Abstract
Ras proteins are proto-oncogenes that are frequently mutated in human cancers. Three closely related isoforms, HRAS, KRAS and NRAS, are expressed in all cells and have overlapping but distinctive functions. Recent work has revealed how differences between the Ras isoforms in their trafficking, localization and protein-membrane orientation enable signalling specificity to be determined. We review the various strategies used to characterize compartmentalized Ras localization and signalling. Localization is an important contextual modifier of signalling networks and insights from the Ras system are of widespread relevance for researchers interested in signalling initiated from membranes.
Keywords: palmitoylation, GTPase, isoforms, organelle, plasma membrane, microdomains, nanoclusters
Introduction
Ras GTPases sit near the top of signalling pathways regulating cell proliferation, differentiation and apoptosis. All cells harbour three Ras isoforms: H-Ras, K-Ras and N-Ras encoded by separate genes. Despite a high degree of sequence homology Ras isoforms are not functionally redundant. This is surprising given that the regions of the proteins that determine interactions with effectors that mediate downstream signalling, with GDP/GTP that mediates Ras activation state and with regulatory proteins that turn on and off Ras, are identical between the Ras isoforms.
Ras proteins are activated by cell surface receptors and their predominant plasma membrane localization has meant that for many years this was believed to be the exclusive site of action of Ras. More recently with the application of improved imaging and protein modification technologies intracellular pools of Ras have been identified and location-specific functions assigned. This is thought to be due to different pools and concentrations of regulators and effectors localized in each compartment that enables different types of signalling to be generated from each location. Since the main difference between Ras isoforms is their overlapping but distinctive subcellular localizations, compartmentalized signalling is believed to contribute to the lack of functional redundancy between the Ras isoforms.
Whilst the last decade has generated detailed mechanistic understanding of compartmentalized Ras signalling there is still debate over the extent to which this is important for cell function. An extreme view is that this is an experimental artifact since most work has had to rely on over-expression of tagged or mutated Ras proteins that distort the balance of normal signalling. In this review we will discuss the evidence for compartment-specific Ras signalling and how it contributes to normal Ras function.
Normal and mutant Ras signalling
Ras proteins are 21kDa molecular switches that cycle between the inactive GDP-bound conformation and the active GTP-bound conformation (Figure 1). Guanine nucleotide exchange factors (GEFs) activate Ras by catalysing the release of GDP, facilitating GTP binding due to its 10-fold higher concentration than GDP in the cytosol [1]. GTP binding induces a conformational change in the Ras switch domains (Switch 1: residues 30-40 and Switch 2: residues 60-76) revealing an effector binding site [2]. Ras has an intrinsic GTPase activity that will return the protein to the inactive GDP-bound state. This process is normally slow however GTPase activating proteins (GAPs) bind GTP-bound Ras and speed this up ≥1000-fold to ensure rapid inactivation.
Figure 1. Ras activation cycle.
Ras is activated when GTP bound. Approximately 20 Ras effectors have been identified that modulate key proliferative, survival and cell migration pathways.
GEFs and GAPs consist of large families that provide varied cell and organelle-specific options for spatio-temporal regulation of Ras signalling. The classic example of a Ras GEF is Sos that is recruited via the adaptor Grb2 to activated receptor tyrosine kinases on the plasma membrane. Cell surface recruitment of Sos enables interaction with and activation of GDP-bound Ras. More recently a second Ras binding site was identified on Sos that recognises GTP-bound Ras enabling a positive feedback loop where active Ras stimulates further Ras activation [3]. An alternative mode of Ras activation via second messengers such as diacylglycerol (DAG) that causes translocation and activation of the GEF RasGRP1 [4]. Whilst Sos activity appears to be limited to the plasma membrane, RasGRP1 plays an important role in determining compartment-specific Ras functions that regulate thymocyte selection [5, 6].
Ras proteins activate signalling pathways by recruiting effectors to membranes; this results in conformational changes in effectors and/or facilitates interactions with activating proteins, cofactors or substrates necessary for signal propagation. Over 20 Ras effectors have been identified although most work has focussed on the Raf protein kinase family (A-Raf, B-Raf and c-Raf-1) and the PtdIns-3 kinase (PI3K) family members that regulate cell proliferation and survival respectively (Figure 1; [7, 8]). Both Ras isoform- and compartment-specific regulation of these effectors have been characterized [9-12].
Mutations that increase Ras activity have been shown to lead to developmental disorders and cancer. Importantly, they also provide some of the best evidence for isoform-specific Ras functions. Whilst K-Ras is the only isoform required for normal mouse development, human developmental disorders reveal a spectrum overlapping syndromes (Noonan, Costello, cardio-facio-cutaneous and autoimmune lymphoproliferative) associated with germ-line mutations of each Ras isoform [13, 14]. Each syndrome is phenotypically distinctive and associated with a specific Ras isoform that has a heterozygous mutation resulting in mild impairment of GDP/GTP binding or GAP interaction.
More potent somatic Ras mutations drive cancer development. On average, 16% of human cancers harbour mutations to codons 12, 13 or 61 that render Ras constitutively active due to significant impairment of GAP stimulation of Ras GTPase activity [15]. Intriguingly, there is a bias in the pattern of Ras isoform mutation rates with K-Ras being the most frequently mutated (20% of human cancers) and H-Ras rarely found mutated (2-3% human cancers). In isogenic cell and mouse models K-Ras is more potent than the other isoforms at supporting transformation and oncogenesis [16, 17]. The reason for this is not understood but may be in part related to relative abundance (K≥N⪢H in many cell types; [18]), differential abilities to induce apoptosis [19], and/or K-Ras specific functions in promoting endodermal cancer stem cell proliferation [20].
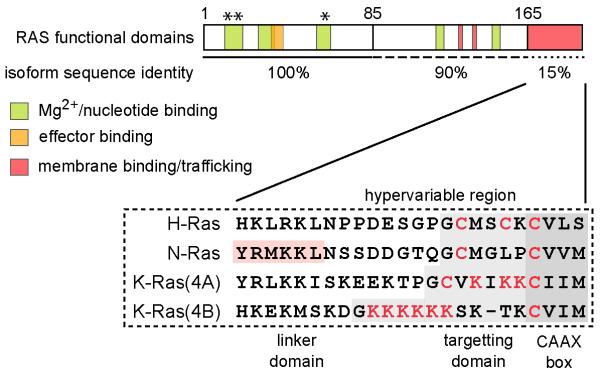
Together these observations point to in vivo functional differences between the Ras isoforms however they are highly homologous sharing 100% sequence identity in the regions that matter for regulating activation state and effector interactions. The only region of significant difference between the isoforms is the C-terminal 23-24 residue hypervariable region (HVR) that has been shown to be important for membrane interactions and determining localization. For this reason compartmentalization has emerged as the most likely mechanism for explaining isoform-specific Ras function.
Ras trafficking and localization
Synthesis, processing and trafficking to the plasma membrane
Ras proteins are synthesized in the cytosol and need subsequent post-translational modifications to enable them to stably associate with membranes where they can function. All three isoforms have a CAAX motif on their extreme C-termini that is sequentially processed; firstly the cysteine is isoprenylated by farnesyl protein transferase. This facilitates endoplasmic reticulum (ER) association where the AAX motif is cleaved off by Ras-converting enzyme 1 (Rce1) and finally the farnesylated cysteine is carboxymethylated by isoprenyl cysteine transferase (Icmt) [9]. Other motifs are necessary to improve the weak membrane binding afforded by the farnesyl moiety. These consist of either stretches of positively charged lysine residues that enable electrostatic interactions with negatively charge phospholipid headgroups or alternatively further lipid modifications with palmitoyl groups (Figure 2) [21].
Figure 2. The Ras Hypervariable Region (HVR) determines functional difference between isoforms.
Ras isoform share sequence identity at all regions regulating activation state and effector interactions. The HVR is post-translationally modified to enable membrane interactions and differential localization (key residues to enable correct localization highlighted in red). Asterixes indicate sites of oncogenic mutations at codons 12, 13 and 61.
The basic hexalysine patch present in the major K-Ras splice variant K-Ras4B (hereafter referred to as K-Ras) together with the farnesyl group is necessary and sufficient for plasma membrane localization of K-Ras via a Golgi-independent cytosolic route [22, 23]. Similarly to the minimal targeting motifs on K-Ras, the farnesyl group and two palmitoyl groups on cysteines 181 and 184 of H-Ras specify targeting to the plasma membrane via the Golgi and transport vesicles [22, 23]. Unexpectedly the monopalmitoylated isoform N-Ras was shown to require additional hydrophobic/basic residues located upstream in the HVR to provide further membrane affinity to enable stable plasma membrane localization [24]. The K-Ras splice variant K-Ras4A is also monopalmitoylated but contains an adjacent basic patch that is sufficient to stablise membrane interactions and allow plasma membrane localization [24].
Reversible membrane interactions
Importantly, both the polybasic electrostatic membrane interactions and the palmitoyl lipid groups enable reversible interactions with membranes and facilitate dynamic associations with multiple subcellular compartments. K-Ras membrane interactions appear to have a very short half life of a few minutes [25].The main reason for accumulation at the plasma membrane is because of its net negative charge versus other intracellular locations [26]. An implication of this short membrane residency is that K-Ras does not have an intrinsic capacity to traffic between compartments via membrane carriers. In contrast, palmitoylated Ras proteins are more stably associated with membranes and traffic post-Golgi via membrane carriers that may include the recycling endosome to reach the plasma membrane [27]. Unlike the farnesyl lipid attachment, the palmitoyl groups can be cleaved off Ras by a thioesterase dramatically reducing membrane avidity and causing translocation to the cytosol. Chemical biology studies and subsequent work using an inhibitor of acylproteinthioesterase 1 (ACPT1) revealed that when the palmitoyl groups of H- or N-Ras cannot be removed the protein end up labelling all membranes [28, 29]. Therefore the dynamics of the acylation/deacylation cycle are critical for ensuring correct localization of the palmitoylated isoforms. When H- and N-Ras become depalmitoylated they return to the Golgi complex for a new round of palmitoylation and trafficking to the surface [30, 31]. This can happen many times in the life of the protein. The number of palmitoyl groups determine the half-life of membrane association and relative Golgi accumulation, with monopalmitoylated N-Ras displaying stronger perinuclear Golgi staining than dipalmitoylated H-Ras in many cell types.
The ability of palmitoyl groups to partition into specific membranes provides a second dynamic feature of this modification. The palmitoyl groups sit next to an adjacent linker domain in the HVR that is highly flexible resulting in significant fluctuations in the depth of insertion of the palmitoyl groups [32, 33]. This is stabilised by the basic residues that sit in the hydrophobic/basic patch within the HVR linker domain [34]. It has been demonstrated that changes in Ras conformation associated with GDP or GTP binding modulate whether these basic residues can interact with the plasma membrane. This has consequent effects on the orientation of the Ras for effector binding and partitioning of Ras in specific signalling domains to enable isoform-specific signalling [35, 36]. This will be discussed in more detail in the section about plasma membrane signalling domains.
Endomembrane trafficking
Whilst all isoforms display their predominant localization at the plasma membrane, a variety of other organelles in addition to the ER/Golgi complex have also been identified as accessible to Ras proteins. Perhaps the least unexpected and most interesting from a signalling point of view is the endosome. Studies in the 1980’s and 1990’s established the role that endocytosis and the endosome plays in modulating signalling from growth factor receptor tyrosine kinases such as EGF receptor [37-39]. Upon activation, the majority of cell surface EGF receptor rapidly internalises via a clathrin-mediated pathway to the endosomal system. Although removed from the external source of activating growth factors the receptors remain ligand bound in the early endosomal system and capable of initiating cytoplasmic signalling until they are sorted into internal vesicles within endosomes. Therefore the endosome is a bona fide signalling platform and contains many key components of Ras signalling pathways.
All Ras isoforms have been localized to the endosomal system although relatively few studies have characterized K-Ras on endosomes compared to H-and N-Ras. Apart from the post-Golgi trafficking described earlier for H- and N-Ras the other route for endosome accumulation of palmitoylated Ras is via conventional clathrin-mediated endocytosis [40]. Whether this is part of a receptor complex or as a passenger during the bulk flow of membrane internalisation during receptor-mediated endocyosis is unclear. Cell surface receptors can be flagged for endocytosis and sorting using ubiquitin; something similar happens for H-, and N-Ras but not K-Ras that are di-ubiquitinated promoting endocytosis and endosomal accumulation [41]. For K-Ras both clathrin-mediated trafficking and a calcium/calmodulin mediated switch causing translocation to the cytosol have been proposed to mediate late endosomal or recycling endosomal accumulation respectively [42, 43]. Given the short half-life of K-Ras on membranes (t1/2 of few minutes; [25]) this conventional clathrin-mediated membrane trafficking is hard to explain and further work is needed to reconcile these observations.
At the endosome various protein-protein and protein-lipid interactions may contribute to retention and sorting of Ras. One recent example proposed involves FERM-like domains that interact with Ras proteins. PX-FERM-like domain containing proteins on endosomes such as SNX17 and SNX27 are suggested to scaffold retention and recycling of trafficked receptors and Ras proteins on this organelle [44].
Finally, Ras has also been observed on the mitochondria. Philips and colleagues showed in a variety of mammalian cell types that protein kinase C (PKC)-dependent phosphorylation of Ser181 on K-Ras antagonises the polybasic domain interaction with the plasma membrane [45]. This promotes translocation to the cytosol and accumulation on the mitochondria where it triggers apoptosis [45, 46]. The reason for the accumulation on mitochondria versus any other organelle is unclear and may be driven by the specific membrane content of this organelle or via protein-protein interactions such as the Bcl-XL interaction characterized in the Philips study.
All of the localization studies described required the over-expression of tagged and mutant forms of Ras to dissect the targeting and trafficking mechanisms. Endogenous Ras localization has been rarely studied; H-Ras is below antibody detection sensitivity whilst K-Ras and N-Ras appear predominantly localized to the plasma membrane with additional limited perinuclear localization associated with N-Ras [47]. Consequently, whilst it is clear which compartments are accessible to Ras proteins, the relative abundance of endogenous Ras isoforms within these locations remains to be accurately determined.
Evidence for compartment-specific Ras signalling
The challenge when studying compartment specific signalling is to separate it from the predominant signalling emanating from other locations such as the plasma membrane. Various methods are used to achieve this that mostly involve either expression of mutant or targetted Ras proteins or reporters of Ras output. An independent indicator of compartment competency to support Ras signalling is the presence of activators, effectors and co-factors for the Ras pathway. In the following section we will discuss how these modes of analysis have been used in complementary studies to provide evidence for compartmentalized Ras signalling.
Evolutionarily ancient origins
Fungal species and Dictyostelium utilise location-specific Ras signalling to control phenotypic outputs. Yeast use Ras signalling via cdc42, MAP kinase and adenylate cyclase/cAMP pathways to regulate many phenotypes including growth, morphogenesis, stress responses and mating [48, 49]. In the budding yeast Saccharomyces cerevisiae there are two Ras isoforms: Ras1 and Ras2, that are both mono-palmitoylated. As in mammalian cells, these Ras isoforms are not functionally redundant and compartment-specific effectors and responses have been proposed to mediate this [50]. The fission yeast Saccharomyces pombe provides a simpler system for Ras study because it harbours only one isoform: Ras1. In S. pombe, Ras1 signalling via Byr2 activates MAP kinase in response to pheromones to control mating whereas Ras1 signalling via Scd1/cdc42 regulates the cytoskeleton to maintain an elongate morphology. Eric Chang and colleagues employed a Ras targetting approach to exclusively restrict Ras to either endomembranes or plasma membrane in S. pombe to investigate these phenotypic responses with striking results. A palmitoylation deficient endomembrane-localized Ras1 stimulated morphogenesis but not the mating pathway whereas the Byr2/MAP kinase mating response was only stimulated from the plasma membrane [51]. Given the potential for signalling artefacts associated with over-expression studies, the authors were careful to optimise exogenous expression of endomembrane and wild type control Ras1 to levels equivalent to endogenous Ras1 however the extent to which these levels compared with plasma membrane Ras was not evident. Despite this omission, this study represents the best example to date of this type of Ras targetting approach because of the simplicity of the system, the mechanistic understanding of compartment influence on the pathways generated and the clear phenotypic differences in output.
Related work with the human fungal pathogen Cryptococcus neoformans also revealed compartment-specific Ras effects on mating and morphogenesis. In this organism Ras1 is dipalmitoylated and endomembrane localized palmitoylation deficient mutants were mating competent but displayed impaired morphogenesis, both observations in direct contrast to the results in S. pombe [52]. This reveals that whilst compartmental Ras signalling appears generic in fungal species, the specific pathways and phenotypes that are regulated from distinct locations may be highly variable.
A smaller scale of compartmentalization is via sub-domains within an organelle. An example of this is the plasma membrane that we now know is organised over various length scales from nanoscale signalling domains to larger regions such as primary cilia and migratory leading edges. Dictyostelium exhibits compartmental Ras signalling within the leading edge to enable directed migration in response to chemical gradients [53, 54]. Of the 11 Ras genes in this organism, RasC and RasG mediate cAMP-dependent chemotaxis [55]. Both of these proteins localize throughout the plasma membrane but GFP-based reporter assays reveal localized activation at the leading edge [54]. Whilst the mechanisms for this are still being defined, a negative feedback loop involving PI3K signalling to the RasGEF Aimless appears to be important [53]. These data highlight a key feature of compartmentalized Ras signalling in that whilst Ras isoforms may display spatially distinctive distributions, spatiotemporal Ras GEF and GAP activity is equally if not more important. Despite this, many of the approaches used to investigate Ras signalling uncouple the experiment from these regulatory mechanisms through use of mutationally active Ras proteins.
ER and Golgi Ras signalling
Whilst all Ras isoforms localize to the ER, Ras localization to the Golgi is well established for isoforms that are palmitoylated en route to the plasma membrane. Localization studies, albeit of over-expressed isoforms reveals the Golgi as the most obvious endomembrane compartment for Ras [9]. Various methods employed by several labs have been used to measure Ras signalling from the Golgi however there remains some debate about whether bona fide signalling occurs from this location. Evidence against is principally based on work using a fluorescent reporter based on the Ras binding domain (RBD) of Raf that recognises GTP-bound Ras. Triplet RBDs attached to GFP enable clear visualisation of endogenous Ras signalling at the plasma membrane in response to T-cell receptor stimulation in Jurkat T-cells [56, 57]. These cells are classically associated with Golgi Ras signalling however, no perinuclear fluorescence indicative of Golgi Ras activation was observed even when employing strategies to further increase endogenous Ras activation. Despite this, a negative result is not evidence of the opposite and questions remain about whether the reporter is sensitive enough to detect endogenous Golgi Ras signalling.
The authors argue that evidence for Golgi Ras activation seen in other studies are a consequence of over-expression however alternative imaging strategies have detected endogenous Golgi Ras activation and the GEFs and GAPs regulating this. Using an innovative bystander FRET approach, localized activation of endogenous Ras on the Golgi of COS-1 cells was observed 10-20 minutes after stimulation with insulin [58]. Therefore whilst plasma membrane Ras activation is transient, endomembranous ER/Golgi Ras activation is sustained. Building on these observations in combination with exogenous expression of H-Ras, it was shown that a variety of growth factors could stimulate Ras in the ER/Golgi with similar kinetics [58]. Differential activation of effectors from ER and Golgi was observed with Golgi-Ras poorly able to active ERK and Akt compared to ER-targetted Ras [59]. In contrast to these data are experiments where Raf was targeted to the ER or Golgi, in this case Golgi-Raf more potently activated ERK compared to ER-Raf [60].
Two compatible modes of ER/Golgi Ras activation have been proposed, one involving in situ GEF activity the other diffusion of activated GTP-bound Ras from the plasma membrane. The relatively slow time-course of endomembrane Ras activation would be consistent with both models. The GEF model is supported by work localising GEFs and revealing the role of second messengers DAG and Ca2+ in activating localized GEF responses. Several GEF families have been localized to the ER [9, 61], and RasGRP1 has been specifically shown to activate Ras on the Golgi following PLC-γ generation of second messengers [58, 62, 63]. Since Ras GRP1 is not ubiquitously expressed and given the known reversible interactions of palmitoylated Ras with membrane the alternative active Ras cytoplasmic diffusion model has been proposed to predominate in some cells [29, 64]. In this case whilst EGF was able to stimulate K-Ras signalling at the plasma membrane, there was no detectable activation of a Golgi-targetted Ras. The only conditions where Golgi-localized Ras activation was observed was with H-Ras that could localize to the plasma membrane and undergo depalmitoylation [64]. Since individual Ras molecules are activated for less than a second [65], this suggests that the activation/depalmitoylation cycle must be very tightly coupled to enable GTP-bound Ras to be released from the membrane. The authors also found that the ER is a second critical regulator of Golgi Ras activity due to local GEF/GAP activity on Ras that transiently localizes to the ER en route to the Golgi [64]. Further support for the translocation of active Ras from the plasma membrane to the Golgi recently came from a combined EM/FRAP investigation of N-Ras signalling. In this case both N-Ras activation and fibronectin induced clustering of cell surface signalling domains was required to promote depalmitoylation and translocation [66].
Using a targetted Ras approach, ER-Ras was found to be capable of stimulating transformation and proliferation equivalent to that seen for oncogenic H-Ras [58, 59]. Despite or perhaps because of similar levels of expression, Golgi Ras was incapable of stimulating these responses. The difficulty with interpreting the relative responses of this type of experiment is that the final concentration of Ras on each organelle cannot be accurately determined or titrated. An interesting alternative approach that can generate localized endogenous Ras activation is through targetting Ras GEF activity. This involves chemically induced dimerisation to acutely induce localization of RasGRF to the plasma membrane or Golgi [67]. Intriguingly, endogenous phospho-ERK labelling was localized as normal to the nucleus in response to plasma membrane Ras but was restricted to the Golgi following Golgi Ras activation – presumably due to interactions with the Golgi localized scaffold Sef [68].
Both positive and negative roles for the Golgi and ER in regulating Ras pathway signalling have been identified. The Raf binding protein RKTG and the MEK/ERK scaffold Sef have both been localized to the Golgi [68, 69]. Whilst Sef modulates MAPK signalling by retaining ERK in the cytosol, RKTG is a negative regulator by sequestering Raf kinases to the Golgi surface in an inactive conformation [70]. Notably, even mutationally active Raf was unable to activate MEK-ERK when bound to RKTG. The mechanism for this inhibition is unclear and seems likely to extend beyond the proposed simple sequestration model proposed by the authors since RKTG is bringing Raf to a compartment that contains the other components of the Ras-MAP kinase cascade.
Finally, there are good examples of ER and Golgi Ras signalling where endogenous Ras has been shown to regulate specific responses. Using the triplet-RBD probe to visualise and cell fractionation to detect endogenous Ras activation and siRNA to knock-down endogenous Ras, Terada and colleagues demonstrated that ER-localized K-Ras stimulates the unfolded protein response (UPR) and autophagy in HUVEC cells in response to oxidative stress [71]. Another example is lymphocyte selection where endogenous Golgi Ras activation results in positive selection whereas plasma membrane Ras activation results in negative selection [5]. Whist the ER/Golgi has been the most intensively studied location for Ras signalling, other endomembranes are also associated with Ras signalling.
Other locations for Ras signalling
Both the endosomal system and mitochondria have been implicated as hosts for Ras signalling. Fluorescence imaging of tagged Ras isoforms and endogenous Ras detection following subcellular fractionation have revealed Ras accumulation on endosomes following growth factor stimulation [38, 72, 73]. Inhibition of dynamin-dependent endocytosis inhibits activation and downstream signalling of endogenous and over-expressed H-Ras and N-Ras but not K-Ras [9, 74]. This is surprising given that K-Ras has been observed on multivesicular bodies/late endosomes [22, 43]; and that K-Ras localization to late endosomes putatively occurs via conventional clathrin-mediated endocytosis and sorting in the early endosome [42, 43]. Dynamin-dependent endocytosis includes both clathrin-dependent and clathrin-independent pathways. An example of a clathrin-independent pathway is the route to Arf6-associated endosomes. Fluorescence microscopy of tagged Ras and effectors revealed accumulation on these structures of both H-Ras and K-Ras together with signalling phosphoinositides and Akt but not Raf indicating specific effector coupling [75, 76].
Ubiquitiination specifies endocytosis of H- and N-Ras although it should be noted that less than 2% of total Ras is ubiquitinated and this is not changed by the GDP/GTP-bound status of Ras [41]. Mutations of H-Ras that abrogate ubiquitination result in enhanced activation of downstream Raf-MAPK [41]. This was interpreted to be due to enhanced plasma membrane association of Ras however there would also be no ubiquitin-dependent sorting of Ras into internal vesicles of endosomes and subsequent degradation in lysosomes that together would normally downregulate signalling.
To what extent endosomal compartments proportionally or uniquely contribute to total Ras signalling is poorly understood. A recent study however utilising targeted Ras constructs came to the provocative conclusion that palmitoylation-deficient endomembane Ras is much more potent at transforming NIH3T3 cells than plasma membrane restricted Ras [77]. This difference in potency was due to endomembrane Ras being able to activate cdc42 that is normally localized on endosomes.
Mitochondria represent the most mysterious location for Ras signalling. Both K-Ras and N-Ras have been localized to mitochondria using fluorescence microscopy and subcellular fractionation. At this location, K-Ras interacts with Bcl-XL triggering apoptosis whilst N-Ras initiates retrograde signalling to the nucleus [45, 78]. Whilst PKC triggers K-Ras translocation to mitochondria to initiate apoptosis in COS-1 and Jurkat T-cells, mitochondria are targets for antiapoptotic Ras signalling in cancer cells. In this case activated ERK in various K-Ras transformed cancer cells localizes to mitochondria to reduce sensitivity of the mitochondrial permeability transition pore that opens to initiate a terminal apoptotic signalling cascade [79]. Therefore whilst in normal and some cancer cells Ras signalling via PI3K and ERK targets the mitochondria to protect against apoptosis, direct interaction of Ras with this organelle subverts this process and triggers cell death.
In this section we have seen how different intracellular locations support Ras signalling to control key cellular phenotypes. Classically, Ras signalling is associated with the cell surface and this represents the predominant location for all Ras isoforms. Work in recent years has revealed how this platform consists of a mosaic of different signalling domains that modulate signal flow across the membrane and regulate isoform-specific interactions.
Plasma membrane signalling domains
The plasma membrane is a complex, dynamic, non-equilibrium mixture of >7000 species of phospholipid, ~30-40mol% cholesterol and ~25% by mass of integral and peripheral membrane proteins. Immediately underneath the lipid bilayer, which itself is asymmetric in composition, is an actin mesh that interacts with and impedes the lateral diffusion of trans-membrane proteins [80-82]. The plasma membrane is therefore not well mixed on many length and time scales, which results in non-random distributions of proteins and lipids within the membrane [83-85]. The lipid anchored Ras proteins provide an excellent example of the outcome of the complex biophysics of diffusion on the plasma membrane.
Ras proteins are arrayed in a mixture of immobile clusters and freely diffusing monomers. Each cluster is estimated to be in the range of 12-24nm in diameter, to comprise 6-7 Ras proteins and to have a lifetime of 0.1-1s [86, 87]. Ras clusters, which because of their size are generally called nanoclusters, are therefore transient, dynamic entities that are constantly forming and disassembling. The formation of Ras nanoclusters involves interplay between the C-terminal lipid anchor, the adjacent hypervariable region and G-domain of Ras with specific phospholipids and cholesterol in the lipid bilayer [88]. In consequence H-, N- and K-ras all operate in spatially distinct, non-overlapping nanoclusters [86]. There is further lateral segregation according to activation state, such that Ras.GDP- and Ras.GTP nanoclusters are spatially distinct for each isoform [89, 90]. Each Ras nanocluster has different structural properties, for example H-ras.GDP and N-ras.GTP nanoclusters are cholesterol dependent whereas all other Ras nanoclusters are not [87, 90-93]. For certain nanoclusters specific scaffold proteins are utilized, to date galectin-1 and galectin-3 have been identified as selective scaffolds for H-Ras.GTP and K-ras.GTP nanoclusters respectively [46, 94-96], whereas nucleolin and nucleophosmin can stabilize K-ras. GTP and K-ras.GDP nanoclusters [97, 98].
A major question has been how the conformational state of the G-domain can regulate membrane Ras / membrane interactions, that in turn drive Ras into distinct types of nanocluster. Initial insights into this problem were gained through molecular dynamic simulations and validated by cell imaging [34, 99, 100]. We now have a fairly clear understanding of the molecular mechanisms that drive H-Ras nanoclustering and the expectation is that similar principles operate for other isoforms [101]. GDP-H-ras membrane anchorage is predominantly stabilized through two basic residues R161 and R164 in the HVR in association with the adjacent membrane anchor [34, 36, 102]. Within the anchor the two palmitates are in an extended, ordered conformation that are able to engage Lo phase lipids in the membrane and assemble into cholesterol-dependent Lo nanoclusters. GTP-loading triggers structural rearrangements in the classical switch regions I and II to operate a novel “switch III” mechanism. Switch III, which comprises the β2-β3 loop and helix α5, pulls R169 and K170 off the membrane [35, 99]. This allows the whole G-domain to rotate and for basic residues in helix-α4 to bind membrane phospholipids and stabilize the new orientation [34, 36, 102]. Simultaneously the palmitates of the lipid anchor become disordered and unable to engage Lo domains. The GTP-orientation is recognized by cytosolic galectin-1, which binds and provides further stability [35, 99]. The stabilized H-Ras.GTP/ galectin-1 complex in turn undergoes homotypic assembly into H-ras.GTP nanoclusters [35, 94, 101]. The RBD of Raf and PI3K also recognize the orientation of the G-domain with respect to the membrane: mutation in the HVR or helix a4 that prevent GTP-driven reorientation abrogate H-ras signal output [35, 36]. Interestingly, certain germline mutations in Noonan’s syndrome that are associated with increased Ras signalling perturb the putative switch III, presumably by stabilizing the effector preferred G-domain orientation [103]. The G-domain orientations of GTP-loaded K-, N-, and H-ras required for optimal effector binding are all different [35]. This observation coupled with the different proteolipid content of the distinct non-overlapping nanoclusters likely accounts for the different signal output and in vivo biochemical diversity of the Ras isoforms [35].
An interesting property of the plasma membrane distribution of Ras proteins is that the ratio of clustered to monomeric Ras is relatively constant over a multilog concentration range [87]. This non-equilibrium property of the Ras distribution is shared by GPI-anchored proteins [104, 105]; the cellular mechanisms that actively maintain these distributions are not understood, but may be related to dynamic turnover of the submembrane actin mesh [106]. Whatever the mechanism it contributes to an important emergent property of the Ras signalling system. The fixed clustered fraction delivers a linear relationship between Ras.GTP levels and number of Ras.GTP nanoclusters on the plasma membrane [107]. Nanoclusters are the sole sites for activation of the Raf/MEK/MAPK cascade on the plasma and scaffolding the MAPK module in nanoclusters renders the biochemistry switch-like, such that the same ERKpp output is generated for all Raf kinase inputs [46, 60, 107, 108]. Each nanocluster therefore operates as a transient, low threshold digital switch that dumps a fixed quantum of ERKpp into the cytosol. The Ras nanocluster system therefore turns the plasma membrane into an analog-digital-analog (ADA) signal converter that transduces the strength of a growth factor signal into a corresponding level of cytosolic activated ERKpp with high fidelity [107-110].
A critical feature of the plasma membrane ADA converter is that the gain of the Ras.GTP to ERKpp signal response is set by the clustered fraction, since this determines what proportion of the total Ras-GTP generated is assemble into signalling nanoclusters. As the clustered fraction is reduced, the ERKpp signal response to a given growth factor stimulation is decreased, even though the total amount of Ras.GTP generated is unchanged [109, 110]. Therefore biological or pharmacological manipulations of the plasma membrane that changes the spatiotemporal organization of Ras could potentially perturb Ras signalling, even without changing the amount of Ras.GTP. Indeed two examples of recent work suggest that this is possible. Growth in n-3 polyunsaturated fatty acids (PUFA) can modulate Ras clustering [111, 112], and a variety of NSAIDs stabilize Lo domains in model and biological membranes [113]. These amphiphilic drugs have pleiotropic activities that extend well beyond COX inhibition, some of which may be explained by perturbation of Ras spatiotemporal dynamics [114]. Novel therapeutic possibilities for targeting Ras nanoclustering in oncogenically transformed cells follow from these studies.
Discussion
We have seen that various mechanisms can ensure signalling specificity of the Ras isoforms. Critical amongst these are their relative abundance and localized concentrations that will determine interaction efficiencies with various GEFs, GAPs and effectors over time. Importantly, the majority of strategies used to date to investigate isoform- or compartment-specific Ras signalling have abrogated the influence of these control mechanisms by overexpressing often constitutively active mutant forms of Ras. There is also the risk that these experiments could scaffold novel signalling in places never seen in vivo or overstate the relative contributions of localized signalling events. Consequently care must be taken when interpreting the results of these experiments, however at the very least many studies have shown the potential of individual compartments for supporting specific signalling pathways.
A final point when considering compartment-specific functions is that whilst organelles are self-contained entities they are frequently observed tethered or closely apposed to each other. A good example of this is the ER that all Ras isoforms initially occupy en route to the plasma membrane and that establishes clear contacts with all of the other organelles in the cell. Contacts between the ER and endosomes enables an ER localized phosphatase, PTP1B, to promote down-regulation of endosomal EGFR [115]. Therefore the boundaries of functionality may be blurred.
In summary, there is general acceptance of the idea that Ras signalling is compartmentalized and that this contributes to specificity within signalling cascades. Compartmentalization can occur over several length scales and whilst this is generic there is variability between organisms and cell types in the phenotypes and pathways that are specifically coupled to individual locations. Ras isoforms represent excellent models for investigating wider questions of signalling compartmentalization whilst insights from this work may also ultimately inform strategies for nullifying aberrant Ras function associated with cancer or developmental disorders.
Acknowledgements
I.A.P is a Royal Society University Research Fellow and work in his laboratory is supported by the NWCRF, BBSRC and the Wellcome Trust. Work in the laboratory of J.F.H is supported by the NIH (GM066717).
References
- [1].Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–77. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- [2].Wittinghofer A, Franken SM, Scheid ig AJ, Rensland H, Lautwein A, Pai EF, et al. Three-dimensional structure and properties of wild-type and mutant H-ras-encoded p21. Ciba Found Symp. 1993;176:6–21. doi: 10.1002/9780470514450.ch2. discussion 21-7. [DOI] [PubMed] [Google Scholar]
- [3].Freedman TS, Sondermann H, Friedland GD, Kortemme T, Bar-Sagi D, Marqusee S, et al. A Ras-induced conformational switch in the Ras activator Son of sevenless. Proc Natl Acad Sci U S A. 2006;103:16692–7. doi: 10.1073/pnas.0608127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ebinu JO, Bottorff DA, Chan EY, Stang SL, Dunn RJ, Stone JC, Ras GRP. a Ras guanyl nucleotide-releasing protein with calcium-and diacylglycerol-binding motifs. Science. 1998;280:1082–6. doi: 10.1126/science.280.5366.1082. [DOI] [PubMed] [Google Scholar]
- [5].Mor A, Campi G, Du G, Zheng Y, Foster DA, Dustin ML, et al. The lymphocyte function-associated antigen-1 receptor costimulates plasma membrane Ras via phospholipase D2. Nat Cell Biol. 2007;9:713–9. doi: 10.1038/ncb1592. [DOI] [PubMed] [Google Scholar]
- [6].Perez de Castro I, Bivona TG, Philips MR. Pellicer A Ras activation in Jurkat T cells following low-grade stimulation of the T-cell receptor is specific to N-Ras and occurs only on the Golgi apparatus. Mol Cell Biol. 2004;24:3485–96. doi: 10.1128/MCB.24.8.3485-3496.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Rajalingam K, Schreck R, Rapp UR, Albert S. Ras oncogenes and their downstream targets. Biochim Biophys Acta. 2007;1773:1177–95. doi: 10.1016/j.bbamcr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- [8].Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer. 2003;3:459–65. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- [9].Omerovic J, Laude AJ, Prior IA. Ras proteins: paradigms for compartmentalised and isoform-specific signalling. Cell Mol Life Sci. 2007;64:2575–89. doi: 10.1007/s00018-007-7133-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Rodriguez-Viciana P, Sabatier C, McCormick F. Signaling specificity by Ras family GTPases is determined by the full spectrum of eff ectors they regulate. Mol Cell Biol. 2004;24:4943–54. doi: 10.1128/MCB.24.11.4943-4954.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Walsh AB, Bar-Sagi D. Differential activation of the Rac pathway by Ha-Ras and K-Ras. J Biol Chem. 2001;276:15609–15. doi: 10.1074/jbc.M010573200. [DOI] [PubMed] [Google Scholar]
- [12].Yan J, Roy S, Apolloni A, Lane A, Hancock JF. Ras isoforms vary in their ab ility to activate Raf-1 and phosphoinositide 3-kinase. J Biol Chem. 1998;273:24052–6. doi: 10.1074/jbc.273.37.24052. [DOI] [PubMed] [Google Scholar]
- [13].Koera K, Nakamura K, Nakao K, Miyoshi J, Toyoshima K, Hatta T, et al. K-ras is essential for the development of the mouse embryo. Oncogene. 1997;15:1151–9. doi: 10.1038/sj.onc.1201284. [DOI] [PubMed] [Google Scholar]
- [14].Tidyman WE, Rauen KA. The RASopathies: developmental syndromes of Ras/MAPK pathway dysregulation. Curr Opin Genet Dev. 2009;19:230–6. doi: 10.1016/j.gde.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–50. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Haigis KM, Kendall KR, Wang Y, Cheung A, Haigis MC, Glickman JN, et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in t he colon. Nat Genet. 2008;40:600–8. doi: 10.1038/ngXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Keller JW, Franklin JL, Graves-Deal R, Friedman DB, Whitwell CW, Coffey RJ. Oncogenic KRAS provides a uniquely powerful and variable oncogenic contribution among RAS family members in the colonic epithelium. J Cell Physiol. 2007;210:740–9. doi: 10.1002/jcp.20898. [DOI] [PubMed] [Google Scholar]
- [18].Omerovic J, Hammond DE, Clague MJ, Prior IA. Ras isoform abundance and signalling in human cancer cell lines. Oncogene. 2008;27:2754–62. doi: 10.1038/sj.onc.1210925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kreeger PK, Wang Y, Haigis KM, Lauffenburger DA. Integration of multiple signaling p athway activities resolves K-RAS/N-RAS mutation paradox in colon epithelial cell response to inflammatory cytokine stimulation. Integr Biol (Camb) 2010;2:202–8. doi: 10.1039/b925935j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Quinlan MP, Settleman J. Explaining the preponderance of Kras mutations in human cancer: A n isoform-specific function in stem cell expansion. Cell Cycle. 2008;7:1332–5. doi: 10.4161/cc.7.10.5927. [DOI] [PubMed] [Google Scholar]
- [21].Hancock JF, Paterson H, Marshall CJ. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell. 1990;63:133–9. doi: 10.1016/0092-8674(90)90294-o. [DOI] [PubMed] [Google Scholar]
- [22].Apolloni A, Prior IA, Lindsay M, Parton RG, Hancock JF. H-ras but not K-ras traffics to the plasma membrane through the exocytic pathway. Mol Cell Biol. 2000;20:2475–87. doi: 10.1128/mcb.20.7.2475-2487.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Choy E, Chiu VK, Silletti J, Feoktistov M, Morimoto T, Michaelson D, et al. Endomembrane trafficking of ras: the CAAX motif targets proteins to the ER and Golgi. Cell. 1999;98:69–80. doi: 10.1016/S0092-8674(00)80607-8. [DOI] [PubMed] [Google Scholar]
- [24].Laude AJ, Prior IA. Palmitoylation and localisation of RAS isoforms are modulated by the hypervariable linker domain. J Cell Sci. 2008;1(21):421–7. doi: 10.1242/jcs.020107. [DOI] [PubMed] [Google Scholar]
- [25].Silvius JR, Bhagatji P, Leventis R, Terrone D. K-ras4B and prenylated proteins lacking “second signals” associate dynamically with cellular membranes. Mol Biol Cell. 2006;17:192–202. doi: 10.1091/mbc.E05-05-0408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Yeung T, Terebiznik M, Yu L, Silvius J, Abidi WM, Philips M, et al. Receptor activation alters inner surface potential during phagocytosis. Science. 2006;313:347–51. doi: 10.1126/science.1129551. [DOI] [PubMed] [Google Scholar]
- [27].Misaki R, Morimatsu M, Uemura T, Waguri S, Miyoshi E, Taniguchi N, et al. Palmitoylated Ras proteins traffic through recycling endosomes to the plasma membrane during exocytosis. J Cell Biol. 2010;191:23–9. doi: 10.1083/jcb.200911143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Dekker FJ, Rocks O, Vartak N, Menninger S, Hedberg C, Balamurugan R, et al. Small-molecule inhibition of APT1 affects Ras localization and signaling. Nat Chem Biol. 2010;6:449–56. doi: 10.1038/nchembio.362. [DOI] [PubMed] [Google Scholar]
- [29].Rocks O, Peyker A, Kahms M, Verveer PJ, Koerner C, Lumbierres M, et al. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science. 2005;307:1746–52. doi: 10.1126/science.1105654. [DOI] [PubMed] [Google Scholar]
- [30].Rocks O, Gerauer M, Vartak N, Koch S, Huang ZP, Pechlivanis M, et al. The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins. Cell. 2010;141:458–71. doi: 10.1016/j.cell.2010.04.007. [DOI] [PubMed] [Google Scholar]
- [31].Goodwin JS, Drake KR, Rogers C, Wright L, Lippincott-Schwartz J, Philips MR, et al. Depalmitoylated Ras traffics to and from the Golgi complex via a nonvesicular pathway. J Cell Biol. 2005;170:261–72. doi: 10.1083/jcb.200502063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Brunsveld L, Waldmann H, Huster D. Membrane binding of lipidated Ras peptides and proteins -- the structural point of view. Biochim Biophys Acta. 2009;1788:273–88. doi: 10.1016/j.bbamem.2008.08.006. [DOI] [PubMed] [Google Scholar]
- [33].Vogel A, Tan KT, Waldmann H, Feller SE, Brown MF, Huster D. Flexibility of ras lipid modifications studied by 2H solid-state NMR and molecular dynamics simulations. Biophys J. 2007;93:2697–712. doi: 10.1529/biophysj.107.104562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Gorfe AA, Hanzal-Bayer M, Abankwa D, Hancock JF, McCammon JA. Structure and dynamics of the full-length lipid-modified H-Ras protein in a 1,2-dimyristoylglycero-3-phosphocholine bilayer. J Med Chem. 2007;50:674–84. doi: 10.1021/jm061053f. [DOI] [PubMed] [Google Scholar]
- [35].Abankwa D, Gorfe AA, Inder K, Hancock JF. Ras membrane orientation and nanodomain localization generate isoform diversity. Proc Natl Acad Sci U S A. 2010;107:1130–5. doi: 10.1073/pnas.0903907107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Abankwa D, Hanzal-Bayer M, Ariotti N, Plowman SJ, Gorfe AA, Parton RG, et al. A novel switch region regulates H-ras membrane orientation and signal output. Embo J. 2008;27:727–35. doi: 10.1038/emboj.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Bergeron JJ, Lai WH, Kay DG, Doherty JJ, 2nd, Khan MN, Posner BI. The endosomal apparatus and transmembrane signalling. Adv Exp Med Biol. 1988;234:213–24. doi: 10.1007/978-1-4757-1980-2_14. [DOI] [PubMed] [Google Scholar]
- [38].Di Guglielmo GM, Baass PC, Ou WJ, Posner BI, Bergeron JJ. Compartmentalization of SHC, GRB2 and mSOS, and hyperphosphorylation of Raf-1 by EGF but not insulin in liver parenchyma. Embo J. 1994;13:4269–77. doi: 10.1002/j.1460-2075.1994.tb06747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Vieira AV, Lamaze C, Schmid SL. Control of EGF receptor signaling by clathrin-mediated endocytosis. Science. 1996;274:2086–9. doi: 10.1126/science.274.5295.2086. [DOI] [PubMed] [Google Scholar]
- [40].Howe CL, Valletta JS, Rusnak AS, Mobley WC. NGF signaling from clathrin-coated vesicles: evidence that signaling endosomes serve as a platform for the Ras-MAPK pathway. Neuron. 2001;32:801–14. doi: 10.1016/s0896-6273(01)00526-8. [DOI] [PubMed] [Google Scholar]
- [41].Jura N, Scotto-Lavino E, Bar-Sagi D. Differential mo dification of Ras proteins by ubiquitination. Mol Cell. 2006;21:679–87. doi: 10.1016/j.molcel.2006.02.011. [DOI] [PubMed] [Google Scholar]
- [42].Fivaz M, Meyer T. Reversible intracellular translocation of KRas but not HRas in hippocampal neurons regulated by Ca2+/calmodulin. J Cell Biol. 2005;170:429–41. doi: 10.1083/jcb.200409157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Lu A, Tebar F, Alvarez-Moya B, Lopez-Alcala C, Calvo M, Enrich C, et al. A clathrin-dependent pathway leads to KRas signaling on late endosomes en route to lysosomes. J Cell Biol. 2009;184:863–79. doi: 10.1083/jcb.200807186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ghai R, Mobli M, Norwood SJ, Bugarcic A, Teasdale RD, King GF, et al. Phox homology band 4.1/ezrin/radixin/moesin-like proteins function as molecular scaffolds that interact with cargo receptors and Ras GTPases. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1017110108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Bivona TG, Quatela SE, Bodemann BO, Ahearn IM, Soskis MJ, Mor A, et al. PK C regulates a farnesyl-electrostatic switch on K-Ras that promotes its association with Bcl-XL on mitochondria and induces apoptosis. Mol Cell. 2006;21:481–93. doi: 10.1016/j.molcel.2006.01.012. [DOI] [PubMed] [Google Scholar]
- [46].Plowman SJ, Ariotti N, Goodall A, Parton RG, Hancock JF. Electrostatic interactions positiv ely regulate K-Ras nanocluster formation and function. Mol Cell Biol. 2008;28:4377–85. doi: 10.1128/MCB.00050-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kranenburg O, Verlaan I, Moolenaar WH. Regulating c-Ras function. cholesterol depletion affects caveolin association, GTP loading, and signaling. Curr Biol. 2001;11:1880–4. doi: 10.1016/s0960-9822(01)00582-6. [DOI] [PubMed] [Google Scholar]
- [48].Toda T, Uno I, Ishikawa T, Powers S, Kataoka T, Broek D, et al. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell. 1985;40:27–36. doi: 10.1016/0092-8674(85)90305-8. [DOI] [PubMed] [Google Scholar]
- [49].Saito H. Regulation of cross-talk in yeast MAPK signaling pathways. Curr Opin Mic robiol. 2010;13:677–83. doi: 10.1016/j.mib.2010.09.001. [DOI] [PubMed] [Google Scholar]
- [50].Chang EC, Philips MR. Spatial segregation of Ras signaling: new evidence from fission yeast. Cell Cycle. 2006;5:1936–9. doi: 10.4161/cc.5.17.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Onken B, Wiener H, Philips MR, Chang EC. Compartmentalized signaling of Ras in fission yeast. Proc Natl Acad Sci U S A. 2006;103:9045–50. doi: 10.1073/pnas.0603318103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Nichols CB, Ferreyra J, Ballou ER, Alspaugh JA. Subcellular localization directs signaling specificity of the Cryptococcus neoformans Ras1 protein. Eukaryot Cell. 2009;8:181–9. doi: 10.1128/EC.00351-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Charest PG, Shen Z, Lakoduk A, Sa saki AT, Briggs SP, Firtel RA. A Ras signaling complex controls the RasC-TORC2 pathway and directed cell migration. Dev Cell. 2010;18:737–49. doi: 10.1016/j.devcel.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Sasaki AT, Chun C, Takeda K, Firtel RA. Localized Ras signaling at the leading edge regulates PI3K, cell polar ity, and directional cell movement. J Cell Biol. 2004;167:505–18. doi: 10.1083/jcb.200406177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Kortholt A, van Haastert PJ. Highlighting the role of Ras and Rap during Dictyostelium chemotaxis. Cell Signal. 2008;20:1415–22. doi: 10.1016/j.cellsig.2008.02.006. [DOI] [PubMed] [Google Scholar]
- [56].Augsten M, Pusch R, Biskup C, Rennert K, Wittig U, Beyer K, et al. Live-cell imaging of endogenous Ras-GTP illustrates predominant Ras activation at the plasma membrane. EMBO Rep. 2006;7:46–51. doi: 10.1038/sj.embor.7400560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Rubio I, Grund S, Song SP, Biskup C, Bandemer S, Fricke M, et al. TCR-induced activation of Ras proceeds at the plasma membrane and requires palmitoylation of N-Ras. J Immunol. 2010;185:3536–43. doi: 10.4049/jimmunol.1000334. [DOI] [PubMed] [Google Scholar]
- [58].Chiu VK, Bivona T, Hach A, Sajous JB, Silletti J, Wiener H, et al. Ras signalling on the endoplasmic reticulum and the Golgi. Nat Cell Biol. 2002;4:343–50. doi: 10.1038/ncb783. [DOI] [PubMed] [Google Scholar]
- [59].Matallanas D, Sanz-Moreno V, Arozarena I, Calvo F, Agudo-Ibanez L, Santos E, et al. Distinct utilization of effectors and biological outcomes resulting from site-specific Ras activation: Ras functions in lipid rafts and Golgi complex are dispensable for pr oliferation and transformation. Mol Cell Biol. 2006;26:100–16. doi: 10.1128/MCB.26.1.100-116.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Inder K, Harding A, Plowman SJ, Philips MR, Parton RG, Hancock JF. Activation of the MAPK module from different spatial locations generates distinct system outputs. Mol Biol Cell. 2008;19:4776–84. doi: 10.1091/mbc.E08-04-0407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Arozarena I, Matallanas D, Berciano MT, Sanz-Moreno V, Calvo F, Munoz MT, et al. Activation of H-Ras in the endoplasmic reticulum by the RasGRF family guanine nucleotide exchange factors. Mol Cell Biol. 2004;24:1516–30. doi: 10.1128/MCB.24.4.1516-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Bivona TG, Perez De Castro I, Ahearn IM, Grana TM, Chiu VK, Lockyer PJ, et al. Phospholipase Cgamma activates Ras on the Golgi apparatus by means of RasGRP1. Nature. 2003;424:694–8. doi: 10.1038/nature01806. [DOI] [PubMed] [Google Scholar]
- [63].Caloca MJ, Zugaza JL, Bustelo XR. Exchange factors of the RasGRP family mediate Ras activation in the Golgi. J Biol Chem. 2003;278:33465–73. doi: 10.1074/jbc.M302807200. [DOI] [PubMed] [Google Scholar]
- [64].Lorentzen A, Kinkhabwala A, Rocks O, Vartak N, Bastiaens PI. Regulation of Ras localization by acylation enables a mode of intracellular signal propagation. Sci Signal. 2010;3:ra68. doi: 10.1126/scisignal.20001370. [DOI] [PubMed] [Google Scholar]
- [65].Murakoshi H, Iino R, Kobayashi T, Fujiwara T, Ohshima C, Yoshimura A, et al. Single-molecule imaging analysis of Ras activation in living cells. Proc Natl Acad Sci U S A. 2004;101:7317–22. doi: 10.1073/pnas.0401354101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Eisenberg S, Beckett AJ, Prior IA, Dekker FJ, Hedberg C, Waldmann H, et al. Raft protein clustering alters N-Ras membrane interactions and activation pattern. Mol Cell Biol. 2011 doi: 10.1128/MCB.05570-11. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Komatsu T, Kukelyansky I, McCaffery JM, Ueno T, Varela LC, Inoue T. Organelle-specific, rapid induction of molecular activities an d membrane tethering. Nat Methods. 2010;7:206–8. doi: 10.1038/nmeth.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Torii S, Kusakabe M, Yamamoto T, Maekawa M, Nishida E. Sef is a spatial regulator for Ras/MAP kinase signaling. Dev Cell. 2004;7:33–44. doi: 10.1016/j.devcel.2004.05.019. [DOI] [PubMed] [Google Scholar]
- [69].Feng L, Xie X, Ding Q, Luo X, He J, Fan F, et al. Spatial re gulation of Raf kinase signaling by RKTG. Proc Natl Acad Sci U S A. 2007;104:14348–53. doi: 10.1073/pnas.0701298104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Fan F, Feng L, He J, Wang X, Jiang X, Zhang Y, et al. RKTG sequesters B-Raf to the Golgi apparatus and inhibits the proliferation and tumorigenicity of human malig nant melanoma cells. Carcinogenesis. 2008;29:1157–63. doi: 10.1093/carcin/bgn119. [DOI] [PubMed] [Google Scholar]
- [71].Wu RF, Ma Z, Liu Z, Terada LS. Nox4-derived H2O2 mediates endoplasmic reticulum signaling through local Ras activation. Mol Cell Biol. 2010;30:3553–68. doi: 10.1128/MCB.01445-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Gomez GA, Daniotti JL. H-Ras dynamically interacts with recycling endosomes in CHO-K1 cells: involvement of Rab5 and Rab11 in the trafficking of H-Ras to this pericentriolar endocytic compartment. J Biol Chem. 2005;280:34997–5010. doi: 10.1074/jbc.M506256200. [DOI] [PubMed] [Google Scholar]
- [73].Pol A, Calvo M, Enrich C. Isolated endosomes from quiescent r at liver contain the signal transduction machinery. Differential distribution of activated Raf-1 and Mek in the endocytic compartment. FEBS Lett. 1998;441:34–8. doi: 10.1016/s0014-5793(98)01517-8. [DOI] [PubMed] [Google Scholar]
- [74].Roy S, Wyse B, Hancock JF. H-Ras signaling and K-Ras signaling are differentially dependen t on endocytosis. Mol Cell Biol. 2002;22:5128–40. doi: 10.1128/MCB.22.14.5128-5140.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].McKay J, Wang X, Ding J, Buss JE, Ambrosio L. H-ras resides on clathrin-independent ARF6 vesicles that harbor little RAF-1, but not on clathrin-dependent endosomes. Biochim Biophys Acta. 2011;1813:298–307. doi: 10.1016/j.bbamcr.2010.11.019. [DOI] [PubMed] [Google Scholar]
- [76].Porat-Shliom N, Kloog Y, Donaldson JG. A unique platform for H-Ras signaling involving clathrin-independent endocytosis. Mol Biol Cell. 2008;19:765–75. doi: 10.1091/mbc.E07-08-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Cheng CM, Li H, Gasman S, Huang J, Schiff R, Chang EC. Compartmentalized Ras proteins tra nsform NIH 3T3 cells with different efficiencies. Mol Cell Biol. 2011;31:983–97. doi: 10.1128/MCB.00137-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Wolfman JC, Planchon SM, Liao J, Wolfman A. Structural and functional consequences of c-N-Ras constitutively associated with intact mitochondria. Biochim Biophys Acta. 2006;1763:1108–24. doi: 10.1016/j.bbamcr.2006.07.015. [DOI] [PubMed] [Google Scholar]
- [79].Rasola A, Sciacovelli M, Chiara F, Pantic B, Brusilow WS, Bernardi P. Activation of mitochondrial ERK protects cancer cells from death through inhibition of the permeability transition. Proc Natl Acad Sci U S A. 107:726–31. doi: 10.1073/pnas.0912742107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Kusumi A, Nakada C, Ritchie K, Murase K, Suzuki K, Murakoshi H, et al. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: high-speed single-molecule tracking of membrane molecules. Annu Rev Biophys B iomol Struct. 2005;34:351–78. doi: 10.1146/annurev.biophys.34.040204.144637. [DOI] [PubMed] [Google Scholar]
- [81].Morone N, Fujiwara T, Murase K, Kasai RS, Ike H, Yuasa S, et al. Three-dimensional reconstruction of the membrane skeleton at the plasma membrane interface by electron tomography. J Cell Biol. 2006;174:851–62. doi: 10.1083/jcb.200606007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Murase K, Fujiwara T, Umemura Y, Suzuki K, Iino R, Yamashita H, et al. Ultrafine membrane compartments for molecular diffusion as revealed by single molecule techniques. Biophys J. 2004;86:4075–93. doi: 10.1529/biophysj.103.035717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Hancock JF. Lipid rafts: contentious only from simplistic standpoints. Nat Rev Mol Cell Biol. 2006;7:456–62. doi: 10.1038/nrm1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Jacobson K, Mouritsen OG, Anderson RG. Lipid rafts: at a crossroad between cell biology and physics. Nature cell biology. 2007;9:7–14. doi: 10.1038/ncb0107-7. [DOI] [PubMed] [Google Scholar]
- [85].Simons K, Vaz WL. Model systems, lipid rafts, and cell membr anes. Annu Rev Biophys Biomol Struct. 2004;33:269–95. doi: 10.1146/annurev.biophys.32.110601.141803. [DOI] [PubMed] [Google Scholar]
- [86].Hancock JF, Parton RG. Ras plasma membrane signalling platforms. Biochem J. 2005;389:1–11. doi: 10.1042/BJ20050231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Plowman SJ, Muncke C, Parton RG, Hancock JF. H-ras, K-ras, and inner plasma membrane raft proteins ope rate in nanoclusters with differential dependence on the actin cytoskeleton. Proc Natl Acad Sci U S A. 2005;102:15500–5. doi: 10.1073/pnas.0504114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Rotblat B, Prior IA, Muncke C, Parton RG, Kloog Y, Henis YI, et al. Three separable domains regulate GTP-dependent association of H-ras with the plasma membrane. Mol Cell Biol. 2004;24:6799–810. doi: 10.1128/MCB.24.15.6799-6810.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Prior IA, Harding A, Yan J, Sluimer J, Parton RG, Hancock JF. GTP-dependent segregation of H-ras from lipid rafts is required for biological activity. Nat Cell Biol. 2001;3:368–75. doi: 10.1038/35070050. [DOI] [PubMed] [Google Scholar]
- [90].Prior IA, Muncke C, Parton RG, Hancock JF. Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J Cell Biol. 2003;160:165–70. doi: 10.1083/jcb.200209091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Eisenberg S, Shvartsman DE, Ehrlich M, Henis YI. Clustering of raft-associated proteins in the external membrane leaflet modulates internal leaflet H-ras diffusion and signaling. Molecular and cellular biology. 2006;26:7190–200. doi: 10.1128/MCB.01059-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Niv H, Gutman O, Kloog Y, Henis YI. Activated K-Ras and H-Ras display different interactions with saturable nonra ft sites at the surface of live cells. J Cell Biol. 2002;157:865–72. doi: 10.1083/jcb.200202009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Roy S, Plowman S, Rotblat B, Prior IA, Muncke C, Grainger S, et al. Individual palmitoyl residues serve distinct roles in H-ras trafficking, microlocalization, and signaling. Mol Cell Biol. 2005;25:6722–33. doi: 10.1128/MCB.25.15.6722-6733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Belanis L, Plowman SJ, Rotblat B, Hancock JF, Kloog Y. Galectin-1 is a novel structural component and a major regulator of h-ras nanoclusters. Mol Biol Cell. 2008;19:1404–14. doi: 10.1091/mbc.E07-10-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Shalom-Feuerstein R, Plowman SJ, Rotblat B, Ariotti N, Tian T, Hancock JF, et al. K-ras nanoclustering is subverted by overexpression of the scaffold protein galectin-3. Cancer Res. 2008;68:6608–16. doi: 10.1158/0008-5472.CAN-08-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Tian T, Plowman SJ, Parton RG, Kloog Y, Hancock JF. Mathematical modeling of K-Ras nanocluster f ormation on the plasma membrane. Biophys J. 2010;99:534–43. doi: 10.1016/j.bpj.2010.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Farin K, Schokoroy S, Haklai R, Cohen-Or I, Elad-Sfadia G, Reyes-Reyes ME, et al. Oncogenic synergism between ErbB1, nucleolin, and mutant Ras. Cancer research. 2011;71:2140–51. doi: 10.1158/0008-5472.CAN-10-2887. [DOI] [PubMed] [Google Scholar]
- [98].Inder KL, Lau C, Loo D, Chaudhary N, Goodall A, Martin S, et al. Nucleophosmin and nucleolin regulate K-Ras plasma membrane interactions and MAPK signal transduction. J Biol Chem. 2009;284:28410–9. doi: 10.1074/jbc.M109.001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Abankwa D, Gorfe AA, Hancock JF. Mechanisms of Ras membrane organization and signalling: Ras on a rocker. Cell Cycle. 2008;7:2667–73. doi: 10.4161/cc.7.17.6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Vogel A, Reuther G, Weise K, Triola G, Nikolaus J, Tan KT, et al. The lipid modifications of Ras that sense membrane environments and induce local enrichment. Angew Chem Int Ed Engl. 2009;48:8784–7. doi: 10.1002/anie.200903396. [DOI] [PubMed] [Google Scholar]
- [101].Abankwa D, Gorfe AA, Hancock JF. Ras nanoclusters: molecular structure and assembly. Semin Cell Dev Biol. 2007;18:599–607. doi: 10.1016/j.semcdb.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Gorfe AA, Babakhani A, McCammon JA. H-ras protein in a bilayer: interaction and structure pertur bation. J Am Chem Soc. 2007;129:12280–6. doi: 10.1021/ja073949v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Cirstea IC, Kutsche K, Dvorsky R, Gremer L, Carta C, Horn D, et al. A restricted spectrum of NRAS mutations causes Noonan syndrome. Nature genetics. 2010;42:27–9. doi: 10.1038/ng.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Mayor S, Rao M. Rafts: scale-dependent, active lipid organization at the cell surface. Traffic. 2004;5:231–40. doi: 10.1111/j.1600-0854.2004.00172.x. [DOI] [PubMed] [Google Scholar]
- [105].Sharma P, Varma R, Sarasij RC, Ira, Gousset K, Krishnamoorthy G, et al. Nanoscale organization of multiple GPI-anchored proteins in living cell membranes. Cell. 2004;116:577–89. doi: 10.1016/s0092-8674(04)00167-9. [DOI] [PubMed] [Google Scholar]
- [106].Goswami D, Gowrishankar K, Bilgrami S, Ghosh S, Raghupathy R, Chadda R, et al. Nanoclusters of GPI-anchored proteins are formed by cortical actin-driven activity. Cell. 2008;135:1085–97. doi: 10.1016/j.cell.2008.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Tian T, Harding A, Inder K, Plowman S, Parton RG, Hancock JF. Plasma membrane nanoswitches generate high-fidelity Ras signal transduction. Nat Cell Biol. 2007;9:905–14. doi: 10.1038/ncb1615. [DOI] [PubMed] [Google Scholar]
- [108].Harding A, Hancock JF. Ras nanoclusters: combining digital and analog signaling. Cell Cycle. 2008;7:127–34. doi: 10.4161/cc.7.2.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Harding AS, Hancock JF. U sing plasma membrane nanoclusters to build better signaling circuits. Trends Cell Biol. 2008;18:364–71. doi: 10.1016/j.tcb.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Kholodenko BN, Hancock JF, Kolch W. Signalling ballet in space and time. Nat Rev Mol Cell Biol. 2010;11:414–26. doi: 10.1038/nrm2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Chapkin RS, Wang N, Fan YY, Lupton JR, Prior IA. Docosahexaenoic acid alters the size and distribution of cell surface microdomains. Biochimica et biophysica acta. 2008;1778:466–71. doi: 10.1016/j.bbamem.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Kim W, Khan NA, McMurray DN, Prior IA, Wang N, Chapkin RS. Regulatory activity of polyunsaturate d fatty acids in T-cell signaling. Progress in lipid research. 2010;49:250–61. doi: 10.1016/j.plipres.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Zhou Y, Hancock JF, Lichtenberger LM. The nonsteroidal anti-inflammatory drug indomethacin induces heterogeneity in lipid membranes: potential implication for its diverse biological action. PLoS One. 2010;5:e8811. doi: 10.1371/journal.pone.0008811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Zhou Y, Plowman SJ, Lichtenberger LM, Hancock JF. The anti-inflammatory drug indomethacin alters nanoclustering in synthetic and cell plasma membranes. J Biol Chem. 2010;285:35188–95. doi: 10.1074/jbc.M110.141200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Eden ER, White IJ, Tsapara A, Futter CE. Membrane contacts between endosomes and ER provide sites for PTP1B-epidermal growth factor receptor interaction. Nat Cell Biol. 2010;12:267–72. doi: 10.1038/ncb2026. [DOI] [PubMed] [Google Scholar]