Abstract
More than a thousand disease susceptibility loci have been identified via genome-wide association studies (GWAS) of common variants; however, the specific genes and full allelic spectrum of causal variants underlying these findings generally remain to be defined. We utilize pooled next-generation sequencing to study 56 genes in regions associated to Crohn’s Disease in 350 cases and 350 controls. Follow up genotyping of 70 rare and low-frequency protein-altering variants (MAF ~ .001-.05) in nine independent case-control series (16054 CD patients, 12153 UC patients, 17575 healthy controls) identifies four additional independent risk factors in NOD2, two additional protective variants in IL23R, a highly significant association to a novel, protective splice variant in CARD9 (p < 1e-16, OR ~ 0.29), as well as additional associations to coding variants in IL18RAP, CUL2, C1orf106, PTPN22 and MUC19. We extend the results of successful GWAS by providing novel, rare, and likely functional variants that will empower functional experiments and predictive models.
Crohn’s disease (CD) and ulcerative colitis (UC) are classified as chronic, idiopathic inflammatory bowel diseases (IBD) of the gastrointestinal tract with unknown etiology (IBD: OMIM #266600). CD is prevalent in roughly 100–150 per 100,000 individuals of European ancestry1. Generally, the disease affects the ileum and colon but can affect any region of the gut. UC has similar population prevalence and although shows some similarities in clinical manifestation, the location of inflammation is limited to the colonic mucosa. Strong familial aggregation has been seen in twin studies of CD and UC. Recent population-based sibling risk is 26-fold greater for CD and 9-fold greater for UC, and overall CD and UC concordance rates in non-selected twin studies is 30% and 15%, respectively among monozygotic (MZ) twins compared to 4% for CD or UC among dizygotic (DZ) twins2,3. Like most complex trait diseases, CD and UC result from a combination of genetic and non-genetic risk factors; each individual factor may be expected to have a relatively modest effect on disease risk4.
There is a clear genetic basis to common immune-mediated diseases such as IBD. However, until recently, identifying disease susceptibility genes was challenging for common, polygenic disease5,6. With the development of HapMap and the GWAS technology, complex trait genetics in general, and IBD in particular, have seen a remarkable increase in the number of bona fide associated loci that have been identified and replicated. In CD, individual genome-wide association scans (GWAS) and a follow-on meta-analysis of those studies have robustly identified over 71 susceptibility loci and have provided significant novel insights beyond the two loci that were established prior to the GWAS era7,8. Similarly in UC, GWAS studies have identified a total of 47 susceptibility loci9,10 and, accounting for the extensive number of alleles associated to both diseases, in total 99 distinct associations have been documented for IBD. While these new findings have already provided novel insight into disease pathways, the common SNPs identified are generally of modest effect and explain only about 23% of the overall variance in CD risk. Moreover, most of the associated variants do not have any known or obvious function and many implicate regions with multiple genes, limiting biological extrapolation.
SNPs implicated by GWAS have tight correlation to other SNPs in the region and are most likely to be in linkage disequilibrium with the causal variant rather than causal themselves. A complete catalog of all variation is required in the search for causal variants11,12. Even with denser reference data from 1000 Genomes Project however, the majority of GWAS hits are not correlated to a coding or obvious functional variant, and therefore do not conclusively implicate a unique gene. Should independently associated rare coding variation be discovered in a gene within a region implicated by GWAS, the gene harboring such variants becomes directly implicated. Furthermore, additional heritability could be explained and specific alleles identified for direct functional experimentation. In CD, multiple independent associated alleles are already documented at NOD2 and IL23R13,14. Exhaustive sequencing of genomic regions has recently become feasible for the first time with the advent of next-generation sequencing (NGS) technologies. Growing collections of genome sequences through international efforts like the 1000 Genomes Project are driving the development of laboratory study designs and analytic methods for utilizing large-scale genomic sequencing in human genetic discovery15.
Targeted sequencing of pooled samples affords the opportunity to efficiently and cost-effectively capture all variation in a more limited target region selectively amplified in multiple DNA samples16,17. Such an approach allows efficient use of NGS technologies, which generate billions of base pairs per experimental unit, yet introduce challenges in data processing and analysis to discover novel variants and assess their potential association to disease. We describe here a pooled NGS study of 350 patients with CD and 350 controls across coding exons of 56 genes contained in regions of confirmed significant association to CD7, and introduce novel SNP calling methods for pooled targeted sequencing projects implemented in the software Syzygy. Novel, potentially functional rare variants identified in the survey are then evaluated in eight independent case-control series, enabling the confirmation of a role for functional, rare variants in CARD9 (Gene ID: 64170), NOD2 (Gene ID: 64127), IL23R (Gene ID: 149233), IL18RAP (Gene ID: 8807) as well as additionally identifying others in MUC19 (Gene ID: 283463), CUL2 (Gene ID: 8453), PTPN22 (Gene ID: 26191), and C1orf106 (Gene ID: 55765) more associated than permitted by chance. The results lend further support to an emerging paradigm seen across both rare diseases (Hirschsprung’s disease, Bardet-Biedl syndrome) and common phenotypes (serum lipids, QT-interval, height, Type 1 Diabetes) where both common, low-penetrance and rarer, often higher penetrance, alleles exist in the same gene and suggest that deep sequencing of regions implicated by GWAS may be effective in extending the heritability and knowledge of specific functional alleles in complex disease16,18,19,20,21.
RESULTS
Discovery of new variants in patients with CD and healthy controls using pooled sequencing
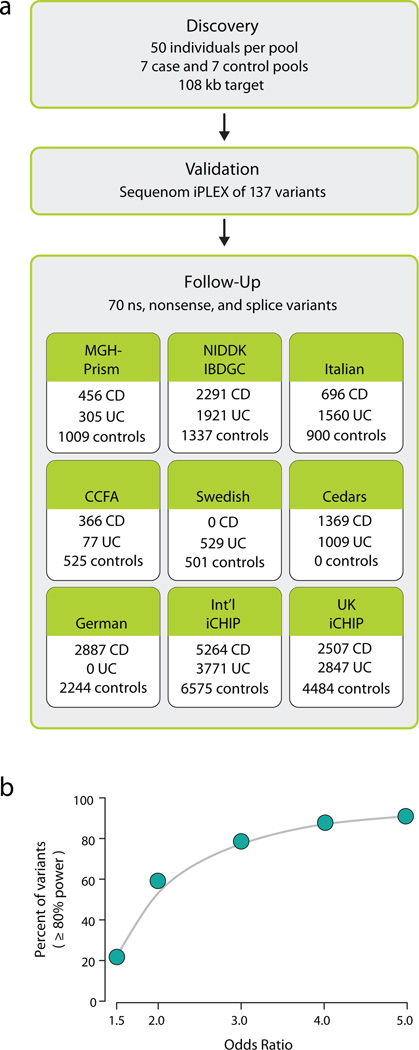
We selected 350 patient with CD and 350 healthy controls of European ancestry from among samples collected by the NIDDK IBD Genetics Consortium (IBDGC) with genome-wide SNP data14,22. Samples were pooled in batches of 50 cases or 50 controls matched for European ancestry using GWAS data. One pool of 50 cases was drawn from self-reported and empirically confirmed (by GWAS data22) Jewish ancestry and was matched with one pool of 50 equivalently defined Jewish controls – remaining pools of cases and controls were selected from the non-Jewish European samples. Pooling of samples was performed only after two rounds of quantification and normalization to insure that the initial DNA pool accurately reflected sample allele frequencies. For each pool we performed PCR amplification to capture the 107.5 kb target of genomic region, which included 645 nuclear-encoded exons (Table S1, S2). We amplified each sample in 593 PCR reactions and the successful PCR amplicons were combined in equimolar amounts, concatenated, and then sheared to construct libraries. The 14 libraries were sequenced using Illumina Genome Analyzer flowcells, with one pool per lane (see Methods) (Figure 1a).
Figure 1. (a) Schematic overview of the Crohn’s disease rare variant phenotype project. (b) Power to detect single-marker rare variant association in follow-up sample sets.
Here we report the results of the Crohn’s pooled resequencing project with follow up genotypes in over 13167 CD patients, 12153 UC patients, 15331 healthy controls. We report that of the 70 markers successfully genotyped 22%,60%,79%,88%,91% have at least 80% power to detect association at minor allele frequency odds ratios of 1.5,2,3,4, and 5 respectively (Figure 1b,S3a,S3b), implying that we are well positioned to address the contribution of rare and low frequency polymorphisms in GWAS loci to IBD.
High-throughput sequencing yielded large amounts of high quality data for each pool. We captured 91% of our nuclear target regions at ≥ 100X coverage and achieved 1500X median coverage per pool (corresponding to 30X per sample/15X per individual chromosome)(Figure S1).
We next aimed to identify rare and low frequency single nucleotide variants/polymorphisms in the pooled samples. We developed a variant calling method (which we named Syzygy) to accommodate the specific pooled study design and confidently identify rare variants (see Supplementary Methods). Through empirical modeling of the sequencing error processes and filters to remove sites with strand inconsistency or clusters of variants suggestive of read misalignment, Syzygy detected 429 putatively high-confidence variants (240 nonsynonymous sites, 169 synonymous sites, and 20 intronic variants within 5 bp of a splice junction) within our 107.5kb targeted region with 45% of the variants already included in dbSNP using dbSNP version 132, nonsynonymous-to-synonymous ratio of 1.42, and transition to transversion ratio of 2.3 (Table 1).
Table 1. Variant Discovery Summary.
Using Syzygy we detected 429 high-confidence variants (240 nonsynonymous sites, 169 synonymous sites, and 20 variants within 5bp of the nearest splice site) within our 107.5kb targeted region with a dbSNP rate of 45%, nonsynonymous-to-synonymous ratio of 1.42, and transition to transversion ratio of 2.3 in the CD pooled sequencing experiment with 350 CD patients and 350 healthy controls.
| Category | High Quality | Moderate Quality |
|---|---|---|
| Variants Identified | 429 | 173 |
| dbSNP % | 45 | 24 |
| NS/S | 1.4 | 1.7 |
| Ts/Tv | 2.3 | 1.4 |
Given that our experimental design aimed to detect variants correctly at the limit of machine quality, we estimated the proposed set of false positive SNPs that would need to be eliminated in subsequent genotyping. Both the proportion of variants in dbSNP and the transition/transversion ratio (Ti/Tv) suggest a relatively high true positive rate in this data set. Specifically, high depth individual level sequencing of 1000 genes performed by the 1000 Genomes project (so-called ‘Pilot 3’) in 697 samples identified a high-quality SNP set with the same %dbSNP (dbSNP version 129), while the Ti/Tv detected here suggests a roughly 90% true positive rate23. To confirm this, a random subset of 137 high-confidence functional nonsynonymous, nonsense, and putative splice variant SNPs was selected for Sequenom iPLEX genotyping of all samples in the sequenced pools and 91.2% validated (Figure 1a). Using a canonical expectation of (theta*SUM(1..1/n)*Nbases), or the rate observed directly in 1000Genomes Pilot 3, we would expect to see ~470 variants across the successfully queried target. Sensitivity for singletons however is incomplete at the lower end of coverage in our experiment (Figure S1) and readily accounts for the modest deficit in our study.
One of the main concerns in any pooled genotyping or sequencing experiment is accurate recovery of allele frequencies. We observed a surprisingly strong correlation between genotype frequencies and sequence level data estimated frequencies (r2 ~ .99) using the method in Syzygy – suggesting the accurate quantitation of DNAs in the pooling steps resulted in good experimental recovery of the pool makeup. A strong correlation is therefore also shown for the case-control test statistic estimated with the pooled data and the test statistic in the genotype data (r2 ~ .925) (Figure S2).
In order to test the role of these rare variants, we identified all nonsynonymous, nonsense or splice site variants which occurred in 2 or more copies up to a frequency of 5% - a total of 115 variants (Table S3). Excluding known GWAS associated low-frequency coding variants at NOD2 IL23R and LRRK2/MUC19, follow-up genotyping was performed for 70 of these markers in eight independent case control samples totaling 16054 CD disease patients, 12153 UC disease patients, and 17575 healthy controls: 1) samples from the MGH-PRISM study, 2) samples assembled from throughout North America and Australia by the NIDDK IBDGC, 3) an Italian-Dutch case-control sample, 4) CCFA Repository Collection, 5) Swedish samples, 6) Cedars samples, 7) German samples as well as Immunochip genotype data provided by 8) the International IBD Genetics Consortium and 9) UK IBD Genetics (n.b., rare coding variants discovered in this study were contributed to Immunochip design) (Figure 1a). Samples 1, 3, 4, and 5 were genotyped for sets of markers using Sequenom iPLEX, Sample 2 genotyping was done as part of a larger IBDGC Illumina GoldenGate study - because of design constraints and assay failures not all markers were examined in all eight follow-up sample sets (see Supplementary Material online for more details of follow-up genotyping). We demonstrate that the current study design is well positioned to address the overall contribution of variants in coding regions of GWAS loci to IBD (Figure 1b, Figure S3, Supplementary Material).
The small number of non-reference alleles expected for many of these variants in each sub-study precludes the use of asymptotic statistics common to association, and the likelihood that population structure becomes an even more significant problem at low frequencies demands a stratified analysis where strict population case-control matching is retained. With this in mind we implemented a mega-analysis of rare variants (MARV) that provides a permutation-based estimate of significance, constraining all permutations to be within each subgroup and thus accommodating arbitrary numbers of sample subsets of diverse population and case-control origin without power loss for single marker and group marker analysis (see Online Methods). Given a target set of 70 variants, in the follow-up analyses we’d expect fewer than 1 SNP to exceed p < .01 by chance and would define traditional experiment-wise significance to be p=.0007. Given both CD and UC are explored in follow-up, to maximize power the primary analysis presented compares all IBD (CD+UC) versus control for genes in which the same common variants have been conclusively associated to both diseases with similar effect (such as CARD9) – for genes specifically associated to only CD (such as NOD2), the UC group is combined with controls (see Anderson et al. 2011 Supplementary Information).
Novel protective splice variant in CARD9
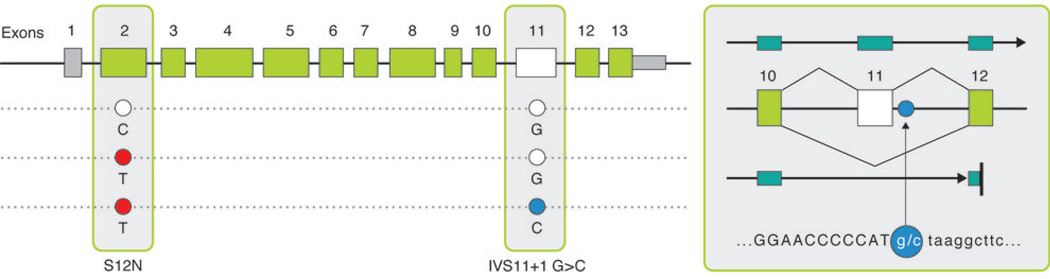
CARD9 has been identified as associated to both CD and UC risk, with a common coding variant (rs4077515 creating substitution S12N – both alleles roughly equifrequent) that represents a ‘typical’ GWAS hit (OR ~ 1.2 in both diseases)8,9. In the pooled sequencing, we identified a splice site variant in CARD9 (Figure 2, Figure S4) altering the first invariant base after exon 11 in 6 controls and 0 disease patients, suggesting a potentially strong protective effect. Follow-up confirms a highly significant association (p<10−16), with the allele appearing at a frequency of roughly .20% of cases and .64% of controls (OR ~ 0.3, Table 2, Table S4). While skipping exon 11 places translation out of frame, the resulting transcript is predicted to escape nonsense mediated decay as premature truncation occurs close to the final splice junction in exon 12. Indeed this hypothetical transcript (Figure 2, Figure S4) has actually been observed in spleen, lymph-node and peripheral blood mononuclear cell (PBMC) derived cDNA libraries. Of note, this rare protective variant actually occurs on a haplotype carrying the risk allele at S12N, indicating that not only are the two associations independent but that the splice variant also completely eliminates the risk normally associated with the common haplotype. Since the CD risk allele at S12N has been associated with higher expression of CARD9, a consistent allelic series may exist if the splice variant is much more substantially low or non-functional and therefore highly protective.
Figure 2. CARD9 protective splice site variant and predicted transcript.
(a) A splice-site variant IVS11+1C>G (OR = 0.29) conferring protection against Crohn’s disease with predicted transcript. This hypothetical transcript has been observed in spleen, lymph-node and peripheral blood mononuclear cell (PBMC) derived CDNA libraries. We predict exon 11 to be skipped and alternative transcript to include exon 9 mRNA sequence continuing to exon 12 including 21 AA before reaching a premature stop.
Table 2. Identification of additional rare and protective variants associated with IBD.
We identify IVS11+1G>C to be protective against IBD with an estimated odds ratio of 0.29 strong (4-fold) protective effect. 5 independent rare mutations in NOD2 are identified to be associated with Crohn’s Disease including R311W, R703C, S431L + V793M, N852S, M863V + fs1007insC. Additional variants conferring protection to IBD are identified in IL23R, and CUL2 and risk missense variants in IL18RAP, C1orf106, MUC19, and PTPN22.
| CD versus UC + HC (CD loci) Gene,mutation chr:positiona |
Targeted Replication | International Immunochip | Combined | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Samples Allele Frequency |
P | Samples Allele Frequency |
P | Samples | OR | P | ||||
| CD | UC + HC | CD | UC + HC | CD | UC+HC | (L95,U95) | ||||
|
NOD2,p.M863V+fs1007insC 16:49308343 |
7969 .0067 |
10179 .00157 |
6.73E-11 | 6544 .0036 |
16126 .0011 |
2.15E-07 | 14523 | 26305 | 4.02 (2.8,5.07) |
<1e-16 |
|
NOD2,p.N852S 16:49308311 |
7962 .0046 |
9590 .0021 |
0.00017 | 6542 .001 |
16121 .000465 |
0.0338 | 14504 | 25711 | 2.47 (1.55,3.93) |
2.90E-05 |
|
NOD2,p.R703C 16:49303430 |
3090 .011 |
4100 .0054 |
0.00025 | 8416 .0079 |
17183 .0052 |
1.59E-04 | 11506 | 21283 | 1.51 (1.12,2.03) |
2.33E-07 |
|
NOD2,p.S431L 16:49302615 |
7949 .0039 |
9569 .0019 |
0.0014 | 6545 .0038 |
16124 .0026 |
0.023 | 14494 | 25693 | 1.45 (1.07,1.95) |
0.00025 |
|
NOD2,p.V793M 16:49303700 |
2227 .0034 |
3252 .0015 |
0.0217 | 6949 .004 |
16156 .0026 |
0.0127 | 9176 | 19408 | 1.45 (1.07,1.95) |
0.002 |
|
NOD2,p.R311W 16:49302254 |
3010 .0017 |
5506 .00099 |
0.118 | 6950 .0014 |
16149 .00073 |
0.029 | 9960 | 21655 | 2.28 (1.37,3.79) |
0.00143 |
|
IL18RAP,p.V527 2:102434852 |
7920 .0036 |
9561 .0015 |
0.0006 | 4131 .00025 |
10336 0 |
0.0456 | 12051 | 19897 | 3.03 (1.95,4.73) |
2.90E-04 |
|
MUC19,p.V56M 12:39226476 |
2227 .0029 |
3253 .00138 |
0.033 | 4963 .0003 |
11324 .00004 |
0.11 | 7190 | 14577 | 4.32 (1.93,9.67) |
0.00546 |
| IBD versus HC (CD + UC loci) Gene,mutation chr:positiona |
Targeted Replication | International Immunochip | Combined | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Samples Allele Frequency |
P | Samples Allele Frequency |
P | Samples | OR | P | ||||
| IBD | HC | IBD | HC | IBD | HC | (L95,U95) | ||||
|
CARD9,c.IVS11+1G>C 9:138379413 |
10439 .002 |
5933 .0058 |
1.9e-08 | 16420 .0024 |
10707 .0071 |
3.33e-16 | 26859 | 16640 | 0.29 (0.22,0.37) |
<1e-16 |
|
IL23R,p.V362I 1:67478488 |
5321 .0131 |
6112 .0127 |
0.27 | 12241 .011 |
10426 .0152 |
2.7e-5 | 17562 | 16538 | 0.72 (0.63,0.83) |
1.18e-5 |
|
IL23R,p.G149R 1:67421184 |
4629 .0026 |
5305 .0045 |
0.064 | 13789 .0025 |
10707 .0043 |
.0013 | 18418 | 16012 | 0.6 (0.45,2.0.79) |
3.2e-4 |
|
CUL2,c.IVS17+5A>G 10:35354137 |
5582 .0056 |
1684 .0063 |
0.2 | 16387 .0065 |
10707 .0092 |
.0004 | 21969 | 12391 | 0.72 (0.6,0.86) |
3.45e-4 |
|
PTPN22,p.H370N 1:114182437 |
5583 .003 |
1682 .002 |
0.3 | 21997 .0031 |
12393 .002 |
.0046 | 21997 | 12393 | 1.6 (1.16,2.24) |
6.2e-3 |
|
C1orf106,p.Y333F 1:19914464 |
13991 .013 |
8486 .01 |
0.009 | NA | NA | NA | 13991 | 8486 | 1.44 (1.02,2.06) |
0.009 |
NCBI human genome build 36 coordinates.
Rare risk variants in NOD2
NOD2, is a member of a family of human cytosolic, non-TIR NACHT-LRR proteins (TIR = Toll/IL-1 receptor; NACHT = neuronal apoptosis inhibitor protein, MHC class 2 transactivator, HET-E, TP1; LRR = leucine-rich repeats)24 first implicated in CD13,25 and later discovered to be involved in Blau Syndrome26. The three previously known causal mutations, R702W, G908R, and fs1007insC, reside in the LRR domain of NOD2, whereas the mutations identified in Blau Syndrome lie on the highly-conserved NACHT nucleotide binding domain (NDB).
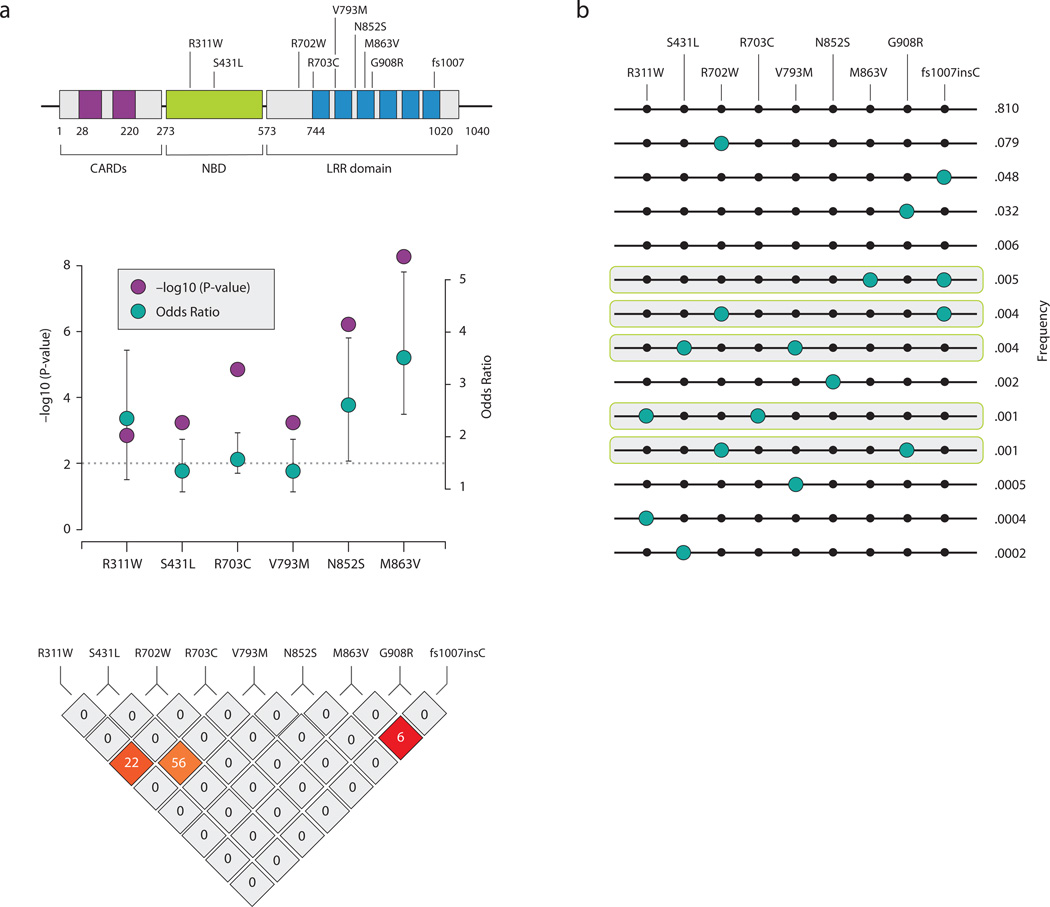
We identify five distinct rare variants (R311W, S431L, R703C, N852S, and M863V), as well as several others in LD with one of these, that are independently associated with CD risk (Table 2, Table S4). S431L (p=.0004) (and the rarer V793M contained on a subset of S431L haplotypes), R703C (p=2.3×10−5) and N852S (p=1.1×10−6) variants are found on distinct haplotypes that do not contain the known causal mutations: R702W, G908R, fs1007insC (Figure 3a, 3b) and are thus completely independent risk variants. R311W shares a subset of haplotypes with R703C (Figure 2), however conditional analysis and haplotype testing indicates both alleles likely contribute independently to risk (Table S5). M863V is a rarer variant that has arisen on the haplotype background of fs1007insC and while the risk estimate of M863V+fs1007insC is stronger (OR=4.02 [2.8,5.7]) is higher than the risk attributable to fs1007insC alone (OR=3.16 [2.9,3.4]), the low frequency of M863V precludes a conclusive statement as to the functionality of M863V at this point – for later calculations of novel variance explained we do not count this an additional risk factor.
Figure 3. (a) Identification of additional rare variants associated with Crohn’s disease and its haplotype structure (b) NOD2 haplotypes observed in 700 individuals with overlapping genotype data (R311W, S431L, R702W, R703C, V793M, N852S, M863V, G908R, fs1007insC).
(a) Five additional risk variants are discovered in NOD2 demonstrating the – log10(P value) and the minor allele odds ratio with 95% CI along with their haplotype block. (b) Note that S431L and V793M are in tight LD and we regard this as one unit S431L + V793M, R703C has a higher frequency than R311W although they share haplotypes conditional analysis (Table S3) demonstrates independent contributions. M863V lies on the background haplotype of fs1007insC.
Functional assessment of additional NOD2 associated alleles
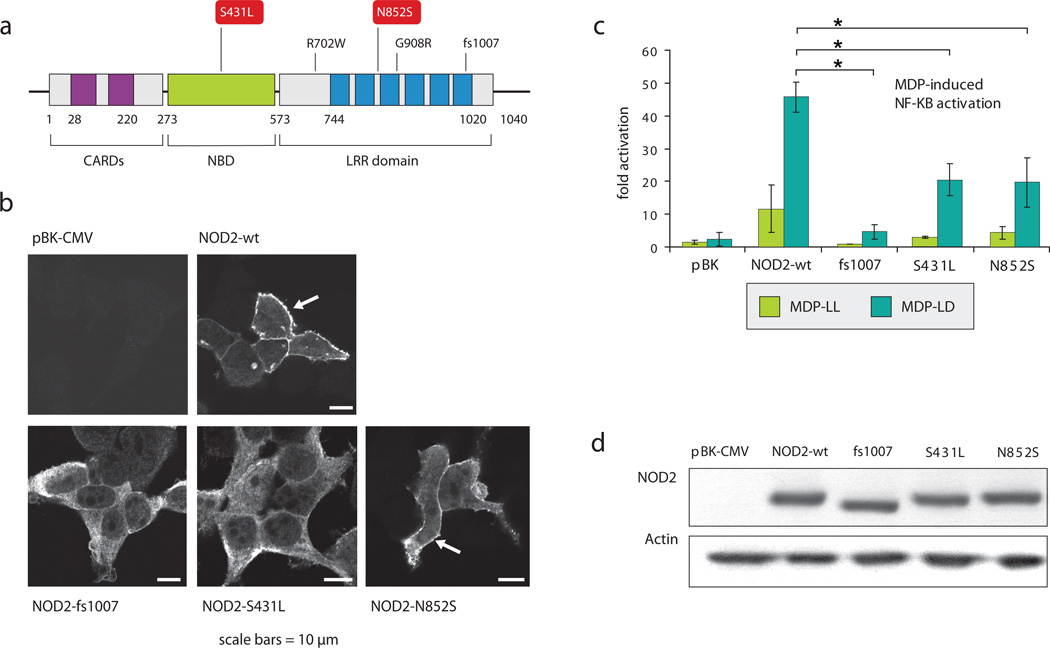
Assays to identify the effect of the mutations on NOD2 intracellular localization demonstrated that S431L and the well-studied insertion mutation (fs1007insC) failed to localize at the membrane area as opposed to N852S (Figure 4). We next determined the abilities of NOD2 mutants S431L and N852S to activate NF-kB in response to NOD2 ligand muramyl dipeptide (MDP). HEK293T cells were transfected with the point mutants as well as wild type NOD2 and the well-studied fs1007 mutant (Figure 4). Western Blot analysis showed that the point mutations did not affect expression level compared to the wild type protein (Figure 4). As published previously, fs1007 mutant failed to induce NF-kB activation after MDP stimulation. The MDP-induced NF-kB activation was also impaired in presence of S431L and N852S (Figure 4).
Figure 4. Functional analyses of NOD2 variants.
HEK293T cells were transfected with NOD2 constructs and fixed using paraformaldehyde 4% at 24h post transfection. Cells were then subjected to immunofluorescent staining to detect NOD2 and fluorescence was collected using a confocal microscope. Image gallery displays a single confocal section.
HEK293T cells were transfected with NOD2 constructs and reporter plasmids encoding firefly luciferase cloned under a promoter containing NF-kB elements and with a plasmid encoding renilla luciferase as a transfection control. After 24h, cells were then stimulated with MDP-LL or MDP-LD (1ug/ml) for 6h. Transcriptional activation was quantified by ratios of firefly luciferase activity to renilla luciferase activity. Data were normalized to the unstimulated condition with empty vector transfection. Statistical analyses were performed using Student t-test. (* p<0.05). Cell lysates were also collected and subjected to western blot analysis to detect NOD2 and actin expression levels.
Together these results indicate that the N852S mutation residing in the LRR domain may perturb MDP recognition without affecting NOD2 intracellular localization, similarly to the common mutations R702W and 908R23. This is opposite to the fs1007insC mutation, which also affects the targeting of NOD2 to the membrane area. Mutation S431L resides in the Nucleotide binding domain (NDB) of the protein and impaired both localization and MDP-induced NF-kB activation. These findings are in line with previous studies demonstrating that critical residues within the NBD region attenuate MDP dependent NF-kB activation24. Further studies are needed to determine the instructive role of NOD2 mutants in coordinating autophagy, control of cellular stress signals and adaptive immune responses.
Asn852Ser and Met863Val rare risk variants are more common in Ashkenazi Jewish individuals
The highest reported prevalence of CD among white individuals is in subjects of Ashkenazi Jewish (AJ) descent, occurring two to four times more frequently than in non-Jewish white populations29. Tukel and Shalata et al (2004)30 screened the NOD2 gene for rare variants and revealed five novel changes (D113N, D357A, I363F, L550V, and N852S) of which N852S occurred only in AJ individuals and was proposed as potentially disease predisposing, with 7 transmissions and only 1 non-transmission from heterozygous parents to affected offspring in an Ashkenazi Jewish family collection – concordant with the case-control observations in this study. In our study, Ashkenazi Jewish individuals had a much higher frequency of both N852S and M863V (4%, 2% cases respectively in Jewish samples and .5% for N852S and M863V in CD non-Jewish case samples) – accounting for the greater incidence of these alleles in the first replication column of table 2 since NIDDK studies in particular had a specific and significant Ashkenazi Jewish ascertainment.
We examined the haplotype carrying N852S in Ashkenazi Jewish individuals (easily determined given the existence of two homozygote cases) and in white non-Jewish individuals in the subset of samples with existing GWAS genotype data8,9,14,22. We found that the N852S mutation in Ashkenazi individuals lies on a unique extended haplotype that extends for several megabases (at least 2 Mb to the left and right). However, N852S mutation in white non-Jewish individuals does not share the extended background haplotype. In Ashkenazi individuals the average shared distance between a pair of AJ 852S chromosomes is at least 4Mb, whereas for a pair of NJ 852S chromosomes is 0.5 Mb (Figure S5) – suggesting that the variant is reasonably old but a single copy was stochastically enriched in the recent Ashkenazi bottleneck ~ 25 generations ago.
Rare protective variants in IL23R
We also identified significant protective effects of amino acid substitutions G149R (P value 3.2×10−4) and V362I (P value 1.2×10−5) in IL23R. This confirms recently published findings32 and is consistent with each of these variants having a protective effect equivalent to that of the more common R381Q substitution (Table 2, Table S4), although they arose on different haplotype backgrounds and are in no LD with R381Q. Despite the large follow-up sample size, we did not find evidence for a protective effect of the previously reported R86Q variant (31747, 0.94). IL23R signaling is attenuated in Th17 cells generated from healthy subjects carrying the R381Q substitution leading to a decrease of IL17A secretion in response to IL-23, indicating that R381Q is associated with reduced Th17 responses33. In addition, recent studies have highlighted a role for IL23 in Th17 cell lineage commitment in the absence of TGF-β. This alternate mode of Th17 differentiation, dependent on IL23R expression, appears to play greater pathogenic role further highlighting the value to discovering protective variants in autoimmunity31. Future therapies for autoimmune disease should consider the phenotypic characters of pathogenic Th17 cells, generated in the absence of TGF-β, and their signaling pathways as possible targets.
Rare risk and protective variants in IL18RAP, C1orf106, CUL2, MUC19, and PTPN22
Although CD and UC do not share an association to the common variant (rs2058660, MAF=.23, OR ~ 1.19, chr2:102.17–102.67 Mb), overlap with celiac disease has recently been documented to rs205866034. We identify a rare risk missense variant, V527L (MAF= .003), in IL18RAP with an estimated minor allele odds ratio of 2.79 to CD. In addition, a low frequency missense variant, Y333F (MAF=.008), in C1orf106 was associated to risk both in CD and UC.
A common CUL2 variant (rs12261843, MAF= 0.30, OR ~ 1.15) has been identified as associated to both CD and UC risk. In the pooled sequencing experiment we identified a splice site variant in CUL2 altering a nucleotide 5 bases downstream exon 17 with an estimated OR of 0.72 in the follow up samples (MAF = .007). Interestingly, several members of the ubiquitin proteosome are present in the autophagy interaction network including CUL2 suggesting cross talk between these processes in intracellular quality control and immunity35.
A common missense variant (risk allele frequency=.90, OR=1.31, rs2476601) in PTPN22 is associated with CD7,8, Type 1 diabetes (T1D)36, Rheumatoid Arthritis (RA)37, and Vitiligo38. This is one of the rare instances where the direction of association differs in different diseases, with the minor allele (W) strongly associated to T1D, RA, and vitiligo but highly protective against CD. Analysis of rare variants in the IBD versus healthy controls comparison demonstrates a modest risk effect (P value = .00026, minor allele odds ratio = 1.6), for a rare (MAF = .003) PTPN22 missense mutation (H370N). Ongoing studies in other autoimmune diseases will help elucidate the overall relevance of H370N and rs2476601 in different conditions.
Examination of haplotype structure (Figure S6) and formal conditional analysis (Table S6) demonstrates that the rare variants highlighted in IL18RAP, MUC19, C1orf106, PTPN22, and CUL2 are independent of the common GWAS variant associated. Specifically, the rare variants at IL18RAP and MUC19 arise on the common higher risk background but confer independently significant risk, the rare variants at PTPN22 and C1orf106 occur on the common low risk background and are therefore obviously independent, and the rare variant at CUL2 is protective and in weak LD with common risk variants at that locus.
Heritability estimates of rare associated variants
We estimated the fraction of additive genetic variance explained using the liability threshold model of Pearson and Lee39 and Fisher40, which assumes an additive effect at each locus and shifts the mean of a normally distributed distribution of disease liability for each genotype class. We assumed a prevalence of CD of 4 per 1,000 and a total narrow-sense heritability of 50%41. We estimate that the discovered rare and low frequency variants associated to CD in this study contribute another 1–2% genetic variance explained over all populations and 2–3% genetic variance explained to the Ashkenazi Jewish population (Table S7).
DISCUSSION
Genomewide association has been remarkably successful in IBD with now 99 confirmed associations already providing important and previously unappreciated views into disease biology. Oddly however, it is quite often what has not yet been discovered or explained (perhaps 75% of the heritability) that consumes much of the debate and focus in human genetics. Next-generation sequencing offers potential insights into both the biology and the heritable component explained by GWAS results through direct ascertainment of a more complete allelic spectrum of functional alleles in cases and controls, including rare variation.
With a targeted, pooled approach, we performed an efficient and cost-effective scan for rare and low frequency polymorphisms in genes in regions identified as relevant in GWAS. After extensive follow-up genotyping, we identify highly significant variants at CARD9, NOD2, CUL2, and IL18RAP that contribute to risk independently from previously defined variants at these loci, and we demonstrate the functionality of the newly implicated NOD2 variants. In addition, we report additional protective variants at IL23R, and identify an excess of additional nominally significant variants in MUC19, PTPN22, and C1orf106.
The results of this experiment are highly relevant to ongoing debates in human genetics. While we found little support for the hypothesis that common variant associations are simply an indirect LD-driven byproduct of higher-penetrance rare alleles, additional independent acting low frequency alleles in genes implicated by common variant association are documented. In the case of the CARD9 splice variant, this novel allele explains more of the overall population variance in risk than does the common S12N associated variant (roughly .3% and .2% respectively). Such observations, should they become commonplace now that technology permits their discovery, may render pointless the strongly worded debates over common versus rare variation. As with many quantitative traits and Mendelian disorders, we observe instances where common alleles of modest effect and rarer alleles with more significant impact peacefully coexist in the same genes – both types of variation providing insight into the same disease biology. In fact the value of these results is likely much more in the realm of functional biology than in nudging the tally of variance explained marginally forward. In addition to the functional confirmation of NOD2 alleles, the identification of a novel CARD9 isoform that is strongly protective against disease development provides a concrete handle with which to study disease biology and potentially a model that could be mimicked therapeutically. Adding .3% to the variance explained and an additional tidbit for the discussion of rare variants and GWAS studies (without which CARD9 would not have been evaluated in this study) are trivial by comparison. Finally, our study validates the principle that additional variants should be routinely searched for by thorough sequencing of genes located within significantly associated regions in GWAS in large sets of cases and appropriate controls.
ONLINE METHODS
DNA preparation and pooling
Crohn’s disease patients and Controls from NIDDK consortium were selected with priority given to samples with adequate amounts of DNA and those with GWAS data available. Samples from the NIDDK consortium undergo rigorous clinical phenotyping and control matching for genetic studies. DNA purification methods are also performed on these samples. The case/control samples selected have already been stringently matched in previous GWAS studies. The baseline concentration of genomic DNA was quantified by Quant-iT™ PicoGreen® dsDNA reagent and detected on the Thermo Scientific Varioskan Flash. All DNAs were normalized to 20ng/µl and repeat quantification was performed to assess accuracy of the normalization step. The quantification and normalization was repeated again to ensure that all samples fell within the desired concentration range. The normalization steps were done with robotic automation using the Packard Multiprobe II HT EX. Once each individual sample is normalized to 10ng/ul, groups of 50 individuals were pooled together using a Multiprobe or Packard Robotic to total 14 pools (700 people).
Target selection and design
Candidate exonic targets from top GWAS published, confirmed genes along with a sample of other highly significant regions of interest were uploaded against HG17 freeze to an in-house database, which houses PRIMER3 software. Amplicons encompassing each target region (coding exons only) were designed using Illumina parameters including a minimum amplicon length of 150bp and maximum amplicon length of 600bp with no buffer sequence added. Additionally, Not1 tails were added to the primer pairs to provide a recognition site for downstream concatenation and shearing step. Amplicons were validated by running PCR product on agarose gels to assess clarity of single bands. Amplicons that had 2/3 clear bands were considered validated. Pfu enzyme, used in Illumina sequencing protocol for PCR, was used in the characterization process. In total, 593 primer pairs passed and covered 95% of the 108 kb target. PCRs contained 20 ng of pooled genomic DNA, 1× HotStar buffer, 0.8 mM dNTPs, 2.5 mM MgCl2, 0.2 units of HotStar Enzyme (Qiagen), and 0.25 µM forward and reverse primers in a 6- or 10-µl reaction volume. PCR cycling parameters were: one cycle of 95°C for 15 min; 35 cycles of 95°C for 20 s, 60°C for 30 s, and 72°C for 1 min; followed by one cycle of 72°C for 3 min. Each PCR product was then treated to similar steps used for the pooling of DNA individuals. The quantification, normalization, and pooling process was again required to ensure that equimolar PCR product went into library construction to have equal representation of all targets. PCR yield was assessed by the same quantification system and the lowest product yield was then used to normalize across PCR plates. Secondary confirmation was ascertained by testing one column of PCR product per plate on 2% agarose E-gel against 1kb DNA ladder to visualize PCR product size. The 593 PCR products were then combined, using the Packard Multiprobe II HT EX, resulting in an amplified target product per sample pool for sequencing.
Sequencing
The PCR products for each pooled sample were concatenated using NotI adapters and sheared into fragments as previously described42. Libraries were constructed by a modified Illumina single-end library protocol, with 225–275 bp gel size selection and PCR enrichment using 14 cycles of PCR, and then single-end sequenced with 76 cycles on an Illumina Genome Analyzer. Each sample pool was sequenced using a single lane of a Illumina GAII analyzer flowcell. 76bp, 36bp and 52bp reads were aligned to the genome using MAQ algorithm43 within the Picard analysis pipeline, and further processed using the SAMtools software44 and custom scripts.
Genotyping
137 high confidence Single Nucleotide Variants (SNVs) were assayed in two phases of genotyping using Sequenom MassARRAY iPLEX GOLD chemistry50. The first phase consisted of 72 SNVs and the second phase of 65 SNVs on 350 NIDDK Crohns samples and 350 NIDDK controls for validation purposes. In each phase of genotyping, oligos were synthesized and mass-spec QCed at Integrated DNA Techologies. All SNVs were genotyped in multiplexed pools of 25–36 assays, designed by AssayDesigner v.3.1 software, starting with 10 ng of DNA per pool. Around 7 nl of reaction was loaded onto each position of a 384-well SpectroCHIP preloaded with 7 nl of matrix (3-hydroxypicolinic acid). SpectroCHIPs were analyzed in automated mode by a MassArray MALDI-TOF Compact system 2with a solid phase laser mass spectrometer (Bruker Daltonics Inc.). We obtained high quality data (>95% genotype call rate, HWE P> 0.001) in all samples that had at least one SNV. Variants were called by real-time SpectroCaller algorithm, analyzed by SpectroTyper v.4.0 software and manually reviewed for rare variants. Additional Sequenom Genotyping was carried out for 9 SNVs in 2887 CD cases and 2244 healthy controls from the German popgen biobank collection. German patients were recruited either at the Department of General Internal Medicine of the Christian-Albrechts-University Kiel, the Charité University Hospital Berlin, through local outpatient services, or nationwide with the support of the German Crohn and Colitis Foundation. German healthy control individuals were obtained from the popgen biobank45.
Beadexpress data generated by the NIDDK IBD consortium on 5549 NIDDK samples aided in validation purposes as well as follow-up of associated variants. Genotyping of IIBDGC samples were done with the Illumina Immunochip where design of SNVs discovered in this experiment were included. Independent Crohn's disease and ulcerative colitis (UC) patients, along with unaffected population controls were genotyped at five genotyping centers (See Supplementary Material on quality control steps).
Cells, Antibodies and Plasmids
HEK293T were obtained from American Type Culture Collection (ATCC) and maintained according to ATCC’s instructions. Anti-β-actin Ab was obtained from SantaCruz. Anti-NOD2 Ab (clone NOD-15) was obtained from BioLegend.
Human wild type NOD2 cDNA was cloned in pBK-CMV vector (stratagene) to express untagged NOD2. Mutated constructs were made using the Quick change site-directed mutagenesis kit (stratagene). Inserts were fully sequenced to check the presence of only the desired mutations.
Immunostaining
HEK293T were seeded on poly-lysine-coated slides and transfected with NOD2 constructs using lipofectamine 2000. The following day, cells were fixed with 4% paraformaldehyde (10 min) and permeabilized PBS-Triton X-100 0.1% (10 min). After washing with PBS, the sections were incubated 15 minutes in PBS containing 1% bovine serum albumin. The sections were then incubated with anti-NOD2 Ab (1:200) for one hour, washed using PBS, incubated with dylight 488 conjugated donkey anti-mouse Ig Ab (Jackson ImmunoResearch) for one hour, washed using PBS and incubated with PBS containing 100µg/ml of DABCO (Sigma) as antifading reagent before mounting in Glycergel medium (Dako). Fluorescence signals were captured using a laser confocal microscope (model Radiance 2000 Bio-Rad).
Luciferase reporter assays
HEK293T cells were co-transfected with 0.025 ng of renilla luciferase plasmid, 2.5 ng of Ig-pIV firefly luciferase reporter and 5 ng of NOD2 plasmids using Lipofectamine 2000 (Invitrogen). After 24h of transfection, cells were stimulated with MDP-LL or MDP-LD (1ug/ml) for 6h. Luciferase activities were measured using the Dual Luciferase reporter assay system (Promega) in a BD moonlight 3010 luminometer (BD Biosciences) and normalized to the internal transfection control of renilla luciferase activity.
Statistical Methods
Variant Discovery Software
Next generation sequencing technology is allowing investigators for the first time to comprehensively survey the full spectrum of genetic variation in large case/control samples. Tools and analytical methods are being developed to address the rapid change in technology and data application capabilities. We have implemented methods in the program Syzygy for analysis of pooled sequencing data generation. The software enables investigators to perform SNP calling on pooled data, estimate allele frequencies of discovered variants, apply single-marker association test in pooled setting, group wise testing of rare and low frequency variants discovered, power evaluation and QC summary, and annotation of variants discovered in regions from primary sequencing data in BAM/SAM format. By doing so allowing researchers to prioritize variants and regions for follow up and dissection of the genetic architecture in the targets of interest.
Mega-Analysis of Rare Variants
One of the goals of the project was to combine data from different groups and subpopulations where samples were carefully matched. We propose the following approach to analyze rare - variants, referred to in this manuscript as M.A.R.V.
- Step 1. Let our random variable
Step 2. The affected/unaffected status is permuted among the individuals within each subgroup, and Step (1) is repeated k times to sample x1*, …, xk* under the null-hypothesis.
- Step 3. The average (μ̂) and sample standard deviation (σ̂) of x1*, …, xk* are calculated and the standardized score is found as
Under the null hypothesis, Z has an approximately standard normal distribution (see Figure S7). Thus, a p-value for the association test can be obtained by comparing Z to the quantiles of the standard normal. Alternatively, a p-value can be obtained by using a standard permutation test, where the p-value is found by (k0+1)/(k+1), and k0 is the number of the k permutations that are at least as extreme as x.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Lauren Solomon for design of figures, R. Onofrio for designing PCR primers, K. Ardlie and S. Mahan for assistance in DNA sample preparation, J. Wilkinson and L. Ambrogio for Illumina sequence project management, Brian Thomson, Gabriel Crenshaw and Andy Taylor for genotyping assistance, J. Flannick and J. Maguire for assistance with pooled sequence analysis, M. dePristo and the Broad GSA team for sequencing analysis help and useful discussions. The work was supported by NHGRI Sequencing Grant, and DK 83756, Funds from the Helmsley Trust to M.J.D & R.J.X. This work was also supported by National Institutes of Health Grants AI062773, DK060049 and DK043351 (to R.J.X.). A.G. was supported by fellowships from La Fondation pour la Recherche Medicale and the Crohn’s and Colitis Foundation of America. JDR holds a Canada Research Chair and is funded by grants from the U.S. National Institutes of Allergy and Infectious Diseases (AI065687; AI067152) from the U.S. National Institute of Diabetes and Digestive and Kidney Diseases (DK064869; DK062432), the Crohn’s and Colitis Foundation of America (SRA512), la Fondation de l’institut de cardiologie de Montréal, the Crohn’s and Colitis Foundation of Canada (CCFC), the Fonds de Recherche en Santé du Québec, and the Canadian Institutes of Health Research. Genotyping of the Italian samples was supported by the Italian Ministry for Health GR-2008-1144485 and unrestricted research grant of Giuliani spa, with case collections provided by the Italian Group for IBD (IG-IBD). The NIDDK IBD Genetics Consortium is funded by the following grants: DK062431 (S.R.B.), DK062422 (J.H.C.), DK062420 (R.H.D.), DK062432 (J.D.R.), DK062423 (M.S.S.), DK062413(D.P.B.M.), and DK062429 (J.H.C.). D.P.B.M. is supported by NCRR grant M01-RR00425 to the Cedars-Sinai General Research Center Genotyping core U01-DK062413(IBD GRC), P01-DK046763 (IBD PPG), Southern California Diabetes Endocrinology Research Center grant, DK063491, R21-DK84554-01, and U01 DK062413. Genotyping of the German samples was supported by the German Ministry of Education and Research through the National Genome Research Network (NGFN) and infrastructure support through the Deutsche Forschungsgemeinschaft cluster of excellence ‘Inflammation at Interfaces’. Genotyping of the Swedish samples was supported by the Swedish Research Council, the Bengt Ihre’s Foundation, and the Örebro University Hospital Research foundation.
Footnotes
CONTRIBUTIONS
M.A.R. and M.J.D. conceived and designed the study. Functional characterization of NOD2 mutants was coordinated and designed by A.G. and R.J.X. Study subject recruitment and phenotyping was supervised by R.H.D., M.D., D.P.B.M., M.D., R.J.X., J.H.C., J.D.R., A.L. Sequenom assay designs were developed by P.G., T.H., J.H., L.K., and A.K. NIDDK IBDGC BeadXpress typing was coordinated and supervised by Y.S. and J.H.C. The pooled sequencing protocol was designed and established at the Broad Institute by M.A.R., C.S., D.A., M.J.D. and S.B.G. IIBDGC contributed sample collection and immunochip genotype data for replication. Project management was performed by M.A.R., G.L., M.S., J.D.R., J.H.C., R.J.X, D.P.B.M., R.H.D., S.B., and M.J.D. C.S., and M.B. performed pooling. C.S., Y.S., P.G., C.L. and M.B. performed the genotyping. M.A.R. and M.J.D. designed and performed the statistical and computational analyses, with assistance from K.S.L., G.B., B.N., J.M.K, T.G., S.R., F.K., T.F, and C.Z. S.R. assisted with QC, PCA, and analysis of ImmunoChip data. Syzygy was developed by M.A.R. and M.J.D. M.J.D. supervised all aspects of the study. The manuscript was written by M.A.R., J.D.R., R.J.X. and M.J.D.
Competing Financial Interests
The author declares no competing financial interest.
Accession codes.
The genomic reference sequence for NOD2 can be found under the GenBank accession number NP_071445.1; for IL23R under NP_653302.2; for CARD9 under NP_434700.2; for CUL2 under NM_003591; for IL18RAP under NP_003844.1; for PTPN22 under NP_057051.3; for C1orf106 under NP_060735.3; for MUC19 under AAP41817.1.
Software availability
Syzygy software is available at: http://www.broadinstitute.org/software/syzygy/
MARV is available at: http://www.broadinstitute.org/ftp/pub/mpg/syzygy/MARV.R
REFERENCES
- 1.Loftus EV., Jr Clinical epidemiology of inflammatory bowel disease: Incidence, prevalence, and environmental influences. Gastroenterology. 2004;126(6):1504–1517. doi: 10.1053/j.gastro.2004.01.063. [DOI] [PubMed] [Google Scholar]
- 2.Bengtson MB, et al. Familial aggregation in Crohn’s disease and ulcerative colitis in a Norwegian population-based cohort followed for ten years. J Crohns Colitis. 2008 Jun;3(2):92–99. doi: 10.1016/j.crohns.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Brant SR. Update on the heritability of inflammatory bowel disease: the importance of twin studies. Inflamm Bowel Dis. 2010 Jan;17(1):1–5. doi: 10.1002/ibd.21385. [DOI] [PubMed] [Google Scholar]
- 4.Rioux JD, Abbas AK. Paths to understanding the genetic basis of autoimmune disease. Nature. 2005;435(7042):584–589. doi: 10.1038/nature03723. [DOI] [PubMed] [Google Scholar]
- 5.Nadeau JH. Single nucleotide polymorphisms: tackling complexity. Nature. 2002;420(6915):517–518. doi: 10.1038/420517a. (2002). [DOI] [PubMed] [Google Scholar]
- 6.Plenge R, Rioux JD. Identifying susceptibility genes for immunological disorders: patterns, power, and proof. Immunol Rev. 2006;210:40–51. doi: 10.1111/j.0105-2896.2006.00372.x. [DOI] [PubMed] [Google Scholar]
- 7.Barrett JC, Hansoul S, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40(8):955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franke A, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGovern DP, et al. Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat Genet. 2010;42(4):332–337. doi: 10.1038/ng.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson CA, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altshuler D, Daly M. Guilt beyond a reasonable doubt. Nat. Genet. 2007;39(7):813–815. doi: 10.1038/ng0707-813. [DOI] [PubMed] [Google Scholar]
- 12.Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322(5903):881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hugot JP, Chamaillard M, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411(6837):599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 14.Duerr RH, Taylor KD, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314(5804):1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nejentsev S, et al. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324(5925):387–389. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calvo SE, Tucker EJ, et al. High-throughput, pooled sequencing identifies mutations in NUBPL and FOXRED1 in human complex I deficiency. Nat Genet. 2010;42(10):851–858. doi: 10.1038/ng.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zaghloul NA, et al. Functional analyses of variants reveal a significant role for dominant negative and common alleles in oligogenic Bardet-Biedl syndrome. Proc Natl Acad Sci U S A. 2010;107(23):10602–10607. doi: 10.1073/pnas.1000219107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emison ES, Garcia-Barcelo M, et al. Differential contributions of rare and common coding and noncoding Ret mutations to multifactorial Hirschsprung disease liability. Am J Hum Genet. 2010;87(1):60–74. doi: 10.1016/j.ajhg.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohen JC, Boerwinkle E, Mosley THJ, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354(12):1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 21.Cohen JC, Pertsemlidis A, et al. Multiple rare variants in NPC1L1 associated with reduced sterol absorption and plasma low-density lipoprotein levels. Proc Natl Acad Sci U S A. 2006;103(6):1810–1815. doi: 10.1073/pnas.0508483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rioux JD, Xavier RJ, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39(5):596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marth G, et al. The functional spectrum of low-frequency coding variation. 2010 doi: 10.1186/gb-2011-12-9-r84. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chamaillard M, Philpott D, et al. Gene-environment interaction modulated by allelic heterogeneity in inflammatory diseases. Proc Natl Acad Sci U S A. 2003;100(6):3455–3460. doi: 10.1073/pnas.0530276100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogura Y, Bonen DK, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411(6837):537–539. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 26.Miceli-Richard C, Lesage S, et al. CARD15 mutations in Blau syndrome. Nat Genet. 2001;29(1):19–20. doi: 10.1038/ng720. [DOI] [PubMed] [Google Scholar]
- 27.Barnich N, Aguirre JE, Reinecker HC, Xavier RJ, Podolsky DK. Membrane recruitment of NOD2 in intenstinal epithelial cells is essential for nuclear factor-kappa B activation in muramyl dipeptide recognition. J Cell Biol. 2005;170(1):21–26. doi: 10.1083/jcb.200502153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanabe T, et al. Regulatory regions and critical residues of NOD2 involved in muramyl dipeptide recognition. EMBO J. 2004;23(7):1587–1597. doi: 10.1038/sj.emboj.7600175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roth MP, et al. Geographic origins of Jewish patients with inflammatory bowel disease. Gastroenterology. 1989;97(4):900–904. doi: 10.1016/0016-5085(89)91495-9. [DOI] [PubMed] [Google Scholar]
- 30.Tukel T, Shalata A, et al. Crohn disease: frequency and nature of CARD15 mutations in Ashkenazi and Sephardi/Oriental Jewish families. Am J Hum Genet. 2004;74(4):623–636. doi: 10.1086/382226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghoreschi K, et al. Generation of pathogenic T(H)17 cells in the absence of TGF-β signalling. Nature. 2010;467(7318):967–971. doi: 10.1038/nature09447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Momozawa Y, et al. Resequencing of positional candidates identifies low frequency IL23R coding variants protecting against inflammatory bowel disease. Nat Genet. 2011;43(1):43–47. doi: 10.1038/ng.733. [DOI] [PubMed] [Google Scholar]
- 33.Meglio PD, et al. The IL23R R381Q Gene Variant Protects against Immune-Mediated Diseases by Impairing IL-23-Induced Th17 Effector Response in Humans. PLoS One. 2011;6(2):e17160. doi: 10.1371/journal.pone.0017160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Festen EAM, Goyette P, et al. A Meta-Analysis of Genome-Wide Association Scans Identifies IL18RAP, PTPN2, TAGAP, and PUS10 as Shared Risk Loci for Crohn’s Disease and Celiac Disease. PLoS Genet. 7(1):e1001283. doi: 10.1371/journal.pgen.1001283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organize of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barrett JC, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41(6):703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stahl E, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42(6):508–514. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin Y, et al. Variant of TYR and autoimmunity susceptibility loci in generalized vitiligo. N Engl J Med. 2010;362(18):1686–1697. doi: 10.1056/NEJMoa0908547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pearson K. Mathematical contributions to the theory of evolution VIII: On the inheritance of characters not capable of exact quantitative measurement. Phil. Trans. R. Soc. Lond. A. 1900;195:79–150. [Google Scholar]
- 40.Fisher RA. The correlation between relatives on the supposition of Mendelian inheritance. Trans R Soc Edinburgh. 1918;52:399–433. (1918). [Google Scholar]
- 41.Ahmad T, Satsangi J, McGovern D, Bunce M, Jewell DP. The genetics of inflammatory bowel disease. Aliment. Pharmacol. Ther. 2001;15:731–748. doi: 10.1046/j.1365-2036.2001.00981.x. [DOI] [PubMed] [Google Scholar]
- 42.Gnirke A, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nature Biotech. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krawczak M, et al. PopGen: population based recruitment of patients and controls for t the analysis of complex genotype phenotype relationships. Community Genet. 2006;9:55–61. doi: 10.1159/000090694. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.