients diagnosed with TNM stage 1 to 3 CRC (n = 52).
b Kaplan–Meier curves showing overall survival of GII high and GII low tumors among patients diagnosed with TNM stage 3 CRC (n = 20)
Low-Level MSI Compared with Patient Outcomes
MSI-L is defined by the Bethesda guidelines as the alteration of repeat sequences in one of five specific microsatellite markers assayed.9 Because mismatch repair errors occur randomly, this commonly used definition is open to considerable statistical error. Studies to more precisely determine MSI-L have analyzed hundreds of microsatellite markers from each tumor sample; such studies have revealed a broad distribution of low-level MSI values ranging from zero to 10% of markers showing alterations, while classical high-level MSI (MSI-H) shows more than 40% of markers altered.13,16
We investigated whether MSI-L cases of CRC have different outcomes, depending on the degree of low-level instability. We followed up on the clinical outcomes of 58 cases we had previously characterized at the genomic level in 2001, where 348 markers were examined for MSI for each tumor specimen.13 The clinicopathologic features of these cases are described in Table 3. We divided these patients into four sets: those showing no microsatellites altered (MSS, n = 6), those with below the median of MSI-L in the study sample (MSI-L1, n = 25), those with above the median low-level MSI (MSI-L2, n = 23), and those with high-level MSI (MSI-H, n = 4). The entire distribution of values is presented in Fig. 2. Sex, race, or age did not vary by MSI status. The four MSI-H tumors were all stage 1 or 2, consistent with the well established superior outcomes for patients with MSI-H tumors. The MSS, MSI-L1, and MSI-L2 tumors showed generally similar distributions among the four stages.
TABLE 3.
Clinicopathological characteristics of 58 patients with CRC according to MSI status of tumor
| MSI statusa,b | MSS n = 6 | MSI-L1 n = 25 | MSI-L2 n = 23 | MSI-H n = 4 |
|---|---|---|---|---|
| Vital statusc | ||||
| Alive | 0 | 8 (32.0%) | 10 (43.5%) | 2 (50.0%) |
| Deaths from CRC | 4 (66.7%) | 11 (44.0%) | 9 (39.1%) | 1 (25.0%) |
| Deaths from other causes | 2 (33.3%) | 6 (24.0%) | 4 (17.4%) | 1 (25.0%) |
| Median follow-up (months)d,e | 0f | 140.1 (14.3) | 150.2 (9.4) | 137.1 (76.7) |
| Disease stage at diagnosis | ||||
| 1 | 0 | 7 (28.0%) | 8 (34.8%) | 1 (25.0%) |
| 2 | 1 (16.7%) | 4 (16.0%) | 3 (13.0%) | 3 (75.0%) |
| 3 | 3 (50.0%) | 8 (32.0%) | 8 (34.8%) | 0 |
| 4 | 2 (33.3%) | 6 (24.0%) | 4 (17.4%) | 0 |
| Age (years) (mean)d | 62.0 | 60.9 | 60.7 | 61.5 |
| Sexc | ||||
| Female | 4 (66.7%) | 12 (48.0%) | 9 (39.1%) | 2 (50.0%) |
| Male | 2 (33.3%) | 13 (52.0%) | 14 (60.9%) | 2 (50.0%) |
| Racec | ||||
| White | 5 (83.3%) | 24 (96.0%) | 22 (100.0%) | 3 (75.0%) |
| Black | 1 (16.7%) | 1 (4.0%) | 0 (0.0%) | 1 (25.0%) |
MSI as determined during genome-wide allelotyping assays.13 Cutpoints followed Bethesda criteria except for MSI-L1 and MSI-L2, which were defined, respectively, as below and above the median within the category of MSI-L, as found in the Bethesda guidelines.9 MSS represents those cases showing no MSI alterations among 348 microsatellites examined
Seven had insufficient tissue for assay
Fisher's exact test for proportions (two-sided); all P > 0.10
Kruskal–Wallis test for continuous values; all P > 0.21
Among only patients alive at the end of the study period
No MSS patients alive at the end of the study period
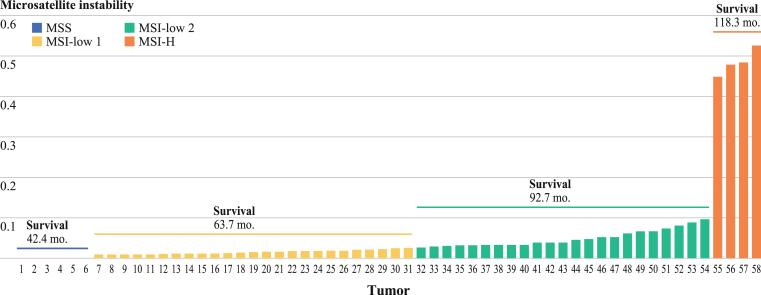
FIG. 2.
Distribution of tumor MSI values and corresponding CRC patient survival times
The outcomes for the four different sets of patients exhibited a gradient of increased survival as MSI level increased (Table 4), though this trend did not reach statistical significance. MSI-H has long been associated with superior outcomes, as it was for our patients where the median survival was 118.3 months. MSI-L2 patients showed a median survival of 92.7 months, while the value for MSI-L1 was 63.7 months. The MSS patients showed the worst outcome, with a median survival of only 42.4 months.
TABLE 4.
Risk of death among 56 patients with CRC
| MSI | n | Survival (months)a,b | Multivariate-adjusted RR of deathb,c,d |
|
|---|---|---|---|---|
| Median | All-cause | 95% CI | ||
| MSS | 6 | 42.4 (11.2) | 1.00 | |
| MSI-L1 | 23 | 63.7 (11.5) | 0.87 | 0.31–2.40 |
| MSI-L2 | 23 | 92.7 (11.9) | 0.71 | 0.24–2.09 |
| MSI-H | 4 | 118.3 (40.5) | 0.46 | 0.05–4.70 |
MSS no microsatellite markers positive for instability, MSI-L1 <10% of markers positive, MSI-L2 ≥10 to <40% of markers positive, MSI-H ≥40% of markers positive
Kruskal–Wallis nonparametric test = 3.93, P = 0.42
Excluded were 2 patients who died less than 6 months after diagnosis (see “Methods” section)
Cox proportional hazard survival analyses
Adjusted for age, race, and TNM stage
Disease stage at diagnosis varied according to MSI status (P = 0.04.) Patients with MSI-high tumors all presented with stage 1 or 2 disease at diagnosis, while 57% of patients with low or no detectable levels of MSI in tumors were diagnosed at stage 3 or 4. MSI status did not appear to vary by sex, race, or age. We did not perform multivariate analyses because of the low number of MSS (n = 6) and MSI-high (n = 4) cases. Hence, we could not statistically assess if MSI level was a favorable prognostic factor independent of stage at diagnosis.
DISCUSSION
For CRC patients after tumor resection, this is the first report of the prognostic value of genomic instability as measured by inter-SSR PCR. We observed a statistically increased risk of death associated with higher levels of inter-SSR PCR instability. In contrast, we observed better outcomes associated with increasing levels of MSI by using a more precise determination with 348 microsatellite markers. By themselves, these results are important; perhaps if combined with genomic instability determinations made with microarray and sequencing approaches or with spectral karyotyping, even more precise associations could be found. Recent reports indicate another manifestation of genomic instability, DNA copy number alteration, is also useful in estimating clinical outcome.10–12 By quantifying the degree of genomic instability, we can get a sense of the likelihood that some cells within the tumor cell population have progressed to a more lethal state. Because all tumors in our study were surgically resected at the time of analysis, such a more lethal state likely relates to undetected distant micrometastases left behind at the time of surgery. Future studies with larger samples are warranted to confirm the prognostic value of genomic instability, and to determine if this effect is maintained with the introduction of additional clinical data in survival models such as treatment type and comorbidities.
MSI is a different case, where higher levels of MSI are associated with increasingly better outcomes. Defects in DNA mismatch repair preferentially alter a small group of genes containing DNA repeat sequences. This in turn causes tumor progression to follow a specific pathway without large genomic events, which generates highly treatable tumors.17,18 Low-level MSI may affect partial aspects of this same system, with higher degrees of low-level instability approaching the superior outcomes observed with MSI-H. Alternatively, recent identification of a microRNA system regulating DNA mismatch repair may relate to the various low-level microsatellite instabilities observed, and a more complex association with clinical outcome.19
In tumor systems such as breast and prostate cancer, where early stage disease is frequently observed but fewer tumors progress further, our approach of analyzing the extent and forms of genomic instability might help provide a useful and practical means for differentiating between those tumors that merit clinical intervention and those that do not.
ACKNOWLEDGMENT
This work was supported by NCI grants R01CA74127 to GRA and P30-CA16056 to the Roswell Park Cancer Institute; by the NCI SEER program contract (N01-PC-35133) and Rapid Response Surveillance Study amendment (HS, 2003) with the Connecticut Tumor Registry in the Connecticut Department of Public Health (DPH); and by the Patrick and Catherine Weldon Donaghue Medial Research Foundation (DF#01-025) and the NCI Program Project Grant (5-P01-CA42101) to BAJ. Certain data used in this publication were obtained from the Connecticut DPH, and the authors assume full responsibility for analyses and interpretation of these data. We thank Nicholas Petrelli for valuable conversations and his role in supplying the specimens, Janet Winston for the pathological examinations, and Roy Heinaman for expert technical assistance. We thank Daniel Rosenberg and staff for microdissection and DNA extraction of the Connecticut subset of tissues.
REFERENCES
- 1.Beerenwinkel N, Antal T, Dingli D, et al. Genetic progression and the waiting time to cancer. PLoS Comp Biol. 2007;3:2239–46. doi: 10.1371/journal.pcbi.0030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sjöblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–74. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 3.Stoler DL, Chen N, Basik M, et al. The onset and extent of genomic instability in sporadic colorectal tumor progression. Proc Natl Acad Sci USA. 1999;96:15121–6. doi: 10.1073/pnas.96.26.15121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stephens PJ, McBride DJ, Lin ML, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–10. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greenman C, Stephens P, Smith R, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–8. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sugarbaker DJ, Richards WG, Gordon GJ, et al. Transcriptome sequencing of malignant pleural mesothelioma tumors. Proc Natl Acad Sci USA. 2008;105:3521–6. doi: 10.1073/pnas.0712399105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benatti P, Gafà R, Barana D, et al. Microsatellite instability and colorectal cancer prognosis. Clin Cancer Res. 2005;11:8332–40. doi: 10.1158/1078-0432.CCR-05-1030. [DOI] [PubMed] [Google Scholar]
- 8.Sinicrope FA, Rego RL, Halling KC, et al. Prognostic impact of microsatellite instability and DNA ploidy in human colon carcinoma patients. Gastroenterology. 2006;131:729–37. doi: 10.1053/j.gastro.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 9.Umar A, Boland CR, Terdiman JP, et al. Revised Bethesda guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–8. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poulogiannis G, Ichimura K, Hamoudi RA, et al. Prognostic relevance of DNA copy number changes in colorectal cancer. J Pathol. 2009;220:338–47. doi: 10.1002/path.2640. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Martens JWM, Yu JX, et al. Copy number alterations that predict metastatic capability of human breast cancer. Cancer Res. 2009;69:3795–801. doi: 10.1158/0008-5472.CAN-08-4596. [DOI] [PubMed] [Google Scholar]
- 12.Pretorius ME, Waehre H, Abeler VM, et al. Large scale genomic instability as an additive prognostic marker in early prostate cancer. Cell Oncol. 2009;31:251–9. doi: 10.3233/CLO-2009-0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson GR, Brenner BM, Swede H, et al. Intrachromosomal genomic instability in human sporadic colorectal cancer measured by genome-wide allelotyping and inter-(simple sequence repeat) PCR. Cancer Res. 2001;61:8274–83. [PubMed] [Google Scholar]
- 14.Jones BA, Christensen AR, Wise JP, Sr, Yu H. Glutathione S-transferase polymorphisms and survival in African-American and white colorectal cancer patients. Cancer Epidemiol. 2009;33:249–56. doi: 10.1016/j.canep.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 15.Basik M, Stoler DL, Kontzoglou KC, Rodriguez-Bigas MA, Petrelli NJ, Anderson GR. Genomic instability in sporadic colorectal cancer quantitated by inter-(simple sequence repeat) PCR analysis. Genes Chromosomes Cancer. 1997;18:19–29. [PubMed] [Google Scholar]
- 16.Laiho P, Launonen V, Lahermo P, et al. Low-level microsatellite instability in most colorectal carcinomas. Cancer Res. 2002;62:1166–70. [PubMed] [Google Scholar]
- 17.Markowitz S, Wang J, Myeroff L, et al. Inactivation of the type II TGF-β receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1276–7. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 18.Vikki S, Launonen V, Karhu A, Sistonen P, Vastrik I, Aaltonen LA. Screening for microsatellite instability target genes in colorectal cancers. J Med Genet. 2002;39:785–9. doi: 10.1136/jmg.39.11.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valeri N, Gasparini P, Fabbri M, et al. Modulation of mismatch repair and genomic stability by miR-155. Proc Natl Acad Sci USA. 2010;107:6982–7. doi: 10.1073/pnas.1002472107. [DOI] [PMC free article] [PubMed] [Google Scholar]