Abstract
The discovery, in 2006, that loss-of-function mutations in the filaggrin gene (FLG) are the cause of ichthyosis vulgaris – the most common disorder of keratinization – and also a strong genetic risk factor for atopic eczema, marked a significant breakthrough in the understanding of eczema pathogenesis. Subsequent investigations of the role of FLG null mutations have identified a series of significant associations with atopic disease phenotypes, including atopic asthma, allergic rhinitis and peanut allergy. However, many questions remain to be answered in relation to the precise mechanisms by which deficiency of an intracellular protein expressed primarily in the differentiating epidermis may contribute to the development of cutaneous and systemic pathology. This review aims to highlight the key milestones in filaggrin research over the past 25 years, to discuss the mechanistic, clinical and therapeutic implications and to consider possible future directions for ongoing investigation.
Keywords: Eczema, ichthyosis vulgaris, keratinizing disorder, atopy, skin barrier
A brief history of filaggrin, from ichthyosis vulgaris to modern molecular genetics
Ichthyosis vulgaris (OMIM #146700) is the most common inherited disorder of keratinisation, with an estimated prevalence of between 1 in 80 and 1 in 250 English schoolchildren (Brown et al., 2008a; Wells and Kerr, 1966). It is an autosomal semidominant condition with incomplete penetrance and variable expressivity, such that disease severity can vary considerably even within affected families (Smith et al., 2006). The ichthyosis is often asymptomatic, although patients may complain of dry, rough skin and cosmetic embarrassment. The disease is characterized by fine scaling which is most apparent on the lower abdomen, extensor aspects of arms and lower legs, with sparing of the flexures (Judge et al., 2004). The semi-adherent scales are white or grey and the ichthyosis varies with environmental conditions, being more severe in cold, dry weather and sometimes resolving completely in a hot, humid environment (DiGiovanni, 2003). The palms and soles are not scaly, but show hyperlinearity, particularly on the thenar eminence (Brown et al., 2009; Judge et al., 2004). Keratosis pilaris is another feature of ichthyosis vulgaris (DiGiovanni, 2003; Mevorah et al., 1985), but since keratosis pilaris is also very common in the normal population, it is not a discriminatory clinical sign (Brown et al., 2008a).
Historically, several convergent lines of reasoning led to a study of the filaggrin gene as a cause for ichthyosis vulgaris. Skin biopsies from patients with ichthyosis vulgaris showed a reduction or absence of keratohyalin granules (the main component of which is profilaggrin) by light microscopy (Manabe et al., 1991; Smith et al., 2006; Sybert et al., 1985) and by electron microscopy the granules were noted to be abnormally shaped or completely absent (Smith et al., 2006; Sybert et al., 1985). Immunostaining of ichthyosis vulgaris skin biopsies showed a reduction in filaggrin protein expression (Fleckman et al., 1987; Sybert et al., 1985) and a reduction in profilaggrin mRNA within keratinocytes was also reported (Manabe et al., 1991; Nirunsuksiri et al., 1995; Nirunsuksiri et al., 1998). The recessive mutant flaky tail mouse, an animal model of ichthyosis vulgaris, demonstrated genetic linkage to the mouse epidermal differentiation complex (Smith et al., 2006) and it expressed abnormal profilaggrin that is not proteolytically processed to filaggrin (Presland et al., 2000). More recently, the phenotype of an absence of granules within the cells of the granular layer was mapped to the epidermal differentiation complex (Mischke et al., 1996) on chromosome 1q21 (Compton et al., 2002).
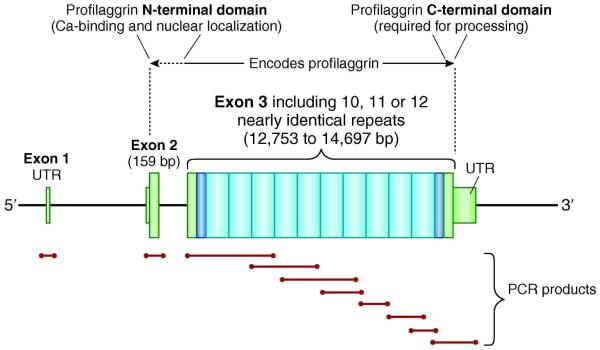
These observations date from the mid-1980s (Figure 1) and the profilaggrin gene was partially sequenced in 1992 (Presland et al., 1992). However the identification of loss-of-function mutations was delayed until 2006 because FLG is such a very large and repetitive gene (Figure 2), making sequencing using conventional polymerase chain reaction (PCR) technically difficult. The formidable challenge of fully sequencing FLG exon 3 was eventually conquered by the use of long range PCR to amplify the whole of exon 3, combined with short specific PCRs to amplify multiple overlapping fragments which could then be used to reconstruct the repetitive sequence as a jig-saw (Figure 2)(Smith et al., 2006). The initial two loss-of-function mutations (R501X and 2282del4) were thus identified in the first repeat of FLG exon 3 (Smith et al., 2006).
Figure 1.
Historical perspective on filaggrin research
Insert shows immunohistochemical staining of human epidermis, with filaggrin in green, basal-specific keratin 5 in red and nuclei stained blue.
Figure 2.
Diagrammatic representation of FLG gene structure and sequencing strategy?
FLG is a large gene located on chromosome 1q21, within the epidermal differentiation complex, a dense cluster of genes involved with keratinocyte terminal differentiation. The gene product is profilaggrin, an insoluble polyprotein which is proteolyzed to release functional filaggrin monomers. Full sequencing of the FLG gene is time-consuming and requires 10 or more PCR reactions and more than 30 sequencing reactions. UTR, untranslated region; bp, base pairs; PCR, polymerase chain reaction
The large and repetitive structure of the FLG gene remains a significant challenge for routine PCR-based sequencing. However, detailed knowledge of selected SNPs, which are unique to each repeat in the extensively studied white European DNA collections, has facilitated the development of repeat-selective PCR primers for sequencing and genotyping assays (Sandilands et al., 2007). The use of next generation sequencing techniques for FLG genotyping is anticipated, but careful application will be required since it will be difficult to accurately align relatively short sequence reads (e.g. 50-100 base pairs) across the highly repetitive third exon, particularly in ethnic populations for which the FLG sequence is not yet well annotated.
The presence of intragenic copy number variation (CNV) in FLG adds a further level of complexity to the sequence analysis. Using Southern blot analysis, three common size alleles encoding 10, 11 or 12 filaggrin repeats were correctly inferred (Gan et al., 1990). These were subsequently confirmed by sequencing and shown to be due to duplication of filaggrin repeat 8 or, duplication of both repeat 8 and repeat 10 (Sandilands et al., 2007). Thus, null mutations aside, the number of filaggrin units varies from 20 to 24 in the human population. Very recently, methods have been developed to allow the genotyping of filaggrin CNV across large populations (Brown et al., 2011b). The CNV allele frequencies in the Irish population were found to be 33.9% 10 repeats, 51.5% 11 repeats and 14.6% 12 repeats. The shortest genotype (10,10), carried by 11.5% of Irish people, was found to infer an eczema risk of 1.67, independent of the loss-of-function mutations. Interestingly, when null mutations are excluded, each additional filaggrin repeat gained decreases the odds ratio for atopic eczema by 0.88 (Brown et al., 2011b). Thus, filaggrin CNV makes a significant, dose-dependent contribution to eczema risk and even a modest 10-20% increase in epidermal filaggrin expression is predicted to be therapeutic for eczema or protective against developing eczema.
It may be pertinent to ask why the strong association of FLG with atopic eczema was not detected by earlier genome-wide association studies? There are a number of possible explanations for this. One early genome-wide genetic study using microsatellite markers did show linkage to the 1q21 locus (Cookson et al., 2001). Factors such as the type of markers used and the study design based on allele transmission (rather than the current more powerful case-control design using high density single-nucleotide polymorphisms – SNPs) led to the importance of this locus being under-recognized. Furthermore, the very strong association of FLG with atopic eczema that we now recognize, results from the combined null genotype reflecting the effect of two or more null mutations, a mechanism that genome-wide association using tagging SNPs is not designed to detect. The most recently published genome-wide study was designed with SNPs tagging the FLG locus and a strong signal was observed (Esparza-Gordillo et al., 2009). It remains to be determined whether this signal is entirely attributable to FLG loss of function mutations or whether other genetic factors nearby in the epidermal differentiation complex of genes also contribute to eczema risk. CNV within the FLG gene contributes to eczema risk, independent of the null mutations (Brown et al., 2011b). This has yet to be factored into interpretation of the genome-wide association data for the 1q21 locus.
Profilaggrin and filaggrin are multi-functional proteins in the maintenance of an optimal skin barrier
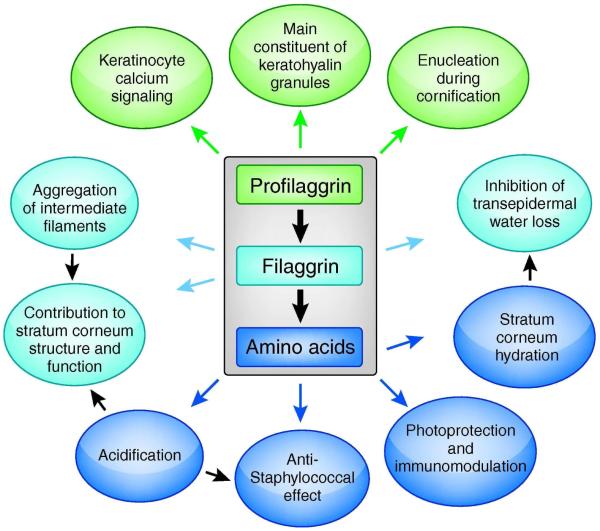
The large (>400kDa), insoluble polyprotein profilaggrin is dephosphorylated and degraded to produce monomeric filaggrin in the stratum corneum and then further proteolyzed to release its component amino acids. Profilaggrin, filaggrin and the amino acids each make different contributions to epidermal structure and barrier function (Figure 3), but the exact mechanisms by which profilaggrin and filaggrin, as intracellular proteins, contribute to what appears to be a paracellular barrier defect (Gruber et al., 2011; Scharschmidt et al., 2009) remain to be defined.
Figure 3.
Profilaggrin, filaggrin and their constituent amino acids are multifunctional proteins contributing to the formation and function of the skin barrier
Diagram summarizing the known and possible functions of profilaggrin, filaggrin and amino acids released by filaggrin proteolysis.
Profilaggrin forms the major component of the keratohyalin granules that are visible by light microscopy within the granular layer of the epidermis. The profilaggrin molecule (Figure 2) is composed of an N-terminal domain which has calcium-binding and nuclear localization components, followed by 10, 11 or 12 nearly identical filaggrin repeats which have keratin-binding properties, plus a C-terminal domain of unknown function (Sandilands et al., 2009). Each of these components has a different function in the differentiating epidermis. The S100-like calcium binding domain (Markova et al., 1993) may play a role in the regulation of calcium-dependent events during terminal epidermal differentiation (Nirunsuksiri et al., 1995; Presland et al., 1992) or conversely, calcium may be involved in the control of profilaggrin processing (Presland et al., 2004; Sandilands et al., 2009). The N-terminal domain is cleaved from profilaggrin and translocates to the nucleus where it may play a role in the enucleation of keratinocytes in the outer stratum corneum (Sandilands et al., 2009). The precise function of the C-terminal domain is unclear but it is known to be required for the processing of profilaggrin to filaggrin. Loss-of-function mutations within FLG exon 3 result in a truncated profilaggrin molecule which lacks the C-terminus, resulting in an almost complete absence of filaggrin monomers (Sandilands et al., 2007). Each of the reported FLG null mutations therefore have an equivalent molecular biological effect, since they each produce biochemically unstable truncated profilaggrin which cannot be processed to release functional filaggrin.
Monomeric filaggrin binds to keratins 1 and 10 and other intermediate filament proteins within the keratinocyte cytoskeleton to form tight bundles (Manabe et al., 1991), facilitating the collapse and flattening of cells in the outermost stratum corneum to produce squames. This process of cell flattening is still seen to occur in the absence of processed filaggrin, but on an ultrastructural level, it appears that loss-of-function mutations in FLG may be associated with disorganized keratin filaments, impaired lamellar body loading and abnormal architecture of the lamellar bilayer (Gruber et al., 2011). A reduction in corneodesmosome density and tight junction protein expression has also been observed in the skin from ichthyosis vulgaris patients (Gruber et al., 2011). It is unclear as to how these observations may be attributable to filaggrin deficiency from our current understanding of filaggrin processing and they may represent secondary effects. Filaggrin contributes to the protein-lipid cornified cell envelope which replaces the plasma membrane of differentiating keratinocytes and is extensively cross-linked by transglutaminases, forming a barrier to water loss and minimizing the entry of allergens and micro-organisms (Candi et al., 2005).
Filaggrin is ultimately deiminated and degraded on the skin surface to release its component amino acids. A mixture of hygroscopic amino acids, at an estimated concentration of 100 millimolar (Candi et al., 2005) is produced, the so-called ‘natural moisturising factor’ (Rawlings and Harding, 2004), which contributes to epidermal hydration and barrier function. FLG null mutations are associated with lower levels of hygroscopic amino acids in the stratum corneum of human subjects, measured using Raman spectroscopy and there is a concomitant increase in transepidermal water loss (Kezic et al., 2008).
Three published studies describing murine models of filaggrin haploinsufficiency have shown a barrier impairment and enhanced percutaneous allergen sensitization (Fallon et al., 2009; Man et al., 2008; Oyoshi et al., 2009). The barrier integrity phenotype associated with FLG null mutations in human skin is currently emerging, with evidence of a significant reduction in natural moisturizing factors in the stratum corneum, to which filaggrin breakdown products contribute (Kezic et al., 2008; Kezic et al., 2011; O’Regan et al., 2010b) as well as FLG genotype-related impairment in stratum corneum integrity and cohesion (Angelova-Fischer et al., 2011). However, eczema severity is itself associated with a reduction in natural moisturizing factor concentrations and the debate is set to continue regarding the relative importance of epidermal defects and immune dysregulation as the key initiating factors in eczema pathogenesis (Elias and Schmuth, 2009; Hudson, 2006; Simpson, 2010; Vercelli, 2009).
Filaggrin is a histidine-rich protein (Lynley and Dale, 1983) and histidine is metabolized to trans-urocanic acid (trans-UCA); pyrrolidone-5-carboxylic acid (PCA) is the other main breakdown product of filaggrin and together these organic acids help to maintain the pH gradient of the epidermis as evidenced by a higher surface pH in FLG null mutation carriers (Jungersted et al., 2010). The ‘acid mantle’ of the stratum corneum has a well-known antimicrobial effect and there is evidence that filaggrin breakdown products at physiological concentrations demonstrate an inhibitory effect on the growth of Staphyloccus aureus (Miajlovic et al., 2010). An acidic pH within the stratum corneum is also important for the functional activity of enzymes involved in ceramide metabolism (Fluhr et al., 2010). The acidic pH also modulates the activity of the serine protease cascade (Ovaere et al., 2009) required for co-ordinated epidermal differentiation and cornified cell envelope formation. Finally, a possible role of filaggrin breakdown products in UV photo-protection has been suggested. The photo-isomerisation of trans- to cis-urocanic acid (Elias and Choi, 2005) produces a molecule with an action spectrum of 280-310nm (McLoone et al., 2005) which is within the UVB range and there is in vitro evidence that cis-UCA has immunomodulatory actions in human keratinocytes and leukocytes (Gilmour et al., 1993; McLoone et al., 2005). There is also recent in vitro evidence from an organotypic siRNA FLG knock-down model that lack of filaggrin leads to a reduction in urocanic acid concentration and an increased sensitivity to UV-induced apoptosis (Mildner et al., 2010).
Filaggrin and filaggrin-related proteins are expressed in the skin of mammals including monotremes, but these molecules are not well conserved and are difficult to identify in lower animal orders. Filaggrin biology has been most extensively studied in mouse and canine species (Chervet et al.; Fallon et al., 2009; Hoste et al.; Man et al., 2008; Marsella and Girolomoni, 2009; Matsui et al.; Moniaga et al.; Oyoshi et al., 2009). These and other experimental models will continue to contribute to our understanding of the functions of this remarkable molecule.
Filaggrin haploinsufficiency plays a key role in phenotypic variation and common disorders of the skin
Filaggrin haploinsufficiency (that is, a lack of filaggrin protein resulting from a heterozyous loss-of-function mutation) contributes to several common dermatological disorders. Ichthyosis vulgaris is a semidominant disorder in which individuals with two FLG null mutations have a more severe phenotype than those with one FLG null mutation. The characteristic skin scaling in ichthyosis vulgaris may logically be attributed to the known functions of FLG, but the mechanisms by which FLG haploinsufficiency contributes to the associated features of keratosis pilaris and palmoplantar hyperlinearity are currently unknown.
Ichthyosis vulgaris is frequently associated with atopic eczema and hence it was a logical next step to investigate the association of FLG null mutations with this disease. A significant association was initially demonstrated in 15 Irish families and then replicated in three separate case/control studies from Irish, Scottish and Danish populations (Palmer et al., 2006). The strong and highly significant association of FLG null mutations with atopic eczema has subsequently been replicated in over 20 independent studies, including case/control studies, family studies and unselected population cohorts. Two recent meta-analyses of these data have estimated the odds ratio of developing atopic eczema to be 4.78 (van den Oord and Sheikh, 2009) and 3.12 (Rodriguez et al., 2009) in association with FLG null genotype. The eczema sub-phenotype that is most strongly associated with FLG null mutations is that of early onset, severe and persistent disease (Barker et al., 2007; Brown et al., 2008b) and with the associated ‘extrinsic’ features of raised total IgE and allergic sensitization (Weidinger et al., 2006; Weidinger et al., 2007). Population cohort studies include atopic eczema of mild-moderate severity and this phenotype has also demonstrated significant association with FLG null mutations (Brown et al., 2008a; Gruber et al., 2010; Henderson et al., 2008). However, mild-moderate eczema shows a lower odds ratio than moderate-severe eczema and its association with FLG null mutations may be significant only in individuals carrying two null mutations (Brown et al., 2008a). Taking into account all the clinical signs that may be attributable to FLG haploinsufficiency (including atopic eczema, ichthyosis, xerosis, palmar hyperlinearity and keratosis pilaris), FLG null mutations are highly penetrant (Brown et al., 2009).
In the light of the very significant association of atopic eczema with FLG null mutations, several other sub-groups of eczema have been investigated for their link with filaggrin. There is a well recognized genetic susceptibility to contact dermatitis (Kezic, 2011) but it is often difficult to distinguish the sub-groups of irritant and allergic contact dermatitis from atopic dermatitis, hence these genetic studies are difficult to interpret (Brown and Cordell, 2008; de Jongh et al., 2008; Lerbaek et al., 2007). Studies on FLG genotype as a risk factor for experimentally-induced irritant dermatitis have given conflicting results (Angelova-Fischer et al., 2011; Jungersted et al., 2010) and this may be due to methodological differences in the concentration of irritant. Studies published to date have reported no association of FLG genotype with allergic contact dermatitis defined by a positive patch test result to one or more substance (Carlsen et al., 2010) nor with a subset of patients having both atopic eczema and contact allergy (Carlsen et al., 2011b). In contrast, FLG null genotype has shown significant association with nickel sensitization, but only in a subgroup of cases reporting intolerance of costume jewellery (Novak et al., 2008) and in a subgroup of women who had not had their ears pierced (Thyssen et al., 2010b). The proposed explanation for this observation is that ear piercing is a stronger risk factor than FLG genotype (Ross-Hansen et al., 2010), emphasising the importance of environmental exposures in allergy development as well as the need for careful phenotype definition in genetic studies.
One small study has investigated FLG genotype as a risk factor for a type I hypersensitivity reaction to latex (Carlsen et al., 2011a) and found no significant association. However this study should be interpreted with caution since it included only three FLG heterozygotes and therefore may not have had sufficient power to exclude a significant association.
Another subgroup of atopic eczema is the severe viral infection eczema herpeticum. Case-control studies from European and African population groups have shown that the loss-of-function mutation R501X confers an even greater risk of eczema herpeticum than atopic eczema (Gao et al., 2009), suggesting a possible role for defective skin barrier in this viral infection.
Two other common cutaneous disorders have been investigated with respect to a hypothetical FLG association: psoriasis, because of its co-association with the 1q21 locus in genome-wide scans (Bowcock et al., 2001); and acne vulgaris, because of a possible protective effect of FLG null mutations (Sergeant et al., 2009). However, adequately powered studies have shown that there is no association of FLG loss-of-function mutations with psoriasis in the UK and Irish populations (Zhao et al., 2007) nor with acne vulgaris in the Singaporean Chinese population (Common et al.).
FLG mutations contribute to genetic risk at each step of the atopic march
The ‘atopic march’ (Hahn and Bacharier, 2005; Spergel, 2010; Wuthrich and Schmid-Grendelmeier, 2003) describes the tendency for atopic eczema to precede the development of food allergies, asthma and allergic rhinitis in a temporal sequence. FLG null mutations have been reported as a risk factor for each step in this march: atopic eczema (Palmer et al., 2006); allergic sensitisation (van den Oord and Sheikh, 2009); the sub-group of asthma in association with eczema (Brown et al., 2008a; Henderson et al., 2008; Marenholz et al., 2006; Weidinger et al., 2008b); allergic rhinitis (Schuttelaar et al., 2009; Weidinger et al., 2008b); and most recently peanut allergy (Brown et al., 2011). Thus, FLG null alleles are a significant risk factor for all aspects of atopy, but with differing odds ratios for each specific phenotype. Overall these studies underscore the role of skin barrier dysfunction as a key driver of allergic disease.
FLG loss-of-function mutations show population specificity
Since the discovery of the first two FLG loss-of-function mutations (R501X and 2282del4) in 2006 (Smith et al., 2006), these two mutations along with the less prevalent S3247X and R2447X have been extensively studied and are present in 7-10% of the white European population. Subsequently a total of more than 20 other rare or family-specific loss-of-function mutations within FLG exon 3 have been discovered in populations of white European ancestry. However, these mutations are population-specific and cannot be used for the basis of genetic epidemiology studies in other populations. The population specificity of FLG null mutations indicates that they have arisen after one population has become split away from another. Thus populations arising from common ancestors share common ancestral mutations, but populations that have not experienced genetic admixture show different mutations. The presence of rare and family-specific mutations indicates that FLG null mutations are still arising within the UK and Irish populations that we have studied.
Asian populations have been shown to have their own mutation spectra, following detailed study of the Japanese population (Enomoto et al., 2008; Hamada et al., 2008; Nemoto-Hasebe et al., 2009b; Nomura et al., 2008; Nomura et al., 2009; Nomura et al., 2007; Osawa et al., 2010) and more recently the Han Chinese (Zhang et al., 2011) and Singaporean Chinese populations (Chen et al., 2011; Chen et al., 2008). In the white European population two prevalent FLG mutations account for over 80% of the FLG null alleles, whereas in the Singaporean Chinese population there are eight different FLG null mutations that account for 80% of the spectrum of FLG mutations (Chen et al., 2011).
The filaggrin story represents an unusual situation in complex trait genetics because the causative variants are either nonsense or frame-shift mutations in a protein-encoding exon and therefore new causative variants can be identified purely from their sequence. In contrast, the vast majority of loci detected in other complex traits are anonymous regions of DNA conferring disease susceptibility. Such loci are not clearly associated with a specific gene and the causative variant(s) either remain unknown or require considerable functional analysis to confirm and understand them. In the case of filaggrin, the readily identifiable causative mutations have revealed the complex genetic architecture of this locus, where each ancestral population has it own unique spectrum of mutations, some rare and some common, each of which are found on a different haplotype of nearby SNPs. Thus, individual SNPs can only reveal part of the signal coming from the FLG locus, further masking its importance in conferring eczema susceptibility.
A smaller number of FLG loss-of-function mutations have so far been reported in other Asian populations, including Korean (Kang et al., 2009), Taiwanese (Hsu et al., 2009), and Bangladeshi (Sinclair et al., 2009) ichthyosis vulgaris and atopic eczema case collections plus one European patient whose mother was from the Philippines (Greisenegger et al., 2010). Until recently, FLG null mutations had not been detected within African populations and an Ethiopian study, including direct sequencing of FLG in 40 cases, has identified only one loss-of-function mutation (Winge et al.). Furthermore, skin biopsies taken from 7 of the Ethiopian cases with palmar hyperlinearity did not show a reduction in filaggrin expression as expected on immunohistochemistry (Winge et al.). These are small numbers from which to draw any firm conclusions, but this study suggests that ichthyosis vulgaris in the Ethiopian population may have a different genetic mechanism from that seen in Europeans.
The prevalence of FLG null mutations varies across Europe, but R501X and 2282del4 are the two most common mutations and they have consistently shown significant association with atopic eczema across the continent (Rodriguez et al., 2009), with the one exception of the Italian population. R501X and 2282del4 are rare in Italian atopic eczema cases (allele frequency <1% for each)(Giardina et al., 2008) and full sequencing of FLG exon 3, exon 2 and the promoter region in a total of 220 Italian atopic dermatitis patients identified only 3 additional mutations and no association with atopic eczema (Cascella et al., 2011). The pattern of FLG mutations in other Mediterranean populations has not yet been examined but the Italian data suggest that different genetic factors may predispose to atopic eczema in these populations and this warrants further investigation.
Filaggrin mutations may act as genetic modifiers in rare genodermatoses
It has been reported that the co-existence of FLG null mutations may be associated with a more severe phenotype of X-linked ichthyosis (Liao et al., 2007) and pachyonychia congenita (Gruber et al., 2009) within an affected family. However, it is not possible to be certain whether FLG is truly acting as a genetic modifier of disease in these single pedigrees because each disease shows considerable inter-individual variation and greater numbers of cases will be needed to clarify this question.
What is the role of FLG-related barrier dysfunction in other inflammatory diseases?
Other inflammatory barrier diseases, including Crohn’s disease, ulcerative colitis and sarcoidosis, share common susceptibility loci with atopic eczema (O’Regan et al., 2010a; Schreiber et al., 2005) but do not show association with FLG null mutations (Ruether et al., 2006; Van Limbergen et al., 2009).
Autoantibody formation to citrullinated profilaggrin is a highly sensitive and specific serological marker for rheumatoid arthritis (Sanmarti et al., 2009) and can predate the onset of joint disease (Perez et al., 2007). Levels of anti-filaggrin antibody may also correlate with arthritis activity (Choi et al., 2005). However, filaggrin is not expressed in articular tissues and these autoantibodies appear to be produced as a result of cross-reaction between deiminated peptide sequences which are present within fibrin chains in the synovium (Mohrenschlager et al., 2006) and deiminated sequences produced during the post-translational processing of profilaggrin (Perez et al., 2007). Anti-filaggrin antibodies are therefore not believed to be pathogenic (Van Steendam et al., 2011). FLG null mutations R501X, 2282del4 and 3702delG, have been studied in rheumatoid arthritis patients and controls (Huffmeier et al., 2008) and FLG mutation carriers showed no increased risk of developing rheumatoid arthritis. However in a separate study FLG heterozygotes did show significantly elevated levels of autoantibodies to citrullinated peptides compared to controls (p=0.018)(Van Steendam et al., 2011) and FLG null mutations may therefore contribute to the development of humoral autoimmunity in early rheumatoid arthritis (Huffmeier et al., 2008) by an as yet undefined mechanism.
Alopecia areata is a tissue-specific autoimmune disease and genetic factors make a significant contribution to its aetiology (Petukhova et al., 2010). Alopecia areata is known to be associated with atopic disease and it has been shown that co-morbidity with atopic eczema as well as the FLG null variants (R501X and 2282del4) predict a more severe form of alopecia areata (Betz et al., 2007; Goh et al., 2006).
FLG demonstrates gene-environment interactions
Eczema is a complex trait, in which multiple genetic and environmental factors contribute to pathogenesis and the interactions of several putative environmental risk factors for atopic disease with FLG genotype have been investigated. It has been hypothesized that FLG haploinsufficiency may weaken the epidermal barrier and hence potentiate the effects of environmental allergens (Bisgaard et al., 2008) resulting in eczema. Birth cohort studies from Denmark and the UK have shown that cat ownership early in life increases the risk of atopic eczema as an additional effect on top of the risk associated with FLG null genotype, whereas dog exposure has no effect (Bisgaard et al., 2008). A separate cohort study from the Netherlands has replicated this finding, showing an increased risk of atopic eczema in FLG null mutation carriers that is further enhanced by early-life exposure to cats (Schuttelaar et al., 2009). However, these data are difficult to interpret mechanistically because, whilst other studies have shown an association of FLG null mutations with raised IgE to cat dander (Henderson et al., 2008) and a significant correlation with atopic eczema severity and specific IgE level for cat dander (Nemoto-Hasebe et al., 2009a), there was no such association with sensitization in the Danish or English cohorts.
Another key environmental influence early in life is the presence or absence of other children, including siblings and contacts in the day-care situation, since these individuals may result in contact with pathogens and allergens. Again these complex epidemiological data are difficult to interpret. A risk analysis of early childhood eczema from a high-risk cohort of children born to mothers with asthma showed no association with time spent in day-care (Bisgaard et al., 2009) but interaction with FLG genotype was not examined. Conversely, two large birth cohort studies from Germany (Cramer et al., 2010) have shown that children with FLG null mutations have a significantly higher risk of eczema if they have an older sibling and attendance at a day-care centre lessened this risk (Cramer et al., 2010). Environmental influences on eczema pathogenesis are therefore not clearly defined and these findings demonstrate the importance of stratification for FLG genotype in future epidemiological studies.
The development of irritant and allergic contact dermatitis is largely dependent on environmental exposure to the irritant or allergic substances, but FLG genotype is also a strong risk factor. However, as discussed above, it is currently unclear whether FLG haploinsufficiency is the risk per se, or whether the risk is mediated via atopic eczema.
The co-existence of FLG mutations and early food sensitization has been shown to improve the positive predictive value for childhood asthma (Marenholz et al., 2009). This observation was interpreted as a representation of two distinct mechanisms interacting in the pathogenesis of asthma (Marenholz et al., 2009). However, it is also possible that the increased risk of asthma occurs as a result of gene-environment interaction between FLG and food allergen exposure. A deeper understanding of the mechanisms within the atopic march will be required for the correct interpretation of these interesting observations.
Filaggrin deficiency forms one portion of a common pathway in eczema pathophysiology
The filaggrin story demonstrates how the study of a monogenic disorder can provide insight into complex trait disease and the striking significance of FLG null mutations as a genetic risk factor for atopic eczema has placed a new focus on the role of barrier impairment in eczema pathogenesis. There is an inherent barrier defect in atopic skin which is present even in the absence of eczema (Flohr et al., 2010; Jakasa et al., 2006; Jakasa et al., 2007) and it is clear that barrier dysfunction can occur as a result of many different molecular mechanisms – independently of filaggrin – but these are outwith the scope of this current review. Focussing on filaggrin, it is apparent that not all filaggrin deficiency is related to FLG null mutations and down-regulation of filaggrin may be brought about by the interplay of other factors. The degree of barrier impairment assessed by transepidermal water loss and stratum corneum hydration shows some correlation with FLG null genotype (Jakasa et al., 2011), although active inflammation further increases the degree of transepidermal water loss (Flohr et al., 2010; Nemoto-Hasebe et al., 2009a). Acute eczematous inflammation itself down-regulates filaggrin expression (Howell et al., 2007) and inflammatory cytokines have been reported to down-regulate filaggrin expression in vitro, including interleukins-4 and −13 (Howell et al., 2007), IL-22 (Gutowska-Owsiak et al., 2011) and IL-25 (Hvid et al., 2011b). Inflammatory cytokines may also down-regulate enzymes required for profilaggrin processing: IL-22 down-regulates cathepsin D in vitro (Gutowska-Owsiak et al., 2011) and Th2-associated cytokines have been shown to reduce caspase 14 mRNA levels in cultured keratinocytes (Hvid et al., 2011a). Similarly, genetic variation in enzymes known to play a role in profilaggrin processing, including caspase 14 (Denecker et al., 2007; Hoste et al., 2011), SASPase (Matsui et al., 2011), calpain I and bleomycin hydrolase (Kamata et al., 2011) may affect the levels of functional filaggrin monomers. In this way inflammatory mediators and enzymatic processing within the stratum corneum may each lead to filaggrin deficiency, forming a common pathway in eczema pathophysiology.
Large collaborative projects are required for the investigation of genetic effects on a population level
One of the hidden stories within the filaggrin field is the fact that many of the significant findings described above have resulted from the collaborative work of national and international groups of skin scientists and clinicians. The work required ‘behind-the-scenes’ to co-ordinate such collaborations is not formally reported, but is clearly an important part of the success of large studies. Similarly, the organization required to collect high quality samples and clinical data from birth cohorts, case collections and population-matched control groups is a very significant undertaking. Within the field of eczema research there are now several large cohorts and case collections which have been used to further our understanding of the role of FLG genotype, including the Irish case collection based in Dublin (Brown et al., 2011; O’Regan et al., 2010a; O’Regan et al., 2010b; Sandilands et al., 2007; Weidinger et al., 2008a), MAS (Marenholz et al., 2006), KORA (Novak et al., 2008; Weidinger et al., 2008a) and ETAC (Muller et al., 2009) in Germany, GENUFAC in Northern Europe (Marenholz et al., 2006), PIAMA in the Netherlands (Schuttelaar et al. 2009), COPSAC in Denmark (Bisgaard et al. 2008; Bisgaard et al. 2009), ALSPAC (Henderson et al., 2008; Weidinger et al., 2008a), NCCGP (Brown et al., 2009; Brown et al., 2008a) and the EAT Study (Flohr et al., 2010) in England, ADVN in the USA (Gao et al., 2009), a Danish population sample (Thyssen et al., 2010a), a Canadian peanut allergy cohort (Brown et al., 2011) and internationally, the ISAAC (Weidinger et al., 2008b). These clinical resources will undoubtedly be useful in future studies and the established collaborative networks may be used to facilitate clinical trials forthcoming from the translational work as a result of new insight into genetic factors in eczema pathogenesis.
What may the next 25 years hold for the field of eczema genetic research?
Key strategic goals for the field of eczema genetics in the next 25 years may be categorized into understanding pathomechanisms, methods for the prevention of atopic disease and therapy development (Figure 1).
There are several rather fundamental questions that remain to be answered in relation to the filaggrin molecule: what are the patterns of filaggrin expression throughout the body; what are the mechanisms by which profilaggrin/filaggrin/amino acids act in healthy tissues; and how exactly does filaggrin deficiency lead to eczema and other atopic diseases? We can be reasonably optimistic that answers to these questions are likely to emerge over the coming years, particularly in view of the worldwide interest in filaggrin research. The field of complex trait genetics is progressing rapidly but one specific challenge is the dissection of the multiple gene-gene interactions that are likely to contribute to the heritability that remains to be explained in atopic eczema. Some genetic interactions with FLG have already been identified: FLG is known to show an independent and multiplicative effect with an eczema risk variant on 11q13.5 (O’Regan et al., 2010a) and with polymorphisms in the genes encoding IL-10 and IL-13 (Lesiak et al., 2011). However many other examples of genes interacting in the pathogenesis of atopic eczema are likely to emerge and these may well provide opportunities for disease prevention and novel therapies. It is hoped and anticipated that the application of next generation sequencing will speed up progress in the discovery of additional eczema genes. An understanding of the importance of FLG mutations in each population group will also facilitate the identification of other important genetic factors. In parallel with this, progress within the field of bioinformatics and statistical genetics is vital in order to make sense of the vast quantities of data produced by the latest high-throughput sequencing.
A clearer understanding of gene-environment interaction is needed in order to explain the dramatic rise in incidence of atopic disease in the industrialized nations over the recent decades. This will require careful epidemiological study of human populations, but the use of mouse models of atopic disease will also be useful in allowing experimental manipulation of both genetic and environmental effects. Gene-environment interactions are key to future preventative strategies, with the ultimate aim of preventing atopic eczema and halting the atopic march.
Another unanswered question is why filaggrin mutations are so prevalent in various human populations? It has been suggested that filaggrin deficiency may confer some heterozygote advantage and that “natural vaccination” against microbial pathogens via the skin barrier may have driven natural selection for these sequence variants during pandemics in our ancient past (Irvine & McLean, 2006). Filaggrin deficient animal models should allow this question to be addressed experimentally in the future (Fallon et al., 2009).
There is potential for the application of filaggrin research to novel therapeutic approaches and personalized medicine
The understanding that has been gained from filaggrin research offers real potential for future improvements in the therapies available for atopic disease. It is clear that the clinical phenotype of ichthyosis vulgaris-related eczema has a poor prognosis in terms of atopic eczema as well as associated atopic asthma, allergic rhinitis and food allergy. We may therefore sub-classify atopic eczema cases according to FLG genotype for the purposes of pharmacogenetic studies. Feasibility studies are currently underway in the UK and USA to investigate the therapeutic potential of barrier enhancement using emollients (BEEP, 2011) and experimental evidence is emerging to show that the FLG gene is amenable to up-regulation. Clarification of the relative importance of the different functions of profilaggrin/filaggrin/amino acids (Figure 3) will allow the further development of strategies to enhance or replace these molecules within human skin.
It is noteworthy that eczema, in contrast to psoriasis, currently lacks any widely effective biological treatment and a clearer understanding of the key functional mechanisms in atopy will be required to identify appropriate biological targets. In the meantime, there is an opportunity to focus on barrier improvement with bespoke emollients such as ceramide-lipid (Elias, 2011) or filaggrin replacement. These barrier improvement therapies have a lower likelihood of undesirable systemic immune effects. Other potential therapeutic strategies include the use of a peroxisome-proliferator-activated receptor-alpha (PPARα) ligand along with topical corticosteroid therapy, since PPARα ligand has been shown in a mouse model to have anti-inflammatory synergy whilst reducing the barrier impairment that results from topical steroids used in isolation (Hatano et al.). TNF-alpha antagonists improve the skin barrier in psoriasis (Kim et al., 2011) and therefore may also be beneficial in eczema.
Concluding remarks
In conclusion, the discovery of loss-of-function mutations within the filaggrin gene represents the single most significant breakthrough in understanding the molecular genetic mechanisms of a wide range of atopic and allergic disorders. Future research into the remarkable filaggrin molecule holds great potential to improve the care of atopic eczema patients and ultimately to prevent the development of atopic disease.
ACKNOWLEDGEMENTS
Sara Brown is supported by a Wellcome Trust Intermediate Clinical Fellowship (086398/Z/08/Z). Filaggrin research in the McLean laboratory is supported by grants from the British Skin Foundation, National Eczema Society, Medical Research Council (G0700314), the Wellcome Trust (090066/B/09/Z and 092530/Z/10/Z) and donations from anonymous families affected by eczema in the Tayside Region of Scotland.
Footnotes
CONFLICT OF INTEREST
Irwin McLean has filed patents on genetic testing and therapy development aimed at the filaggrin gene. Sara Brown has no conflict of interest to declare.
REFERENCES
- Angelova-Fischer I, Mannheimer AC, Hinder A, et al. Distinct barrier integrity phenotypes in filaggrin-related atopic eczema following sequential tape stripping and lipid profiling. Exp Dermatol. 2011;20:351–356. doi: 10.1111/j.1600-0625.2011.01259.x. [DOI] [PubMed] [Google Scholar]
- Barker JN, Palmer CN, Zhao Y, et al. Null mutations in the filaggrin gene (FLG) determine major susceptibility to early-onset atopic dermatitis that persists into adulthood. J Invest Dermatol. 2007;127:564–567. doi: 10.1038/sj.jid.5700587. [DOI] [PubMed] [Google Scholar]
- BEEP Feasibility Study of Barrier Enhancement for Eczema Prevention (BEEP) 2011 http://www.beepstudy.org/ and http://clinicaltrials.gov/ct2/show/NCT01142999.
- Betz RC, Pforr J, Flaquer A, et al. Loss-of-function mutations in the filaggrin gene and alopecia areata: strong risk factor for a severe course of disease in patients comorbid for atopic disease. J Invest Dermatol. 2007;127:2539–2543. doi: 10.1038/sj.jid.5700915. [DOI] [PubMed] [Google Scholar]
- Bisgaard H, Halkjaer LB, Hinge R, et al. Risk analysis of early childhood eczema. J Allergy Clin Immunol. 2009;123:1355–1360. e1355. doi: 10.1016/j.jaci.2009.03.046. [DOI] [PubMed] [Google Scholar]
- Bisgaard H, Simpson A, Palmer CN, et al. Gene-environment interaction in the onset of eczema in infancy: filaggrin loss-of-function mutations enhanced by neonatal cat exposure. PLoS Med. 2008;5:e131. doi: 10.1371/journal.pmed.0050131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowcock AM, Shannon W, Du F, et al. Insights into psoriasis and other inflammatory diseases from large-scale gene expression studies. Hum Mol Genet. 2001;10:1793–1805. doi: 10.1093/hmg/10.17.1793. [DOI] [PubMed] [Google Scholar]
- Brown SJ, Asai Y, Cordell HJ, et al. Loss-of-function variants in the filaggrin gene are a significant risk factor for peanut allergy. J Allergy Clin Immunol. 2011a;127:661–667. doi: 10.1016/j.jaci.2011.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SJ, Cordell HJ. Are filaggrin mutations associated with hand eczema or contact allergy?--we do not know. Br J Dermatol. 2008;158:1383–1384. doi: 10.1111/j.1365-2133.2008.08551.x. [DOI] [PubMed] [Google Scholar]
- Brown SJ, Kroboth KE, Sandilands A, Campbell LE, Pohler E, Kezic S, et al. Intragenic copy number variation within filaggrin contributes to risk of atopic dermatitis with a dose-dependent effect. J Invest Dermatol. 2011b doi: 10.1038/jid.2011.342. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SJ, Relton CL, Liao H, et al. Filaggrin haploinsufficiency is highly penetrant and is associated with increased severity of eczema: further delineation of the skin phenotype in a prospective epidemiological study of 792 school children. Br J Dermatol. 2009;161:884–889. doi: 10.1111/j.1365-2133.2009.09339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SJ, Relton CL, Liao H, et al. Filaggrin null mutations and childhood atopic eczema: a population-based case-control study. J Allergy Clin Immunol. 2008a;121:940–946. e943. doi: 10.1016/j.jaci.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SJ, Sandilands A, Zhao Y, et al. Prevalent and low-frequency null mutations in the filaggrin gene are associated with early-onset and persistent atopic eczema. J Invest Dermatol. 2008b;128:1591–1594. doi: 10.1038/sj.jid.5701206. [DOI] [PubMed] [Google Scholar]
- Candi E, Schmidt R, Melino G. The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol. 2005;6:328–340. doi: 10.1038/nrm1619. [DOI] [PubMed] [Google Scholar]
- Carlsen BC, Johansen JD, Menne T, et al. Filaggrin null mutations and association with contact allergy and allergic contact dermatitis: results from a tertiary dermatology clinic. Contact Dermatitis. 2010;63:89–95. doi: 10.1111/j.1600-0536.2010.01748.x. [DOI] [PubMed] [Google Scholar]
- Carlsen BC, Meldgaard M, Hamann D, et al. Latex allergy and filaggrin null mutations. J Dent. 2011a;39:128–132. doi: 10.1016/j.jdent.2010.11.001. [DOI] [PubMed] [Google Scholar]
- Carlsen BC, Thyssen JP, Menne T, et al. Association between filaggrin null mutations and concomitant atopic dermatitis and contact allergy. Clin Exp Dermatol. 2011b;36:467–472. doi: 10.1111/j.1365-2230.2010.03994.x. [DOI] [PubMed] [Google Scholar]
- Cascella R, Foti Cuzzola V, et al. Full sequencing of the FLG gene in Italian patients with atopic eczema: evidence of new mutations, but lack of an association. J Invest Dermatol. 2011;131:982–984. doi: 10.1038/jid.2010.398. [DOI] [PubMed] [Google Scholar]
- Chen H, Common JE, Haines RL, et al. Wide spectrum of filaggrin-null mutations in atopic dermatitis highlights differences between Singaporean Chinese and European populations. Br J Dermatol. 2011 doi: 10.1111/j.1365-2133.2011.10331.x. [DOI] [PubMed] [Google Scholar]
- Chen H, Ho JC, Sandilands A, et al. Unique and recurrent mutations in the filaggrin gene in Singaporean Chinese patients with ichthyosis vulgaris. J Invest Dermatol. 2008;128:1669–1675. doi: 10.1038/sj.jid.2008.2. [DOI] [PubMed] [Google Scholar]
- Chervet L, Galichet A, McLean WH, et al. Missing C-terminal filaggrin expression, NFkappaB activation and hyperproliferation identify the dog as a putative model to study epidermal dysfunction in atopic dermatitis. Exp Dermatol. 19:e343–346. doi: 10.1111/j.1600-0625.2010.01109.x. [DOI] [PubMed] [Google Scholar]
- Choi KH, Lee EB, Yoo CD, et al. Clinical significance of anti-filaggrin antibody recognizing uncitrullinated filaggrin in rheumatoid arthritis. Exp Mol Med. 2005;37:546–552. doi: 10.1038/emm.2005.67. [DOI] [PubMed] [Google Scholar]
- Common JE, Brown SJ, Haines RL, et al. Filaggrin null mutations are not a protective factor for acne vulgaris. J Invest Dermatol. 131:1378–1380. doi: 10.1038/jid.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton JG, DiGiovanna JJ, Johnston KA, et al. Mapping of the associated phenotype of an absent granular layer in ichthyosis vulgaris to the epidermal differentiation complex on chromosome 1. Exp Dermatol. 2002;11:518–526. doi: 10.1034/j.1600-0625.2002.110604.x. [DOI] [PubMed] [Google Scholar]
- Cookson WO, Ubhi B, Lawrence R, et al. Genetic linkage of childhood atopic dermatitis to psoriasis susceptibility loci. Nat Genet. 2001;27:372–373. doi: 10.1038/86867. [DOI] [PubMed] [Google Scholar]
- Cramer C, Link E, Horster M, et al. Elder siblings enhance the effect of filaggrin mutations on childhood eczema: results from the 2 birth cohort studies LISAplus and GINIplus. J Allergy Clin Immunol. 2010;125:1254–1260. e1255. doi: 10.1016/j.jaci.2010.03.036. [DOI] [PubMed] [Google Scholar]
- de Jongh CM, Khrenova L, Verberk MM, et al. Loss-of-function polymorphisms in the filaggrin gene are associated with an increased susceptibility to chronic irritant contact dermatitis: a case-control study. Br J Dermatol. 2008;159:621–627. doi: 10.1111/j.1365-2133.2008.08730.x. [DOI] [PubMed] [Google Scholar]
- Denecker G, Hoste E, Gilbert B, et al. Caspase-14 protects against epidermal UVB photodamage and water loss. Nat Cell Biol. 2007;9:666–674. doi: 10.1038/ncb1597. [DOI] [PubMed] [Google Scholar]
- DiGiovanni JJ. Ichthyosiform dermatoses. In: Freedberg IM, Eisen AZ, Wolff K, Austen KF, Goldsmith LA, Katz SI, editors. Fitzpatrick’s Dermatology in General Medicine. 6th ed. Vol. 1. McGraw-Hill; 2003. pp. 481–505. [Google Scholar]
- Elias PM. Therapeutic Implications of a Barrier-based Pathogenesis of Atopic Dermatitis. Ann Dermatol. 2011;22:245–254. doi: 10.5021/ad.2010.22.3.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias PM, Choi EH. Interactions among stratum corneum defensive functions. Exp Dermatol. 2005;14:719–726. doi: 10.1111/j.1600-0625.2005.00363.x. [DOI] [PubMed] [Google Scholar]
- Elias PM, Schmuth M. Abnormal skin barrier in the etiopathogenesis of atopic dermatitis. Curr Opin Allergy Clin Immunol. 2009;9:437–446. doi: 10.1097/ACI.0b013e32832e7d36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto H, Hirata K, Otsuka K, et al. Filaggrin null mutations are associated with atopic dermatitis and elevated levels of IgE in the Japanese population: a family and case-control study. J Hum Genet. 2008;53:615–621. doi: 10.1007/s10038-008-0293-z. [DOI] [PubMed] [Google Scholar]
- Esparza-Gordillo J, Weidinger S, Folster-Holst R, et al. A common variant on chromosome 11q13 is associated with atopic dermatitis. Nat Genet. 2009;41:596–601. doi: 10.1038/ng.347. [DOI] [PubMed] [Google Scholar]
- Fallon PG, Sasaki T, Sandilands A, et al. A homozygous frameshift mutation in the mouse Flg gene facilitates enhanced percutaneous allergen priming. Nat Genet. 2009;41:602–608. doi: 10.1038/ng.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleckman P, Holbrook KA, Dale BA, et al. Keratinocytes cultured from subjects with ichthyosis vulgaris are phenotypically abnormal. J Invest Dermatol. 1987;88:640–645. doi: 10.1111/1523-1747.ep12470251. [DOI] [PubMed] [Google Scholar]
- Flohr C, England K, Radulovic S, et al. Filaggrin loss-of-function mutations are associated with early-onset eczema, eczema severity and transepidermal water loss at 3 months of age. Br J Dermatol. 2010;163:1333–1336. doi: 10.1111/j.1365-2133.2010.10068.x. [DOI] [PubMed] [Google Scholar]
- Fluhr JW, Elias PM, Man MQ, et al. Is the filaggrin-histidine-urocanic acid pathway essential for stratum corneum acidification? J Invest Dermatol. 2010;130:2141–2144. doi: 10.1038/jid.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan SQ, McBride OW, Idler WW, Markova N, Steinert PM. Organization, structure, and polymorphisms of the human profilaggrin gene. Biochemistry. 1990;29:9432–40. doi: 10.1021/bi00492a018. [DOI] [PubMed] [Google Scholar]
- Gao PS, Rafaels NM, Hand T, et al. Filaggrin mutations that confer risk of atopic dermatitis confer greater risk for eczema herpeticum. J Allergy Clin Immunol. 2009;124:507–513. doi: 10.1016/j.jaci.2009.07.034. 513 e501-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardina E, Paolillo N, Sinibaldi C, et al. R501X and 2282del4 filaggrin mutations do not confer susceptibility to psoriasis and atopic dermatitis in Italian patients. Dermatology. 2008;216:83–84. doi: 10.1159/000109365. [DOI] [PubMed] [Google Scholar]
- Gilmour JW, Vestey JP, George S, et al. Effect of phototherapy and urocanic acid isomers on natural killer cell function. J Invest Dermatol. 1993;101:169–174. doi: 10.1111/1523-1747.ep12363652. [DOI] [PubMed] [Google Scholar]
- Goh C, Finkel M, Christos PJ, et al. Profile of 513 patients with alopecia areata: associations of disease subtypes with atopy, autoimmune disease and positive family history. J Eur Acad Dermatol Venereol. 2006;20:1055–1060. doi: 10.1111/j.1468-3083.2006.01676.x. [DOI] [PubMed] [Google Scholar]
- Greisenegger E, Novak N, Maintz L, et al. Analysis of four prevalent filaggrin mutations (R501X, 2282del4, R2447X and S3247X) in Austrian and German patients with atopic dermatitis. J Eur Acad Dermatol Venereol. 2010;24:607–610. doi: 10.1111/j.1468-3083.2009.03469.x. [DOI] [PubMed] [Google Scholar]
- Gruber R, Elias PM, Crumrine D, et al. Filaggrin genotype in ichthyosis vulgaris predicts abnormalities in epidermal structure and function. Am J Pathol. 2011;178:2252–2263. doi: 10.1016/j.ajpath.2011.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber R, Janecke AR, Grabher D, et al. Lower prevalence of common filaggrin mutations in a community sample of atopic eczema: is disease severity important? Wien Klin Wochenschr. 2010;122:551–557. doi: 10.1007/s00508-010-1449-3. [DOI] [PubMed] [Google Scholar]
- Gruber R, Wilson NJ, Smith FJ, et al. Increased pachyonychia congenita severity in patients with concurrent keratin and filaggrin mutations. Br J Dermatol. 2009;161:1391–1395. doi: 10.1111/j.1365-2133.2009.09471.x. [DOI] [PubMed] [Google Scholar]
- Gutowska-Owsiak D, Schaupp AL, Salimi M, et al. IL-22 down-regulates filaggrin expression and affects expression of profilaggrin processing enzymes. Br J Dermatol. 2011 doi: 10.1111/j.1365-2133.2011.10400.x. [DOI] [PubMed] [Google Scholar]
- Hahn EL, Bacharier LB. The atopic march: the pattern of allergic disease development in childhood. Immunol Allergy Clin North Am. 2005;25:231–246. doi: 10.1016/j.iac.2005.02.004. v. [DOI] [PubMed] [Google Scholar]
- Hamada T, Sandilands A, Fukuda S, et al. De novo occurrence of the filaggrin mutation p.R501X with prevalent mutation c.3321delA in a Japanese family with ichthyosis vulgaris complicated by atopic dermatitis. J Invest Dermatol. 2008;128:1323–1325. doi: 10.1038/sj.jid.5701164. [DOI] [PubMed] [Google Scholar]
- Hatano Y, Elias PM, Crumrine D, et al. Efficacy of Combined Peroxisome Proliferator-Activated Receptor-alpha Ligand and Glucocorticoid Therapy in a Murine Model of Atopic Dermatitis. J Invest Dermatol. doi: 10.1038/jid.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson J, Northstone K, Lee SP, et al. The burden of disease associated with filaggrin mutations: a population-based, longitudinal birth cohort study. J Allergy Clin Immunol. 2008;121:872–877. e879. doi: 10.1016/j.jaci.2008.01.026. [DOI] [PubMed] [Google Scholar]
- Hoste E, Kemperman P, Devos M, et al. Caspase-14 Is Required for Filaggrin Degradation to Natural Moisturizing Factors in the Skin. J Invest Dermatol. doi: 10.1038/jid.2011.153. [DOI] [PubMed] [Google Scholar]
- Hoste E, Kemperman P, Devos M, et al. Caspase-14 Is Required for Filaggrin Degradation to Natural Moisturizing Factors in the Skin. J Invest Dermatol. 2011 doi: 10.1038/jid.2011.153. [DOI] [PubMed] [Google Scholar]
- Howell MD, Kim BE, Gao P, et al. Cytokine modulation of atopic dermatitis filaggrin skin expression. J Allergy Clin Immunol. 2007;120:150–155. doi: 10.1016/j.jaci.2007.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CK, Akiyama M, Nemoto-Hasebe I, et al. Analysis of Taiwanese ichthyosis vulgaris families further demonstrates differences in FLG mutations between European and Asian populations. Br J Dermatol. 2009;161:448–451. doi: 10.1111/j.1365-2133.2009.09112.x. [DOI] [PubMed] [Google Scholar]
- Hudson TJ. Skin barrier function and allergic risk. Nat Genet. 2006;38:399–400. doi: 10.1038/ng0406-399. [DOI] [PubMed] [Google Scholar]
- Huffmeier U, Boiers U, Lascorz J, et al. Loss-of-function mutations in the filaggrin gene: no contribution to disease susceptibility, but to autoantibody formation against citrullinated peptides in early rheumatoid arthritis. Ann Rheum Dis. 2008;67:131–133. doi: 10.1136/ard.2007.073239. [DOI] [PubMed] [Google Scholar]
- Hvid M, Johansen C, Deleuran B, et al. Regulation of caspase 14 expression in keratinocytes by inflammatory cytokines - a possible link between reduced skin barrier function and inflammation? Exp Dermatol. 2011a doi: 10.1111/j.1600-0625.2011.01280.x. [DOI] [PubMed] [Google Scholar]
- Hvid M, Vestergaard C, Kemp K, et al. IL-25 in Atopic Dermatitis: A Possible Link between Inflammation and Skin Barrier Dysfunction? J Invest Dermatol. 2011b doi: 10.1038/jid.2010.277. [DOI] [PubMed] [Google Scholar]
- Jakasa I, de Jongh CM, Verberk MM, et al. Percutaneous penetration of sodium lauryl sulphate is increased in uninvolved skin of patients with atopic dermatitis compared with control subjects. Br J Dermatol. 2006;155:104–109. doi: 10.1111/j.1365-2133.2006.07319.x. [DOI] [PubMed] [Google Scholar]
- Jakasa I, Koster ES, Calkoen F, et al. Skin barrier function in healthy subjects and patients with atopic dermatitis in relation to filaggrin loss-of-function mutations. J Invest Dermatol. 2011;131:540–542. doi: 10.1038/jid.2010.307. [DOI] [PubMed] [Google Scholar]
- Jakasa I, Verberk MM, Esposito M, et al. Altered penetration of polyethylene glycols into uninvolved skin of atopic dermatitis patients. J Invest Dermatol. 2007;127:129–134. doi: 10.1038/sj.jid.5700582. [DOI] [PubMed] [Google Scholar]
- Judge MR, McLean WHI, Munro CS. Disorders of keratinization. In: Burns T, Cox N, Griffiths C, editors. Rook’s Textbook of Dermatology. 7th ed. Vol. 34. Blackwell Publishing; 2004. pp. 37–34. 10. [Google Scholar]
- Jungersted JM, Scheer H, Mempel M, et al. Stratum corneum lipids, skin barrier function and filaggrin mutations in patients with atopic eczema. Allergy. 2010;65:911–918. doi: 10.1111/j.1398-9995.2010.02326.x. [DOI] [PubMed] [Google Scholar]
- Kamata Y, Yamamoto M,, Kawakami F,, et al. Bleomycin hydrolase is regulated biphasically in a differentiation- and cytokine-dependent manner: relevance to atopic dermatitis. J Biol Chem. 2011;286:8204–8212. doi: 10.1074/jbc.M110.169292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang TW, Lee JS, Oh SW, et al. Filaggrin mutation c.3321delA in a Korean patient with ichthyosis vulgaris and atopic dermatitis. Dermatology. 2009;218:186–187. doi: 10.1159/000163083. [DOI] [PubMed] [Google Scholar]
- Kezic S. Genetic susceptibility to occupational contact dermatitis. Int J Immunopathol Pharmacol. 2011;24:73S–78S. [PubMed] [Google Scholar]
- Kezic S, Kemperman PM, Koster ES, et al. Loss-of-function mutations in the filaggrin gene lead to reduced level of natural moisturizing factor in the stratum corneum. J Invest Dermatol. 2008;128:2117–2119. doi: 10.1038/jid.2008.29. [DOI] [PubMed] [Google Scholar]
- Kezic S, O’Regan GM, Yau N, et al. Levels of filaggrin degradation products are influenced by both filaggrin genotype and atopic dermatitis severity. Allergy. 2011 doi: 10.1111/j.1398-9995.2010.02540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BE, Howell MD, Guttman E, et al. TNF-alpha downregulates filaggrin and loricrin through c-Jun N-terminal kinase: role for TNF-alpha antagonists to improve skin barrier. J Invest Dermatol. 2011;131:1272–1279. doi: 10.1038/jid.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerbaek A, Bisgaard H, Agner T, et al. Filaggrin null alleles are not associated with hand eczema or contact allergy. Br J Dermatol. 2007;157:1199–1204. doi: 10.1111/j.1365-2133.2007.08252.x. [DOI] [PubMed] [Google Scholar]
- Lesiak A, Kuna P, Zakrzewski M, et al. Combined occurrence of filaggrin mutations and IL-10 or IL-13 polymorphisms predisposes to atopic dermatitis. Exp Dermatol. 2011;20:491–495. doi: 10.1111/j.1600-0625.2010.01243.x. [DOI] [PubMed] [Google Scholar]
- Liao H, Waters AJ, Goudie DR, et al. Filaggrin mutations are genetic modifying factors exacerbating X-linked ichthyosis. J Invest Dermatol. 2007;127:2795–2798. doi: 10.1038/sj.jid.5700971. [DOI] [PubMed] [Google Scholar]
- Lynley AM, Dale BA. The characterization of human epidermal filaggrin. A histidine-rich, keratin filament-aggregating protein. Biochim Biophys Acta. 1983;744:28–35. doi: 10.1016/0167-4838(83)90336-9. [DOI] [PubMed] [Google Scholar]
- Man MQ, Hatano Y, Lee SH, et al. Characterization of a hapten-induced, murine model with multiple features of atopic dermatitis: structural, immunologic and biochemical changes following single versus multiple oxazolone challenges. J Invest Dermatol. 2008;128:79–86. doi: 10.1038/sj.jid.5701011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manabe M, Sanchez M, Sun TT, et al. Interaction of filaggrin with keratin filaments during advanced stages of normal human epidermal differentiation and in ichthyosis vulgaris. Differentiation. 1991;48:43–50. doi: 10.1111/j.1432-0436.1991.tb00241.x. [DOI] [PubMed] [Google Scholar]
- Marenholz I, Kerscher T, Bauerfeind A, et al. An interaction between filaggrin mutations and early food sensitization improves the prediction of childhood asthma. J Allergy Clin Immunol. 2009;123:911–916. doi: 10.1016/j.jaci.2009.01.051. [DOI] [PubMed] [Google Scholar]
- Marenholz I, Nickel R, Ruschendorf F, et al. Filaggrin loss-of-function mutations predispose to phenotypes involved in the atopic march. J Allergy Clin Immunol. 2006;118:866–871. doi: 10.1016/j.jaci.2006.07.026. [DOI] [PubMed] [Google Scholar]
- Markova NG, Marekov LN, Chipev CC, et al. Profilaggrin is a major epidermal calcium-binding protein. Mol Cell Biol. 1993;13:613–625. doi: 10.1128/mcb.13.1.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsella R, Girolomoni G. Canine models of atopic dermatitis: a useful tool with untapped potential. J Invest Dermatol. 2009;129:2351–2357. doi: 10.1038/jid.2009.98. [DOI] [PubMed] [Google Scholar]
- Matsui T, Miyamoto K, Kubo A, et al. SASPase regulates stratum corneum hydration through profilaggrin-to-filaggrin processing. EMBO Mol Med. 2011;3:320–333. doi: 10.1002/emmm.201100140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLoone P, Simics E, Barton A, et al. An action spectrum for the production of cis-urocanic acid in human skin in vivo. J Invest Dermatol. 2005;124:1071–1074. doi: 10.1111/j.0022-202X.2005.23731.x. [DOI] [PubMed] [Google Scholar]
- Mevorah B, Marazzi A, Frenk E. The prevalence of accentuated palmoplantar markings and keratosis pilaris in atopic dermatitis, autosomal dominant ichthyosis and control dermatological patients. Br J Dermatol. 1985;112:679–685. doi: 10.1111/j.1365-2133.1985.tb02336.x. [DOI] [PubMed] [Google Scholar]
- Miajlovic H, Fallon PP, Irvine AD, et al. Effect of filaggrin breakdown products on growth of and protein expression by Staphylococcus aureus. J Allergy Clin Immunol. 2010 doi: 10.1016/j.jaci.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mildner M, Jin J, Eckhart L, et al. Knockdown of filaggrin impairs diffusion barrier function and increases UV sensitivity in a human skin model. J Invest Dermatol. 2010;130:2286–2294. doi: 10.1038/jid.2010.115. [DOI] [PubMed] [Google Scholar]
- Mischke D, Korge BP, Marenholz I, et al. Genes encoding structural proteins of epidermal cornification and S100 calcium-binding proteins form a gene complex (“epidermal differentiation complex”) on human chromosome 1q21. J Invest Dermatol. 1996;106:989–992. doi: 10.1111/1523-1747.ep12338501. [DOI] [PubMed] [Google Scholar]
- Mohrenschlager M, Schafer T, Huss-Marp J, et al. The course of eczema in children aged 5-7 years and its relation to atopy: differences between boys and girls. Br J Dermatol. 2006;154:505–513. doi: 10.1111/j.1365-2133.2005.07042.x. [DOI] [PubMed] [Google Scholar]
- Moniaga CS, Egawa G, Kawasaki H, et al. Flaky tail mouse denotes human atopic dermatitis in the steady state and by topical application with Dermatophagoides pteronyssinus extract. Am J Pathol. 176:2385–2393. doi: 10.2353/ajpath.2010.090957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Marenholz I, Lee YA, et al. Association of Filaggrin loss-of-function-mutations with atopic dermatitis and asthma in the Early Treatment of the Atopic Child (ETAC) population. Pediatr Allergy Immunol. 2009;20:358–361. doi: 10.1111/j.1399-3038.2008.00808.x. [DOI] [PubMed] [Google Scholar]
- Nemoto-Hasebe I, Akiyama M, Nomura T, et al. Clinical severity correlates with impaired barrier in filaggrin-related eczema. J Invest Dermatol. 2009a;129:682–689. doi: 10.1038/jid.2008.280. [DOI] [PubMed] [Google Scholar]
- Nemoto-Hasebe I, Akiyama M, Nomura T, et al. FLG mutation p.Lys4021X in the C-terminal imperfect filaggrin repeat in Japanese patients with atopic eczema. Br J Dermatol. 2009b;161:1387–1390. doi: 10.1111/j.1365-2133.2009.09406.x. [DOI] [PubMed] [Google Scholar]
- Nirunsuksiri W, Presland RB, Brumbaugh SG, et al. Decreased profilaggrin expression in ichthyosis vulgaris is a result of selectively impaired posttranscriptional control. J Biol Chem. 1995;270:871–876. doi: 10.1074/jbc.270.2.871. [DOI] [PubMed] [Google Scholar]
- Nirunsuksiri W, Zhang SH, Fleckman P. Reduced stability and bi-allelic, coequal expression of profilaggrin mRNA in keratinocytes cultured from subjects with ichthyosis vulgaris. J Invest Dermatol. 1998;110:854–861. doi: 10.1046/j.1523-1747.1998.00208.x. [DOI] [PubMed] [Google Scholar]
- Nomura T, Akiyama M, Sandilands A, et al. Specific filaggrin mutations cause ichthyosis vulgaris and are significantly associated with atopic dermatitis in Japan. J Invest Dermatol. 2008;128:1436–1441. doi: 10.1038/sj.jid.5701205. [DOI] [PubMed] [Google Scholar]
- Nomura T, Akiyama M, Sandilands A, et al. Prevalent and rare mutations in the gene encoding filaggrin in Japanese patients with ichthyosis vulgaris and atopic dermatitis. J Invest Dermatol. 2009;129:1302–1305. doi: 10.1038/jid.2008.372. [DOI] [PubMed] [Google Scholar]
- Nomura T, Sandilands A, Akiyama M, et al. Unique mutations in the filaggrin gene in Japanese patients with ichthyosis vulgaris and atopic dermatitis. J Allergy Clin Immunol. 2007;119:434–440. doi: 10.1016/j.jaci.2006.12.646. [DOI] [PubMed] [Google Scholar]
- Novak N, Baurecht H, Schafer T, et al. Loss-of-function mutations in the filaggrin gene and allergic contact sensitization to nickel. J Invest Dermatol. 2008;128:1430–1435. doi: 10.1038/sj.jid.5701190. [DOI] [PubMed] [Google Scholar]
- O’Regan GM, Campbell LE, Cordell HJ, et al. Chromosome 11q13.5 variant associated with childhood eczema: an effect supplementary to filaggrin mutations. J Allergy Clin Immunol. 2010a;125:170–174. e171–172. doi: 10.1016/j.jaci.2009.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Regan GM, Kemperman PM, Sandilands A, et al. Raman profiles of the stratum corneum define 3 filaggrin genotype-determined atopic dermatitis endophenotypes. J Allergy Clin Immunol. 2010b;126:574–580. e571. doi: 10.1016/j.jaci.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa R, Konno S, Akiyama M, et al. Japanese-specific filaggrin gene mutations in Japanese patients suffering from atopic eczema and asthma. J Invest Dermatol. 2010;130:2834–2836. doi: 10.1038/jid.2010.218. [DOI] [PubMed] [Google Scholar]
- Ovaere P, Lippens S, Vandenabeele P, et al. The emerging roles of serine protease cascades in the epidermis. Trends Biochem Sci. 2009;34:453–463. doi: 10.1016/j.tibs.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Oyoshi MK, Murphy GF, Geha RS. Filaggrin-deficient mice exhibit TH17-dominated skin inflammation and permissiveness to epicutaneous sensitization with protein antigen. J Allergy Clin Immunol. 2009;124:485–493. doi: 10.1016/j.jaci.2009.05.042. 493 e481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer CN, Irvine AD, Terron-Kwiatkowski A, et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet. 2006;38:441–446. doi: 10.1038/ng1767. [DOI] [PubMed] [Google Scholar]
- Perez ML, Gomara MJ, Ercilla G, et al. Antibodies to citrullinated human fibrinogen synthetic peptides in diagnosing rheumatoid arthritis. J Med Chem. 2007;50:3573–3584. doi: 10.1021/jm0701932. [DOI] [PubMed] [Google Scholar]
- Petukhova L, Duvic M, Hordinsky M, et al. Genome-wide association study in alopecia areata implicates both innate and adaptive immunity. Nature. 2010;466:113–117. doi: 10.1038/nature09114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presland RB, Boggess D, Lewis SP, et al. Loss of normal profilaggrin and filaggrin in flaky tail (ft/ft) mice: an animal model for the filaggrin-deficient skin disease ichthyosis vulgaris. J Invest Dermatol. 2000;115:1072–1081. doi: 10.1046/j.1523-1747.2000.00178.x. [DOI] [PubMed] [Google Scholar]
- Presland RB, Coulombe PA, Eckert RL, et al. Barrier function in transgenic mice overexpressing K16, involucrin, and filaggrin in the suprabasal epidermis. J Invest Dermatol. 2004;123:603–606. doi: 10.1111/j.0022-202X.2004.23226.x. [DOI] [PubMed] [Google Scholar]
- Presland RB, Haydock PV, Fleckman P, et al. Characterization of the human epidermal profilaggrin gene. Genomic organization and identification of an S-100-like calcium binding domain at the amino terminus. J Biol Chem. 1992;267:23772–23781. [PubMed] [Google Scholar]
- Rawlings AV, Harding CR. Moisturization and skin barrier function. Dermatol Ther. 2004;17(Suppl 1):43–48. doi: 10.1111/j.1396-0296.2004.04s1005.x. [DOI] [PubMed] [Google Scholar]
- Rodriguez E, Baurecht H, Herberich E, et al. Meta-analysis of filaggrin polymorphisms in eczema and asthma: robust risk factors in atopic disease. J Allergy Clin Immunol. 2009;123:1361–1370. e1367. doi: 10.1016/j.jaci.2009.03.036. [DOI] [PubMed] [Google Scholar]
- Ross-Hansen K, Menne T, Johansen JD, et al. Nickel reactivity and filaggrin null mutations--evaluation of the filaggrin bypass theory in a general population. Contact Dermatitis. 2010;64:24–31. doi: 10.1111/j.1600-0536.2010.01815.x. [DOI] [PubMed] [Google Scholar]
- Ruether A, Stoll M, Schwarz T, et al. Filaggrin loss-of-function variant contributes to atopic dermatitis risk in the population of Northern Germany. Br J Dermatol. 2006;155:1093–1094. doi: 10.1111/j.1365-2133.2006.07500.x. [DOI] [PubMed] [Google Scholar]
- Sandilands A, Sutherland C, Irvine A, et al. Filaggrin in the frontline: role in skin barrier function and disease. J Cell Sci. 2009;122:1285–1294. doi: 10.1242/jcs.033969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandilands A, Terron-Kwiatkowski A, Hull PR, et al. Comprehensive analysis of the gene encoding filaggrin uncovers prevalent and rare mutations in ichthyosis vulgaris and atopic eczema. Nat Genet. 2007;39:650–654. doi: 10.1038/ng2020. [DOI] [PubMed] [Google Scholar]
- Sanmarti R, Graell E, Perez ML, et al. Diagnostic and prognostic value of antibodies against chimeric fibrin/filaggrin citrullinated synthetic peptides in rheumatoid arthritis. Arthritis Res Ther. 2009;11:R135. doi: 10.1186/ar2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharschmidt TC, Man MQ, Hatano Y, et al. Filaggrin deficiency confers a paracellular barrier abnormality that reduces inflammatory thresholds to irritants and haptens. J Allergy Clin Immunol. 2009;124:496–506. doi: 10.1016/j.jaci.2009.06.046. 506 e491-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber S, Rosenstiel P, Albrecht M, et al. Genetics of Crohn disease, an archetypal inflammatory barrier disease. Nat Rev Genet. 2005;6:376–388. doi: 10.1038/nrg1607. [DOI] [PubMed] [Google Scholar]
- Schuttelaar ML, Kerkhof M, Jonkman MF, et al. Filaggrin mutations in the onset of eczema, sensitization, asthma, hay fever and the interaction with cat exposure. Allergy. 2009;64:1758–1765. doi: 10.1111/j.1398-9995.2009.02080.x. [DOI] [PubMed] [Google Scholar]
- Sergeant A, Campbell LE, Hull PR, et al. Heterozygous null alleles in filaggrin contribute to clinical dry skin in young adults and the elderly. J Invest Dermatol. 2009;129:1042–1045. doi: 10.1038/jid.2008.324. [DOI] [PubMed] [Google Scholar]
- Simpson E. Are epidermal defects the key initiating factors in the development of atopic dermatitis? Br J Dermatol. 2010;163:1147–1148. doi: 10.1111/j.1365-2133.2010.10115.x. [DOI] [PubMed] [Google Scholar]
- Sinclair C, O’Toole EA, Paige D, et al. Filaggrin mutations are associated with ichthyosis vulgaris in the Bangladeshi population. Br J Dermatol. 2009;160:1113–1115. doi: 10.1111/j.1365-2133.2009.09050.x. [DOI] [PubMed] [Google Scholar]
- Smith FJ, Irvine AD, Terron-Kwiatkowski A, et al. Loss-of-function mutations in the gene encoding filaggrin cause ichthyosis vulgaris. Nat Genet. 2006;38:337–342. doi: 10.1038/ng1743. [DOI] [PubMed] [Google Scholar]
- Spergel JM. From atopic dermatitis to asthma: the atopic march. Ann Allergy Asthma Immunol. 2010;105:99–106. doi: 10.1016/j.anai.2009.10.002. quiz 107-109, 117. [DOI] [PubMed] [Google Scholar]
- Sybert VP, Dale BA, Holbrook KA. Ichthyosis vulgaris: identification of a defect in synthesis of filaggrin correlated with an absence of keratohyaline granules. J Invest Dermatol. 1985;84:191–194. doi: 10.1111/1523-1747.ep12264813. [DOI] [PubMed] [Google Scholar]
- Thyssen JP, Carlsen BC, Menne T, et al. Filaggrin null mutations increase the risk and persistence of hand eczema in subjects with atopic dermatitis: results from a general population study. Br J Dermatol. 2010a;163:115–120. doi: 10.1111/j.1365-2133.2010.09822.x. [DOI] [PubMed] [Google Scholar]
- Thyssen JP, Johansen JD, Linneberg A, et al. The association between null mutations in the filaggrin gene and contact sensitization to nickel and other chemicals in the general population. Br J Dermatol. 2010b doi: 10.1111/j.1365-2133.2010.09708.x. [DOI] [PubMed] [Google Scholar]
- van den Oord RA, Sheikh A. Filaggrin gene defects and risk of developing allergic sensitisation and allergic disorders: systematic review and meta-analysis. BMJ. 2009;339:b2433. doi: 10.1136/bmj.b2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Limbergen J, Russell RK, Nimmo ER, et al. Filaggrin loss-of-function variants are associated with atopic comorbidity in pediatric inflammatory bowel disease. Inflamm Bowel Dis. 2009;15:1492–1498. doi: 10.1002/ibd.20926. [DOI] [PubMed] [Google Scholar]
- Van Steendam K, Tilleman K, Deforce D. The relevance of citrullinated vimentin in the production of antibodies against citrullinated proteins and the pathogenesis of rheumatoid arthritis. Rheumatology (Oxford) 2011;50:830–837. doi: 10.1093/rheumatology/keq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercelli D. Of flaky tails and itchy skin. Nat Genet. 2009;41:512–513. doi: 10.1038/ng0509-512. [DOI] [PubMed] [Google Scholar]
- Weidinger S, Baurecht H, Wagenpfeil S, et al. Analysis of the individual and aggregate genetic contributions of previously identified serine peptidase inhibitor Kazal type 5 (SPINK5), kallikrein-related peptidase 7 (KLK7), and filaggrin (FLG) polymorphisms to eczema risk. J Allergy Clin Immunol. 2008a;122:560–568. e564. doi: 10.1016/j.jaci.2008.05.050. [DOI] [PubMed] [Google Scholar]
- Weidinger S, Illig T, Baurecht H, et al. Loss-of-function variations within the filaggrin gene predispose for atopic dermatitis with allergic sensitizations. J Allergy Clin Immunol. 2006;118:214–219. doi: 10.1016/j.jaci.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Weidinger S, O’Sullivan M, Illig T, et al. Filaggrin mutations, atopic eczema, hay fever, and asthma in children. J Allergy Clin Immunol. 2008b;121:1203–1209. e1201. doi: 10.1016/j.jaci.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Weidinger S, Rodriguez E, Stahl C, et al. Filaggrin mutations strongly predispose to early-onset and extrinsic atopic dermatitis. J Invest Dermatol. 2007;127:724–726. doi: 10.1038/sj.jid.5700630. [DOI] [PubMed] [Google Scholar]
- Wells RS, Kerr CB. Clinical features of autosomal dominant and sex-linked ichthyosis in an English population. Br Med J. 1966;1:947–950. doi: 10.1136/bmj.1.5493.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winge MC, Bilcha KD, Lieden A, et al. Novel filaggrin mutation but no other loss-of-function variants found in ethiopian atopic-dermatitis patients. Br J Dermatol. doi: 10.1111/j.1365-2133.2011.10475.x. [DOI] [PubMed] [Google Scholar]
- Wuthrich B, Schmid-Grendelmeier P. The atopic eczema/dermatitis syndrome. Epidemiology, natural course, and immunology of the IgE-associated (“extrinsic”) and the nonallergic (“intrinsic”) AEDS. J Investig Allergol Clin Immunol. 2003;13:1–5. [PubMed] [Google Scholar]
- Zhang H, Guo Y, Wang W, et al. Mutations in the filaggrin gene in Han Chinese patients with atopic dermatitis. Allergy. 2011 doi: 10.1111/j.1398-9995.2010.02493.x. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Terron-Kwiatkowski A, Liao H, et al. Filaggrin null alleles are not associated with psoriasis. J Invest Dermatol. 2007;127:1878–1882. doi: 10.1038/sj.jid.5700817. [DOI] [PubMed] [Google Scholar]