Abstract
For locomotion, vertebrate animals use the force generated by contractile skeletal muscles. These muscles form an actin/myosin-based bio-machinery that is attached to skeletal elements to effect body movement and maintain posture. The mechanics, physiology, and homeostasis of skeletal muscles in normal and disease states are of significant clinical interest. How muscles originate from progenitors during embryogenesis has attracted considerable attention from developmental biologists. How skeletal muscles regenerate and repair themselves after injury by the use of stem cells is an important process to maintain muscle homeostasis throughout lifetime. In recent years, much progress has been made towards uncovering the origins of myogenic progenitors and stem cells as well as the regulation of these cells during development and regeneration.
Introduction
In humans, more than 600 skeletal muscle groups are anatomically defined. Despite their complexity in shape and function, each muscle group is made up of hundreds to thousands of fundamental structural units called myofibers. The myofiber is unique in its constitution as it is a multi-nucleated syncytium containing tens to hundreds of nuclei resulting from cellular fusion of differentiated single muscle cells, the myocytes. Progenitors that give rise to these differentiated myocytes are a subject of this review. Stem cells that repair damaged myofibers or regenerate new myofibers after trauma in the adult are also evaluated. In particular, we contrast similarities and differences of cellular and molecular events that orchestrate muscle development and regeneration.
I. Cell origin and lineage of myogenic progenitors and stem cells
The embryonic origin of skeletal muscles and their progenitors
The entire trunk and limb skeletal muscles arise from a transient embryonic mesodermal structure called the somite (Fig. 1). Somites are segmented mesodermal units flanking both sides of the spinal cord that were first visualized by Marcello Malpighi in the chick embryo1. It is therefore fitting that chick embryos have been a primary experimental system for investigating skeletal muscle development since the 1970s2. In particular, chick-quail chimera experiments3, in which surgically combined host and donor cells can be distinguished by nucleolar morphology or quail-specific antigen, were performed to demonstrate a somitic origin of the limb musculature4,5. The dorsal epithelial portion of the somite, the dermomyotome, contains the myogenic progenitors6. Furthermore, limb and ventral body wall muscles only come from the ‘lateral’ half of the somite, while the dorsal axial muscles derive from the ‘medial’ half7. Focal labeling of somitic cells with fluorescent dyes was also used to evaluate the morphology of emerging myogenic cells8,9. Live imaging of such labeled cells revealed that cells near the medial and lateral borders (or ‘lips’) of the dermomyotome, represent the primary wave of myogenic cells 10. The myogenic contribution of the central portion of the dermomyotome was not addressed in these studies.
Figure 1. Developmental progression of myogenesis and myogenic gene expression.
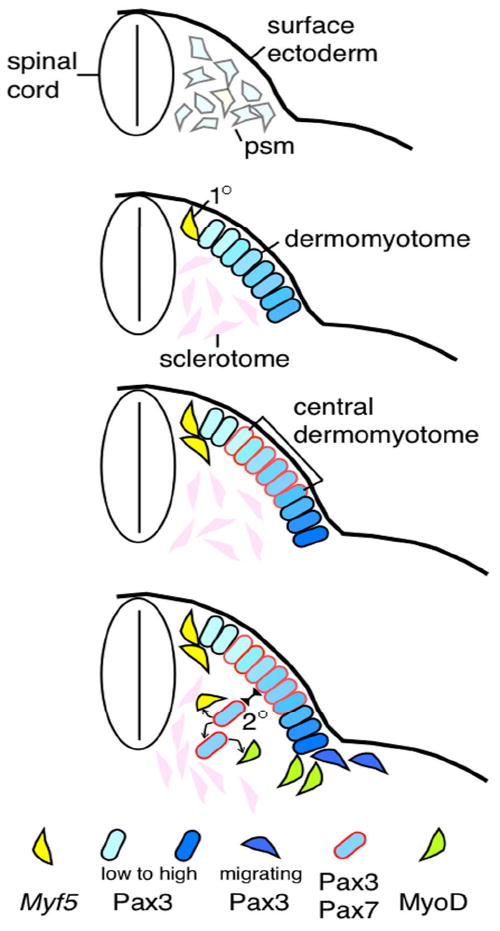
Top panel: presomitic mesoderm cells express Pax3 (pale blue) and low levels of Myf5 (pale yellow) transcripts. Epithelial somite stage is omitted. Second panel: appearance of sclerotome, dermomyotome (Pax3+), and primary (1°) myogenic cells (Myf5+) at the dorsal medial edge. Pax3 expression is at a higher level at the lateral edge. Third panel: Pax7 expression emerges and overlaps with Pax3+ cells in the central dermomyotome in more mature somites. Bottom panel: Vertical division of the central dermomyotomal cells, which give rise to the secondary (2°) myogenic progenitors. These cells are presumed to give rise to more progenitors and Myf5+ or MyoD+ myogenic cells. The lateral Pax3+ cells give rise to migrating myoblasts entering the ventral body wall and limbs. Keys to cells with specific gene expression are at the bottom.
En1 is specifically expressed in the central dermomyotome of the mouse. Using loxP-recombination-based LacZ reporter expression for cell marking/tracing via En1 gene-directed Cre (En1-Cre) activity, Atit et al.11 showed that marked central dermomyotome cells (at embryonic day 9.5; E9.5) become a triangular domain underneath the dermomyotome a day later (at E10.5). Descendant cells become incorporated not only into dorsal trunk muscles, but also interscapular brown fat and dorsal dermis at E16.511 (Table 1). A chick-quail chimera study has long since determined that embryonic muscles and their progenitors have the same somitic origin12. A recent revisit of this issue agrees with trunk muscle and dermis fates deriving from central dermomyotome cells13, though chick has no brown fat for comparison. Gros et al.13 further showed by live imaging that GFP-expressing central dermomyotome cells divided vertically (relative to the epithelial plane) with one daughter cell moving inward, explaining the triangular domain observed in En1-Cre marked somites. Using reporter gene knock-in alleles of two dermomyotome-expressing genes, Pax3 and Pax7 (encoding related transcription factors), Relaix et al.14 concluded that the vertically dividing cells were indeed Pax3+Pax7+ central dermomyotome cells that give rise to a new population of inner cells. As Pax3;Pax7 double mutants failed to generate additional myogenic cells after the primary wave of myogenesis, Pax3+Pax7+ cells represent the secondary progenitors for continuous expansion of muscle mass (Fig. 1).
Table 1.
| Knock-in allele | Cell types labeled | References | Comments |
|---|---|---|---|
|
| |||
| Pax3-Cre | Endothelium | Hutchenson et al. | possible Cre activity in presomitic mesoderm |
| M-Cre | Trunk and limb muscle | Shienda et al. | |
| Limb satellite cells | |||
| Side population cells | |||
|
| |||
| En1-Cre | Trunk Muscle | Atit et al. | no limb muscle labeled |
| En1-Cre-ERT | Dorsal dermis | ||
| Brown fat | |||
|
| |||
| Pax7-Cre-ERT2 | Trunk Muscle | Lepper and Fan Keller et al. | cell types labeled dependent on timing of tamoxifen administration |
| Pax7-IRES-Cre-ERT | Limb muscle | ||
| Dorsal Dermis | |||
| Brown fat | |||
| Fetal muscle progenitors | |||
| Adult satellite cells | |||
| Adult muscles | |||
|
| |||
| Myf5-Cre | Trunk muscle | Seale et al. | Cre activty in presomitic mesoderm *also as committed muscle progenitors |
| Limb muscle | Kuang et al. | ||
| Brown fat | Gensch et al. | ||
| Cartilage | |||
| Endothelium | |||
| Adult satellite cells* | |||
|
| |||
| MyoD-Cre | Trunk muscle | Kanisicak et al. | only myogenic cells were characterized |
| Limb muscle | |||
| Perinatal satellite cells | |||
|
| |||
| Myogenin-Cre | Trunk Muscle | Gensch et al. | only muscles are labeled |
| Limb muscle | |||
Central dermomytome cells do not contribute to ventral body wall or limb muscles. These two populations originate from the lateral half of the somite7, presumably the lateral dermomyotome. This region expresses high levels of Pax3 and mice mutant for Pax3 alone lack these muscles15. Because Pax3 is also expressed in the presomitic mesoderm16,17, Pax3-Cre-mediated lineage tracing precludes assignment of limb and ventral body wall muscle progenitors specifically to the lateral dermoymotome18 (Table 1). A transgenic line called M-Cre was used to help define the lateral dermomyotome as a source of limb muscle progenitors18. However, constitutive Cre mediated lineage-tracing marks all cells expressing Cre ‘at any one time’ prior to the assay time point, thus negating temporal specificity. As a gene often possesses a dynamic expression pattern, analysis of constitutive Cre-based lineage tracing must include all expression patterns prior to the assay time point for accurate interpretation.
The tamoxifen inducible forms of Cre, the Cre-ER fusion and its successive improved versions Cre-ERT and Cre-ERT2, offer an opportunity for temporally controlled cell marking19. Using a Pax7-Cre-ERT2 allele for inducible lineage tracing, it was found that Pax7-expressing cells marked at different time points contribute to different sets of cell fates and muscle groups20 (Table 1). When central dermomyotome cells were marked at E9.5, they contribute to the three fates revealed by the En1-Cre and En1-Cre-ERT study11. Pax7+ cells marked at E10.5 contribute to ventral and proximal forelimb muscles, and brown fat, but less so to dermis. E11.5 marked cells do not contribute to dermis, but they can be traced to distal fore- and hind-limb muscles and some brown fat. By E12.5, Pax7-descendant cells become restricted to the myogenic lineage and are found even in the most distal limb muscles. How three fates become segregated from a simple epithelium (the dermomyotome) and how Pax7+ cells become fate restricted to the myogenic lineage are fundamental cell fate determination issues that have yet to be experimentally explored.
Myogenic specification and differentiation
Myogenic differentiation has been extensively studied and reviewed21,22. Following is a brief summary. Cloning of MyoD changed the landscape of the myogenic field23. Forced expression of this transcription factor can convert various cultured cell types to the myogenic fate, earning its reputation as the master regulator of myogenesis. MyoD has three related family members, Myf5, Myf6 (also called Mrf4), and Myogenin24. Their expression is largely restricted to the myogenic lineage. MyoD and Myf5 expression prefigures the differentiated myocytes and defines the myogenic domain (Fig. 1). Myogenin is turned on in the myocytes prior to their fusion into myofibers. Myf6 expression is the last to be detected. Myf5 mutants lack the initial myotome, but this defect is compensated and mutants eventually develop with negligible muscle defects25. Single mutants for MyoD or Myf6 are also viable with no severe muscle defects at birth26,27. However, Myf5/Myf6/MyoD triple mutants have drastically reduced embryonic muscles28. Myogenin mutants have no defects in myogenic specification and differentiation, but a defect in myocyte fusion in the trunk and limbs29. MyoD;Myf6 double mutants are also defective in myocyte fusion30. Importantly, MyoD;Myf6;Myogenin triple mutants are devoid of any myocyte fusion, despite normal Myf5 expression30. It therefore appears that Myf5 acts primarily at the early step of specification and Myogenin at the late step of myocyte fusion, while MyoD and/or Myf6 can participate in both steps in a semi-redundant fashion. These four genes’ function and regulation are complex and not necessarily agreed upon by all investigators in the field. Nevertheless, embryonic myogenesis involves intricate interplays between these myogenic regulatory factors (MRFs).
Surprisingly, Myf5-Cre marked cells contribute to brown fat31, in addition to muscles and committed myogenic progenitors32, suggesting that its expression does not commit cells to myogenic differentiation (Table 1). However, the Myf5-Cre knock-in construct used a constitutive Cre and an exogenous polyA signal, which precludes endogenous 3’UTR usage and miRNA regulation. It is therefore not completely clear whether there was ‘aberrant’ or ‘unintended’ expression of Cre in an earlier progenitor stage. For example, Myf5-Cre is expressed in the presomitic mesoderm, where Myf5 protein is not detected33. Therefore, not surprisingly, Myf5-Cre also directs lineage labeling in cartilage and dermis in this study. If a subpopulation of Myf5+ cells is indeed multi-potential, it will be important to determine their temporal and spatial distribution. By contrast, MyoD-Cre34 and Myogenin-Cre33 have thus far only been reported to mark the myogenic lineage (Table 1).
Muscle progenitors in the peri-natal period
The Pax7+ cell source represents a major contributor to myofibers during the first 3 weeks of birth based on temporally controlled labeling and tracing of hind limb muscles using the Pax7-Cre-ERT2 allele35. In other muscle groups, notably most trunk and the diaphragm muscles, myogenic progenitors also express Pax336. Whether all Pax7+ myogenic progenitor cells detected in the perinatal period are descendants of embryonic Pax7+ cells has not been conclusively determined as tamoxifen-induced labeling efficiency in the embryo is low20. There is a recent report that peri-natal Pax7+ cells can arise from PW1+Pax7- muscle interstitial cells37. Using CD34 and Sca1 surface antigens for fluorescent-activated cell sorting (FACS), the investigators described ~56% of the CD34+Sca1high fraction expresses PW1 but not Pax7, and these cells can generate muscles and Pax7+ cells after transplantation into adult animals. Because these cells never express Pax3 during embryogenesis (not marked in Pax3-Cre mice), they are presumably not of somitic origin. However, at least some embryonically and perinatally marked Pax7+ cells can directly contribute to adult muscle stem cells20,35. The relative contributions of embryonic versus peri-natal de novo Pax7+ cells to muscle fiber formation and adult muscle stem cells are currently unclear.
Although the Pax7 mutant has no discernable embryonic muscle defect (unless combined with the Pax3 mutation), Pax7 is essential for perinatal myogenesis. Pax7 mutants are born with a normal number of cells actively transcribing the Pax7 locus in hind limb muscles (scored by knock-in reporter expression or by Pax7-Cre-ERT2-mediated reporter expression35). This number declines in ensuing weeks; consequently, surviving mutant adults have barely any satellite cell and are extremely compromised in injury induced muscle regeneration38,39. Mitotic defect38, cell death40, and reduced myogenic potential39 have been implicated in the loss of satellite cells. Inducible lineage tracing of mutant cells showed that they do incorporate into myofibers more readily and over a longer period relative to control cells35. Thus, Pax7 likely maintains the progenitor pool size by counter-balancing the tendency to differentiate. How Pax7 acts, how its expression is regulated, and how the number of Pax7+ cells is controlled during this period are critical questions pertaining to the establishment of a normal pool of muscle stem cells for use throughout the active life of a vertebrate animal.
As Pax7+ cells continuously incorporate into myofibers during the perinatal period, one predicts that their differentiation depends on the MyoD gene family. However, as mentioned above, Myf5, MyoD, and Myf6 single mutants can develop to adulthood with relatively normal skeletal muscles, presumably due to redundancy between these genes. Intriguingly, MyoD-Cre was shown to direct lineage labeling of perinatal Pax7+ muscle progentiors at a high rate34. Myf5-Cre also directs marking of postnatal Pax7+ cells32. Since neither protein has been detected in muscle stem cells, these knock-in Cre genes must be transcribed in muscle progenitor/stem cells sometime in the history of their making. Whether these two genes play a redundant role in this process is unknown. Systematic combinations of conditional gene inactivation among Myf5, MyoD, Mrf4, and Myogenin during the peri-natal period will help to define their roles, relative to those defined in embryogenesis by the respective germ line mutants.
Adult myogenic stem cells in injury-induced skeletal muscle regeneration
In adulthood, skeletal muscle possesses a tremendous capacity to repair itself after extensive injury, which is exemplified by full tissue restoration after mincing of an entire skeletal muscle41. The tissue’s regenerative potential is maintained over up to 50 toxin-induced injury/regeneration cycles42. Such regenerative capacity has been largely attributed to muscle stem cells endogenous to the skeletal muscle. One such cell type is the satellite cell, which was first identified by Alexander Mauro exactly fifty years ago using transmission electron microscopy (TEM) performed on frog tibialis anticus (i.e. tibialis anterior) muscle43. These cells are named after their physical location: attached to the sarcolemma and beneath the basal lamina ‘orbiting’ the muscle fiber. In the same year, in vitro culture experiments with myoblasts obtained by enzymatic digestion of skeletal muscle tissue established that mononucleated muscle progenitor cells could give rise to multi-nucleated myofibers via fusion44,45. Whether in vitro isolated myoblasts corresponded to TEM-defined satellite cells in vivo was not certain at the time. Years later, elegant transplantation experiments of muscles, in which satellite cell nuclei pulse-labeled with tritiated thymidine were chased into myonuclei, demonstrated that satellite cells could proliferate and potentially differentiate into muscle fibers after transplantation-induced injury46. Further progress in the satellite cell field was impeded in large part due to a lack of satellite cell specific molecular markers and consequently, satellite cell research was limited to laborious ultra-anatomical studies by TEM.
Identification of Pax7 being specifically expressed in skeletal myofiber associated cells that resemble satellite cells has greatly accelerated research in the field47. An EM study of the frog leg muscle has detected Pax7 immuno-reactivity in the satellite cell48. Similarly, we found Pax7 immuno-reactivity and β-gal enzymatic activity driven by Pax7-Cre-ERT2;LacZ reporter in satellite cells of mouse tibialis anterior muscles by TEM (Fig. 2). These findings provided evidence that Pax7+ cells are bona fide satellite cells as described by Alexander Mauro. However, Pax7 is not the only marker used for identifying satellite cells. For example, the Myf5nLacZ knock-in allele directs β-gal activity in a large proportion of satellite cells. Transplantation of single muscle fibers along with their myofiber-associated cells from Myf5+/nLacZ mice into immuno-compromised and irradiated dystrophic muscles gave rise to hundreds of genetically marked myonuclei and satellite cells49. Montarras et al.36 obtained similar results via transplantation of Pax3+ cells isolated by FACS on the basis of fluorescent reporter gene expression from the Pax3 locus. More recently, it has become possible to isolate adult muscle progenitor cells via FACS using a combination of positive and negative selections of cell surface markers32,50,51. When transplanted in bulk into dystrophic muscles of immune-deficient mice, these various populations of cells proficiently contributed myonuclei to myofibers, as well as satellite cells to the host muscle fiber niches. Importantly, Sacco et al.50 found all cells sorted by CD45-Sca1-CD34+β1integrin+ criteria positive for Pax7 by single cell RT-PCR assay, and transplantation of such single cells results in the generation of new muscles and stem cells, fulfilling the ‘gold standard’ for demonstrating their stemness52. It remains to be determined whether all or only some in vitro defined transplantation competent muscle stem cell populations contain Pax7+ cells.
Figure 2. Pax7 expression is detected in satellite cells of the mouse tibialis anterior muscle.
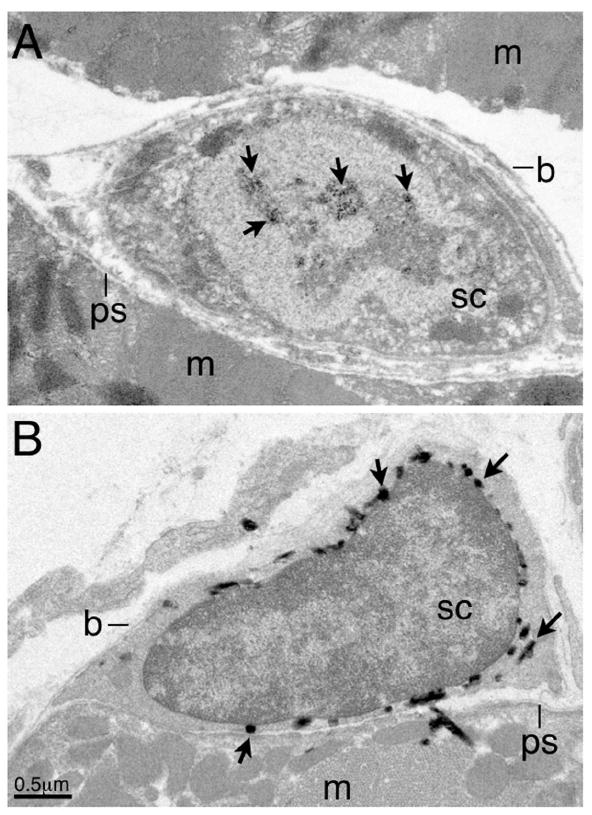
(A) Immuno-EM detection of endogenous Pax7 by a monoclonal antibody (DSHB), followed by a HRP-conjugated goat anti-mouse IgG1 antibody (Molecular Probes) and enzymatic reaction using the DAB substrate (Vecta Lab). The sample was fixed in Zamboni’s fixative following the protocol in Chen et al. (2007). (B) TEM of samples from tamoxifen-treated Pax7-Cre-ERT2;LacZ reporter mice, fixed in 4% paraformaldehyde, reacted with X-gal substrate, following the procedure in Kanisick et al. (2009). Abbreviations: m, muscle fiber, b, basement membrane; ps, plasmalemmal surface; sc, satellite cell. Arrows in (A) indicate reacted DAB deposits in the nucleus, in (B), reacted X-gal precipitates in the cytoplasm. Some X-gal precipitates are often seen next to satellite cells, likely due to substrate diffusion during enzymatic reaction. Scale bar in (B).
The above evidence for various sources of muscle stem cells is based upon in vitro isolation followed by transplantation, a long time tradition for studying stem cells. Using the Pax7-Cre-ERT2 mice for adult-specific cell marking and lineage tracing, Lepper et al.35 provided direct in vivo evidence for Pax7+ satellite cells being the major source of muscle stem cells: after toxin-induced injury to the tibialis anterior muscle, lineage-labeled Pax7+ descendant cells robustly contributed to all regenerative myofibers and self-renewed to replenish the pool of quiescent satellite cells35. While this unequivocally demonstrates that Pax7+ cells are a major source of muscle stem cells in acute injury induced muscle regeneration, it will be important to determine the role of these cells in other physiological contexts, including long-term skeletal muscle tissue maintenance, exercise-induced muscle growth, and muscle wasting diseases. Moreover, a critical question concerns whether Pax7+ cells are the exclusive muscle stem cell source. As new myofibers form as a syncytium by fusion of multiple myocytes, lineage marking of the Pax7+ cell population alone cannot be used to exclude the input from other cell sources within the same myofiber due to diffusion of the lineage tracer. Indeed, numerous transplantation-based studies have reported several different Pax7- cell types contributing to both myofibers as well as the satellite cell compartment53. To determine the physiological relevance of these clinically relevant cells for curing muscle degenerative diseases, it will be important to ascertain the role of them via direct lineage-tracing and/or genetic cell ablation of Pax7+ satellite cells to reveal if there is any remaining capacity for adult myogenesis.
Quite unexpectedly, tamoxifen-induced adult-specific inactivation of Pax7 alone (via Pax7-Cre-ERT2 allele35) or Pax3 and Pax7 together (via a widely expressed Cre-ERT allele54) does not cause an obvious defect in injury-induced muscle regeneration. The conditional mutant Pax7 cell is not only able to regenerate myofibers effectively, but also proliferate, re-occupy the architectural satellite cell niche, and support an additional round of muscle regeneration. One could not help asking what is the function of Pax7 in adult satellite cells? Whether other transcription factors compensate for or replace its role in adult satellite cell function is an open question. Along the same line of comparing gene function at different stages of myogenesis, the MyoD mutant develops with embryonic muscles relatively normally, but in adulthood muscle regeneration is compromised due to defects in myoblast proliferation and differentiation55,56. Myf5 mutants also develop muscles normally, but are defective in muscle regeneration due to compromised myoblast proliferation57,58. Conversely, while Myogenin is required for embryonic myocyte fusion, this function of Myogenin is dispensable post-natally59. Instead Myogenin controls a distinct transcriptional program59, in particular the response to denervation-induced muscle atrophy60. The significance of such temporal switching of myogenic gene functions may lie in the fundamental differences between development and regeneration.
II. Signaling during myogenesis
Myogenic induction by Wnt proteins
In the embryo, the spinal cord and surface ectoderm surrounding the somite are necessary and sufficient for inducing myogenesis17,61-63. Several Wnt genes are expressed in these two tissues. Co-cultures of presomitic cells and cells producing selective Wnt proteins induce expression of Pax3 and Pax764, as well as Myf5 and MyoD65,66. In accordance, Wnt1;Wnt3a (both normally expressed in the dorsal spinal cord) double mutant embryos have reduced expression of Pax3 and Myf5 in the medial dermomyotome67. The myogenic inducing potential of Wnt genes expressed by the surface ectoderm has not been tested genetically. In vitro pharmacological studies suggest that Wnts activate the Gα - adenylyl cyclase - protein kinase A signaling cascade to phosphorylate the transcription factor CREB, which in turn activates myogenic genes. Consistently, both germ line inactivation of Creb and dominant negative Creb over-expression in the somite lead to severely reduced expression of Pax3, Myf5, and MyoD in vivo66. In addition, the PKC pathway, which can be activated via Gα signaling, has also been implicated in myogenic induction by modulating Pax3 activity and MyoD expression68.
The canonical Wnt/β-catenin pathway also plays a role in myogenesis. Inactivation of β-catenin in presomitic mesoderm cells leads to disorganized somites due to segmentation defects69, disallowing a firm conclusion about its role in the primary wave of myogenesis70. En1-Cre directed β-catenin inactivation in the central dermomyotome leads to an increase in the domain of myogenic cells at the expense of dermal cells, indicating an inhibitory role of β-catenin during the secondary wave of myogenesis11. By contrast, β-catenin inactivation by Pax7-Cre caused only a modest defect in muscle architecture, represented by a change in muscle fiber sub-type distribution70. Since both Pax7 and En1 are expressed in the central dermoymtome, the discrepancies likely reflect different timing of expression and/or efficiency of Cre between the two alleles. Analysis of the role of β-catenin via gene inactivation is further complicated by an accompanying cell adhesion defect in mutant cells. Inactivation of BCL9, which regulates β-catenin’s nuclear activity but not its adhesion function, should in principle eliminate this issue. Yet, conditional inactivation of BCL9 by Myf5-Cre, causes no developmental defects71. The timing of Myf5-Cre activity and BCL9 protein perdurance may obscure a potential role of the canonical pathway in embryonic myogenesis.
The role of Wnt signaling in adult regenerative myogenesis is multifaceted (Fig. 3). A selective combination of Wnt5a, 5b, 7a, and 7b can convert muscle resident CD45+Sca1+ cells (i.e. the side-population cells) to express Pax772, suggesting that CD45+Sca1+ cells are a source of de novo Pax7+ stem cells for muscle regeneration. Although these Wnt genes were originally reported to be up-regulated during regeneration72, subsequent re-evaluation did not substantiate such claim73. Thus, the physiological significance of the side-population cells and whether their conversion to Pax7+ cells occurs in vivo are not yet known. On the other hand, FGF6 and FGFR4 are expressed during the early phase of the regenerative response73,74, and both Fgf6 and Fgfr4 mutants display compromised muscle regeneration74,75, supporting the importance of FGF signaling pathway for proficient muscle repair.
Figure 3. Gene expression and signaling regulation during adult muscle regeneration.
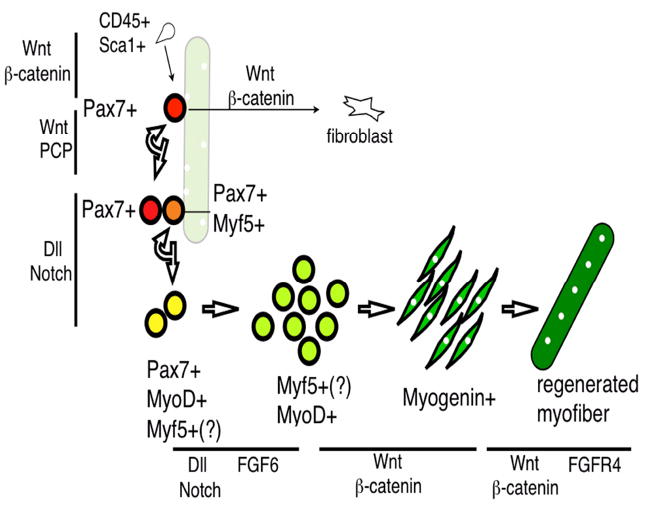
CD34+Sca1+ side population cells can be converted into Pax7+ cells via Wnt/β-catenin signaling. Pax7+ satellite cells (red) are associated with the myofiber (pale green) with peripheral myonuclei (white). Upon injury, Pax7+ cells either go through symmetric division for self-renewal via the Wnt/PCP pathway or asymmetric division to give rise to committed progenitors (Pax7+Myf5+, orange) involving Dll/Notch signaling. These cells expand (as transit amplifying cells) and express MyoD (yellow). Myf5 is presumed (Myf5 (?)) to be expressed in transit amplifying cells and Pax7-MyoD+ myoblasts (yellowish green). Myogenin (green) is turned on in differentiated myocytes, which eventually fuse to form the new myofiber (dark green), which has the characteristic of centrally located myonuclei. Wnt/β-catenin can also convert Pax7+ cells into fibroblasts. There are at least two steps of muscle regeneration involving FGF signaling: FGF6 affects MyoD+ cells, while FGF4R4 affects myofiber number and size. Gene expression for each stage is confined to those covered in the text and does not represent a full list. Wnt, FGF, and Notch pathways that regulate specific steps of myogenic progression are represented by black lines next to the defined steps.
For symmetric division of Pax7+ muscle stem cells, Wnt7a has been shown to employ the planar cell polarity (PCP) pathway: Wnt7a gene delivery into muscles via electroporation causes increased muscle stem cell number, and consequently muscle hypertrophy, and the PCP component Vangl-2 is a likely effector mediating this function76. Although β-catenin nuclear localization has been documented in cultured proliferating myoblasts, it may act in the transit amplifying progenitors rather than in the self-renewing stem cells77. β-catenin nuclear localization is also detected in differentiating myogenic cells71. Importantly, BCL9 inactivation or pharmacological intervention of β-catenin activity in vivo caused delayed myogenic differentiation but did not disturb progenitor proliferation during regeneration, evidence that the canonical pathway participates in the differentiation step71,78. Wnt3a has also been shown to employ the canonical pathway to accelerate myocyte fusion using the in vitro C2C12 myoblast model79. As myocyte fusion immediately follows differentiation, separating Wnt/β-catenin functions in these two steps in unsynchronized populations of cells may prove to be difficult.
Lastly, Wnt/β-catenin pathway activation also induces muscle fibrosis and thus, is implicated in the decline of muscle function in aging. Gene delivery of Wnt3a via intramuscular eletroporation leads to fibrotic accumulation76. In vitro, recombinant Wnt3a causes direct conversion of myoblasts into fibroblasts78. Importantly, a β-catenin transcriptional reporter is activated in aging but not in young muscles after injury, and depletion of Wnt proteins via soluble Frz receptors can rejuvenize aged myoblasts to prevent fibrogenesis71. While Wnt acts locally to divert aging muscle stem cells to the fibrogenic fate, TGFβ1 has been reported to be a candidate systemic factor that may attenuate muscle stem cell activation80,81 as serum levels of TGFβ1 increase in aging humans and mice82. Most of the above cited effects of Wnt and TGFβ1 in adult and aging muscle regeneration are primarily based on in vitro culture or in vivo overexpression, pharmacology and RNAi studies; the genetic evidence remains to be obtained.
Maintenance of myogenic progenitors and stem cells by Notch signaling
The Notch-Delta receptor-ligand mediated signaling pathway has been shown to be instrumental in the regulation of various progenitor/stem cells and their differentiated daughters, including the myogenic progenitor and their differentiating progeny. Delta like 1 (Dll-1) mutant embryos initially have a normal number of Pax3+ and Pax7+ cells; over time, these cells become diminished83. Similarly, somitic conditional inactivation of RBP-Jκ, a transcriptional mediator of Notch signaling, also leads to a gradual depletion of Pax7+ cells84. Consequently, by mid- to late embryogenesis, both Dll-1 and conditional RBP-Jκ mutants have much reduced musculature. These data reinforce the positive role of Notch signaling in controlling the myogenic progenitor pool size.
One prominent negative regulator of Notch signaling is Numb. Asymmetric distribution of Numb during cell division helps determine distinct developmental paths of daughter cells by affecting differential Notch activities85. Curiously, forced expression of a Numb-GFP fusion protein in mouse embryos86 resulted in an increased progenitor pool, seemingly contradicting the positive role for Notch signaling in progenitor maintenance. Because this particular Numb-GFP transgene did not cause down-regulation of Notch reporter expression, it is still unresolved how Numb acts in regulating embryonic muscle progenitor cell number. When Numb protein distribution was followed by in vitro live imaging of post-natal myoblasts, either isolated in bulk87 or from dissociated myofibers88, it was found to be distributed both asymmetrically and symmetrically during cell division. The proportion of asymmetrically segregated Numb in daughter cells declined over successive divisions, possibly reflecting the absence of in vivo regulators in culture. While Shinin et al.88 proposed that Numb co-segregates with the progenitor, Conboy et al.87 suggested that Numb associates with the differentiating daughter cell. As of yet, the precise role of Numb in the myogenic lineage is not resolved.
Without examining Numb localization, symmetric and asymmetric satellite cell divisions can be observed in cultured myofibers and their associated satellite cells isolated from Myf5-Cre;YFP reporter mice32. In this experimental paradigm, YFP was used as an indicator for a sub-population of satellite cells at one point in time expressing Myf5 (Pax7+YFP+), relative to those expressing only Pax7 (Pax7+YFP-)32. Two types of cell division were found. Type 1, planar symmetric cell division (along the plane of the myofiber) resulted in the majority of daughter cells either both maintaining the Pax7+YFP- stem cell fate or both displaying Myf5 expression (Pax7+YFP+). Type 2, asymmetric division along the apical-basal plane mostly gave rise to cells with different fates: the cell that divided away from the basal lamina gained Myf5 expression (Pax7+YFP+), while the basal sister stayed in the satellite cell compartment and did not express Myf5 (Pax7+YFP-). Furthermore, the cell that remained basally displayed elevated levels of Notch3 while the apical cell expressed the Dll-1, suggesting that activation of Notch3 by Dll-1 positively selects the basal cells to maintain the stem cell fate. However, in response to repetitive muscle injury, Notch3-deficient mice regenerated better and had more satellite cells than control animals, suggesting that Notch3 is instead a negative regulator of the muscle stem cell pool89. By contrast, over-expression of an intracellular domain of Notch1 (NICD, an activated form of Notch1) increased proliferation of cultured myoblasts87, supporting its positive role in stem cell expansion. It is interesting to note that while Notch1 is expressed in nearly all satellite cells, Notch3 is expressed only in a subset. One may therefore speculate that the Notch pathway serves to regulate the number of satellite cells through the balanced activity between Notch1 (positive) and Notch3 (negative). Whether different Delta like ligands are involved in mediating differential Notch activities is yet to be explored.
Although Dll-1 expression is up-regulated in injured muscles of young adults87, aging muscles lose the ability to activate Dll-1 after injury and are compromised in the regenerative process80. Pharmacological inhibition of Notch signaling can indeed block muscle regeneration in young adult mice, while forced activation of Notch signaling is sufficient to restore efficient muscle regeneration in old mice80. These results provide strong support for the continuous participation of Notch signaling in the maintenance of the skeletal muscle stem cell activity throughout lifetime. For studying the aging process, gene over-expression or functional blocking reagents, rather than genetic means, are often used. As such, the nuances of differential roles between gene family members may be overlooked. Restricting knockouts of each Delta and Notch gene to the muscle progenitors versus their surrounding cells will help to delineate the specific role of each gene member.
III. Regeneration recapitulates development?
Studies of embryonic muscle development and the muscle regeneration have primarily been two distinct disciplines. It has been largely by inferential extensions that these processes were compared. From the above descriptions, it should be clear that in many cases, the molecular mechanisms that control adult muscle regeneration do not recapitulate those that control embryonic myogenesis. During skeletal muscle progenitor and stem cell expansion and maintenance, there is a progressive change of genetic requirements for Pax3 and Pax7, to Pax7 only, to neither, from embryo, to neonate, to adult, respectively. For muscle differentiation, there is also a temporal shift of genetic contributions among the MRF members, for example, the roles of MyoD and Myf5 during embryonic muscle development versus adult muscle regeneration reviewed above. The most striking shift of function is exemplified by Myogenin, which is required for fusion of embryonic myocytes but dispensable for fusion of regenerating adult myocytes90. Lastly, the Wnt/β-catenin pathway also appears to act differently during embryonic myogenesis, adult muscle regeneration, and aging muscle fibrosis. The pathway that is thus far continuously required for maintaining embryonic muscle progenitors and adult muscle stem cells is Notch signaling. Yet, Notch3 is only required for maintaining muscle stem cell pool size after repeated injuries, but not for regulating the embryonic myogenic progenitor pool. There are many other signaling and transcriptional regulators of muscle development and regeneration that have only been examined in one context but not in the other, but they are not covered here due to space. A comprehensive evaluation of these regulators in both contexts will help to conceptualize the differences between these two processes. Because embryonic development relies on active progenitors cycling continuously but adult muscle stem cells are relatively quiescent in the absence of injury, these two processes are fundamentally distinct. In particular, the muscle regenerative process involves stem cell activation and a return to a homeostatic state in a chaotic inflammatory environment not seen during embryogenesis. Therefore, it is only reasonable that these adult stem cells utilize different strategies from those employed by embryonic progenitors to adapt to unpredictable circumstances such as injury-induced regeneration.
Acknowledgments
We thank Mike Sepanski for help with electron microscopy of the satellite cells. C.-F. Fan and C. Lepper are supported by an NIH grant (AR060042). L. Li and M. E. Rozo are supported by Carnegie Endowment.
References
- 1.Malpighi M. 1672 [Google Scholar]
- 2.Sanders EJ, Lash JW, Ordahl CP. Origin and Fate of Somites. IOS Press; 2001. [Google Scholar]
- 3.Le Douarin N, Barq G. Sur l’utilisation des cellules de la caille japonaise comme ‘marqueurs biologiques’en embryologie expérimentale. C r hebd Séanc Acad Sei Paris D. 1969;269:1543–1546. [PubMed] [Google Scholar]
- 4.Chevallier A, Kieny M, Mauger A. Limb-somite relationship:origin of the limb musculature. Embryol exp Morph. 1977;41:245–258. [PubMed] [Google Scholar]
- 5.Christ B, Jacob HJ, Jacob M. Experimental analysis of the origin of the wing musculature in avian embryos. Anat Embryol. 1977;150:171–186. doi: 10.1007/BF00316649. [DOI] [PubMed] [Google Scholar]
- 6.Christ B, Brand-Saberi B, Grim M, Wilting J. Local signalling in dermomyotomal cell type specification. Anat Embryol. 1992;186:505–510. doi: 10.1007/BF00185464. [DOI] [PubMed] [Google Scholar]
- 7.Ordahl CP, Le Douarin NM. Two myogenic lineages within the developing somite. Development (Cambridge, England) 1992;114:339–353. doi: 10.1242/dev.114.2.339. [DOI] [PubMed] [Google Scholar]
- 8.Kaehn K, Jacob HJ, Christ B, Hinrichsen K, Poelmann RE. The onset of myotome formation in the chick. Anatomy and embryology. 1988;177:191–201. doi: 10.1007/BF00321131. [DOI] [PubMed] [Google Scholar]
- 9.Denetclaw WF, Jr, Christ B, Ordahl CP. Location and growth of epaxial myotome precursor cells. Development (Cambridge, England) 1997;124:1601–1610. doi: 10.1242/dev.124.8.1601. [DOI] [PubMed] [Google Scholar]
- 10.Denetclaw WF, Jr, Berdougo E, Venters SJ, Ordahl CP. Morphogenetic cell movements in the middle region of the dermomyotome dorsomedial lip associated with patterning and growth of the primary epaxial myotome. Development (Cambridge, England) 2001;128:1745–1755. doi: 10.1242/dev.128.10.1745. [DOI] [PubMed] [Google Scholar]
- 11.Atit R, et al. Beta-catenin activation is necessary and sufficient to specify the dorsal dermal fate in the mouse. Developmental biology. 2006;296:164–176. doi: 10.1016/j.ydbio.2006.04.449. [DOI] [PubMed] [Google Scholar]
- 12.Armand O, Boutineau AM, Mauger A, Pautou MP, Kieny M. Origin of satellite cells in avian skeletal muscles. Arch Anat Microsc Morphol Exp. 1983;72:163–181. [PubMed] [Google Scholar]
- 13.Gros J, Manceau M, Thome V, Marcelle C. A common somitic origin for embryonic muscle progenitors and satellite cells. Nature. 2005;435:954–958. doi: 10.1038/nature03572. [DOI] [PubMed] [Google Scholar]
- 14.Relaix F, Rocancourt D, Mansouri A, Buckingham M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature. 2005;435:948–953. doi: 10.1038/nature03594. [DOI] [PubMed] [Google Scholar]
- 15.Bober E, Franz T, Arnold HH, Gruss P, Tremblay P. Pax-3 is required for the development of limb muscles: a possible role for the migration of dermomyotomal muscle progenitor cells. Development (Cambridge, England) 1994;120:603–612. doi: 10.1242/dev.120.3.603. [DOI] [PubMed] [Google Scholar]
- 16.Goulding MD, Chalepakis G, Deutsch U, Erselius JR, Gruss P. Pax-3, a novel murine DNA binding protein expressed during early neurogenesis. The EMBO journal. 1991;10:1135–1147. doi: 10.1002/j.1460-2075.1991.tb08054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan CM, Tessier-Lavigne M. Patterning of mammalian somites by surface ectoderm and notochord: evidence for sclerotome induction by a hedgehog homolog. Cell. 1994;79:1175–1186. doi: 10.1016/0092-8674(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 18.Schienda J, et al. Somitic origin of limb muscle satellite and side population cells. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:945–950. doi: 10.1073/pnas.0510164103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feil R, Wagner J, Metzger D, Chambon P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochemical and biophysical research communications. 1997;237:752–757. doi: 10.1006/bbrc.1997.7124. [DOI] [PubMed] [Google Scholar]
- 20.Lepper C, Fan CM. Inducible lineage tracing of Pax7-descendant cells reveals embryonic origin of adult satellite cells. Genesis. 2010;48:424–436. doi: 10.1002/dvg.20630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buckingham M. Skeletal muscle development and the role of the myogenic regulatory factors. Biochem Soc Trans. 1996;24:506–509. doi: 10.1042/bst0240506. [DOI] [PubMed] [Google Scholar]
- 22.Buckingham M, et al. The formation of skeletal muscle: from somite to limb. J Anat. 2003;202:59–68. doi: 10.1046/j.1469-7580.2003.00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lassar AB, Paterson BM, Weintraub H. Transfection of a DNA locus that mediates the conversion of 10T1/2 fibroblasts to myoblasts. Cell. 1986;47:649–656. doi: 10.1016/0092-8674(86)90507-6.0092-8674(86)90507-6 [DOI] [PubMed] [Google Scholar]
- 24.Olson EN. MyoD family: a paradigm for development? Genes & development. 1990;4:1454–1461. doi: 10.1101/gad.4.9.1454. [DOI] [PubMed] [Google Scholar]
- 25.Kaul A, Koster M, Neuhaus H, Braun T. Myf-5 revisited: loss of early myotome formation does not lead to a rib phenotype in homozygous Myf-5 mutant mice. Cell. 2000;102:17–19. doi: 10.1016/s0092-8674(00)00006-4.S0092-8674(00)00006-4 [DOI] [PubMed] [Google Scholar]
- 26.Rudnicki MA, Braun T, Hinuma S, Jaenisch R. Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell. 1992;71:383–390. doi: 10.1016/0092-8674(92)90508-a.0092-8674(92)90508-A [DOI] [PubMed] [Google Scholar]
- 27.Yoon JK, Olson EN, Arnold HH, Wold BJ. Different MRF4 knockout alleles differentially disrupt Myf-5 expression: cis-regulatory interactions at the MRF4/Myf-5 locus. Developmental biology. 1997;188:349–362. doi: 10.1006/dbio.1997.8670.S0012-1606(97)98670-X [DOI] [PubMed] [Google Scholar]
- 28.Kassar-Duchossoy L, et al. Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature. 2004;431:466–471. doi: 10.1038/nature02876.nature02876 [DOI] [PubMed] [Google Scholar]
- 29.Hasty P, et al. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature. 1993;364:501–506. doi: 10.1038/364501a0. [DOI] [PubMed] [Google Scholar]
- 30.Valdez MR, Richardson JA, Klein WH, Olson EN. Failure of Myf5 to support myogenic differentiation without myogenin, MyoD, and MRF4. Developmental biology. 2000;219:287–298. doi: 10.1006/dbio.2000.9621. [DOI] [PubMed] [Google Scholar]
- 31.Seale P, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182.nature07182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuang S, Kuroda K, Le Grand F, Rudnicki MA. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007;129:999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gensch N, Borchardt T, Schneider A, Riethmacher D, Braun T. Different autonomous myogenic cell populations revealed by ablation of Myf5-expressing cells during mouse embryogenesis. Development (Cambridge, England) 2008;135:1597–1604. doi: 10.1242/dev.019331. [DOI] [PubMed] [Google Scholar]
- 34.Kanisicak O, Mendez JJ, Yamamoto S, Yamamoto M, Goldhamer DJ. Progenitors of skeletal muscle satellite cells express the muscle determination gene, MyoD. Developmental biology. 2009;332:131–141. doi: 10.1016/j.ydbio.2009.05.554.S0012-1606(09)00873-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lepper C, Conway SJ, Fan CM. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature. 2009 doi: 10.1038/nature08209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Montarras D, et al. Direct isolation of satellite cells for skeletal muscle regeneration. Science (New York, N Y) 2005;309:2064–2067. doi: 10.1126/science.1114758. [DOI] [PubMed] [Google Scholar]
- 37.Mitchell KJ, et al. Identification and characterization of a non-satellite cell muscle resident progenitor during postnatal development. Nature cell biology. 2010;12:257–266. doi: 10.1038/ncb2025.ncb2025 [DOI] [PubMed] [Google Scholar]
- 38.Kuang S, Charge SB, Seale P, Huh M, Rudnicki MA. Distinct roles for Pax7 and Pax3 in adult regenerative myogenesis. The Journal of cell biology. 2006;172:103–113. doi: 10.1083/jcb.200508001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oustanina S, Hause G, Braun T. Pax7 directs postnatal renewal and propagation of myogenic satellite cells but not their specification. The EMBO journal. 2004;23:3430–3439. doi: 10.1038/sj.emboj.7600346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Relaix F, et al. Pax3 and Pax7 have distinct and overlapping functions in adult muscle progenitor cells. The Journal of cell biology. 2006;172:91–102. doi: 10.1083/jcb.200508044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Studitsky AN. Free Auto- and Homografts of Muscle Tissue in Experiments on Animals. Ann N Y Acad Sci. 1964;120:789–801. doi: 10.1111/j.1749-6632.1964.tb34771.x. [DOI] [PubMed] [Google Scholar]
- 42.Luz MA, Marques MJ, Santo Neto H. Impaired regeneration of dystrophin-deficient muscle fibers is caused by exhaustion of myogenic cells. Braz J Med Biol Res. 2002;35:691–695. doi: 10.1590/s0100-879x2002000600009.S0100-879X2002000600009 [DOI] [PubMed] [Google Scholar]
- 43.Mauro A. Satellite cell of skeletal muscle fibers. The Journal of biophysical and biochemical cytology. 1961;9:493–495. doi: 10.1083/jcb.9.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stockdale FE, Holtzer H. DNA synthesis and myogenesis. Exp Cell Res. 1961;24:508–520. doi: 10.1016/0014-4827(61)90450-5. [DOI] [PubMed] [Google Scholar]
- 45.Cooper WG, Konigsberg IR. Dynamics of myogenesis in vitro. The Anatomical record. 1961;140:195–205. doi: 10.1002/ar.1091400305. [DOI] [PubMed] [Google Scholar]
- 46.Snow MH. An autoradiographic study of satellite cell differentiation into regenerating myotubes following transplantation of muscles in young rats. Cell and tissue research. 1978;186:535–540. doi: 10.1007/BF00224941. [DOI] [PubMed] [Google Scholar]
- 47.Seale P, et al. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102:777–786. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- 48.Chen Y, Lin G, Slack JM. Control of muscle regeneration in the Xenopus tadpole tail by Pax7. Development (Cambridge, England) 2006;133:2303–2313. doi: 10.1242/dev.02397.dev.02397 [DOI] [PubMed] [Google Scholar]
- 49.Collins CA, et al. Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell. 2005;122:289–301. doi: 10.1016/j.cell.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 50.Sacco A, Doyonnas R, Kraft P, Vitorovic S, Blau HM. Self-renewal and expansion of single transplanted muscle stem cells. Nature. 2008 doi: 10.1038/nature07384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cerletti M, et al. Highly efficient, functional engraftment of skeletal muscle stem cells in dystrophic muscles. Cell. 2008;134:37–47. doi: 10.1016/j.cell.2008.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Osawa M, Hanada K, Hamada H, Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science (New York, N Y) 1996;273:242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- 53.Peault B, et al. Stem and progenitor cells in skeletal muscle development, maintenance, and therapy. Mol Ther. 2007;15:867–877. doi: 10.1038/mt.sj.6300145. [DOI] [PubMed] [Google Scholar]
- 54.Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Developmental biology. 2002;244:305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- 55.Megeney LA, Kablar B, Garrett K, Anderson JE, Rudnicki MA. MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes & Dev. 1996;10:1173–1183. doi: 10.1101/gad.10.10.1173. [DOI] [PubMed] [Google Scholar]
- 56.Cornelison DD, Olwin BB, Rudnicki MA, Wold BJ. MyoD(-/-) satellite cells in single-fiber culture are differentiation defective and MRF4 deficient. Developmental biology. 2000;224:122–137. doi: 10.1006/dbio.2000.9682.S0012-1606(00)99682-9 [DOI] [PubMed] [Google Scholar]
- 57.Ustanina S, Carvajal J, Rigby P, Braun T. The myogenic factor Myf5 supports efficient skeletal muscle regeneration by enabling transient myoblast amplification. Stem Cells. 2007;25:2006–2016. doi: 10.1634/stemcells.2006-0736. [DOI] [PubMed] [Google Scholar]
- 58.Gayraud-Morel B, et al. A role for the myogenic determination gene Myf5 in adult regenerative myogenesis. Developmental biology. 2007;312:13–28. doi: 10.1016/j.ydbio.2007.08.059. [DOI] [PubMed] [Google Scholar]
- 59.Meadows E, Cho JH, Flynn JM, Klein WH. Myogenin regulates a distinct genetic program in adult muscle stem cells. Developmental biology. 2008;322:406–414. doi: 10.1016/j.ydbio.2008.07.024.S0012-1606(08)01074-9 [DOI] [PubMed] [Google Scholar]
- 60.Moresi V, et al. Myogenin and class II HDACs control neurogenic muscle atrophy by inducing E3 ubiquitin ligases. Cell. 2010;143:35–45. doi: 10.1016/j.cell.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rong PM, Teillet MA, Ziller C, Le Douarin NM. The neural tube/notochord complex is necessary for vertebral but not limb and body wall striated muscle differentiation. Development (Cambridge, England) 1992;115:657–672. doi: 10.1242/dev.115.3.657. [DOI] [PubMed] [Google Scholar]
- 62.Kuratani S, et al. The expression pattern of the chick homeobox gene gMHox suggests a role in patterning of the limbs and face and in compartmentalization of somites. Developmental biology. 1994;161:357–369. doi: 10.1006/dbio.1994.1037.S0012-1606(84)71037-2 [DOI] [PubMed] [Google Scholar]
- 63.Cossu G, et al. Activation of different myogenic pathways: myf-5 is induced by the neural tube and MyoD by the dorsal ectoderm in mouse paraxial mesoderm. Development (Cambridge, England) 1996;122:429–437. doi: 10.1242/dev.122.2.429. [DOI] [PubMed] [Google Scholar]
- 64.Fan CM, Lee CS, Tessier-Lavigne M. A role for WNT proteins in induction of dermomyotome. Developmental biology. 1997;191:160–165. doi: 10.1006/dbio.1997.8713. [DOI] [PubMed] [Google Scholar]
- 65.Tajbakhsh S, et al. Differential activation of Myf5 and MyoD by different Wnts in explants of mouse paraxial mesoderm and the later activation of myogenesis in the absence of Myf5. Development (Cambridge, England) 1998;125:4155–4162. doi: 10.1242/dev.125.21.4155. [DOI] [PubMed] [Google Scholar]
- 66.Chen AE, Ginty DD, Fan CM. Protein kinase A signalling via CREB controls myogenesis induced by Wnt proteins. Nature. 2005;433:317–322. doi: 10.1038/nature03126.nature03126 [DOI] [PubMed] [Google Scholar]
- 67.Ikeya M, Takada S. Wnt signaling from the dorsal neural tube is required for the formation of the medial dermomyotome. Development (Cambridge, England) 1998;125:4969–4976. doi: 10.1242/dev.125.24.4969. [DOI] [PubMed] [Google Scholar]
- 68.Brunelli S, Relaix F, Baesso S, Buckingham M, Cossu G. Beta catenin-independent activation of MyoD in presomitic mesoderm requires PKC and depends on Pax3 transcriptional activity. Developmental biology. 2007;304:604–614. doi: 10.1016/j.ydbio.2007.01.006.S0012-1606(07)00020-6 [DOI] [PubMed] [Google Scholar]
- 69.Dunty WC, Jr, et al. Wnt3a/beta-catenin signaling controls posterior body development by coordinating mesoderm formation and segmentation. Development (Cambridge, England) 2008;135:85–94. doi: 10.1242/dev.009266.dev.009266 [DOI] [PubMed] [Google Scholar]
- 70.Hutcheson DA, Zhao J, Merrell A, Haldar M, Kardon G. Embryonic and fetal limb myogenic cells are derived from developmentally distinct progenitors and have different requirements for beta-catenin. Genes & development. 2009;23:997–1013. doi: 10.1101/gad.1769009.gad.1769009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brack AS, et al. BCL9 is an essential component of canonical Wnt signaling that mediates the differentiation of myogenic progenitors during muscle regeneration. Developmental biology. 2009;335:93–105. doi: 10.1016/j.ydbio.2009.08.014.S0012-1606(09)01137-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Polesskaya A, Seale P, Rudnicki MA. Wnt signaling induces the myogenic specification of resident CD45+ adult stem cells during muscle regeneration. Cell. 2003;113:841–852. doi: 10.1016/s0092-8674(03)00437-9.S0092867403004379 [DOI] [PubMed] [Google Scholar]
- 73.Zhao P, Hoffman EP. Embryonic myogenesis pathways in muscle regeneration. Developmental dynamics : an official publication of the American Association of Anatomists. 2004;229:380–392. doi: 10.1002/dvdy.10457. [DOI] [PubMed] [Google Scholar]
- 74.Floss T, Arnold HH, Braun T. A role for FGF-6 in skeletal muscle regeneration. Genes & development. 1997;11:2040–2051. doi: 10.1101/gad.11.16.2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhao P, et al. Fgfr4 is required for effective muscle regeneration in vivo. Delineation of a MyoD-Tead2-Fgfr4 transcriptional pathway. The Journal of biological chemistry. 2006;281:429–438. doi: 10.1074/jbc.M507440200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Le Grand F, Jones AE, Seale V, Scime A, Rudnicki MA. Wnt7a activates the planar cell polarity pathway to drive the symmetric expansion of satellite stem cells. Cell stem cell. 2009;4:535–547. doi: 10.1016/j.stem.2009.03.013.S1934-5909(09)00148-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Otto A, et al. Canonical Wnt signalling induces satellite-cell proliferation during adult skeletal muscle regeneration. Journal of cell science. 2008;121:2939–2950. doi: 10.1242/jcs.026534.jcs.026534 [DOI] [PubMed] [Google Scholar]
- 78.Brack AS, et al. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science (New York, N Y) 2007;317:807–810. doi: 10.1126/science.1144090. [DOI] [PubMed] [Google Scholar]
- 79.Pansters NA, et al. Segregation of myoblast fusion and muscle-specific gene expression by distinct ligand-dependent inactivation of GSK-3beta. Cell Mol Life Sci. 2011;68:523–535. doi: 10.1007/s00018-010-0467-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Conboy IM, et al. Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature. 2005;433:760–764. doi: 10.1038/nature03260.nature03260 [DOI] [PubMed] [Google Scholar]
- 81.Carlson ME, Hsu M, Conboy IM. Imbalance between pSmad3 and Notch induces CDK inhibitors in old muscle stem cells. Nature. 2008;454:528–532. doi: 10.1038/nature07034.nature07034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Carlson ME, et al. Relative roles of TGF-beta1 and Wnt in the systemic regulation and aging of satellite cell responses. Aging Cell. 2009;8:676–689. doi: 10.1111/j.1474-9726.2009.00517.x.ACE517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schuster-Gossler K, Cordes R, Gossler A. Premature myogenic differentiation and depletion of progenitor cells cause severe muscle hypotrophy in Delta1 mutants. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:537–542. doi: 10.1073/pnas.0608281104.0608281104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vasyutina E, et al. RBP-J (Rbpsuh) is essential to maintain muscle progenitor cells and to generate satellite cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:4443–4448. doi: 10.1073/pnas.0610647104.0610647104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Spana EP, Doe CQ. Numb antagonizes Notch signaling to specify sibling neuron cell fates. Neuron. 1996;17:21–26. doi: 10.1016/s0896-6273(00)80277-9.S0896-6273(00)80277-9 [DOI] [PubMed] [Google Scholar]
- 86.Jory A, et al. Numb promotes an increase in skeletal muscle progenitor cells in the embryonic somite. Stem Cells. 2009;27:2769–2780. doi: 10.1002/stem.220. [DOI] [PubMed] [Google Scholar]
- 87.Conboy IM, Rando TA. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev Cell. 2002;3:397–409. doi: 10.1016/s1534-5807(02)00254-x.S153458070200254X [DOI] [PubMed] [Google Scholar]
- 88.Shinin V, Gayraud-Morel B, Gomès D, Tajbakhsh S. Asymmetric division and cosegregation of template DNA strands in adult muscle satellite cells. Nature cell biology. 2006;8:677–687. doi: 10.1038/ncb1425. [DOI] [PubMed] [Google Scholar]
- 89.Kitamoto T, Hanaoka K. Notch3 null mutation in mice causes muscle hyperplasia by repetitive muscle regeneration. Stem Cells. 2010;28:2205–2216. doi: 10.1002/stem.547. [DOI] [PubMed] [Google Scholar]
- 90.Meadows E, Flynn JM, Klein WH. Myogenin regulates exercise capacity but is dispensable for skeletal muscle regeneration in adult mdx mice. PLoS One. 2011;6:e16184. doi: 10.1371/journal.pone.0016184. [DOI] [PMC free article] [PubMed] [Google Scholar]


