Abstract
Lysosomes are ubiquitous intracellular organelles that have an acidic internal pH, and play crucial roles in cellular clearance. Numerous functions depend on normal lysosomes, including the turnover of cellular constituents, cholesterol homeostasis, downregulation of surface receptors, inactivation of pathogenic organisms, repair of the plasma membrane and bone remodeling. Lysosomal storage disorders (LSDs) are characterized by progressive accumulation of undigested macromolecules within the cell due to lysosomal dysfunction. As a consequence, many tissues and organ systems are affected, including brain, viscera, bone and cartilage. The progressive nature of phenotype development is one of the hallmarks of LSDs. In recent years biochemical and cell biology studies of LSDs have revealed an ample spectrum of abnormalities in a variety of cellular functions. These include defects in signaling pathways, calcium homeostasis, lipid biosynthesis and degradation and intracellular trafficking. Lysosomes also play a fundamental role in the autophagic pathway by fusing with autophagosomes and digesting their content. Considering the highly integrated function of lysosomes and autophagosomes it was reasonable to expect that lysosomal storage in LSDs would have an impact upon autophagy. The goal of this review is to provide readers with an overview of recent findings that have been obtained through analysis of the autophagic pathway in several types of LSDs, supporting the idea that LSDs could be seen primarily as “autophagy disorders.”
Keywords: Mucolipidosis Type IV, autophagy, glycogenosis, lysosomal storage disorders, lysosomes, mucopolysaccharidoses, sphingolipidoses
Introduction
Lysosomes are ubiquitous intracellular organelles that have an acidic internal pH, and have crucial roles in cellular clearance. Numerous functions have been shown to depend on normal lysosomes, including turnover of cellular constituents, cholesterol homeostasis, downregulation of surface receptors, inactivation of pathogenic organisms, repair of the plasma membrane and bone remodeling. Lysosomal biogenesis involves maturation of early endosomes to form multivesicular bodies (i.e., late endosomes), followed by fusion to the lysosomes and subsequent lysosome reformation.1-3 Lysosomes are involved in the degradation of a wide variety of structurally diverse substances into their basic building blocks, such as proteins, glycosaminoglycans (GAGs), sphingolipids, glycogen, nucleic acids, oligosaccharides and complex lipids. These are either recycled through biosynthetic pathways or further degraded to generate energy. Cellular and foreign material destined for degradation reach lysosomes via endocytosis, phagocytosis, autophagy, or direct transport.
Lysosomal storage disorders (LSDs) are characterized by progressive accumulation of undigested macromolecules within the cell due to lysosomal dysfunction. LSDs were first defined as lysosomal enzyme deficiency states in 1965 by H.G. Hers based on his discovery that the glycogen storage disorder known as Pompe disease exhibits an absence of acidic α-glucosidase activity.4 Hers’ conceptual breakthrough provided the foundation for understanding literally dozens of additional so-called “storage” disorders, including the gangliosidoses and other sphingolipidoses, the mucopolysaccharidoses, the glycoproteinoses, and so forth. What became readily apparent in time, however, was that nonlysosomal enzymes, as well as soluble and transmembrane proteins of late endosomes and lysosomes—when defective—could also cause lysosomal storage defects essentially identical to conditions lacking a specific lysosomal hydrolase. Hence the emergence of an understanding of the latter type of LSDs, which include I-cell disease, multiple sulfatase deficiency, Niemann-Pick type C disease, mucolipidosis IV, Danon disease, juvenile neuronal ceroid lipofuscinosis and others.5 Today, LSDs are recognized as a cohort of nearly 60 different inherited disorders, with each sharing a genetic defect that renders the lysosomal system dysfunctional and unable to degrade specific materials normally processed within the cell. As a consequence, many tissues and organ systems are affected, including brain, viscera, bone and cartilage, with early onset central nervous system (CNS) dysfunction predominating. Whereas clinical features of these disorders vary widely, most are fatal within the first two decades of life following many years of worsening disease. The progressive nature of phenotype development is one of the hallmarks of LSDs.
In spite of the clarity brought about by modern genetic and biochemical studies of storage diseases, a persisting conundrum is why defects in the lysosomal system per se ultimately cause cell and organ dysfunction, particularly for the brain. This question grows in part out of the realization that early on, cells and organs in most storage disorders appear to develop and function normally, and only later is this followed by insidious disease onset and progression. In recent years biochemical and cell biology studies of LSDs have revealed an ample spectrum of abnormalities in a variety of cellular functions. These include defects in signaling pathways, calcium homeostasis, lipid biosynthesis and degradation and intracellular trafficking (see ref. 6 for a review). Therefore, an emerging view7 to explain the disease cascade is one focused on the importance of the multiple endosomal and autophagosomal streams flowing into the lysosomal system for processing. Failure to degrade and recycle sequestered materials in lysosomal disease may also lead to deficiency states in which precursors for metabolic pathways in the cell are diminished, followed by altered synthetic pathways and an increased metabolic demand on affected cells. The lysosomal system has thus emerged from being considered only an end-organelle, to being at the very hub of metabolic regulatory control. Perhaps the best example of the interconnection of the lysosome with other cellular systems is autophagy. Lysosomes play a fundamental role in the autophagic pathway by fusing with autophagosomes and digesting their content. Recent evidence of the cooperative and integrated roles of lysosomes and autophagosomes comes from the discovery of an overarching regulatory gene network (CLEAR) and a master gene, transcription factor EB (TFEB), controlling the biogenesis and function of both lysosomes and autophagosomes.8-10
Considering the highly integrated function of lysosomes and autophagosomes it was reasonable to expect that lysosomal storage in LSDs would have an impact upon autophagy. The goal of the present review is to provide readers with an overview of recent findings that have been obtained through analysis of the autophagic pathway in several types of LSDs. The main results of the studies on autophagy in LSDs are reported in Table 1.
Table 1. The main results of the studies on autophagy in LSDs.
| Disease | AV accumulation† | Defective AV degradation‡ | Increased AV formation° | Increased poly-ub proteins¶ | Increased dysfunctional mitochondria§ | Increased p62• | Refs. |
|---|---|---|---|---|---|---|---|
|
GLYCOGENOSES |
|
|
|
|
|
|
|
|
Pompe disease |
Y |
Y |
Y |
Y |
NT |
Y |
11,12 |
|
Danon disease |
Y |
Y |
NT |
NT |
NT |
NT |
13 |
|
MUCOPOLYSACCHARIDOSES |
|
|
|
|
|
|
|
|
MSD |
Y |
Y |
N |
Y |
Y |
Y |
14,15 |
|
MPSIII A |
Y |
Y |
N |
Y |
Y |
Y |
14,15 |
|
MPS VI |
Y |
Y |
NT |
Y |
Y |
Y |
16 |
|
SPHINGOLIPIDOSES |
|
|
|
|
|
|
|
|
NPC1, NPC2 |
Y |
Y |
Y |
Y |
Y |
Y |
17,18 |
|
Gaucher disease |
Y |
NT |
NT |
NT |
NT |
Y |
19 |
|
Fabry disease |
Y |
Y |
NT |
Y |
NT |
Y |
20 |
|
GM1 gangliosidosis |
Y |
NT |
Y |
NT |
Y |
NT |
21 |
|
MUCOLIPIDOSES |
|
|
|
|
|
|
|
|
MLII |
Y |
NT |
N |
Y |
Y |
Y |
22 |
|
MLIII |
Y |
NT |
N |
Y |
Y |
Y |
22,23 |
|
MLIV |
Y |
Y |
Y |
Y |
Y |
Y |
24–26 |
|
CEROID LIPOFUSCINOSES |
|
|
|
|
|
|
|
|
CLN10 |
Y |
NT |
NT |
NT |
NT |
NT |
27 |
| CLN 3 | Y | NT | Y | NT | NT | NT | 28 |
†Number of autophagic vesicles (AV) quantified by electron microcopy or LC3-immunofluorescence, amounts of LC3-II by western blotting. ‡Impaired autophagosome-lysosome fusion, defective degradation of long-lived proteins. °MTOR downregulation, BECN1 activation ¶Poly-ubiquitinated proteins (poly-ub) revealed by immunofluorescence or western blotting using anti-ubiquitin antibodies. §Dysfunctional mitochondria revealed by western blotting using mitochondrial markers. •p62/SQSTM1 protein revealed by immunofluorescence or western blotting using anti-p62 antibodies.
Autophagy in Glycogenosis Type II (Pompe Disease) and Danon Disease
Pompe disease
Pompe disease was the first recognized lysosomal storage disorder. The disease is caused by deficiency of acid maltase, also known as acid α-glucosidase (GAA). Clinical heterogeneity of the disease is a well-established phenomenon.29,30 In the most serious infantile form, the disease leads to profound weakness, heart failure, and if left untreated causes death within one year. Even in milder late-onset form the illness is relentless and debilitating: patients eventually become confined to a wheelchair or a bed and many die prematurely from respiratory failure.
It took more than 30 y to define the cause of Pompe disease after its discovery in 1932,31,4 and even longer to develop a therapy. Only recently, enzyme replacement therapy (ERT) using recombinant human acid α-glucosidase designed to supplement the defective enzyme was approved for all forms of the disease. This therapy stemmed from a straightforward hypothesis to explain the disease pathogenesis, namely that progressive enlargement of glycogen-filled lysosomes would lead to lysosomal rupture and to release of glycogen and toxic substances into the cytosol.32,33 The assumption was that early treatment, initiated before lysosomal integrity was compromised, would reverse this pathogenic cascade. Now, after years of preclinical and clinical experience with this therapy it has become clear that this assumption is only partially correct: while cardiac muscle responds very well to therapy, skeletal muscle does not. The poor muscle response to therapy led to the need to revisit the pathogenesis of the disease, with increased attention to an involvement of the autophagic pathway in disease pathogenesis.
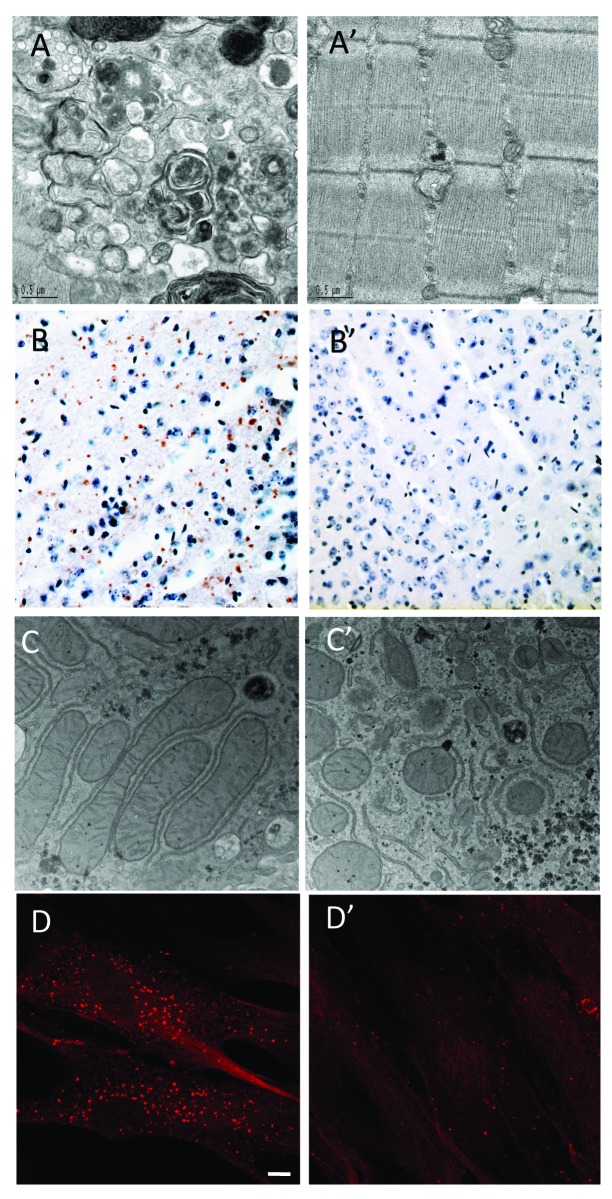
Poor skeletal muscle response to ERT was initially shown in long-term preclinical studies in a mouse model of Pompe disease [GAA-knockout (GAA-KO)].34 The presence of large areas of autophagic accumulation detected by electron microscopy in the therapy-resistant fast muscle suggested that abnormal autophagy might be the culprit (Fig. 1A). Surprisingly, huge clusters of LAMP1 (a lysosomal marker)- and LC3 (an autophagosomal marker)-positive vesicles are seen in the core of virtually every muscle fiber even in young Pompe mice. Only outside the core is the expected pathology of Pompe disease observed, namely individual or isolated groups of expanded lysosomes with clearly defined borders. The autophagic mass grows in size as the animals age, and from a morphological perspective, it is evident that abnormal autophagy, rather than lysosomal expansion, is what eventually causes muscle fiber destruction and the loss of muscle force.35 It appears that both ends of the autophagic process—the generation of autophagosomes and their resolution by lysosomal fusion36—are impaired in Pompe skeletal muscle. The components of the system, such as BECN1, GABARAP, ATG7 and LC3, are present in excess, but this excess is associated with functional deficiency of autophagy. The phenotypic defect in fast muscle of the GAA-KO mice appears to arise not from an induction of autophagy but rather from a block in autophagosomal turnover (a defect in autophagosome-lysosome fusion) in the diseased muscle. Autophagy substrates, such as p62/SQSTM1 and ubiquitinated proteins37-39 accumulate in the core of Pompe muscle fibers.11 The formation of ubiquitin-positive protein aggregates in the GAA-KO mice precedes the development of clinical symptoms and parallels the progression of the disease.11
Figure 1. Representative examples of autophagy defects observed in LSDs. (A) Electron microscopy (EM) provides evidence for the presence of autophagic accumulation in the fast muscles (white part of the gastocnemius muscle) of a 5-mo-old Pompe knockout mouse. (A’) Control. (B) Immuno-histochemical staining of ubiquitin-positive inclusions in the cerebral cortex of 3-mo old MSD mice. (B’) Control. (C) EM showing aberrant mitochondria in the liver of 1-mo-old MSD mice. (C’) Control. (D) Accumulation of autophagosomes in MLIV. Fibroblasts from MLIV patients were grown in complete media, fixed, permeabilized and immunostained with a polyclonal antibody to LC3. Scale bar, 10 µm. (D’) Control.
Autophagic buildup poses a significant problem for the lysosomal delivery of the therapeutic enzyme. The enzyme traffics to its destination, lysosomes, via mannose-6-phosphate receptor-mediated endocytosis. Autophagic and endocytic pathways converge at several steps along the way: autophagosomes fuse not only with lysosomes, but also with late and even early endosomes.40 Therefore, it is perhaps not surprising that the recombinant enzyme ends up in the area of autophagosome accumulation.41–43 Thus, the removal of autophagic buildup seemed a reasonable approach to improve the therapy.
Indeed, ERT in muscle-specific Atg5- or Atg7-deficient Pompe mice results in near-complete glycogen clearance.12 This outcome observed in both young and older mice is never seen in Pompe mice in which autophagy is not tampered with. Of note, ERT plus suppression of autophagy converts Pompe mice into muscle-specific autophagy-deficient wild-type mice; the health (longevity, mobility and single fiber contractility) of these mice is far better than the health of Pompe mice. The accumulation of dysfunctional mitochondria, mild atrophy and age-dependent decrease in force have been reported in muscle-specific autophagy-deficient wild-type mice,44,45 but these abnormalities seem to be a reasonable price to pay for the reversal of pathology in Pompe disease.
In humans, autophagic buildup is present in many muscle cells in late-onset patients (both juvenile and adults), thus making the observations in the mouse model relevant to the human study. Furthermore, in many fibers autophagic accumulation is the overwhelming (and in some fibers the only) pathology, because the lysosomes that lie outside the autophagic region appear essentially normal.46,47 In contrast, the role of autophagy in the pathogenesis of infantile Pompe disease is much less obvious. Unexpectedly, the autophagic component which is so prominent in late-onset cases is insignificant in infants whose biopsies were analyzed within days after birth.47 Instead, the major characteristic of these fibers is the presence of hugely expanded lysosomes without clear borders, a finding consistent with the hypothesis of lysosomal rupture as a cause of muscle destruction.32,33
The difference between the relative contribution of the lysosomal and autophagic pathologies in untreated infants and adults presents a conundrum in Pompe disease. One thing appears to be clear: infants lacking the enzyme are born with already severely damaged muscle fibers filled with giant lysosomes with ruptured membranes and massive glycogen deposits. The lack of autophagic buildup in such infants suggests that the role of autophagy in muscle development is minimal, perhaps because of a constant supply of nutrients through the umbilical cord. This hypothesis is consistent with the data in mice showing a low level of autophagy throughout the embryonic period.48 Analysis of biopsies from infants receiving ERT shows that lysosomes shrink in some patients, but remarkably, autophagic buildup resembling that found in skeletal muscle from adults emerges.47 A long-term study and a larger number of samples are needed to evaluate the fate of autophagic buildup in ERT-treated infants.
Glycogen passage from the cytosol to lysosomes must involve some type of autophagic pathway since by definition autophagy is a process of transport and lysosomal degradation of any intracellular material. Early morphological data showing the presence of glycogen particles inside autophagic vacuoles in skeletal muscle, liver and heart of the newborn rats implied the involvement of macroautophagy.49,50 If this process operates later in life, then inhibition of macroautophagy would alleviate glycogen burden in lysosomes by preventing its transport from the cytosol. Indeed, suppression of autophagosome formation by inactivation of Atg5 or Atg7 in skeletal muscle in untreated Pompe mice results in reduction of accumulated glycogen; for reasons that are unclear, this reduction is much more pronounced in ATG7-deficient Pompe mice.43 Nevertheless, data from both strains suggest that at least some glycogen is delivered to the lysosomes via the macroautophagic pathway. However, it remains unclear how the remaining glycogen is transported to the lysosome. Yet another recent hypothesis suggests that the intracellular glycogen trafficking to lysosomes may proceed via a nonclassical LC3-independent autophagy mediated by glycogen-binding protein STBD1 (starch binding domain-containing protein 1), which anchors glycogen molecules to vesicular membranes.51
Danon disease
Danon disease (also called “glycogen storage disease due to LAMP2 deficiency” or “lysosomal glycogen storage disease with normal acid maltase activity”) is a lysosomal glycogen storage disease due to LAMP2 (lysosomal-associated membrane protein 2) deficiency.52 The disease is inherited as an X-linked trait and is extremely rare. The disease phenotype is characterized by severe cardiomyopathy and variable skeletal muscle weakness often associated with mental retardation.
Danon disease was the first LSD in which an involvement of autophagy was reported.52 Accumulation of autophagic vacuoles is detected in several tissues, particularly the muscle, from a mouse model of the disease.13
Autophagy in Mucopolysaccharidoses
Multiple sulfatase deficiency (MSD) and mucopolysaccharidosis type IIIA (MPS IIIA)
The mucopolysaccharidoses (MPSs) are a group of lysosomal storage disorders caused by deficiency of enzymes catalyzing the degradation of glycosaminoglycans (GAGs), which are long, repeating chains of complex sugar molecules. When one of several enzymes is absent, a progressive buildup of GAGs occurs. Depending on the enzyme deficiency, catabolism of dermatan sulfate, heparan sulfate, keratan sulfate, chondroitin sulfate, or hyaluronan may be blocked, singly or in combination.53 Lysosomal accumulation of GAGs results in cell, tissue and organ dysfunction. There are 11 known enzyme deficiencies that give rise to 7 distinct MPSs. MPS III, also referred to as Sanfilippo syndrome, is caused by a deficiency of enzymes that are needed to break down heparan sulfate. MPS III is classified into four types (A,B,C and D), with each type caused by a deficiency of a different enzyme. MPSIIIA is due to deficiency of the heparan N-sulfatase enzyme, and is characterized by profound mental deterioration, hyperactivity, and relatively mild somatic manifestations.
Multiple sulfatase deficiency (MSD) is a rare but very severe disorder in which affected individuals present a complex multisystemic phenotype due to the impaired activity of all sulfatases. It is caused by mutations in the Sulfatase Modifying Factor 1 (SUMF1) gene, which is responsible for an essential post-translational modification of all sulfatases.54,55 Lack of sulfatase activities in MSD patients leads to accumulation of sulfated lipids and mucopolysaccharides resulting in a clinical phenotype that combines the features of at least seven diseases, five of which are mucopolysaccharidoses.56 The sumf1−/− KO mouse model of MSD recapitulates most of the features of the human disease.57
As for other types of LSDs, the pathogenic mechanisms that lead from enzyme deficiency to cell death in MSD and MPS IIIA are not completely understood. An impairment of autophagy is postulated to play a major role in disease pathogenesis.14 LC3 immunofluorescence reveals a higher number of autophagosomes in several brain regions of mouse models of MSD and MPS IIIA compared with wild-type mice. A decreased colocalization of LAMP1 with the autophagosomal marker LC3 in MSD and MPS IIIA cells compared with wild-type cells in vitro suggests the possibility of impaired lysosome-autophagosome fusion. A defective autophagic flux was also demonstrated by the accumulation of autophagy substrates, such as polyubiquitinated proteins and dysfunctional mitochondria (Fig. 1B and C), both of which are significantly increased in tissues from MSD and MPS IIIA mice.14 In addition, MSD cells show a decreased ability to degrade exogenously expressed aggregate-prone proteins, such as expanded huntingtin and mutated α-synuclein, which are responsible for Huntington and Parkinson disease, respectively.14 Similarly, during skeletal development, chondrocytes from MSD mice suffer from a severe lysosomal storage defect and display a defective autophagosome digestion leading to a defect in energy metabolism and to cell death.58
Studies aimed at the identification of the mechanisms underlying the impairment of the fusion between lysosomes and autophagosomes in MSD and MPS IIIA point to abnormalities of membrane lipid composition and SNARE (Soluble NSF Attachment protein REceptor) protein distribution.15 In particular, a significant accumulation of cholesterol is found in the lysosomal membranes of both MSD and MPS IIIA cells. This is associated with a disorganization of the endolysosomal membrane and with disruption of “lipid rafts.” Interestingly, wild-type cells “loaded” with cholesterol in vitro mirror the fusion defects observed in LSD cells. Conversely, lowering cholesterol by the use of methyl β-cyclodextrin in MSD and MPS IIIA cells rescues normal lysosomal function. The SNARE proteins, which are key components of the cellular membrane fusion machinery, are found aberrantly sequestered within cholesterol-rich regions of LSD lysosomal membranes. This abnormal distribution appears to lock these proteins in complexes and to impair their function and recycling, thereby directly affecting lysosomal membrane fusion capacities.15
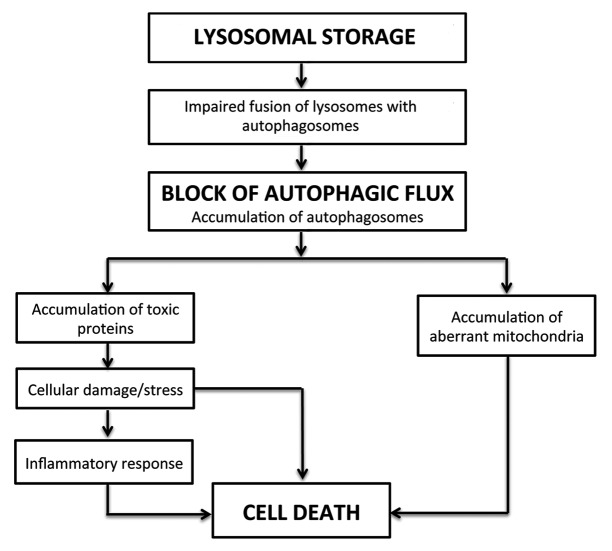
These studies provided evidence that a global lysosomal dysfunction leads to the impaired autophagy observed in the pathogenesis of MSD and MPS IIIA and possibly in other types of LSDs. A possible model for disease pathogenesis is depicted in Figure 2. According to this model, lysosomal accumulation of undegraded substrates results in impaired lysosome-autophagosome fusion and a block of the autophagic flux. This figure depicts a generalized mechanism that may apply to several LSDs, even though a reduced efficiency of autophagic degradation may not necessarily be due to impaired lysosomal fusion. The secondary accumulation of autophagic substrates, such as polyubiquitinated proteins and dysfunctional mitochondria, are a consequence of this block. This model places the emphasis on the secondary accumulation of autophagy substrates as a key determinant of cell death, rather than on the primary accumulation of lysosomal substrates.
Figure 2. Model depicting disease pathogenesis in LSDs.
Mucopolysaccharidosis type VI (MPS VI)
Mucopolysaccharidosis VI (MPS VI), also known as Maroteaux-Lamy syndrome, is caused by deficiency of the lysosomal enzyme N-acetylgalactosamine-4-sulfatase (arylsulfatase B, ARSB).59 ARSB hydrolyzes sulfate esters from GAGs, mainly dermatan sulfate (DS), and a deficiency of the enzyme prevents the sequential degradation of DS leading to its accumulation in various cells and tissues.53 Clinically, MPS VI is characterized by coarse faces, short stature, dysostosis multiplex, stiffness and functional impairment of joints, hepatosplenomegaly, cardiac valve anomalies and corneal clouding.53 No evidence of CNS involvement is evident in clinically severe MPS VI,59 although scattered neuronal storage is evident in animal models lacking ARSB activity.60
Studies using fibroblasts from MPS VI patients demonstrated that lysosomal storage in these cells results in impaired autophagy, accumulation of polyubiquitinated proteins and mitochondrial dysfunction.16 These studies also demonstrated that the extent of LAMP2-LC3 colocalization is similar between MPS VI and normal fibroblasts, indicating that that autophagosome-lysosome fusion is not completely blocked in MPS VI fibroblasts. Since glycosaminoglycans are reported to inhibit cathepsin activity61 and cathepsin-activity deficiency results in impaired autophagy,62 Tessitore et al. suggest that this mechanism might play a role in autophagy impairment in MPS VI. Impaired autophagy is also observed in vivo in a rat model of MPS VI.16 Electron microscopy in livers from affected rats shows an accumulation of autophagic vacuoles, abnormal ubiquitination and abnormal mitochondrial function. Interestingly, gene transfer of wild-type ARSB results in the prevention of derman sulfate accumulation and in the rescue of the defects observed in the autophagic pathway, ubiquitination and mitochondrial function.16
Autophagy in Sphingolipidoses
The sphingolipidoses are a heterogeneous group of inherited disorders of sphingolipid metabolism that often affect the nervous system. These diseases predominantly occur in the pediatric population, and the resulting neurodegeneration produces psychomotor retardation and myoclonus due to diffuse and progressive involvement of neurons, or weakness and spasticity due to involvement of white matter tracts. The underlying genetic defects are diverse and variably result in accumulation of sphingomyelin, glycolipids, glucocerebrosides, gangliosides, unesterified cholesterol and sulfatide compounds.63,64 In some instances, this storage material results from hydrolytic enzyme deficiency, whereas in others its occurrence is less well understood and may be secondary to the accumulation of other lipids. In either case glycosphingolipids can alter macroautophagy. Recent studies demonstrate that loading cells with glycosphingolipids, simply by adding them to the culture media, leads to autophagy induction, decreased clearance of autophagosomes, and an accumulation of autophagy intermediates.65 Strikingly, many of these same changes are observed in model systems used to study the sphingolipidoses, suggesting that an accumulation of one or more glycosphingolipids may alter the function of the autophagic pathway in these disorders.
Niemann-Pick type C disease (NPC)
Niemann-Pick type C disease (NPC) is caused by mutations in the NPC1 or NPC2 genes,66,67 whose protein products are thought to act cooperatively in the efflux of cholesterol from late endosomes and lysosomes.68 NPC1 encodes a multipass transmembrane protein that contains a sterol sensing domain, similar to that found in the regulators of cholesterol biosynthesis, HMG-CoA reductase, SCAP and 7DHCR;69 an N-terminal cholesterol binding domain on the lumenal side of the membrane, and several lumenal loops of unknown function.70,71 NPC2 is a soluble protein primarily localized to the late endosome/lysosome lumen,67 where it binds cholesterol in an orientation opposite that of the NPC1 N-terminal domain. NPC2 is capable of extracting cholesterol from lipid bilayers, and then transferring it to another bilayer or to the NPC1 N-terminal domain.70,72 As a result of NPC1 or NPC2 deficiency, unesterified cholesterol derived from receptor-mediated endocytosis of low-density lipoprotein (LDL)-cholesterol accumulates widely in cells throughout the body. A broad array of glycosphingolipids accumulates as well73 although it is uncertain whether this latter phenomenon is attributable to a direct role for NPC1-NPC2 in glycosphingolipid trafficking or is secondary to cholesterol accumulation.74
A marked accumulation of autophagosomes occurs in the brains of NPC mice and in skin fibroblasts from NPC patients.17,75,76 This is attributable, in part, to the induction of autophagy through BECN1,75 a critical regulator of macroautophagy that binds class III phosphatidylinositol 3-kinase and is both required for the initiation of autophagosome formation and contributes to autophagosome maturation.77 Notably, BECN1 and LC3-II levels in wild-type fibroblasts are increased by U18666A, a small molecule that induces NPC-like lipid trafficking defects, suggesting that an accumulation of unesterified cholesterol or sphinogolipids may act as a proximal trigger.71 This induction is accompanied by a relatively modest increase in autophagic flux as measured by the degradation of long-lived proteins, as well as by the accumulation of the autophagic substrate p62/SQSTM1 in RIPA-insoluble fractions of brain lysates18,75 and by an accumulation of ubiquitinated proteins,78 preferentially in the endosomal/lysosomal fraction of npc1−/− mouse brain lysates.76 Taken together, these findings suggest that NPC1 deficiency leads to both an induction of autophagy and an impairment of autophagic flux, similar to the effects triggered by loading cells with glycosphingolipids. The impairment in degradation of autophagic substrates may contribute to several aspects of NPC neuropathology, including the accumulation of ubiquitinated proteins and the generation of reactive oxygen species.
Gaucher disease
As in NPC, models of Gaucher disease show both an induction of autophagy and an accumulation of autophagosomes and autophagic substrates. Gaucher disease is caused by mutations in either glucocerebrosidase, the lysosomal enzyme that degrades glucosylceramide and glucosylsphingosine, or its activator protein saposin C. Fibroblasts from some patients deficient in saposin C show increased autophagy induction.79 Mice homozygous for mutant V394L glucocerebrosidase and deficient in saposin C, used to model neuronopathic Gaucher disease, exhibit punctate p62/SQSTM1 accumulations in neurons and astrocytes, and sequester undigested material in vesicles within axons, consistent with an accumulation of autophagic substrates.19 These observations suggest that degradation of autolysosome cargo is deficient in Gaucher disease cells. Interestingly, an impairment of lysosomal protein degradation due to glucocerebrosidase haploinsufficiency was recently suggested to occur in carriers of the Gaucher disease mutation, leading to increased risk for Parkinson disease due to diminished α-synuclein degradation.80
Fabry disease and GM1 gangliosidosis
Increased basal expression of the autophagosome marker LC3-II is observed in models of other sphingolipidoses as well, including GM1-gangliosidosis21 and Fabry disease.20 GM1-gangliosidosis is an autosomal recessive lysosomal lipid storage disorder caused by mutations of the lysosomal β-galactosidase (β-gal) and results in the accumulation of GM1 ganglioside. The disease phenotype is characterized by severe CNS dysfunction, visceromegaly and skeletal dysplasia. An increased number of autophagosomes, as detected by the LC3 marker, elevated BECN1 levels, and dysfunctional mitochondria are observed in brains from GM1 gangliosidosis mice.21 In Fabry disease, deficiency of the lysosomal enzyme α-galactosidase A leads to an accumulation of its substrate globotriaosylceramide (Gb3) throughout the body. Measurement of LC3 in cultured cells from Fabry patients reveals increased basal levels compared with wild-type cells and a larger increase in response to starvation. Treatment of starved Fabry disease fibroblasts and lymphocytes with lysosomal protease inhibitors reveals a block in autophagic flux, demonstrating a more severe disruption of degradation through macroautophagy than that observed in other sphingolipidoses. Furthermore, increased staining of p62/SQSTM1 and ubiquitin in renal tissues and in cultured fibroblasts from Fabry patients further supports impairment of autophagic flux.20 For Fabry disease and other sphingolipid storage diseases, defining where and how the impairment of autophagic flux occurs, and establishing the extent to which alterations in macroautophagy contribute to the disease phenotype remain important research objectives.
Autophagy in Mucolipidosis
Mucolipidosis type IV
Mucolipidosis type IV (MLIV) is an autosomal recessive disorder characterized by acute psychomotor delays, achlorydria and visual abnormalities including retinal degeneration, corneal clouding, optic atrophy and strabismus.81-83 Lysosomal inclusions are found in most tissues in MLIV patients. The composition of the storage material is heterogeneous and includes lipids and mucopolysaccharides forming characteristic multiconcentric lamellae, as well as soluble, granulated proteins.84-87 MLIV is caused by mutations in MCOLN1 (mucolipin 1, also known as TRPML1), an endolysosomal cation channel belonging to the transient receptor potential (TRP) superfamily of ion channels.87-90 Whole cell patch clamp, as well as recording of native endolysosomal membranes, have led to the conclusion that MCOLN1 functions as an inwardly (from lumen to cytoplasm) rectifying channel permeable to Ca2+, Na+, K+ and Fe2+/Mn2+ whose activity is potentiated by low pH.91-93
Although the cellular role of MCOLN1 is still under investigation, the current model suggests that this protein mediates Ca2+ efflux from late endosomes and lysosomes.94,95 Localized Ca2+ release from such acidic stores is required for fusion between endocytic vesicles and to maintain organelle homeostasis, thus suggesting that MCOLN1 is a key regulator of membrane trafficking along the endosomal pathway. In agreement with this model, delivery of cargo from the cell surface to the lysosome and fusion of lysosomes with the plasma membrane are impaired in MCOLN1-deficient cells.24,96-99 Autophagosome turnover is also defective in fibroblasts derived from MLIV patients. The degradation of the autophagosome content requires fusion of autophagosomes with the late endocytic pathway. In MCOLN1-deficient fibroblasts both degradation of the autophagosome content and fusion of autophagosomes with late endosomes/lysosomes are delayed compared with control cells.24 This leads to a dramatic accumulation of autophagosomes in the cytosol of MLIV cells as demonstrated by indirect immunofluorescence (Fig. 1D), LC3-II/LC3-I immunoblot and electron microscopy.24 Impairment of the autophagic pathway has detrimental consequences for the cell leading to inefficient degradation of protein aggregates and damaged organelles. In particular, accumulation of p62/SQSTM1 inclusions and abnormal mitochondria has been described in MLIV fibroblasts.24,100
Findings suggest additional roles for MCOLN1 in the regulation of autophagy. MCOLN1 interacts with HSPA8 (HSC70) and DNAJB1 (HSP40), two components of the chaperone-mediated autophagy (CMA) molecular machinery.25 MLIV fibroblasts show defective CMA and decreased levels of LAMP2A resulting in increased levels of oxidized proteins.25 Finally, MCOLN1 has the ability to form heteroligomers with the two other members of the mucolipin family, MCOLN2 and MCOLN3.101,102 Depletion of endogenous MCOLN3 or expression of a channel-dead dominant negative MCOLN3 mutant inhibits starvation-induced autophagy.102,103 Therefore, MCOLN1 might also play an indirect role in the initiation of autophagy.
Autophagy has also been shown to be defective in primary neurons cultured from a murine model of MLIV. The mcoln1−/− mice provide an excellent phenotypic model of the human disease, and all of the hallmarks of MLIV are present in the mice with the exception of corneal clouding.104 At birth, the knockout animals display no obvious behavioral or morphological phenotypes when compared with wild-type littermates, but as they age they show progressive limb weakening, eventual limb paralysis, and death at approximately eight months. Analysis of the brain at 8 mo shows lysosomal inclusions in multiple cell types, including neurons, astrocytes, oligodendrocytes, microglia and endothelial cells, with larger inclusions present in neurons. The MLIV inclusions are unusual in their combination of dense lamella and granular matrices, and they do not resemble those typically seen in the glycosphingolipidoses, the mucopolysaccharidoses, or most other types of lysosomal disease.104,105 Electron microscopy of primary cerebellar neurons from Mcoln1-deficient mouse embryos demonstrates significant membranous intracytoplasmic storage bodies, despite the lack of gross phenotype at birth.26 Evaluation of macroautophagy in neurons by LC3-II/LC3-I immunoblot shows increased levels of LC3-II, similar to what is seen in human MLIV fibroblasts. LC3-II clearance is also defective, as treatment of the mcoln1−/− neuronal cultures with protease inhibitors to stimulate autophagy does not result in increased LC3-II levels.26 Demonstration of defective autophagy in MCOLN1-deficient neurons suggests a possible mechanism underlying neurodegeneration, whereby increased protein aggregation and organelle damage lead to autophagic stress and eventual neuronal death.24 The MLIV mouse model provides an important tool for evaluating the complicated interplay between chaperone-mediated autophagy and macroautophagy and their role in neurodegenerative disease.
Mucolipidosis type II and Mucolipidosis type III
Mucolipidosis type II (MLII) and Mucolipidosis type III (MLIII) are autosomal recessive diseases caused by deficiency of the enzyme N-acetylglucosamine 1-phosphotransferase (GlcNAc-phosphotransferase).106,107 This enzyme modifies newly synthesized lysosomal hydrolases by attaching a molecule of mannose-6-phosphate (M6P) that functions as a tag for delivery to lysosomes.108 Mutations in GlcNAc-phosphotransferase result in the missorting and cellular loss of lysosomal enzymes and lysosomal accumulation of storage material.109,110 MLII is characterized by skeletal abnormalities, short stature, cardiomegaly and developmental delays. MLIII is a late onset, milder form of MLII.111 Alterations in autophagy have been reported recently in MLII and MLIII fibroblasts. This includes accumulation of autophagosomes, p62/SQSTM1, ubiquitin and fragmented mitochondria. In contrast, no variations in the levels of BECN1 are observed, suggesting that formation of autophagosomes is not increased in these disorders.22 Accumulation of LC3-positive structures and ubiquitin aggregates is also reported in neuronal cells of dorsal root ganglions in a MLIII patient.23 Importantly, inhibition of autophagy restores mitochondrial alterations in MLII and MLIII cells, thus suggesting that increased autophagy might be detrimental for proper mitochondrial function.22
Autophagy in Neuronal Ceroid-Lipofuscinoses
The neuronal ceroid lipofuscinoses (NCLs) are the most common cause of neurodegeneration among children. These disorders typically manifest with blindness, seizures, progressive cognitive defects and motor failure. NCLs are both genetically and phenotypically heterogeneous.112 The juvenile onset NCL (JNCL), also known as Batten disease, is the most common form of NCL and is due to mutations in the CLN3 gene.113 The congenital form of NCL is due to mutations in the cathepsin D gene (CTSD/CLN10).114 A significant increase in the number of autophagosomes is observed in patients affected by these two types of NCLs and in the murine models of these diseases.27,28 In addition, accumulation of dysfunctional mitochondria and downregulation of the MTOR pathway, a sign of activation of autophagosome formation, are detected in JNCL due to mutations of the Cln3 gene.28
Concluding Remarks
Autophagy has been analyzed in a variety of LSDs with different severities of the phenotype, different tissues involved, and different types of storage molecules. Table 1 shows the main findings obtained and Figure 1 displays some examples of the results obtained by these studies. In spite of all the above-mentioned differences among the diseases and samples analyzed, a common theme can be recognized (see also ref. 115 for a further review). In most cases there is an impairment of autophagic flux, causing a secondary accumulation of autophagy substrates such as polyubiquitinated proteins, p62/SQSTM1 and dysfunctional mitochondria, on one end, and an increase in factors involved in autophagosome formation, such as BECN1, as an attempt to compensate for the impaired autophagic flux, on the other. Accordingly, LSDs can be seen primarily as “autophagy disorders.”10 Interestingly, a defect in autophagic lysosome formation, due to abnormal MTOR activation, is reported in several LSDs.3
Impairment of autophagy and accumulation of autophagy substrates in LSDs suggest that at least some mechanisms underlying the LSD phenotype may be similar to other diseases in which defective autophagy has been observed. In particular, many neurodegenerative diseases that affect the aging population including Alzheimer, Parkinson and Huntington diseases, exhibit the presence of intraneuronal accumulations of protein aggregates as key elements of their pathogenic cascades. Yet the potential importance of macroautophagy as a critical player in the formation of these aggregates was largely unappreciated prior to studies in which key autophagy regulatory proteins were conditionally knocked out in mouse models. Genetic ablation in the CNS of Atg5 and Atg7, each critical for the formation of autophagosomes in cells, was found to cause progressive neurological disease in mice accompanied the accumulation of protein aggregates and inclusion bodies, and by neuron death.116,117 Subsequent studies further revealed the critical role of ATG5 and ATG7 in maintaining normal integrity of axons, particularly for Purkinje cells, which develop early and progressive neuroaxonal dystrophy characterized by accumulation of autophagosome-like membrane structures in swellings along the length of the axon.117,118
A further link between LSDs and neurodegenerative disorders comes from studies showing accumulation of phosphorylated-tau aggregates and neurofibrillary tangles, which are typically detected in patients with Alzheimer disease, in LSDs such as Niemann-Pick type C119 and mucopolysaccharidoses,120 and deficient degradation of amyloid precursor protein (APP) in sphingolipidoses.65 Parallels between the neuropathology of Niemann-Pick C and several age-dependent protein aggregation neurodegenerative disorders are striking, and include the accumulation of phosphorylated tau and α-synuclein,121-124 and in some cases amyloid β.125 The recent identification of a defect of lysosomal acidification in a murine model of Alzheimer disease126 and of pathogenic lysosomal depletion in a mouse model of Parkinson disease127 further support a connection between lysosomal dysfunction and neurodegenerative diseases.
The acknowledged importance of autophagy in maintaining normal bulk protein recycling in these diseases has led to interest in whether pharmacological stimulation of autophagy could provide therapeutic benefit by delaying or preventing protein aggregate accumulations in disorders such as Huntington disease.128 Rapamycin was demonstrated to protect against neurodegeneration in a fly model of Huntington and to improve neuropathology in a mouse model of Huntington disease by enhancing autophagy-mediated clearance of huntingtin accumulation.129 Rapamycin was also shown to have a clearing effect on a variety of aggregate-prone proteins with polyglutamine or polyalanine expansions, leading to an improvement in cellular phenotype.130 These data suggest that enhancement of autophagy can be an effective approach to reduce toxic protein accumulation and cell death also in LSDs. Trehalose, a disaccharide present in many nonmammalian species, is an mTOR-independent autophagy activator, that has an anti-apoptotic effect in cultured neurons and accelerates the clearance of mutant hungtintin and α-synuclein.131 However, one should also take into consideration that the defect observed in most LSDs is not a dysfunction in the formation of autophagosomes, but a block in autophagic flux. Therefore, activation of autophagy in this scenario may have detrimental effects. This would be consistent with the observation that suppression of autophagy has a beneficial effects on ERT for Pompe disease.43
Recently, it was shown that cholesterol accumulation in the endolysosomal membranes in LSDs changes their organization and composition and reduces their fusion capacity.15 It is possible that drugs aimed at reducing cholesterol in membranes, such as methyl-β-cyclodextrin (MβCD), can restore the fusion capacity of lysosomal membranes and release the block of autophagy in LSDs, as recently described for MSD and MPS IIIA.15 This compound is toxic in vivo, but FDA approved cyclodextrins (e.g., Kleptose, Trappsol and Captisol) may be used. Indeed, chronic treatment using hydroxypropyl β-cyclodextrin in mice with NPC disease results in cholesterol and ganglioside storage reduction, normalization in the autophagy marker LC3-II and significantly increased survival, although similar studies in MPS IIIA and GM1 gangliosidosis mice do not show similar benefit.132 Finally, a potentially attractive possibility would be to induce a global enhancement of both the lysosomal and autophagic pathways by acting on the master gene TFEB.9,99 Further studies are needed to explore the effects of modulators of autophagy, which operate at different steps of the autophagic flux, on the LSD phenotype in cell culture and in animal models. Hopefully, these studies will lead to the development of effective treatments for several LSDs.
Acknowledgments
We thank Dr. G. Diez-Roux for manuscript preparation, helpful discussions and critical reading of the manuscript and C. Settembre for critical reading of the manuscript. A.P.L. is supported by the NIH grant R01 NS063967; R.P. is supported by the Intramural Research Program of the NIH, National Heart, Lung, and Blood Institute (NHLBI); S.U.W. is supported by NIH grant HD045561; A.B. is supported by the Beyond Batten Disease Foundation, the Telethon Foundation and the European Research Council (ERC) grant #250154.
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/19469
References
- 1.Luzio JP, Pryor PR, Bright NA. Lysosomes: fusion and function. Nat Rev Mol Cell Biol. 2007;8:622–32. doi: 10.1038/nrm2217. [DOI] [PubMed] [Google Scholar]
- 2.Piper RC, Luzio JP. Ubiquitin-dependent sorting of integral membrane proteins for degradation in lysosomes. Curr Opin Cell Biol. 2007;19:459–65. doi: 10.1016/j.ceb.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465:942–6. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hers HG. alpha-Glucosidase deficiency in generalized glycogenstorage disease (Pompe’s disease) Biochem J. 1963;86:11–6. doi: 10.1042/bj0860011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Platt FM, Walkley SU. Lysosomal Disorders of the Brain. Oxford University Press, 2004. [Google Scholar]
- 6.Ballabio A, Gieselmann V. Lysosomal disorders: from storage to cellular damage. Biochim Biophys Acta. 2009;1793:684–96. doi: 10.1016/j.bbamcr.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Walkley SU. Pathogenic cascades in lysosomal disease-Why so complex? J Inherit Metab Dis. 2009;32:181–9. doi: 10.1007/s10545-008-1040-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sardiello M, Ballabio A. Lysosomal enhancement: a CLEAR answer to cellular degradative needs. Cell Cycle. 2009;8:4021–2. doi: 10.4161/cc.8.24.10263. [DOI] [PubMed] [Google Scholar]
- 9.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–33. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Settembre C, Fraldi A, Rubinsztein DC, Ballabio A. Lysosomal storage diseases as disorders of autophagy. Autophagy. 2008;4:113–4. doi: 10.4161/auto.5227. [DOI] [PubMed] [Google Scholar]
- 11.Raben N, Hill V, Shea L, Takikita S, Baum R, Mizushima N, et al. Suppression of autophagy in skeletal muscle uncovers the accumulation of ubiquitinated proteins and their potential role in muscle damage in Pompe disease. Hum Mol Genet. 2008;17:3897–908. doi: 10.1093/hmg/ddn292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raben N, Schreiner C, Baum R, Takikita S, Xu S, Xie T, et al. Suppression of autophagy permits successful enzyme replacement therapy in a lysosomal storage disorder--murine Pompe disease. Autophagy. 2010;6:1078–89. doi: 10.4161/auto.6.8.13378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanaka Y, Guhde G, Suter A, Eskelinen EL, Hartmann D, Lüllmann-Rauch R, et al. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature. 2000;406:902–6. doi: 10.1038/35022595. [DOI] [PubMed] [Google Scholar]
- 14.Settembre C, Fraldi A, Jahreiss L, Spampanato C, Venturi C, Medina D, et al. A block of autophagy in lysosomal storage disorders. Hum Mol Genet. 2008;17:119–29. doi: 10.1093/hmg/ddm289. [DOI] [PubMed] [Google Scholar]
- 15.Fraldi A, Annunziata F, Lombardi A, Kaiser HJ, Medina DL, Spampanato C, et al. Lysosomal fusion and SNARE function are impaired by cholesterol accumulation in lysosomal storage disorders. EMBO J. 2010;29:3607–20. doi: 10.1038/emboj.2010.237. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Tessitore A, Pirozzi M, Auricchio A. Abnormal autophagy, ubiquitination, inflammation and apoptosis are dependent upon lysosomal storage and are useful biomarkers of mucopolysaccharidosis VI. Pathogenetics. 2009;2:4. doi: 10.1186/1755-8417-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ko DC, Milenkovic L, Beier SM, Manuel H, Buchanan J, Scott MP. Cell-autonomous death of cerebellar purkinje neurons with autophagy in Niemann-Pick type C disease. PLoS Genet. 2005;1:81–95. doi: 10.1371/journal.pgen.0010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pacheco CD, Elrick MJ, Lieberman AP. Tau deletion exacerbates the phenotype of Niemann-Pick type C mice and implicates autophagy in pathogenesis. Hum Mol Genet. 2009;18:956–65. doi: 10.1093/hmg/ddn423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun Y, Liou B, Ran H, Skelton MR, Williams MT, Vorhees CV, et al. Neuronopathic Gaucher disease in the mouse: viable combined selective saposin C deficiency and mutant glucocerebrosidase (V394L) mice with glucosylsphingosine and glucosylceramide accumulation and progressive neurological deficits. Hum Mol Genet. 2010;19:1088–97. doi: 10.1093/hmg/ddp580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chévrier M, Brakch N, Céline L, Genty D, Ramdani Y, Moll S, et al. Autophagosome maturation is impaired in Fabry disease. Autophagy. 2010;6:589–99. doi: 10.4161/auto.6.5.11943. [DOI] [PubMed] [Google Scholar]
- 21.Takamura A, Higaki K, Kajimaki K, Otsuka S, Ninomiya H, Matsuda J, et al. Enhanced autophagy and mitochondrial aberrations in murine G(M1)-gangliosidosis. Biochem Biophys Res Commun. 2008;367:616–22. doi: 10.1016/j.bbrc.2007.12.187. [DOI] [PubMed] [Google Scholar]
- 22.Otomo T, Higaki K, Nanba E, Ozono K, Sakai N. Inhibition of autophagosome formation restores mitochondrial function in mucolipidosis II and III skin fibroblasts. Mol Genet Metab. 2009;98:393–9. doi: 10.1016/j.ymgme.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi H, Takahashi-Fujigasaki J, Fukuda T, Sakurai K, Shimada Y, Nomura K, et al. Pathology of the first autopsy case diagnosed as mucolipidosis type III α/β suggesting autophagic dysfunction. Mol Genet Metab. 2011;102:170–5. doi: 10.1016/j.ymgme.2010.09.014. [DOI] [PubMed] [Google Scholar]
- 24.Vergarajauregui S, Connelly PS, Daniels MP, Puertollano R. Autophagic dysfunction in mucolipidosis type IV patients. Hum Mol Genet. 2008;17:2723–37. doi: 10.1093/hmg/ddn174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Venugopal B, Mesires NT, Kennedy JC, Curcio-Morelli C, Laplante JM, Dice JF, et al. Chaperone-mediated autophagy is defective in mucolipidosis type IV. J Cell Physiol. 2009;219:344–53. doi: 10.1002/jcp.21676. [DOI] [PubMed] [Google Scholar]
- 26.Curcio-Morelli C, Charles FA, Micsenyi MC, Cao Y, Venugopal B, Browning MF, et al. Macroautophagy is defective in mucolipin-1-deficient mouse neurons. Neurobiol Dis. 2010;40:370–7. doi: 10.1016/j.nbd.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koike M, Shibata M, Waguri S, Yoshimura K, Tanida I, Kominami E, et al. Participation of autophagy in storage of lysosomes in neurons from mouse models of neuronal ceroid-lipofuscinoses (Batten disease) Am J Pathol. 2005;167:1713–28. doi: 10.1016/S0002-9440(10)61253-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao Y, Espinola JA, Fossale E, Massey AC, Cuervo AM, MacDonald ME, et al. Autophagy is disrupted in a knock-in mouse model of juvenile neuronal ceroid lipofuscinosis. J Biol Chem. 2006;281:20483–93. doi: 10.1074/jbc.M602180200. [DOI] [PubMed] [Google Scholar]
- 29.van der Ploeg AT, Reuser AJ. Pompe’s disease. Lancet. 2008;372:1342–53. doi: 10.1016/S0140-6736(08)61555-X. [DOI] [PubMed] [Google Scholar]
- 30.Hirschhorn R, Reuser A.Glycogen storage disease type II: acid a-glucosidase (acid maltase) deficiency. In: Scriver C, Beaudet A, Sly W, Valle D, eds. The metabolic and molecular bases of inherited disease. New York, NY: McGraw-Hill; 2001:3389-420 [Google Scholar]
- 31.Pompe JC. Over idiopatische hypertrophie van het hart. Ned Tijdschr Geneeskd. 1932;76:304. [Google Scholar]
- 32.Griffin JL. Infantile acid maltase deficiency. I. Muscle fiber destruction after lysosomal rupture. Virchows Arch B Cell Pathol Incl Mol Pathol. 1984;45:23–36. doi: 10.1007/BF02889849. [DOI] [PubMed] [Google Scholar]
- 33.Thurberg BL, Lynch Maloney C, Vaccaro C, Afonso K, Tsai AC, Bossen E, et al. Characterization of pre- and post-treatment pathology after enzyme replacement therapy for Pompe disease. Lab Invest. 2006;86:1208–20. doi: 10.1038/labinvest.3700484. [DOI] [PubMed] [Google Scholar]
- 34.Raben N, Fukuda T, Gilbert AL, de Jong D, Thurberg BL, Mattaliano RJ, et al. Replacing acid alpha-glucosidase in Pompe disease: recombinant and transgenic enzymes are equipotent, but neither completely clears glycogen from type II muscle fibers. Mol Ther. 2005;11:48–56. doi: 10.1016/j.ymthe.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 35.Xu S, Galperin M, Melvin G, Horowits R, Raben N, Plotz P, et al. Impaired organization and function of myofilaments in single muscle fibers from a mouse model of Pompe disease. J Appl Physiol. 2010;108:1383–8. doi: 10.1152/japplphysiol.01253.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–26. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–14. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 39.Ichimura Y, Kumanomidou T, Sou YS, Mizushima T, Ezaki J, Ueno T, et al. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem. 2008;283:22847–57. doi: 10.1074/jbc.M802182200. [DOI] [PubMed] [Google Scholar]
- 40.Berg TO, Fengsrud M, Strømhaug PE, Berg T, Seglen PO. Isolation and characterization of rat liver amphisomes. Evidence for fusion of autophagosomes with both early and late endosomes. J Biol Chem. 1998;273:21883–92. doi: 10.1074/jbc.273.34.21883. [DOI] [PubMed] [Google Scholar]
- 41.Fukuda T, Roberts A, Ahearn M, Zaal K, Ralston E, Plotz PH, et al. Autophagy and lysosomes in Pompe disease. Autophagy. 2006;2:318–20. doi: 10.4161/auto.2984. [DOI] [PubMed] [Google Scholar]
- 42.Fukuda T, Ahearn M, Roberts A, Mattaliano RJ, Zaal K, Ralston E, et al. Autophagy and mistargeting of therapeutic enzyme in skeletal muscle in Pompe disease. Mol Ther. 2006;14:831–9. doi: 10.1016/j.ymthe.2006.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raben N, Wong A, Ralston E, Myerowitz R. Autophagy and mitochondria in Pompe disease: nothing is so new as what has long been forgotten. Am J Med Genet C Semin Med Genet. 2012;160:13–21. doi: 10.1002/ajmg.c.31317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu JJ, Quijano C, Chen E, Liu H, Cao L, Fergusson MM, et al. Mitochondrial dysfunction and oxidative stress mediate the physiological impairment induced by the disruption of autophagy. Aging (Albany NY) 2009;1:425–37. doi: 10.18632/aging.100038. [AlbanyNY] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Masiero E, Sandri M. Autophagy inhibition induces atrophy and myopathy in adult skeletal muscles. Autophagy. 2010;6:307–9. doi: 10.4161/auto.6.2.11137. [DOI] [PubMed] [Google Scholar]
- 46.Raben N, Takikita S, Pittis MG, Bembi B, Marie SKN, Roberts A, et al. Deconstructing Pompe disease by analyzing single muscle fibers: to see a world in a grain of sand. Autophagy. 2007;3:546–52. doi: 10.4161/auto.4591. [DOI] [PubMed] [Google Scholar]
- 47.Raben N, Ralston E, Chien YH, Baum R, Schreiner C, Hwu WL, et al. Differences in the predominance of lysosomal and autophagic pathologies between infants and adults with Pompe disease: implications for therapy. Mol Genet Metab. 2010;101:324–31. doi: 10.1016/j.ymgme.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–6. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 49.Schiaffino S, Hanzlíková V. Autophagic degradation of glycogen in skeletal muscles of the newborn rat. J Cell Biol. 1972;52:41–51. doi: 10.1083/jcb.52.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kondomerkos DJ, Kalamidas SA, Kotoulas OB, Hann AC. Glycogen autophagy in the liver and heart of newborn rats. The effects of glucagon, adrenalin or rapamycin. Histol Histopathol. 2005;20:689–96. doi: 10.14670/HH-20.689. [DOI] [PubMed] [Google Scholar]
- 51.Jiang S, Heller B, Tagliabracci VS, Zhai L, Irimia JM, DePaoli-Roach AA, et al. Starch binding domain-containing protein 1/genethonin 1 is a novel participant in glycogen metabolism. J Biol Chem. 2010;285:34960–71. doi: 10.1074/jbc.M110.150839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nishino I, Fu J, Tanji K, Yamada T, Shimojo S, Koori T, et al. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease) Nature. 2000;406:906–10. doi: 10.1038/35022604. [DOI] [PubMed] [Google Scholar]
- 53.Neufeld E, Muenzer J. The mucopolysaccharidoses New York: McGraw-Hill, 2001. [Google Scholar]
- 54.Cosma MP, Pepe S, Annunziata I, Newbold RF, Grompe M, Parenti G, et al. The multiple sulfatase deficiency gene encodes an essential and limiting factor for the activity of sulfatases. Cell. 2003;113:445–56. doi: 10.1016/S0092-8674(03)00348-9. [DOI] [PubMed] [Google Scholar]
- 55.Dierks T, Schmidt B, Borissenko LV, Peng J, Preusser A, Mariappan M, et al. Multiple sulfatase deficiency is caused by mutations in the gene encoding the human C(alpha)-formylglycine generating enzyme. Cell. 2003;113:435–44. doi: 10.1016/S0092-8674(03)00347-7. [DOI] [PubMed] [Google Scholar]
- 56.Hopwood JJ, Ballabio A. Multiple Sulfatase Deficiency and the Nature of the Sulfatase Family. In: Scriver CR, Beaudet AL, Valle D, Sly WS, Childs B, Kinzler KW, et al., eds. The Metabolic & Molecular Bases of Inherited Disease. New York: McGraw Hill, 2001:3725-32. [Google Scholar]
- 57.Settembre C, Annunziata I, Spampanato C, Zarcone D, Cobellis G, Nusco E, et al. Systemic inflammation and neurodegeneration in a mouse model of multiple sulfatase deficiency. Proc Natl Acad Sci U S A. 2007;104:4506–11. doi: 10.1073/pnas.0700382104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Settembre C, Arteaga-Solis E, McKee MD, de Pablo R, Al Awqati Q, Ballabio A, et al. Proteoglycan desulfation determines the efficiency of chondrocyte autophagy and the extent of FGF signaling during endochondral ossification. Genes Dev. 2008;22:2645–50. doi: 10.1101/gad.1711308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hopwood JJ, Morris CP. The mucopolysaccharidoses. Diagnosis, molecular genetics and treatment. Mol Biol Med. 1990;7:381–404. [PubMed] [Google Scholar]
- 60.Walkley SU, Thrall MA, Haskins ME, Mitchell TW, Wenger DA, Brown DE, et al. Abnormal neuronal metabolism and storage in mucopolysaccharidosis type VI (Maroteaux-Lamy) disease. Neuropathol Appl Neurobiol. 2005;31:536–44. doi: 10.1111/j.1365-2990.2005.00675.x. [DOI] [PubMed] [Google Scholar]
- 61.Li Z, Yasuda Y, Li W, Bogyo M, Katz N, Gordon RE, et al. Regulation of collagenase activities of human cathepsins by glycosaminoglycans. J Biol Chem. 2004;279:5470–9. doi: 10.1074/jbc.M310349200. [DOI] [PubMed] [Google Scholar]
- 62.Boland B, Kumar A, Lee S, Platt FM, Wegiel J, Yu WH, et al. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer’s disease. J Neurosci. 2008;28:6926–37. doi: 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eckhardt M. Pathology and current treatment of neurodegenerative sphingolipidoses. Neuromolecular Med. 2010;12:362–82. doi: 10.1007/s12017-010-8133-7. [DOI] [PubMed] [Google Scholar]
- 64.Staretz-Chacham O, Lang TC, LaMarca ME, Krasnewich D, Sidransky E. Lysosomal storage disorders in the newborn. Pediatrics. 2009;123:1191–207. doi: 10.1542/peds.2008-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tamboli IY, Hampel H, Tien NT, Tolksdorf K, Breiden B, Mathews PM, et al. Sphingolipid storage affects autophagic metabolism of the amyloid precursor protein and promotes Abeta generation. J Neurosci. 2011;31:1837–49. doi: 10.1523/JNEUROSCI.2954-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carstea ED, Morris JA, Coleman KG, Loftus SK, Zhang D, Cummings C, et al. Niemann-Pick C1 disease gene: homology to mediators of cholesterol homeostasis. Science. 1997;277:228–31. doi: 10.1126/science.277.5323.228. [DOI] [PubMed] [Google Scholar]
- 67.Naureckiene S, Sleat DE, Lackland H, Fensom A, Vanier MT, Wattiaux R, et al. Identification of HE1 as the second gene of Niemann-Pick C disease. Science. 2000;290:2298–301. doi: 10.1126/science.290.5500.2298. [DOI] [PubMed] [Google Scholar]
- 68.Kwon HJ, Abi-Mosleh L, Wang ML, Deisenhofer J, Goldstein JL, Brown MS, et al. Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol. Cell. 2009;137:1213–24. doi: 10.1016/j.cell.2009.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ioannou YA. Multidrug permeases and subcellular cholesterol transport. Nat Rev Mol Cell Biol. 2001;2:657–68. doi: 10.1038/35089558. [DOI] [PubMed] [Google Scholar]
- 70.Infante RE, Wang ML, Radhakrishnan A, Kwon HJ, Brown MS, Goldstein JL. NPC2 facilitates bidirectional transfer of cholesterol between NPC1 and lipid bilayers, a step in cholesterol egress from lysosomes. Proc Natl Acad Sci U S A. 2008;105:15287–92. doi: 10.1073/pnas.0807328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Scott C, Ioannou YA. The NPC1 protein: structure implies function. Biochim Biophys Acta. 2004;1685:8–13. doi: 10.1016/j.bbalip.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 72.Xu Z, Farver W, Kodukula S, Storch J. Regulation of sterol transport between membranes and NPC2. Biochemistry. 2008;47:11134–43. doi: 10.1021/bi801328u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Karten B, Peake KB, Vance JE. Mechanisms and consequences of impaired lipid trafficking in Niemann-Pick type C1-deficient mammalian cells. Biochim Biophys Acta. 2009;1791:659–70. doi: 10.1016/j.bbalip.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 74.Zhou S, Davidson C, McGlynn R, Stephney G, Dobrenis K, Vanier MT, et al. Endosomal/lysosomal processing of gangliosides affects neuronal cholesterol sequestration in Niemann-Pick disease type C. Am J Pathol. 2011;179:890–902. doi: 10.1016/j.ajpath.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pacheco CD, Kunkel R, Lieberman AP. Autophagy in Niemann-Pick C disease is dependent upon Beclin-1 and responsive to lipid trafficking defects. Hum Mol Genet. 2007;16:1495–503. doi: 10.1093/hmg/ddm100. [DOI] [PubMed] [Google Scholar]
- 76.Liao G, Yao Y, Liu J, Yu Z, Cheung S, Xie A, et al. Cholesterol accumulation is associated with lysosomal dysfunction and autophagic stress in Npc1 -/- mouse brain. Am J Pathol. 2007;171:962–75. doi: 10.2353/ajpath.2007.070052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–76. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Higashi Y, Murayama S, Pentchev PG, Suzuki K. Cerebellar degeneration in the Niemann-Pick type C mouse. Acta Neuropathol. 1993;85:175–84. doi: 10.1007/BF00227765. [DOI] [PubMed] [Google Scholar]
- 79.Vaccaro AM, Motta M, Tatti M, Scarpa S, Masuelli L, Bhat M, et al. Saposin C mutations in Gaucher disease patients resulting in lysosomal lipid accumulation, saposin C deficiency, but normal prosaposin processing and sorting. Hum Mol Genet. 2010;19:2987–97. doi: 10.1093/hmg/ddq204. [DOI] [PubMed] [Google Scholar]
- 80.Mazzulli JR, Xu YH, Sun Y, Knight AL, McLean PJ, Caldwell GA, et al. Gaucher disease glucocerebrosidase and α-synuclein form a bidirectional pathogenic loop in synucleinopathies. Cell. 2011;146:37–52. doi: 10.1016/j.cell.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Altarescu G, Sun M, Moore DF, Smith JA, Wiggs EA, Solomon BI, et al. The neurogenetics of mucolipidosis type IV. Neurology. 2002;59:306–13. doi: 10.1212/wnl.59.3.306. [DOI] [PubMed] [Google Scholar]
- 82.Bonavita S, Virta A, Jeffries N, Goldin E, Tedeschi G, Schiffmann R. Diffuse neuroaxonal involvement in mucolipidosis IV as assessed by proton magnetic resonance spectroscopic imaging. J Child Neurol. 2003;18:443–9. doi: 10.1177/08830738030180070701. [DOI] [PubMed] [Google Scholar]
- 83.Wakabayashi K, Gustafson AM, Sidransky E, Goldin E. Mucolipidosis type IV: an update. Mol Genet Metab. 2011;104:206–13. doi: 10.1016/j.ymgme.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Berman ER, Livni N, Shapira E, Merin S, Levij IS. Congenital corneal clouding with abnormal systemic storage bodies: a new variant of mucolipidosis. J Pediatr. 1974;84:519–26. doi: 10.1016/S0022-3476(74)80671-2. [DOI] [PubMed] [Google Scholar]
- 85.Riedel KG, Zwaan J, Kenyon KR, Kolodny EH, Hanninen L, Albert DM. Ocular abnormalities in mucolipidosis IV. Am J Ophthalmol. 1985;99:125–36. doi: 10.1016/0002-9394(85)90220-x. [DOI] [PubMed] [Google Scholar]
- 86.Goldin E, Cooney A, Kaneski CR, Brady RO, Schiffmann R. Mucolipidosis IV consists of one complementation group. Proc Natl Acad Sci U S A. 1999;96:8562–6. doi: 10.1073/pnas.96.15.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Slaugenhaupt SA, Acierno JS, Jr., Helbling LA, Bove C, Goldin E, Bach G, et al. Mapping of the mucolipidosis type IV gene to chromosome 19p and definition of founder haplotypes. Am J Hum Genet. 1999;65:773–8. doi: 10.1086/302549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bargal R, Avidan N, Ben-Asher E, Olender Z, Zeigler M, Frumkin A, et al. Identification of the gene causing mucolipidosis type IV. Nat Genet. 2000;26:118–23. doi: 10.1038/79095. [DOI] [PubMed] [Google Scholar]
- 89.Bassi MT, Manzoni M, Monti E, Pizzo MT, Ballabio A, Borsani G. Cloning of the gene encoding a novel integral membrane protein, mucolipidin-and identification of the two major founder mutations causing mucolipidosis type IV. Am J Hum Genet. 2000;67:1110–20. doi: 10.1016/s0002-9297(07)62941-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sun M, Goldin E, Stahl S, Falardeau JL, Kennedy JC, Acierno JS, Jr., et al. Mucolipidosis type IV is caused by mutations in a gene encoding a novel transient receptor potential channel. Hum Mol Genet. 2000;9:2471–8. doi: 10.1093/hmg/9.17.2471. [DOI] [PubMed] [Google Scholar]
- 91.Dong XP, Cheng X, Mills E, Delling M, Wang F, Kurz T, et al. The type IV mucolipidosis-associated protein TRPML1 is an endolysosomal iron release channel. Nature. 2008;455:992–6. doi: 10.1038/nature07311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Puertollano R, Kiselyov K. TRPMLs: in sickness and in health. Am J Physiol Renal Physiol. 2009;296:F1245–54. doi: 10.1152/ajprenal.90522.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dong XP, Shen D, Wang X, Dawson T, Li X, Zhang Q, et al. PI(3,5)P(2) controls membrane trafficking by direct activation of mucolipin Ca(2+) release channels in the endolysosome. Nat Commun. 2010;1:38. doi: 10.1038/ncomms1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cheng X, Shen D, Samie M, Xu H. Mucolipins: Intracellular TRPML1-3 channels. FEBS Lett. 2010;584:2013–21. doi: 10.1016/j.febslet.2009.12.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Abe K, Puertollano R. Role of TRP channels in the regulation of the endosomal pathway. Physiology (Bethesda) 2011;26:14–22. doi: 10.1152/physiol.00048.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fares H, Greenwald I. Regulation of endocytosis by CUP-5, the Caenorhabditis elegans mucolipin-1 homolog. Nat Genet. 2001;28:64–8. doi: 10.1038/ng0501-64. [DOI] [PubMed] [Google Scholar]
- 97.LaPlante JM, Sun M, Falardeau J, Dai D, Brown EM, Slaugenhaupt SA, et al. Lysosomal exocytosis is impaired in mucolipidosis type IV. Mol Genet Metab. 2006;89:339–48. doi: 10.1016/j.ymgme.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 98.Thompson EG, Schaheen L, Dang H, Fares H. Lysosomal trafficking functions of mucolipin-1 in murine macrophages. BMC Cell Biol. 2007;8:54. doi: 10.1186/1471-2121-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Medina DL, Fraldi A, Bouche V, Annunziata F, Mansueto G, Spampanato C, et al. Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev Cell. 2011;21:421–30. doi: 10.1016/j.devcel.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jennings JJ, Jr., Zhu JH, Rbaibi Y, Luo X, Chu CT, Kiselyov K. Mitochondrial aberrations in mucolipidosis Type IV. J Biol Chem. 2006;281:39041–50. doi: 10.1074/jbc.M607982200. [DOI] [PubMed] [Google Scholar]
- 101.Curcio-Morelli C, Zhang P, Venugopal B, Charles FA, Browning MF, Cantiello HF, et al. Functional multimerization of mucolipin channel proteins. J Cell Physiol. 2010;222:328–35. doi: 10.1002/jcp.21956. [DOI] [PubMed] [Google Scholar]
- 102.Zeevi DA, Lev S, Frumkin A, Minke B, Bach G. Heteromultimeric TRPML channel assemblies play a crucial role in the regulation of cell viability models and starvation-induced autophagy. J Cell Sci. 2010;123:3112–24. doi: 10.1242/jcs.067330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kim HJ, Soyombo AA, Tjon-Kon-Sang S, So I, Muallem S. The Ca(2+) channel TRPML3 regulates membrane trafficking and autophagy. Traffic. 2009;10:1157–67. doi: 10.1111/j.1600-0854.2009.00924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Venugopal B, Browning MF, Curcio-Morelli C, Varro A, Michaud N, Nanthakumar N, et al. Neurologic, gastric, and opthalmologic pathologies in a murine model of mucolipidosis type IV. Am J Hum Genet. 2007;81:1070–83. doi: 10.1086/521954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Micsenyi MC, Dobrenis K, Stephney G, Pickel J, Vanier MT, Slaugenhaupt SA, et al. Neuropathology of the Mcoln1(-/-) knockout mouse model of mucolipidosis type IV. J Neuropathol Exp Neurol. 2009;68:125–35. doi: 10.1097/NEN.0b013e3181942cf0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tiede S, Storch S, Lübke T, Henrissat B, Bargal R, Raas-Rothschild A, et al. Mucolipidosis II is caused by mutations in GNPTA encoding the alpha/beta GlcNAc-1-phosphotransferase. Nat Med. 2005;11:1109–12. doi: 10.1038/nm1305. [DOI] [PubMed] [Google Scholar]
- 107.Kudo M, Brem MS, Canfield WM. Mucolipidosis II (I-cell disease) and mucolipidosis IIIA (classical pseudo-hurler polydystrophy) are caused by mutations in the GlcNAc-phosphotransferase alpha / beta—subunits precursor gene. Am J Hum Genet. 2006;78:451–63. doi: 10.1086/500849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hickman S, Neufeld EF. A hypothesis for I-cell disease: defective hydrolases that do not enter lysosomes. Biochem Biophys Res Commun. 1972;49:992–9. doi: 10.1016/0006-291X(72)90310-5. [DOI] [PubMed] [Google Scholar]
- 109.Vladutiu GD, Rattazzi MC. Abnormal lysosomal hydrolases excreted by cultured fibroblasts in I-cell disease (mucolipidosis II) Biochem Biophys Res Commun. 1975;67:956–64. doi: 10.1016/0006-291X(75)90768-8. [DOI] [PubMed] [Google Scholar]
- 110.Kornfeld S, Sly WS. I-cell disease and pseudo-hurler polydystrophy: disorders of lysosomal enzyme phosphorylation and localization. In: Scriver CR, Beaudet AL, Sly WS, Valle D, eds. The Metabolic and Molecular Bases of Inherited Disease. New York: McGraw-Hill, 2001:3469-82. [Google Scholar]
- 111.Paik KH, Song SM, Ki CS, Yu HW, Kim JS, Min KH, et al. Identification of mutations in the GNPTA (MGC4170) gene coding for GlcNAc-phosphotransferase alpha/beta subunits in Korean patients with mucolipidosis type II or type IIIA. Hum Mutat. 2005;26:308–14. doi: 10.1002/humu.20205. [DOI] [PubMed] [Google Scholar]
- 112.Wisniewski KE, Zhong N, Philippart M. Pheno/genotypic correlations of neuronal ceroid lipofuscinoses. Neurology. 2001;57:576–81. doi: 10.1212/wnl.57.4.576. [DOI] [PubMed] [Google Scholar]
- 113.The International Batten Disease Consortium Isolation of a novel gene underlying Batten disease, CLN3. Cell. 1995;82:949–57. doi: 10.1016/0092-8674(95)90274-0. [DOI] [PubMed] [Google Scholar]
- 114.Siintola E, Partanen S, Strömme P, Haapanen A, Haltia M, Maehlen J, et al. Cathepsin D deficiency underlies congenital human neuronal ceroid-lipofuscinosis. Brain. 2006;129:1438–45. doi: 10.1093/brain/awl107. [DOI] [PubMed] [Google Scholar]
- 115.Schultz ML, Tecedor L, Chang M, Davidson BL. Clarifying lysosomal storage diseases. Trends Neurosci. 2011;34:401–10. doi: 10.1016/j.tins.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 117.Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–63. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 118.Nishiyama J, Miura E, Mizushima N, Watanabe M, Yuzaki M. Aberrant membranes and double-membrane structures accumulate in the axons of Atg5-null Purkinje cells before neuronal death. Autophagy. 2007;3:591–6. doi: 10.4161/auto.4964. [DOI] [PubMed] [Google Scholar]
- 119.Walkley SU, Suzuki K. Consequences of NPC1 and NPC2 loss of function in mammalian neurons. Biochim Biophys Acta. 2004;1685:48–62. doi: 10.1016/j.bbalip.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 120.Ohmi K, Kudo LC, Ryazantsev S, Zhao HZ, Karsten SL, Neufeld EF. Sanfilippo syndrome type B, a lysosomal storage disease, is also a tauopathy. Proc Natl Acad Sci U S A. 2009;106:8332–7. doi: 10.1073/pnas.0903223106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Auer IA, Schmidt ML, Lee VM, Curry B, Suzuki K, Shin RW, et al. Paired helical filament tau (PHFtau) in Niemann-Pick type C disease is similar to PHFtau in Alzheimer’s disease. Acta Neuropathol. 1995;90:547–51. doi: 10.1007/BF00318566. [DOI] [PubMed] [Google Scholar]
- 122.Bu B, Li J, Davies P, Vincent I. Deregulation of cdk5, hyperphosphorylation, and cytoskeletal pathology in the Niemann-Pick type C murine model. J Neurosci. 2002;22:6515–25. doi: 10.1523/JNEUROSCI.22-15-06515.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Spillantini MG, Tolnay M, Love S, Goedert M. Microtubule-associated protein tau, heparan sulphate and alpha-synuclein in several neurodegenerative diseases with dementia. Acta Neuropathol. 1999;97:585–94. doi: 10.1007/s004010051034. [DOI] [PubMed] [Google Scholar]
- 124.Saito Y, Suzuki K, Hulette CM, Murayama S. Aberrant phosphorylation of alpha-synuclein in human Niemann-Pick type C1 disease. J Neuropathol Exp Neurol. 2004;63:323–8. doi: 10.1093/jnen/63.4.323. [DOI] [PubMed] [Google Scholar]
- 125.Saito Y, Suzuki K, Nanba E, Yamamoto T, Ohno K, Murayama S. Niemann-Pick type C disease: accelerated neurofibrillary tangle formation and amyloid beta deposition associated with apolipoprotein E epsilon 4 homozygosity. Ann Neurol. 2002;52:351–5. doi: 10.1002/ana.10266. [DOI] [PubMed] [Google Scholar]
- 126.Lee JH, Yu WH, Kumar A, Lee S, Mohan PS, Peterhoff CM, et al. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell. 2010;141:1146–58. doi: 10.1016/j.cell.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dehay B, Bové J, Rodríguez-Muela N, Perier C, Recasens A, Boya P, et al. Pathogenic lysosomal depletion in Parkinson’s disease. J Neurosci. 2010;30:12535–44. doi: 10.1523/JNEUROSCI.1920-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–6. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 129.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–95. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 130.Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11:1107–17. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 131.Sarkar S, Ravikumar B, Floto RA, Rubinsztein DC. Rapamycin and mTOR-independent autophagy inducers ameliorate toxicity of polyglutamine-expanded huntingtin and related proteinopathies. Cell Death Differ. 2009;16:46–56. doi: 10.1038/cdd.2008.110. [DOI] [PubMed] [Google Scholar]
- 132.Davidson CD, Ali NF, Micsenyi MC, Stephney G, Renault S, Dobrenis K, et al. Chronic cyclodextrin treatment of murine Niemann-Pick C disease ameliorates neuronal cholesterol and glycosphingolipid storage and disease progression. PLoS One. 2009;4:e6951. doi: 10.1371/journal.pone.0006951. [DOI] [PMC free article] [PubMed] [Google Scholar]