Abstract
Globin gene switching is a complex, highly regulated process allowing expression of distinct globin genes at specific developmental stages. Here, for the first time, we have characterized all of the zebrafish globins based on the completed genomic sequence. Two distinct chromosomal loci, termed major (chromosome 3) and minor (chromosome 12), harbor the globin genes containing α/β pairs in a 5′-3′ to 3′-5′ orientation. Both these loci share synteny with the mammalian α-globin locus. Zebrafish globin expression was assayed during development and demonstrated two globin switches, similar to human development. A conserved regulatory element, the locus control region (LCR), was revealed by analyzing DNase I hypersensitive sites, H3K4 trimethylation marks and GATA1 binding sites. Surprisingly, the position of these sites with relation to the globin genes is evolutionarily conserved, despite a lack of overall sequence conservation. Motifs within the zebrafish LCR include CACCC, GATA, and NFE2 sites, suggesting functional interactions with known transcription factors but not the same LCR architecture. Functional homology to the mammalian α-LCR MCS-R2 region was confirmed by robust and specific reporter expression in erythrocytes of transgenic zebrafish. Our studies provide a comprehensive characterization of the zebrafish globin loci and clarify the regulation of globin switching.
Keywords: zebrafish, erythropoiesis, globin switching, locus control region
Introduction
In organisms dependent on functional hemoglobin for oxygen transport, regulation of its production is vital, and misregulation can have catastrophic effects. An elaborate regulatory mechanism has evolved to govern globin production and globin switching, the process by which precise changes occur in α- and β-globin production in an organism throughout development. globin switching occurs in many species across ontogeny, including humans and other mammals, and relies on both highly conserved and unique cis- and trans-regulatory elements (Gumucio et al., 1996; Hardison, 1998; Higgs et al., 2008; Li et al., 2002; Noordermeer and de Laat, 2008; Sankaran et al., 2010). A driving force of globin switching at the organismal level is a series of waves of hematopoiesis defined by the production of erythrocyte precursors in different anatomical locations (McGrath and Palis, 2008)“Maturational” globin switching, or the switch in globin production of an individual cell as it matures through erythropoiesis, also plays a critical role in defining the globin expression signature of the organism as a whole (Kingsley et al., 2006).
Genetic disorders in which this process is disrupted, either through the mutation of regulatory regions or globin coding sequences themselves, are collectively known as hemoglobinopathies (Johnson et al., 2002; Noordermeer and de Laat, 2008; Stamatoyannopoulos, 2005). These disorders include the thalassemias and sickle cell disease (Galanello and Origa, 2010; Harteveld and Higgs, 2010; Mousa and Qari, 2010), which remain a major health concern worldwide (Orkin and Higgs, 2010; World Health and Thalassemia International, 2008). The molecular complexity and clinical relevance of globin switching has made it an area of intense basic and clinical research; sickle cell anemia was the first disease for which the molecular basis was described (Eaton, 2003; Orkin and Higgs, 2010; Pauling et al., 1949).
Fundamental mechanisms of gene regulation (Fritsch et al., 1980; Leder et al., 1980), particularly those of long-range regulatory elements, were initially discovered and studied in this system (Grosveld et al., 1987; Li et al., 2002). Clinical observations, including that higher levels of persistent fetal Globin in sickle cell patients ameliorates symptoms, have focused further research towards influencing this globin switch as a treatment option (Orkin and Higgs, 2010; Watson, 1948). Despite these substantial efforts and even breakthroughs, aspects of the mechanism of globin regulation remain unclear and a cure for the hemoglobinopathies elusive (Orkin and Higgs, 2010).
Conserved long-range regulatory elements play a critical role in globin expression and switching. These enhancer regions are typically, as in the globin locus, characterized by DNase I hypersensitive sites (HS) annotated by their position (in kilobases) upstream of the globin coding sequences (Li et al., 2002). The DNase I hypersensitive site ~40kb upstream (HS-40) of the α-globin locus in humans has been demonstrated to be essential for proper globin expression, with other hypersensitive sites spanning as much as 150kb playing roles in the process (Higgs et al., 2008; Noordermeer and de Laat, 2008; Palstra et al., 2008). The conservation in coding, noncoding regulatory and overall synteny of the globin loci can be traced to the ancestral globin locus present in early jawed vertebrates, which contained both α- and β-globins. The locus has diverged over time and segregated into separate α and β loci after the divergence of amphibians (Gillemans et al., 2003; Hardison, 1998), but essential functions have been shown to be conserved (Anguita et al., 2001; Flint et al., 2001; Gillemans et al., 2003; Goodman et al., 1975; Gumucio et al., 1996; Hardison, 1998; Hughes et al., 2005). Analysis of this conservation in noncoding regions reveals the presence of multispecies conserved sequences (MCS), which when aligned with DNase I HS often define a functional regulatory sequence. This has allowed for the definition of homologous regions in different species, as well as guided identification of new regulatory elements (Higgs et al., 2008).
The zebrafish, Danio rerio has already been established as an important model to study developmental hematopoiesis (Bahary and Zon, 1998; Shafizadeh and Paw, 2004). Advantages include high fecundity, accelerated development, external fertilization, and transparent embryos, allowing for the real-time in vivo visualization. Importantly, the embryos do not require hemoglobin or red blood cells through at least the first 15 days of development (Rombough and Drader, 2009), allowing for detailed loss-of-function studies not possible in other organisms. Similarly, these features allow for large-scale chemical screening not feasible in mammals (Trompouki and Zon, 2010). Of particular interest is the ability to generate transgenic organisms quickly and perform whole-organism, live fluorescent imaging.
The adult (Chan et al., 1997) and embryonic (Brownlie et al., 2003) globins in the zebrafish have been characterized by identifying some of the adult and embryonic globins, both α and β, within both globin loci. Here, the first detailed elucidation of the globin expression pattern changes allowed the observation and characterization of embryonic-to-larval and larval-to-adult globin switches during development, consistent with humans (Stamatoyannopoulos, 2005). A conserved regulatory element has also been revealed by DNase I hypersensitivity mapping and GATA1 binding data, which we show functionally drives robust and specific expression in red blood cells, validating these techniques for identifying regulatory regions. Additional putative enhancer elements in the major locus and minor locus were also observed and warrant further investigation. These data fully characterize the zebrafish as a model of globin switching by defining the coding regions, synteny, genomic structure, regulatory regions and expression pattern of the zebrafish globin gene loci as well as demonstrate the level of conservation at both the molecular and functional level between zebrafish and humans.
Materials and Methods
Zebrafish maintenance
Zebrafish were staged, raised, and maintained as described (Kimmel et al., 1995; Westerfield, 2000). All zebrafish experiments and procedures were performed as approved by the Children’s Hospital Boston institutional Animal Care and Use Committee.
RNA isolation and cDNA preparation
Pools of fifty zebrafish from the 18 somite stage (ss) through 32 days postfertilization (dpf) were collected and homogenized in Trizol reagent (Invitrogen, Carlsbad, CA). Small pools of adult fish were also homogenized in Trizol, and total RNA was isolated using the manufacturer’s protocol and further purified with the RNeasy kit (Qiagen, Germantown, MD). Subsequently, 1 μg of total RNA was used to generate cDNA using the Superscript III system (Invitrogen).
Quantitative real-time PCR
Quantitative real-time PCR was performed on an iCycler IQ5 Real-Time PCR detection system (Bio-Rad, Hercules, CA) using SYBR Green Supermix (Bio-Rad), and fold change was determined using the Gene Expression Analysis for iCycler iQ Macro (Bio-Rad). Additional details located in Supplemental Methods.
Generation of zebrafish transgenics
The LCR and α/β adult globin 2 (α/βa2) bidirectional promoter were, respectively, isolated by restriction enzyme and PCR amplified and cloned into pEGFP-1 vector to create the α/βa2-GFP construct. Additional LCR fragments were amplified by PCR and assembled into expression vectors containing transposon elements using the Gateway system (Invitrogen). Additional details located in Supplemental Methods.
FACS Analysis of zebrafish peripheral blood
Zebrafish peripheral blood was isolated from deeply anesthetized adult α-LCR-α/βa2-eGFP zebrafish as previously described (Lin et al., 2005), with the following modifications. Peripheral blood cells were placed into 200-300 uL of 0.9% PBS containing 5% fetal calf serum and 100 U/mL heparin, then filtered through a 40 μm nylon mesh to ensure a single cell suspension. Propidium iodide at 1 ug/mL (Sigma) was added as a marker to exclude dead cells and debris. Fluorescence activated cell sorter (FACS) analysis was performed based on PI exclusion, forward scatter, side scatter, and GFP fluorescence using a FACS Vantage flow cytometer (Becton Dickinson, San Jose, CA). FACS data were analyzed using FloJo software.
DNase I hypersensitivity assay
Zebrafish peripheral blood was collected by cardiac puncture in anesthetized adult zebrafish. As the mature erythrocytes in lower vertebrates retain their nuclei, we are able to isolate erythrocyte nuclei from peripheral blood. Zebrafish liver tissues were dissected from anesthetized adult fish and cell suspensions were made using a glass tissue homogenizer in 1X PBS. DNase I hypersensitivity assays of both erythrocytes and liver cells were carried out as previously described (Sabo et al., 2006). Bowtie (Langgmead et al., 2009) was used to map DNase I sequence reads onto Zv9 and formatted into BAM files. MACS (Zhang et al., 2008) was used to compute peaks and create browser tracks.Additional details located in Supplemental Methods.
ChIP-seq of zebrafish red cells
ChIP was performed (Lee et al., 2006), the sample sequenced and bioinformatic analysis performed (Zhang et al., 2008) as previously described. Briefly, red cells from 10 adult zebrafish were isolated for each ChIP-seq reaction, cross-linked, prepared with the Illumina/Solexa Genomic DNA kit (Illumina-IP-102-1001), sequenced and analyzed using Model-based Analysis of ChIP-seq (MACS) (Zhang et al., 2008). Additional details located in Supplemental Methods.
In situ hybridization
The whole-mount in situ hybridization protocol was carried out as previously described (Thisse and Thisse, 2008) using antisense probes amplified from digested plasmids.
Accession numbers
The Dnase I and ChIP-seq data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE35895 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE35895).
Results
Analysis of the genomic regions of the globin gene loci
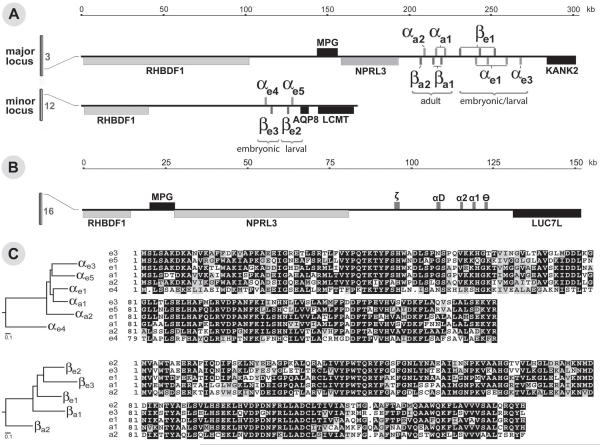
The genes encoding the zebrafish globins reside on two separate chromosomes. Chromosome 3 contains the major globin locus with 13 globin genes and chromosome 12 houses the minor globin locus with four globin genes (Figure 1A). The sequence from the major globin locus was assembled from fully sequenced BACs and phage artificial chromosomes (PACs) available from the zebrafish genome sequencing project at the Sanger Genome Sequencing Center (GenBank ID: AC103581, GenBank ID: AL953863, GenBank ID: BX004811 and GenBank ID: CU464181) based on overlapping sequences among these genomic clones and their encoded globins. Additional sequenced BACs contain different numbers of globin genes, such as AL845551 and AL929176, indicating the presence of various globin locus haplotypes within the population. Figure 1A likely represents a summary of more than one haplotype of the globin region in individual zebrafish that were used to make BAC and PAC libraries (BUSM1, CHORI, CH73, and DanioKey). The major globin locus is syntenic to human chromosome 16 which contains the α-globin locus (Figure 1B). The adjacent genes around the zebrafish major locus share many similarities with the human α-globin locus (Gillemans et al., 2003; Hardison, 1998). These include rhbdf1 (c16orf8), mpg, nprl3 (c16orf35), and the kank2 (flj20004; an ankyrin-like gene). The LCR is found within an intron of the adjacent nprl3 gene (Higgs et al., 2005). There are several differences in the zebrafish major locus compared to mammals. First, genes are primarily found in α/β-globin pairs with a head-to-head orientation. This gene distribution implicates a primitive mechanism to coordinate equal expression of the two globin genes: utilization of a single common promoter and shared enhancer. Second, in the zebrafish the adult globin cluster is located closer to the putative locus control region (“The Putative α-LCR and Proximal globin Promoter Confers Erythroid Specific Expression”) than the embryonic/larval cluster. This is in contrast to humans and mice where the globin genes are expressed in the order of their position within the locus, with the earliest expressed globins closest to the LCR (Noordermeer and de Laat, 2008).
Figure 1. Major and minor zebrafish chromosomal loci and protein sequences.
(A) The major and minor zebrafish globin loci are located on chromosome 3 and 12 respectively. The major locus was assembled by aligning bacterial artificial chromosomes (BAC) and phage artificial chromosomes (PAC), AC103581, AL953863, BX004811 and CU464181, and contains 13 globin genes. The minor locus was assembled by aligning 2 BACs, CR352324 and BX572076, and contains 4 globin genes. The timing of expression is denoted by “embryonic,” “embryonic/larval,” and “adult.”The surrounding regions included are syntenic with other teleost and mammalian species (“Results”). The scale indicated the distance of the region from the beginning of rhbdf1 on both loci. (B) The human α-globin locus, adapted from Higgs & Wood (2008), is syntenic with both the major and minor zebrafish globin loci. (C) Analysis of the similarity of the various globins, broken into α- and β-globins, by protein sequence. Phylogenic analysis was performed using the Clustal W method (MegAlign; DNASTAR). Shades of gray indicate level of conservation at the amino acid level.
The minor locus is covered on two BACs, CR352324 and BX572076, and each contains the same four globins. This locus is less syntenic with the mammalian globin loci, while remaining syntenic with other fish globin loci. Similar to Fugu it shares the rhbdf1 gene that is also present at the major globin locus (Gillemans et al., 2003). This locus is flanked on the other side by the aqp8 and lcmt genes (Figure 1), with the lcmt gene also being a conserved flanking gene with Fugu (Gillemans et al., 2003). Like the major locus, genes are also arranged in pairs; however, only the intermediate expressed globins αe5 and βe2 are arranged in a head-to-head orientation. The αe4 and βe3 pair is arranged in a tail-to-tail fashion.
Genome wide survey of globin coding loci, on Zv9 and available BAC sequences, has identified 17 coding regions that harbor conserved globin genes (Table S1). Through sequence similarity analysis at both the nucleotide and amino acid levels (Figure 1C), 11 unique globin genes were identified with evidence from existing cDNA sequences. Several of the genes have been duplicated or triplicated and share identical sequence at the nucleotide and amino acid level. Comparative analysis of their proximal promoters also shows that they are identical (data not shown). The majority of these gene species have been previously identified in other studies for embryonic (Brownlie et al., 2003) and adult zebrafish globins (Chan et al., 1997). Based on phylogenetic analysis, these globins are grouped into α- and β-globin branches. Based on sequence diversity of BACs containing globin genes, comparison of different strains reveals the presence of different haplotypes of adult globins (data not shown). New globins have been identified on the minor locus using a sequence search. These globin genes, αe4, αe5, βe2 and βe3, were not previously analyzed for their sequence similarity and gene expression. Both βe2 and βe3 group with the previously known zebrafish β-globin genes based on amino acid sequence. Similarly, αe5 groups with the previously known zebrafish α-globin genes. αe4 is the most divergent of all the newly analyzed globin genes and, apart from teleosts, is the most similar to the αD globin found in chickens. The unique number of globins present and similar organization of the zebrafish globin loci is mostly likely the result of genome duplication(s).
Globin gene expression during development identifies two globin switches
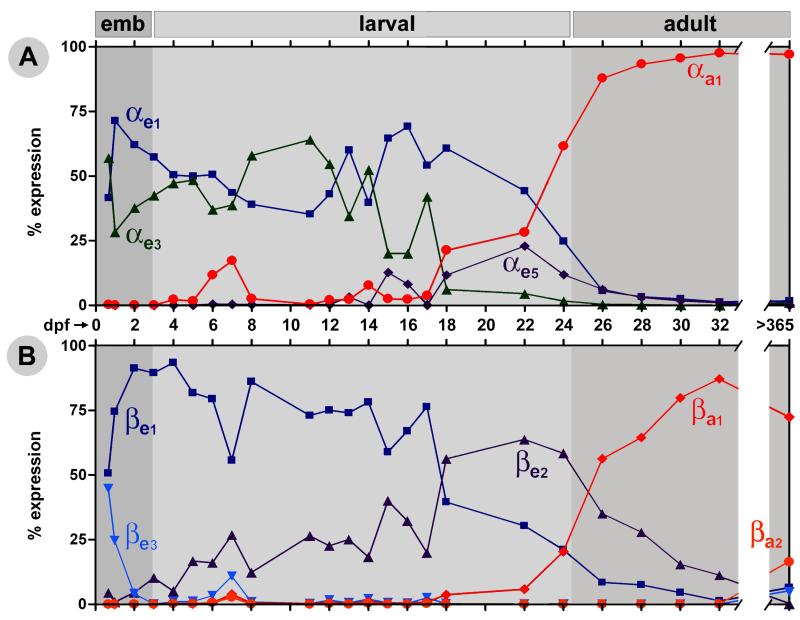
To characterize the developmental globin switching process, the expression of the globin genes was followed in embryos, juveniles and adult fish up to 32 days post fertilization (dpf), and then again assessed at one year of age. Total RNA samples were analyzed with quantitative real-time PCR (qPCR) primers designed to amplify specific globin species.
The globin expression studies delineate three stages of expression (Figure 2). The embryonic stage is defined by the expression of βe3, the only exclusively embryonic globin, with substantial contribution from αe1, αe3 and βe1 (Figure 2 “emb” panel). The first switch then appears to begin between 24 hpf and 36 hpf, marked by the sharp decrease in βe3. The larval stage is characterized by increasing expression of βe2 and, in the later portion, αe5, two genes oriented head-to-head in the genome in the minor locus (Figure 1), as well as the maintained expression of many of the globins from the embryonic stage. These embryonic globins begin to decrease as the larval globins peak near the end of this period. The second switch that establishes the mature, adult globin expression, as defined by the expression pattern observed at 1 year of age, is characterized by the decreasing expression of the embryonic and larval globins and the increasing expression of the nearly exclusively adult globins αa1 and βa1 (Figure 2). This switch begins around 22 dpf, with the continued decline of the embryonic/larval stage globins and the start of the decline of the larval globins αe5 and βe2. The adult globin expression pattern is nearly completely established by 32 dpf. The genes from the cluster closest to the Locus Control Region (LCR), αa1 and βa1, contributing the majority of hemoglobin for the adult fish and a smaller contribution from βa2.
Figure 2. Relative globin expression levels throughout development.
The relative expression level changes of the α-globin (A) genes and the β-globin genes (B) in the zebrafish are shown. In both A and B, the approximate embryonic (emb), larval and adult stages of globin expression are denoted by shades of gray. The first time point depicted is the 16 somite stage. Relative expression levels were determined by quantitative real-time PCR and normalized to band3. The globin genes αe4 and αa2 are not depicted as no significant expression was detected at any of these time points.
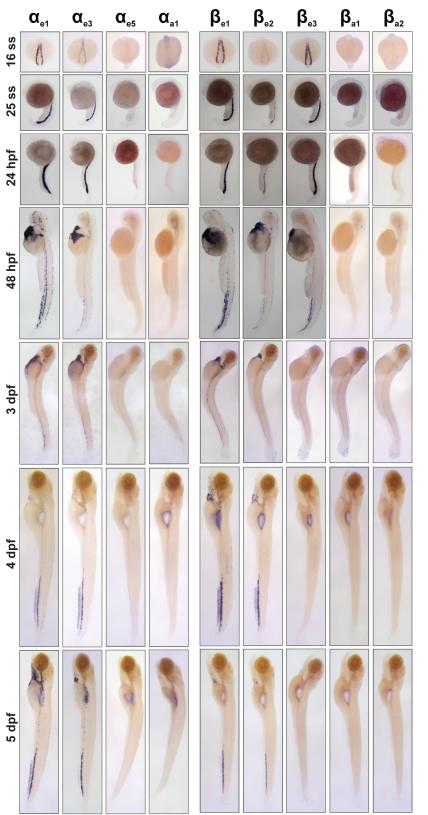
For the first 5 dpf, we examined the expression of the globin genes by in situ hybridization (Figure 3). These results support the qPCR results and provide further evidence there is a globin switch from the embryonic to larval stages in the zebrafish. The decrease of βe3 expression between 48 hpf and 3 dpf observed in the qPCR data (Figure 2) is also observed by in situ hybridization as a loss of the staining between these two time points (Figure 3). The increase in the expression of βe2 between the 16 s.s. and 5 dpf identified by qPCR is also evident in the increased intensity of the staining over the course of these time points by in situ hybridization. Together, these gene expression studies demonstrate that the zebrafish has an embryonic to larval switch and a larval to adult globin switch.
Figure 3.
globin expression by in situ hybridization through 5 dpf. Expression patterns of the α- (A) and β-globin (B) genes. For detected genes, expression is seen in bilateral stripes at the 16ss, in the ICM at the 25ss and 24 hpf stage. With the onset of circulation at 24 hpf, globin positive cells can be seen throughout the vasculature with probes for expressed globin genes, particularly in the vascular plexus of the caudal hematopoietic tissue.
Analysis of the Genomic Structure of the globin Loci
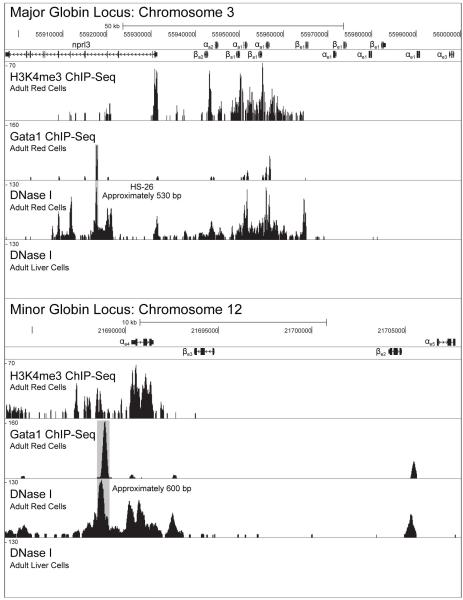
To define long-range globin enhancers and promoters, DNase I hypersensivity mapping, the active H3K4 trimethylation (H3K4me3) mark was mapped and ChIP-seq analysis for the canonical erythrocyte transcription factor GATA1 (Higgs et al., 2008) was carried out in mature adult erythrocytes, which are nucleated in the zebrafish. DNase I and H3K4me3 peaks were observed at expected genes such as GATA1, SCL and KLF4 in comparison to the control liver cells (Figure 4). As expected, the GATA1 ChIP-seq showed distinct peaks corresponding to the proximal promoters of the transcriptionally active adult globin genes. Within both the major and minor globin loci, the HS are highly correlated with H3K4me3 within gene bodies, and the GATA1 sites show a restricted binding correlated with strong HS signals (Figure 4). HS and H3k4me3 signals are evident through the body of the adult globins in the major locus and generally absent from those of the embryonic globins, consistent with the, respectively, high and low levels of expression of these gene in adult red cells (Figure 2). These data indicate an open genomic structure at the adult globin promoters. Within the globin gene cluster, the binding of GATA1 appears to be localized to the proximal promoters of the highly expressed adult globin genes in conjunction with elevated HS signal and H3K4me3 signal. An additional strong HS peak that does not correspond with a GATA1 signal appears near the proximal promoter for βe1, the embryonic gene closest to the transcriptionally active adult globins. As observed in other organisms, this HS may be playing a role in repressing the subsequent globins (Giles et al., 2010). A similar pattern is observed in the minor locus (Figure 4); the only coding region with high levels of H3K4me3 and HS is αe4, for which no expression was detected. A peak within the repressed region, as determined by qPCR expression (Figure 2), of βe2 and αe5 is also observed. These results are specific to red blood cells as the control liver cells do not exhibit any of these patterns.
Figure 4.
Analysis of the chromosomal state of the major and minor globin loci. All tracks were mapped to Zv9 on the UCSU genome browser (http://genome.ucsc.edu/). In some cases annotated genes were renamed in order to adhere to the naming convention, and in cases where a globin gene was not annotated, the UCSC BLAT tool was used to locate the ORFs included in the figure. The shaded gray areas indicate the confirmed and putative regulatory regions in the major and minor loci respectively.
In addition to the HS, H3K4me3 and GATA1 sites within the globin gene cluster itself, a number of putative upstream regulatory elements in both loci were identified. The gene adjacent to the globin coding sequences of the major locus, nprl3, which is also expressed in red cells, contains a number of HS. One of these peaks spans a region of approximately 530 bp and contains a core region of approximately 200 bp that features NFE2, CACCC and functional GATA1 motif binding sites. This region is located approximately 26 kb upstream of the nearest globin coding sequence (HS-26; gray box, Figure 4 “Major Globin Locus”)). This region shares similarity to MCS-R2 (HS-26 in mice and HS-40 in humans) (Hughes et al., 2005). The overall genomic structure of the minor locus appears to be less homologous to globin loci in higher vertebrates, but a number of putative regulatory sites were able to be detected. Three peaks, two with strong H3K4me3 and HS signal and one HS without H3K4me3 signal, are spread across the locus (Figure 4). The first is located upstream of αe4, the second spanning the coding sequence of the gene and the third between βe2 and αe5. All three contain numerous GATA1-, NF-E2- and CACC-binding motifs. As in the major locus, the GATA1 binding is mainly localized to proximal promoters, though the genes are not active (Figure 4). The the strongest DNase hypersensitive peak in the minor locus (gray box, Figure 4 “Minor Globin Locus”), located upstream of all the coding sequences, contains the characteristic binding motifs and coordinates with the strongest GATA1 peak, and it is therefore most likely the LCR for the minor locus. These measures of the genomic structure of the globin locus strongly indicate a role for these features in the overall regulation of globin expression and demarcating putative regulatory elements.
The Putative α-LCR and Proximal globin Promoter Confers Erythroid Specific Expression
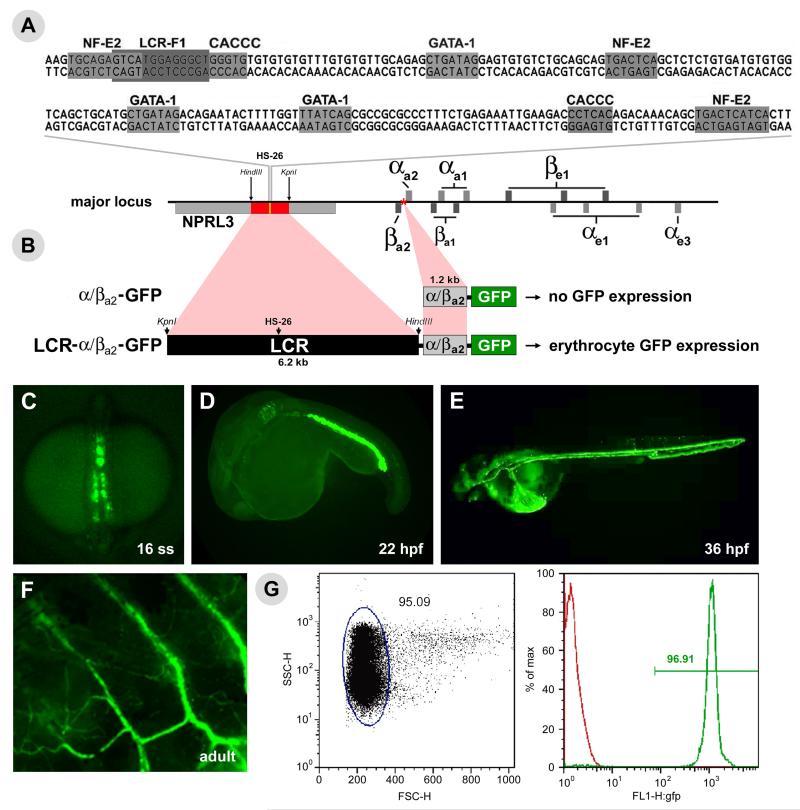
Due to the open chromatin and high degree of sequence homology to known globin regulatory regions (MCS-R2) the role of the HS-26 peak in the regulation of globin gene expression in zebrafish was further investigated. The MCS-R2 regulatory site has been mapped in humans to a 300 bp region within the 5th intron of the nprl3, that contains GATA(1)-, NF-E2/AP1- and CACC-binding motifs (Higgs et al., 2008; Noordermeer and de Laat, 2008); both the synteny and binding motifs are conserved in the zebrafish homologue, HS-26 (Figure 5A). A similar region was identified through comparison to additional species (Maruyama et al., 2007; Hughes et al., 2005). This region has been identified as being sufficient for robust and specific reporter expression in red blood cells (Higgs et al., 2008).
Figure 5.
Generation and characterization of LCR-GFP transgenic zebrafish line. (A) Sequence and location of the HS-26 DNase I hypersensitivity site (HS-26; yellow rectangle) and locus control region (LCR; red rectangle) within the major globin locus. The proximal globin promoter used is indicated with a red star. Shaded boxes indicate binding motifs for canonical erythrocyte transcription factors. (B) Representation of the vectors assembled from genomic fragments and the resulting GFP expression patterns. Fluorescent images of LCR-GFP transgenic zebrafish embryos at 16ss, 22 hpf and 36 hpf respectively (C-E). (F) Fluorescent image of the flank of an adult LCR-GFP transgenic zebrafish. (G) Red blood cell gate as determined by forward and side scatter for peripheral blood, and the analysis of the percent of GFP positive red blood cells in an LCR-GFP transgenic adult (green) versus in a wild-type adult (red).
In order to functionally test the ability of the putative zebrafish regulatory region HS-26 to confer robust, erythroid-specific expression in vivo, a fragment from a bacterial artificial chromosome (CH211-113F11; AL953863) containing HS-26 from the 5th intron of nprl3 as well as a portion of the α/βa2 bidirectional proximal promoter were cloned into GFP reporter constructs and assessed their expression in vivo (Figure 5B). Transgenic embryos were grown to adulthood and transgenic lines established. Multiple lines were monitored and found to have robust erythroid specific visible GFP expression beginning at the 16 somite stage in bilateral stripes in the posterior mesoderm (Figure 5C). At 22 hpf, the ICM region, the site of primitive hematopoiesis, is label with GFP (Figure 5D). At 24 hpf, these GFP positive cells enter circulation and persist through adulthood (Figure 5E-F). The specificity of the GFP expression to mature erythrocytes was confirmed by FACS analysis of adult zebrafish peripheral blood (Figure 5G). GFP expression in erythrocytes is visible from the 16 somite stage to adulthood, demonstrating that the reporter construct does not “switch.” In addition, expression of the GFP mRNA was assessed in order to directly compare the expression of the endogenous globins versus that of the transgene. GFP mRNA expression can be detected beginning at approximately the 12 somite stage, which coincides with the onset of endogenous globin expression (Brownlie et al., 2003) (Supp. Figure 1).
Additional constructs were assembled to confirm HS-26 as the functional globin enhancer. The various constructs were injected at the one cell stage, the embryos were allowed to develop until the 24 hpf stage and observed under a fluorescent microscope. The ability of the α/βa2-bidirectional promoter to drive erythroid expression in the absence of the enhancer was tested by injecting the reporter construct without the putative LCR region. None (0%; 0/50) of the injected embryos expressed visible GFP expression, while 92% (22/25) of those injected with the construct containing the full LCR expressed GFP (data not shown). Two constructs were designed with a truncated LCR region containing either HS-26, as defined by the bounds of the DNase I peak (Figure 4), or the fraction of HS-26 located under the GATA1 peak (Figure 4) driving GFP under the control of the α/βa2-bidirectional promoter. Injection of each construct, in parallel with an mCherry reporter plasmid to control for injection efficiency, showed robust ICM GFP expression in 100% (50/50; 50/50) of productively injected embryos. Replacing the zebrafish α/βa2-bidirectional promoter with a minimal promoter derived from the mouse β-globin locus confirmed the enhancer ability of both the DNase and GATA1 peaks. GFP expression was observed in the ICM of 100% (50/50) of embryos productively injected with the GATA1 peak enhancer/minimal promoter construct and 94% (47/50) of embryos injected with the DNase peak/minimal promoter construct (Supp. Figure 2). Conversely, 82% (41/50) of embryos productively injected with LCRP1/P2-MinPro-GFP, which contains the full LCR except for the DNase I peak, did not express GFP in the ICM. This demonstrates that the functional component of the full LCR region is contained within the DNase I peak. The ability of HS-26 to confer robust, specific expression in vivo confirms that the region is the LCR for the major globin locus and its functional homology to MSC-R2.
Discussion
Globin gene regulation serves as a paradigm for the study of gene expression. The zebrafish offers a genetic model to study globin gene switching in vivo. Although the erythroid program in zebrafish utilizes many of the same transcription factors as mammals, the unique structure of the fish globin loci ensures globin chain balance by regulating pairs of globin genes. Our analysis of the globin locus in zebrafish demonstrates a high level of synteny between the teleost globin loci and the mammalian α-globin locus. The LCR is very well conserved based on functional studies. The switching patterns include a switch from embryonic to larval, and from larval to adult globins. Surprisingly, the genomic structure of the loci and the binding of a canonical erythrocyte transcription factor are very similar between mammals and fish, despite the overall lack of primary DNA sequence conservation. This suggests that the overall genomic structure, including the binding of transcription factors, is more critical for globin gene expression than strict sequence conservation.
Our work supports the hypothesis that the ancestral locus is an α-globin locus (Gillemans et al., 2003; Hardison, 1998). The presence of both α- and β-like globin genes on both the major and minor loci may place the zebrafish closer to this ancestral locus than the puffer fish (Fugu rubripes), where one locus contains only α-like genes, more similar to the organizational structure found in mammals. The arrangement of the globin genes within the loci also implies a bi-directional promoter located between an α- and β-like gene as an ancestral mechanism for obtaining comparable levels of α- and β-like Globin protein. This is also supported by the localization of the GATA1 peaks (Figure 4) to these putative promoter regions in major locus. The mammalian globin loci are arranged such that, for the predominant globins, the genes are temporally expressed in the order in which they are present in the genome (Higgs et al., 2008; Hughes et al., 2005). Within the zebrafish genome, the genes are grouped into clusters of embryonic, embryonic/larval, larval and adult gene expression, and while the importance of physical location with respect to the LCR is conserved, the orientation is reversed for the major locus (Figures 1-2). Globin switching occurs in fish and utilizes the LCR elements to interact with specific regulatory elements near the individual globin genes.
The conservation of the regulatory network responsible for controlling globin switching, including the conservation of the transcription factors, the primary sequence of their binding site, the location of these site and their affects on transcriptional effect on other regulators and the globin gene themselves, establishes the similarity between the overall process of globin production in zebrafish and higher mammals. At approximately 16 s.s. in the bilateral stripes of the developing embryo, the primitive erythrocytes have a high expression of the embryonic globin genes αe1, αe3, βe1 and βe3 (Figure 2, Figure 3). Around 24 hours post fertilization (hpf), primitive proerythroblasts enter circulation (Chen and Zon, 2009), expressing the same embryonic globins, but with ratios different than those present at earlier time points. These cells presumably undergo “maturational” globin switching (Kingsley et al., 2006), as the globin expression of the embryo continues to change prior to the emergence of red blood cells derived from the next wave. These cells continue to mature in circulation and are the only circulating red cell population through 4 dpf (Weinstein et al., 1996), but can contribute past 7 dpf (Chen and Zon, 2009). Between 1 dpf and 2 dpf, the ratios of the embryonic globins previously expressed continue to change, and through 4 dpf βe3’s contribution drops while βe2 expression increases. These changes coincide with the emergence of the erythromyeloid progenitor (EMP) population, which is responsible for generating the next wave of red blood cells to enter circulation (Bertrand et al., 2007). Mature cells begin to enter circulation about 36 hpf, but contribute to the globin expression of the whole embryo prior to this. Through this time period maturational switching as well as the evolving ratio of primitive to EMP cells are most likely affecting the global globin profile. The definitive hematopoietic stem cells are specified as early as 24 hpf but do not contribute to the mature red blood cell pool until approximately 10-14 dpf, having already migrated from the AGM to the caudal hematopoietic tissue (CHT), but may be contributing to the whole embryo globin profile prior to entering circulation. HSCs arriving from the CHT begin to seed the pronephros beginning around 4 dpf, with the definitive hematopoietic activity detectable short thereafter. This will provide full multilineage hematopoietic support for the remainder of the animal’s life (Murayama et al., 2006). Therefore, it is likely that the precipitous decrease in the embryonic globins between approximately 17 dpf and 26 dpf (Figure 2) is the result of a decreasing contribution from the EMP wave erythrocytes, and an unknown contribution from maturational switching. The larval globins αe5 and βe2 peak at a distinct time period. Definitive erythrocytes may not undergo maturational globin switching as the increase in αa1 and βa1 coincides approximately with the increasing contribution of definitive erythrocytes to circulation. The overlapping of these distinct cell populations in the embryo as well as remaining gaps in our understanding of zebrafish hematopoiesis do not allow us to fully resolve the nature of each observed switch. Our data support that both maturational and cellular switching processes contribute to the changes in the overall globin expression observed throughout development.
The functional identification of the LCR in the zebrafish through the use of DNase I hypersensitivity mapping and GATA1 ChIP demonstrates the power of this technique to quickly identify key hematopoietic regulatory sequences. The identification of novel co-localized peaks within mature red cells may indicate the importance of such regions. In addition, the comparison of such dataset between red cell from multiple waves or maturational stages could provide insights into the dynamics of particular regions and their putative relevance to the functional changes occurring. These techniques are complementary to and not redundant with sequence analysis techniques as the rearrangement of elements can obscure sequence similarity despite functional similarity. In addition, regions identified by sequence similarity may not be identified by the genomic state of the region (MCS-E2) (Higgs et al., 2005), and regions identified by sequence homology may not be as robust regulatory regions as those identified by genomic state (Maruyama et al., 2007).
The well documented (Gillemans et al., 2003; Hardison, 1998) high level of syntenic conservation of the organization of the globin locus throughout evolution was able to both inform our work and is highly suggestive that there can be a fluid translation of information between zebrafish and mammalian systems. The sequence conservation observed through our analysis and annotation of both the coding and some non-coding regions of the loci concur with the assessment of previous work that both the synteny and regulatory regions have changed little over 500 million years of evolution (Higgs et al., 2008). In particular, the sequence conservation of the cis-regulatory elements, including GATA-1, NF-E2 and SCL binding motifs, suggest functional conservation in the trans-regulatory networks interacting with the cis elements and demonstrate the evolutionary constraints on this essential process. The syntenic conservation of flanking genes furthers this point, including nprl3 which contains the LCR in mouse, humans, pufferfish, medaka (Maruyama et al., 2007; Hughes et al., 2005) as well as the regulatory element identified here.
The identification of the functional LCR in zebrafish can facilitate the dissection of the functional role(s) of surrounding regions in the process of globin switching. With the relative ease of transgenics in the zebrafish, fragments of DNA can be coupled to GFP and analyzed in vivo in the context of wildtype or hematopoietic mutants or morphants. The transgenic construct generated here recapitulates endogenous globin expression (Figure 5C; Supp. Figure 1) and mimics the temporal expression changes observed for analogous constructs in the mammalian system (Behringer et al., 1990; Enver et al., 1990). Assessing the affects of alterations to the construct can, as shown here, be more easily tested in the zebrafish than a mammalian model. The loci’s genomic context, presence of “intervening” sequence between the LCR and proximal promoter/coding sequence and the absence of additional genes in the region have all been investigated and/or shown to have an effect on globin switching (de Laat and Grosveld, 2003; Flint et al., 2001; Higgs et al., 2008; Li et al., 2002; Noordermeer and de Laat, 2008; Palstra et al., 2008; Tang et al., 2006). Determining the role(s) of these additional regulatory elements can provide a better understanding of globin switching. For instance, it is known that the zinfandel mutation (zin), which has been linked to the major globin locus, alters embryonic globin expression from both loci (Brownlie et al., 2003). This mutation is likely in a critical regulatory element, and identifying the mutation and its role in the regulatory network using this information and these tools will shed light on the broader process of globin switching.
Supplementary Material
Highlights for.
Zebrafish globin Switching Occurs in Two Developmental Stages and is Controlled by the LCR
The zebrafish globin loci and sequences were fully identified.
Two globin switches occur in the zebrafish development, similar to humans.
Analysis of the chromatin state of globin loci identifies the LCR.
The zebrafish LCR functions in vivo.
Table 1. Sequences of primers used in quantitative real-time PCR.
| Gene | PCR primer | Sequence 5′→ 3′ |
|---|---|---|
| αe1a/b/c | forward | CCAGGATGTTGATTGTCTAC |
| αe1a/b/c | reverse | CAGTCTTGCCGTGTTTC |
| αe3 | forward | CCTAAGCCCCAACTCTC |
| αe3 | reverse | CTCCCTTCAGGTCATCC |
| αe4 | forward | GTGGCGGAAGAAATTGG |
| αe4 | reverse | ATGGAACCTGCTAAGTGG |
| αe5 | forward | CCTCAGACCAAGACCTAC |
| αe5 | reverse | TCAGACAGAGCCAAAGC |
| αa1a/b | forward | CAAGGCTGTTGTTAAGGC |
| αa1a/b | reverse | ATTCTGGCGAGGGCTTC |
| αa2 | forward | GCGAATGCTTACTGTCTAC |
| αa2 | reverse | TGTCACGAGGATGTTATGG |
| βe1a/b/c | forward | CTTGACCATCGTTGTTG |
| βe1a/b/c | reverse | GATGAATTTCTGGAAAGC |
| βe2 | forward | ACTATGAGGAGGCTGGAC |
| βe2 | reverse | CGGCGTAGGTGTTCTTG |
| βe3 | forward | ATGCTTGGTCGTCTATCC |
| βe3 | reverse | ATGATTGCCTCTGTGTTG |
| βa1a/b | forward | ATATTTCGCCACATTCG |
| βa1a/b | reverse | GCATAGGTGTTCTTGA |
| βa2 | forward | ATTGCGAGTGTCTGGAG |
| βa2 | reverse | CCGTGTTCTGAAACTTTGG |
| slc4A1 | forward | AGTTAGAGCCTGTTGTATCC |
| slc4A1 | reverse | TCCATCACCACCGAATCC |
Acknowledgements
We would like to thank Owen Tamplin for providing pME-MinPro-GFP. We would like to thank Hsuan-Ting Huang for reviewing the manuscript. The authors acknowledge the help of the UW High-Throughput Genomic Center staff. The authors would like to thank members of the Zon laboratory for suggestions on the manuscript. This work was supported by grants from the National Institutes Health (NIDDK 5RO1 DK53298-14 to L.I.Z.; NIDDK 2P30 DK49216-17, NIDDK 5R37 DK55381-11 and NHLBI 5R01HL048801-18 to Y.Z.; R01GM71923 to J.A.S.; NHLBI 5PO1HL32262-29 to E.T.) and the Howard Hughes Medical Institute (L.I.Z.). J.A.S. was also supported by UW internal sources.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contact Emails: jganis@fas.harvard.edu; nelhsia@gmail.com; etrompouki@enders.tch.harvard.edu; jdejong@peds.bsd.uchicago.edu; adibiase@enders.tch.harvard.edu; zyjia2010@163.com; psabo@stamlab.org; mweaver@stamlab.org; sull@uw.edu; jstam@u.washington.edu; yzhou@enders.tch.harvard.edu; zon@enders.tch.harvard.edu
Authorship
Contributions: J.J.G and N.H. designed and performed experiments, analyzed data and wrote the manuscript; E.T., J.L.O.d., P.J.S., M.W. and R.S. performed experiments and analyzed data; A.D. analyzed data; J.S.L and Z.J. performed experiments; J.A.S and Y.Z designed experiments and analyzed data; L.I.Z. designed experiments, analyzed data and wrote the manuscript.
Conflict-of-interest disclosure: L.I.Z. is a founder and stock holder of Fate, Inc. and a scientific advisor for Stemgent.
References
- Anguita E, Johnson CA, Wood WG, Turner BM, Higgs DR. Identification of a conserved erythroid specific domain of histone acetylation across the alpha-globin gene cluster. Proc. Natl. Acad. Sci. U. S. A. 2001;98:12114–12119. doi: 10.1073/pnas.201413098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahary N, Zon LI. Use of the zebrafish (Danio rerio) to define hematopoiesis. Stem Cells. 1998;16:89–98. doi: 10.1002/stem.160089. [DOI] [PubMed] [Google Scholar]
- Behringer RR, Ryan TM, Palmiter RD, Brinster RL, Townes TM. Human gamma- to beta-globin gene switching in transgenic mice. Genes Dev. 1990;4:380–389. doi: 10.1101/gad.4.3.380. [DOI] [PubMed] [Google Scholar]
- Bertrand JY, Kim AD, Violette EP, Stachura DL, Cisson JL, Traver D. Definitive hematopoiesis initiates through a committed erythromyeloid progenitor in the zebrafish embryo. Development. 2007;134:4147–4156. doi: 10.1242/dev.012385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlie A, Hersey C, Oates AC, Paw BH, Falick AM, Witkowska HE, Flint J, Higgs D, Jessen J, Bahary N, Zhu H, Lin S, Zon L. Characterization of embryonic globin genes of the zebrafish. Dev. Biol. 2003;255:48–61. doi: 10.1016/s0012-1606(02)00041-6. [DOI] [PubMed] [Google Scholar]
- Chan FY, Robinson J, Brownlie A, Shivdasani RA, Donovan A, Brugnara C, Kim J, Lau BC, Witkowska HE, Zon LI. Characterization of adult alpha- and beta-globin genes in the zebrafish. Blood. 1997;89:688–700. [PubMed] [Google Scholar]
- Chen AT, Zon LI. Zebrafish blood stem cells. J. Cell. Biochem. 2009;108:35–42. doi: 10.1002/jcb.22251. [DOI] [PubMed] [Google Scholar]
- de Laat W, Grosveld F. Spatial organization of gene expression: the active chromatin hub. Chromosome Res. 2003;11:447–459. doi: 10.1023/a:1024922626726. [DOI] [PubMed] [Google Scholar]
- Eaton WA. Linus Pauling and sickle cell disease. Biophys. Chem. 2003;100:109–116. doi: 10.1016/s0301-4622(02)00269-7. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enver T, Raich N, Ebens AJ, Papayannopoulou T, Costantini F, Stamatoyannopoulos G. Developmental regulation of human fetal-to-adult globin gene switching in transgenic mice. Nature. 1990;344:309–313. doi: 10.1038/344309a0. [DOI] [PubMed] [Google Scholar]
- Flint J, Tufarelli C, Peden J, Clark K, Daniels RJ, Hardison R, Miller W, Philipsen S, Tan-Un KC, McMorrow T, Frampton J, Alter BP, Frischauf AM, Higgs DR. Comparative genome analysis delimits a chromosomal domain and identifies key regulatory elements in the alpha globin cluster. Hum. Mol. Genet. 2001;10:371–382. doi: 10.1093/hmg/10.4.371. [DOI] [PubMed] [Google Scholar]
- Fritsch EF, Lawn RM, Maniatis T. Molecular cloning and characterization of the human beta-like globin gene cluster. Cell. 1980;19:959–972. doi: 10.1016/0092-8674(80)90087-2. [DOI] [PubMed] [Google Scholar]
- Galanello R, Origa R. Beta-thalassemia. Orphanet J. Rare Dis. 2010;5:11. doi: 10.1186/1750-1172-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles KE, Gowher H, Ghirlando R, Jin C, Felsenfeld G. Chromatin boundaries, insulators, and long-range interactions in the nucleus. Cold Spring Harb. Symp. Quant. Biol. 2010;75:79–85. doi: 10.1101/sqb.2010.75.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillemans N, McMorrow T, Tewari R, Wai AW, Burgtorf C, Drabek D, Ventress N, Langeveld A, Higgs D, Tan-Un K, Grosveld F, Philipsen S. Functional and comparative analysis of globin loci in pufferfish and humans. Blood. 2003;101:2842–2849. doi: 10.1182/blood-2002-09-2850. [DOI] [PubMed] [Google Scholar]
- Goodman M, Moore GW, Matsuda G. Darwinian evolution in the genealogy of haemoglobin. Nature. 1975;253:603–608. doi: 10.1038/253603a0. [DOI] [PubMed] [Google Scholar]
- Grosveld F, van Assendelft GB, Greaves DR, Kollias G. Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- Gumucio DL, Shelton DA, Zhu W, Millinoff D, Gray T, Bock JH, Slightom JL, Goodman M. Evolutionary strategies for the elucidation of cis and trans factors that regulate the developmental switching programs of the beta-like globin genes. Mol. Phylogenet. Evol. 1996;5:18–32. doi: 10.1006/mpev.1996.0004. [DOI] [PubMed] [Google Scholar]
- Hardison R. Hemoglobins from bacteria to man: evolution of different patterns of gene expression. J. Exp. Biol. 1998;201:1099–1117. doi: 10.1242/jeb.201.8.1099. [DOI] [PubMed] [Google Scholar]
- Harteveld CL, Higgs DR. Alpha-thalassaemia. Orphanet J. Rare Dis. 2010;5:13. doi: 10.1186/1750-1172-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs DR, Garrick D, Anguita E, De Gobbi M, Hughes J, Muers M, Vernimmen D, Lower K, Law M, Argentaro A, Deville MA, Gibbons R. Understanding alpha-globin gene regulation: Aiming to improve the management of thalassemia. Ann. N. Y. Acad. Sci. 2005;1054:92–102. doi: 10.1196/annals.1345.012. [DOI] [PubMed] [Google Scholar]
- Higgs DR, Vernimmen D, Wood B. Long-range regulation of alpha-globin gene expression. Adv. Genet. 2008;61:143–173. doi: 10.1016/S0065-2660(07)00005-3. [DOI] [PubMed] [Google Scholar]
- Hughes JR, Cheng JF, Ventress N, Prabhakar S, Clark K, Anguita E, De Gobbi M, de Jong P, Rubin E, Higgs DR. Annotation of cis-regulatory elements by identification, subclassification, and functional assessment of multispecies conserved sequences. Proc. Natl. Acad. Sci. U. S. A. 2005;102:9830–9835. doi: 10.1073/pnas.0503401102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RM, Gumucio D, Goodman M. Globin gene switching in primates. Comp. Biochem. Physiol. A. Mol. Integr. Physiol. 2002;133:877–883. doi: 10.1016/s1095-6433(02)00205-2. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kingsley PD, Malik J, Emerson RL, Bushnell TP, McGrath KE, Bloedorn LA, Bulger M, Palis J. Maturational” globin switching in primary primitive erythroid cells. Blood. 2006;107:1665–1672. doi: 10.1182/blood-2005-08-3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leder P, Hansen JN, Konkel D, Leder A, Nishioka Y, Talkington C. Mouse globin system: a functional and evolutionary analysis. Science. 1980;209:1336–1342. doi: 10.1126/science.7414319. [DOI] [PubMed] [Google Scholar]
- Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Peterson KR, Fang X, Stamatoyannopoulos G. Locus control regions. Blood. 2002;100:3077–3086. doi: 10.1182/blood-2002-04-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HF, Traver D, Zhu H, Dooley K, Paw BH, Zon LI, Handin RI. Analysis of thrombocyte development in CD41-GFP transgenic zebrafish. Blood. 2005;106:3803–3810. doi: 10.1182/blood-2005-01-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama K, Ishikawa Y, Yasumasu S, Iuchi I. Globin gene enhancer activity of a DNase-I hypersensitive site-40 homolog in medaka, Oryzias latipes. Zoolog. Sci. 2007;24:997–1004. doi: 10.2108/zsj.24.997. [DOI] [PubMed] [Google Scholar]
- McGrath K, Palis J. Ontogeny of erythropoiesis in the mammalian embryo. Curr. Top. Dev. Biol. 2008;82:1–22. doi: 10.1016/S0070-2153(07)00001-4. [DOI] [PubMed] [Google Scholar]
- Mousa SA, Qari MH. Diagnosis and management of sickle cell disorders. Methods Mol. Biol. 2010;663:291–307. doi: 10.1007/978-1-60761-803-4_13. [DOI] [PubMed] [Google Scholar]
- Murayama E, Kissa K, Zapata A, Mordelet E, Briolat V, Lin HF, Handin RI, Herbomel P. Tracing hematopoietic precursor migration to successive hematopoietic organs during zebrafish development. Immunity. 2006;25:963–975. doi: 10.1016/j.immuni.2006.10.015. [DOI] [PubMed] [Google Scholar]
- Noordermeer D, de Laat W. Joining the loops: beta-globin gene regulation. IUBMB Life. 2008;60:824–833. doi: 10.1002/iub.129. [DOI] [PubMed] [Google Scholar]
- Orkin SH, Higgs DR. Medicine. Sickle cell disease at 100 years. Science. 2010;329:291–292. doi: 10.1126/science.1194035. [DOI] [PubMed] [Google Scholar]
- Palstra RJ, de Laat W, Grosveld F. Beta-globin regulation and long-range interactions. Adv. Genet. 2008;61:107–142. doi: 10.1016/S0065-2660(07)00004-1. [DOI] [PubMed] [Google Scholar]
- Pauling L, Itano HA, et al. Sickle cell anemia a molecular disease. Science. 1949;110:543–548. doi: 10.1126/science.110.2865.543. [DOI] [PubMed] [Google Scholar]
- Rombough P, Drader H. Hemoglobin enhances oxygen uptake in larval zebrafish (Danio rerio) but only under conditions of extreme hypoxia. J. Exp. Biol. 2009;212:778–784. doi: 10.1242/jeb.026575. [DOI] [PubMed] [Google Scholar]
- Sabo PJ, Kuehn MS, Thurman R, Johnson BE, Johnson EM, Cao H, Yu M, Rosenzweig E, Goldy J, Haydock A, Weaver M, Shafer A, Lee K, Neri F, Humbert R, Singer MA, Richmond TA, Dorschner MO, McArthur M, Hawrylycz M, Green RD, Navas PA, Noble WS, Stamatoyannopoulos JA. Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat. Methods. 2006;3:511–518. doi: 10.1038/nmeth890. [DOI] [PubMed] [Google Scholar]
- Sankaran VG, Xu J, Orkin SH. Advances in the understanding of haemoglobin switching. Br. J. Haematol. 2010;149:181–194. doi: 10.1111/j.1365-2141.2010.08105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafizadeh E, Paw BH. Zebrafish as a model of human hematologic disorders. Curr. Opin. Hematol. 2004;11:255–261. doi: 10.1097/01.moh.0000138686.15806.71. [DOI] [PubMed] [Google Scholar]
- Stamatoyannopoulos G. Control of globin gene expression during development and erythroid differentiation. Exp. Hematol. 2005;33:259–271. doi: 10.1016/j.exphem.2004.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Wang Z, Huang Y, Liu DP, Liu G, Shen W, Tang X, Feng D, Liang CC. Gene order in human alpha-globin locus is required for their temporal specific expressions. Genes Cells. 2006;11:123–131. doi: 10.1111/j.1365-2443.2006.00923.x. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B. High-resolution in situ hybridizatrion to whole-mount zebrafish embryos. Nat. Protoc. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- Trompouki E, Zon LI. Small molecule screen in zebrafish and HSC expansion. Methods Mol. Biol. 2010;636:301–316. doi: 10.1007/978-1-60761-691-7_19. [DOI] [PubMed] [Google Scholar]
- Watson J. The significance of the paucity of sickle cells in newborn Negro infants. Am. J. Med. Sci. 1948;215:419–423. doi: 10.1097/00000441-194804000-00008. [DOI] [PubMed] [Google Scholar]
- Weinstein BM, Schier AF, Abdelilah S, Malicki J, Solnica-Krezel L, Stemple DL, Stainier DY, Zwartkruis F, Driever W, Fishman MC. Hematopoietic mutations in the zebrafish. Development. 1996;123:303–309. doi: 10.1242/dev.123.1.303. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book: A guide for the laboratory use of zebrafish (Danio rerio) 4th ed Oregon; Eugene: 2000. [Google Scholar]
- World Health, O. Thalassemia International, F. Management of Haemoglobin Disorders: Report of Joint WHO-TIF Meeting, Nicosia, Cyprus; World Health Organization, Geneva, Switzerland. 2008. [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.