Abstract
Somatic missense mutations in PIK3CA, which encodes the p110α catalytic subunit of phosphoinositide 3-kinases (PI3Ks), occur frequently in human cancers. Activating mutations spread across multiple domains, some of which are located at inhibitory contact sites formed with the regulatory subunit p85α. PIK3R1, which encodes p85α, also has activating somatic mutations. We find a strong correlation between lipid kinase and lipid binding activities, for both wild-type (WT) and a representative set of oncogenic mutant complexes of p110α/p85α. Lipid binding involves both electrostatic and hydrophobic interactions. Activation caused by a phosphorylated receptor tyrosine kinase (RTK) peptide binding to the p85α N-terminal SH2 domain (nSH2) induces lipid binding. This depends on the polybasic activation loop as well as a conserved hydrophobic motif in the C-terminal region of the kinase domain. The hotspot E545K mutant largely mimics the activated WT p110α. It shows the highest basal activity and lipid binding, and is not significantly activated by an RTK phosphopeptide. Both the hotspot H1047R mutant and rare mutations (C420R, M1043I, H1047L, G1049R and p85α-N564D) also show increased basal kinase activities and lipid binding. However, their activities are further enhanced by an RTK phosphopeptide to levels markedly exceeding that of activated WT p110α. Phosphopeptide binding to p110β/p85α and p110δ/p85α complexes also induces their lipid binding. We present a crystal structure of WT p110α complexed with the p85α inter-SH2 domain (iSH2) and the inhibitor PIK-108. Additional to the ATP-binding pocket, an unexpected, second PIK-108 binding site is observed in the kinase C-lobe. We show a global conformational change in p110α consistent with allosteric regulation of the kinase domain by nSH2. These findings broaden our understanding of the differential biological outputs exhibited by distinct types of mutations regarding growth factor dependence, and suggest a two-tier classification scheme relating p110α and p85α mutations with signalling potential.
Keywords: PIK3CA mutations, p110alpha, lipid binding, crystal structure, inhibitor, growth factor signalling
Introduction
Class IA PI3Ks transduce growth factor signalling by phosphorylating phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2) to produce PtdIns(3,4,5)P3 on the plasma membrane. Recruitment to membrane receptors and enzyme activation are coordinated by engagement of the p85 SH2 domains to regions bearing phosphorylated ‘YXXM’ motifs on activated RTKs or RTK-associated adaptors (Backer et al., 1992; Carpenter et al., 1993). PtdIns(3,4,5)P3 serves as docking sites for downstream signalling molecules that harbour pleckstrin homology domains. Central among these are PDK1 and PKB/AKT, which orchestrate signalling cascades promoting cell growth, proliferation, survival, metabolism and migration (Manning and Cantley, 2007; Franke, 2008; Bayascas, 2010). Cellular levels of PtdIns(3,4,5)P3 are tightly regulated. PTEN down-regulates p110 signalling by removing the 3′-phosphate of PtdIns(3,4,5)P3 (Carracedo and Pandolfi, 2008). Up-regulation of PI3K signalling, through amplification or gain-of-function mutations of RTKs, p110α and AKTs, as well as loss of PTEN function and expression, occurs frequently in human cancers (Yuan and Cantley, 2008; Chalhoub and Baker, 2009).
Uniquely among the four class I PI3K isoforms that produce PtdIns(3,4,5)P3, p110α has been identified with activating cancer-linked somatic mutations (Samuels et al., 2004). Over 80% of the missense mutations cluster in two ‘hotspots’ commonly represented by E545K in the helical domain, and H1047R in the kinase domain. Both display transforming activities in cell culture (Zhao et al., 2005; Gymnopoulos et al., 2007). The ability to initiate tumorigenesis in transgenic mouse models has been demonstrated for the H1047R mutant (Engelman et al., 2008; Adams et al., 2011; Meyer et al., 2011).
Crystal structures of p110α/85α complexes have provided molecular details of p85-mediated inhibition (Huang et al., 2007; Mandelker et al., 2009), revealing that many activating mutations, including ones occurring at low frequencies (Ikenoue et al., 2005; Gymnopoulos et al., 2007; Oda et al., 2008; Zhang et al., 2008; Jaiswal et al., 2009; Rudd et al., 2011), are located in inter-domain contact sites. These sites include the inhibitory interfaces between the p110α C2/helical/kinase domains and the p85α nSH2 domain (Miled et al., 2007; Mandelker et al., 2009), and between the p110α C2 domain and the p85α iSH2 domain (Wu et al., 2009). Phosphopeptide-binding to nSH2 competes with its inhibitory interaction with p110, constituting the dominant activation mechanism for p110α, and E545K mimics this activation (Yu et al., 1998; Miled et al., 2007; Mandelker et al., 2009). Since p110α adaptor binding domain (ABD) binds to p85α iSH2 tightly, the often-mutated interface between ABD and the kinase domain can be viewed as an extension of the p85 inhibitory network. Mutations in the kinase C-terminal region do not appear to interfere with p85 inhibition since p85α cSH2 does not play an inhibitory role in p110α regulation (Yu et al., 1998), although it does for p110β and p110δ via direct contact with this region (Burke et al., 2011; Zhang et al., 2011).
Some of the cancer-linked mutations confer a positive charge, such as C420R and H1047R in p110α, and have been suggested to enhance membrane binding (Gymnopoulos et al., 2007; Mandelker et al., 2009). However, direct membrane-binding measurements are lacking. A common assumption is that these mutations directly facilitate electrostatic interaction with phospholipid headgroups. However, this has limited applicability to other activating mutations, notably H1047L. The E545K and H1047R mutants were first reported to have similar biological activities, in terms of promoting cell growth and resisting apoptosis under growth factor limiting conditions (Samuels et al., 2005). More recent studies showed that they can have differential functional outcomes, in terms of chemotactic and metastatic phenotypes (Pang et al., 2009) and transforming potential (Chakrabarty et al., 2010) of isogenic human breast cancer cells. Transforming ability of chicken embryonic fibroblasts differs between the E545K and the H1047R mutants, invoking the suggestion that these two mutants operate via different activation mechanisms (Zhao and Vogt, 2008).
Previously, the E545K and H1047R mutants were found to be more active than the WT enzyme, but their similar affinities for ATP did not explain the differences in lipid kinase activities (Carson et al., 2008). Here, we investigated the premise that enhanced lipid binding forms a general mechanism for p110 activation, particularly regarding cancer mutations. We dissected the structural elements important for lipid binding. Our results show that p85α nSH2, a key regulatory element for p110α lipid kinase activity, controls access of the catalytic subunit lipid binding sites to membrane. We examined a set of p110α/p85α cancer-linked mutants of diverse structural and chemical types, and find a strong correlation linking their elevated lipid kinase activities to their lipid binding levels. We present a crystal structure of WT p110α/p85α-iSH2 in complex with an inhibitor. Its structural features in the kinase domain resemble those of the H1047R mutant (Mandelker et al., 2009), instead of the WT apo structure (Huang et al., 2007). We also noted unusual structural features of the kinase C-terminal tail and tested their function. We observe global conformational changes that might be of relevance to allosteric regulation of p110α, and provide a structural context to understand the functional data presented here.
Results
Structure of a wildtype p110α/p85α-iSH2 complex
A crystal structure of mouse WT p110α in complex with human p85α niSH2 fragment and the p110β/p110δ selective inhibitor PIK-108 has been determined and refined to 3.5 Å (Rwork/Rfree=0.184/0.228) (acronyms of p110α and p85α domain structures and mutations are illustrated in Figure 1). Details of crystallographic statistics are provided in Supplementary Table S1. Although other compounds that inhibit p110α more specifically were surveyed for co-crystallization, the p110β/δ selective PIK-108 produced the best crystals. As in the structure of human WT p110α/p85α-iSH2 (Huang et al., 2007), the nSH2 of the p85α niSH2 fragment is not observed in the electron density map. The high salt concentration in the crystallization cocktail might have competed off nSH2 binding to p110α. As such, our structure represents an alternative view of p110α not constrained by nSH2 binding. Unlike previous structures of p110α/p85α complexes, our structure shows clear electron density for the entire activation loop (Figure 2a). However, key conserved activation loop residues, K942 and R949, previously identified to be important for p110γ recognising the substrate PtdIns(4,5)P2 head group (Pirola et al., 2001), point away from the ATP binding site (Figure 2d). Hence, although structure of a p110α/p85α-iSH2 complex should mimic an RTK-activated state (see below), the observed conformation of this loop does not appear to be compatible with positioning the lipid headgroup for phosphoryl transfer. The activation loop is also involved in crystal contacts (Supplementary Figure S1), which likely influenced the conformation we observe.
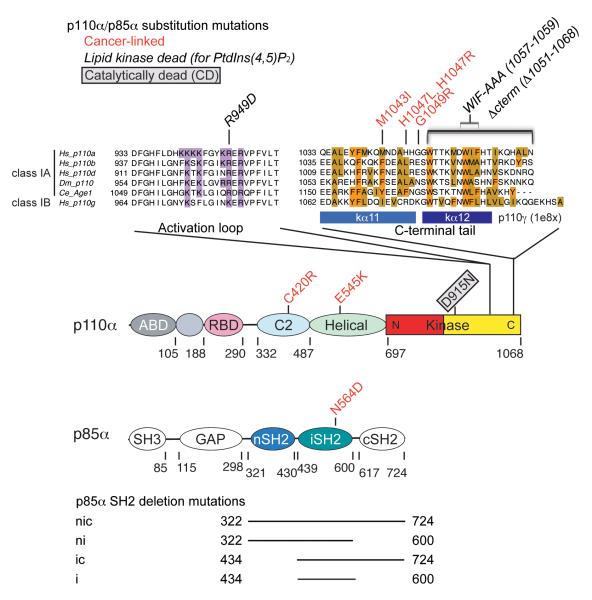
Figure 1.
Schematics of p110α and p85α domain structures. Substitution and deletion mutants used in this study are illustrated. Sequence alignment display was prepared with Jalview (Waterhouse et al., 2009). Basic residues in the activation loop (which binds the lipid substrate headgroup) and hydrophobic residues in the C-terminal tail are highlighted.
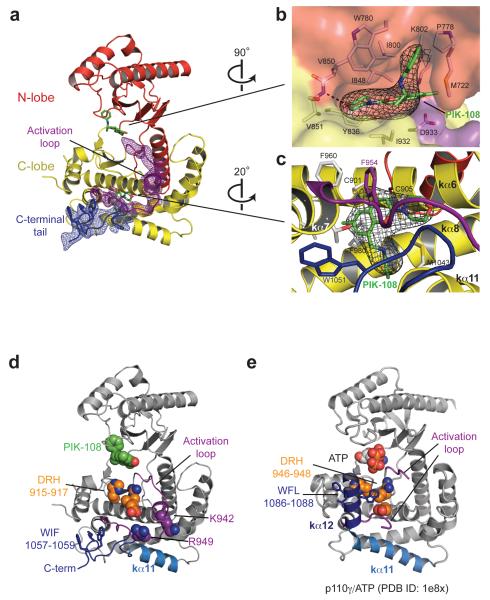
Figure 2.
Structure of the kinase domain in WT p110α/p85α-iSH2 complexed with the inhibitor PIK-108. (a) Omit maps. The σA weighted Fo-Fc electron density maps (contoured at 3.5σ) were calculated separately with the activation loop and the C-terminal tail omitted from the refined model. (b,c) PIK-108 binding sites in the ATP-binding pocket of the kinase domain (b) and a novel site in the kinase C-lobe (c). The PIK-108 omit maps are contoured at 3.5σ. PIK-108 interacting residues (≤ 3.8 Å inter-atomic distances) are shown as stick models. (d) Functional elements in the kinase domain. (e) Kinase domain of p110γ catalytic core, shown for comparison with respect to secondary structure in the C-terminal tail. Note the interactions between the conserved W1086 in helix kα12 and the conserved catalytic residues DRH.
PIK-108 belongs to the class of propeller-shaped PI3K inhibitors. In our structure, the PIK-108 phenyl substituent occupies what has been coined the ‘specificity pocket’, which is gated by the P-loop M772, in the kinase ATP-binding site (Figure 2b). Opening of this pocket by propeller-shaped compounds appears to be more favourable in p110δ than in other p110 isotypes (Berndt et al., 2010). This could be the reason for the PIK-108 selectivity for p110β and p110δ. There is also complete density for what we interpret as a second PIK-108, snuggling in a hydrophobic pocket in the kinase C-lobe, surrounded by helices kα6-8, kα11 and the C-terminal end of the activation loop (Figure 2c). This pocket is partially occupied by the activation loop residues 954-956 (or their equivalents) in other p110 structures. The presence of the compound in this induced pocket may have contributed to the ordering of the activation loop, which is involved in crystal packing, and might explain why this crystal form is specific to PIK-108, out of twenty tested p110 selective inhibitors.
The kinase C-terminal region of p110α exhibits intrinsic flexibility
Our structure is for the WT p110α, but the kinase C-terminal tail more closely resembles those in the structures of the oncogenic mutant H1047R p110α/p85α-niSH2 (Mandelker et al., 2009), than that in the WT apo p110α/p85α-iSH2 structure (Huang et al., 2007). In the WT apo p110α/p85α-iSH2 structure, helix kα11 is complete to residue 1048, and the rest of the C-terminal sequence is largely disordered (Huang et al., 2007) [hereafter all p110 secondary structure nomenclature is according to that of p110γ (Walker et al., 1999)]. The kinase C-terminal tail in our structure adopts an extended conformation from residue 1045 to 1061, with the last helical turn of kα11 appearing to be distorted (Figure 2a). A striking feature in this region is the WIF motif (1057-1059), whose side-chains stack against each other (Figure 2d). This motif is conserved as a triplet of hydrophobic residues in class I and class II PI3Ks, with W1057 being invariant in class I p110 (Figure 1). The equivalent ‘WFL’ (1086-1088) in p110γ is part of helix kα12 (Figure 2e), which corresponds to residues 1050-1061 in p110α. Helix kα12 also exists in the crystal structures of p110β and Vps34 (Miller et al., 2010; Zhang et al., 2011). This C-terminal region in our structure, and also in the H1047R p110α/p85α-niSH2 structures, is involved in crystal packing with the Ras-binding domain (RBD) of a symmetry-related molecule (Supplementary Figure S1). Therefore, the conformations of the C-terminal tail among the current p110α structures appear to be dictated by the crystalline environments. It is likely fortuitous that the RBD and the kinase domain pack against each other in different crystal forms of p110α. We would not ascribe any functional significance to these crystal contacts, particularly with regard to Ras activation.
Deletion of helix kα12 in Vps34, p110β and p110δ abrogated lipid kinase activity and lipid binding (Miller et al., 2010; Burke et al., 2011; Zhang et al., 2011). This region is also of functional importance for p110α (see below). Deletion of kα12 in Vps34 and in p110β resulted in increased intrinsic ATPase activity in the absence of lipid substrate (Miller et al., 2010; Zhang et al., 2011), an observation replicated by deletion of residues 1051-1068 in p110α (Supplementary Figure S2, Δcterm mutant). Interactions between helix kα12 and the catalytic loop (illustrated for p110γ in Figure 2e) were suggested to shield the conserved catalytic DRH motif from performing futile ATP hydrolysis (Miller et al., 2010; Zhang et al., 2011). These interactions are not observed in the current p110α crystal structures, suggesting that in solution, p110α could also have a helix kα12, similar to p110γ, p110β and Vps34. The extended conformations observed in current crystal structures of p110α suggest an intrinsic flexibility in this region.
Correlation of lipid kinase activity with lipid binding
To test whether lipid binding forms the basis of p110 activation, we compared lipid kinase with lipid binding activities for three sets of p110α/p85α complexes: SH2 deletions in p85α, engineered mutations in the p110α kinase domain and cancer-linked mutations in both p110α and p85α (Figure 3). We evaluated activation by RTK phosphopeptide for all complexes, using the doubly tyrosine-phosphorylated peptide (pY2-peptide) derived from PDGFRβ. We used liposomes of defined compositions generally mimicking that of plasma membranes (Narayan and Lemmon, 2006). The substrate PtdIns(4,5)P2 was used at 2% (close to the physiological concentration), and the anionic lipid phosphatidylserine (PS) was tested at two concentrations, 10% and 20%. Cancer-linked mutations and nSH2 deletion in p85α severely suppressed expression of p110/p85 in insect cells. To obtain sufficient materials for lipid binding analyses, we produced activating p110α/p85α proteins in the catalytically dead (CD) background, bearing the D915N mutation in the catalytic DRH motif of p110α.
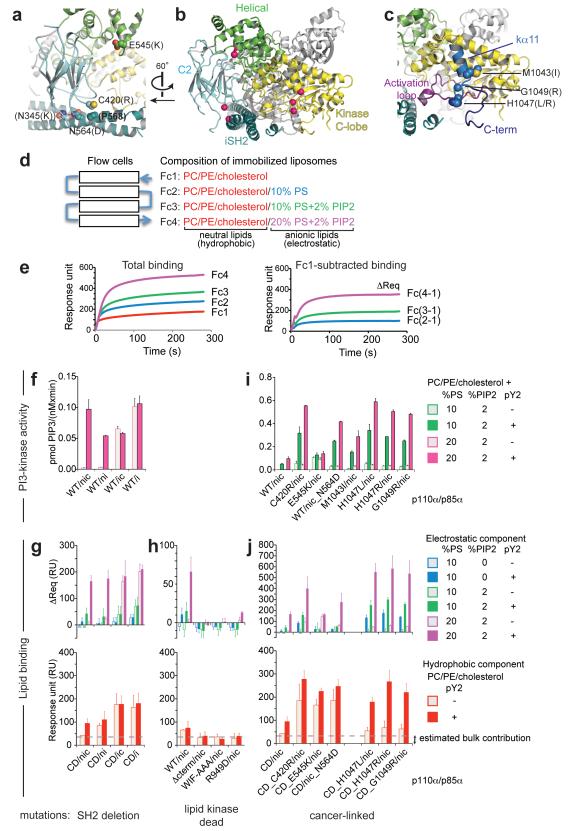
Figure 3.
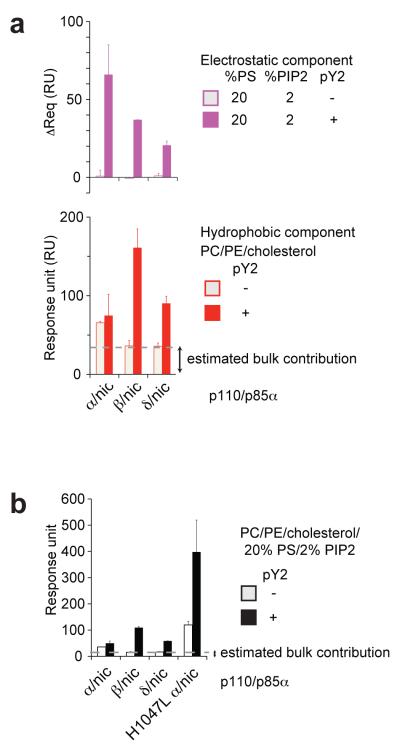
Lipid kinase and lipid binding activities. (a-c) Structural views of the mutant positions in our WT p110α/p85α-iSH2 complex. A global view is shown in (b) with C-alpha atoms of the mutated residues displayed as pink spheres. (a,c) Detailed views of mutant positions at the C2 domain/iSH2 interface and the helical domain (a) and in the kinase C-lobe (c). (a) The C2 C420 contacts the iSH2 P568, and the iSH2 N564 contacts the C2 N345 (N345K is a cancer-linked mutant, not analysed in this study). (d) Schematics of the SPR lipid binding experiment. Liposome vesicles are captured via the lipophilic groups attached to the dextran matrix on the flow cells. (e) Example (C420R/nic + pY2-peptide) of a steady-state binding sensogram before (left) and after (right) signal subtraction from flow cell 1 (Fc1). (f,i) Lipid kinase activities. The initial rates of PtdIns(3,4,5)P3 production were measured with fixed substrate concentration (50 μM PtdIns(4,5)P2 and 100 μM ATP) using liposomes of the indicated compositions. No lipid kinase activity was detected for the lipid kinase-dead mutants, which were active in hydrolysing ATP in the absence of lipid substrate. (g,h,j) Lipid binding measured by SPR. Binding levels were compared at identical protein concentration in each set (g,j: 500 nM, h: 1 μM). Averaged binding levels of the lipid kinase-dead mutants on Fc1, ~35 RU, were taken as the estimated bulk contribution, i.e. refractive index change due to the presence of proteins in the buffer (h). Data in f-j represent the mean ± s.d. of two to three independent experiments. (Abbreviations: PC= phosphatidylcholine; PE=phosphatidylethanolamine; PS=phosphatidylserine; PIP2=PtdIns(4,5)P2; PIP3=PtdIns(3,4,5)P3; pY2=pY2-peptide with two pYXXM motifs from PDGFRβ; CD=’catalytically dead’ D915N mutation in p110α)
We employed surface plasmon resonance (SPR) to measure lipid binding (schematics shown in Figure 3d,e). We examined the electrostatic and the hydrophobic components of lipid binding explicitly, since both are important properties for many membrane-binding proteins (Cho and Stahelin, 2005; Lemmon, 2008). We immobilized liposomes of different compositions in each of the four serially connected flow cells on the sensor chip. Flow cell one (Fc1) of the chip contained only neutral lipids phosphatidylcholine (PC), phosphatidylethanolamine (PE) and cholesterol, while Fc2, Fc3 and Fc4 contained additionally negatively-charged lipids PS (Fc2), or PS with PtdIns(4,5)P2 (Fc3 and Fc4). As such, Fc1 subtracted signals represent the electrostatic component of binding. We attribute any binding signals on Fc1 that are significantly above background (estimated as the averaged signal on Fc1 produced by the lipid kinase dead mutants) to be the hydrophobic component of lipid binding.
p85α nSH2, but not cSH2, regulates both lipid kinase activity and membrane binding of p110α
The nSH2 plays a dominant role in regulating the lipid kinase activity of p110α. The basal activities of p110α/p85α-nicSH2 and p110α/p85α-niSH2 complexes are very low using liposome substrate mimicking physiological concentrations of PS and PtdIns(4,5)P2. Activation, whether by binding of RTK phosphopeptide to nSH2-containing complexes, or by nSH2 deletion, increases the activity by 25-50 fold (Figure 3f). Qualitatively, our results recapitulate those of previous studies performed with pure phosphatidylinositol (PtdIns) as substrate (Yu et al., 1998). Correspondingly, RTK phosphopeptide binding to nSH2-containing complexes induces lipid binding (Figure 3g). Binding of RTK phosphopeptide to cSH2 does not affect either the kinase activity or membrane binding (Figure 3f,g). Total negative charge on the liposomes positively influenced lipid binding. Liposome compositions differing by 10% in PS confer up to two-fold increases in lipid binding levels. Hence, it appears that for p110α, the nSH2-p110α inhibitory contact, which is relieved by phosphopeptide binding, suppresses the basal lipid kinase activity by inhibiting membrane binding.
Both kinase activation loop and C-terminal tail are necessary for binding to anionic lipids
Using PtdIns(4,5)P2-containing liposomes, no kinase activity could be detected for three engineered kinase domain mutants: (i) the activation loop mutant R949D, (ii) the kinase helix kα12 deletion mutant (Δcterm), nor (iii) a helix kα12 triple mutant with much reduced hydrophobicity (WIF-AAA(1057-1059)), with protein concentration as high as 500 nM and incubation time up to an hour. These mutants, however, are catalytically active in terms of intrinsic ATPase activity (Supplementary Figure S2). In lipid binding experiments, correspondingly, the R949D mutant showed much reduced binding to anionic lipids (Figure 3h). The same result was observed for K942Q (data not shown). These results are consistent with the observation that mutations removing the positive charges of the equivalent residues in p110γ prevent PtdIns(4,5)P2, but not PtdIns or PtdIns4P, from acting as a substrate (Pirola et al., 2001). The kinase C-terminal mutants, Δcterm and WIF-AAA, did not show any detectable binding to anionic lipids at concentration up to 1 μM. This establishes the WIF motif as a key determinant for lipid binding in the C-terminal region. These results indicate that there are at least two lipid-binding sites in the kinase domain: one is in the polybasic activation loop, electrostatic in nature, and the other in the C-terminal region, hydrophobic in nature.
Effects of cancer-linked mutations in p110α/p85α on lipid binding
To gain a broader view of the effects of cancer-linked mutations, we studied seven mutations that have previously been characterized to be activating (Ikenoue et al., 2005; Gymnopoulos et al., 2007; Zhang et al., 2008; Jaiswal et al., 2009). They cover three structurally distinct regions (Figure 3a-c), namely, the C2-iSH2 interface (p110α C420R and p85α N564D), the helical-nSH2 interface (p110α E545K) and the kinase domain (M1043I, H1047L, H1047R and G1049R), and represent different chemical property changes.
Overall, there is a strong correlation between lipid kinase activity and lipid binding (Figure 3i,j, total lipid binding on Fc4 is shown in Supplementary Figure S3), underscoring enhanced lipid binding as a general mechanism for cancer-linked activating mutations. All mutants have increased basal activity, and other than E545K, can be significantly further activated by phosphopeptide binding. This affirms that the C2-iSH2 inhibitory contact site does not function identically to the nSH2 inhibitory site, which includes both C2 and helical domains in the crystal structure (Mandelker et al., 2009). The degree of activation is enhanced by increase in negative charges on the liposomes. Both anionic lipids PS and PtdIns(4,5)P2 enhance lipid binding.
There is a pattern correlating the location of the mutations with their hydrophobic component of lipid binding (Figure 3j): non-kinase domain mutants (C420R, E545K and p85α-N564D) display high levels of hydrophobic interaction with neutral lipids in the basal state, whereas the kinase C-terminal mutants require phosphopeptide activation to achieve the same level of hydrophobic binding. We will consider the two groups separately.
Despite their different chemical properties, the kinase domain mutants, H1047L, H1047R and G1049R, exhibit similarly high levels of hydrophobic binding to neutral lipids, and electrostatic binding to PS/PtdIns(4,5)P2-containing lipids upon phosphopeptide activation. These binding levels are a few fold higher than those of the activated WT enzyme. We have shown that the kinase helix kα12 constitutes a lipid-binding site. It is likely that helix kα11 and the kα11-kα12 “elbow”, on which these mutations reside, is an extension of the same site.
For non-kinase domain mutants, the nature of the effects of hydrophobicity is not immediately obvious. Disruption of the C2-iSH2 (C420R and p85α-N564D) or the C2/helical-nSH2 contacts (E545K) could expose hydrophobic regions in the C2 domain that are normally inaccessible to lipid binding. The iSH2 N564 forms polar contacts with the C2 N345, which is in the C2β1-β2 loop. The C2 C420, which is in the C2β5-β6 loop, forms van der Waals contacts with iSH2 P568 (Figure 3a). These two C2 loops in p110α are equivalent to the lipid-binding loops in the C2 domains of PKCα, which binds PS (Guerrero-Valero et al., 2009), and of cPLA2, which binds PC (Perisic et al., 1999; Stahelin et al., 2003). Superposition of the PKCα C2 structure, which has a bound short acyl-chain PS molecule (PDB ID: 1dsy), onto the C2 domain of our p110α/p85α-iSH2 structure reveals that both N564D and C420R would be compatible with accessing phospholipid headgroups (Supplementary Figure S4a). This interpretation would account for the observed increase in electrostatic interaction with anionic lipids (Figure 3j).
Global conformational change in p110α accompanies nSH2 disengagement
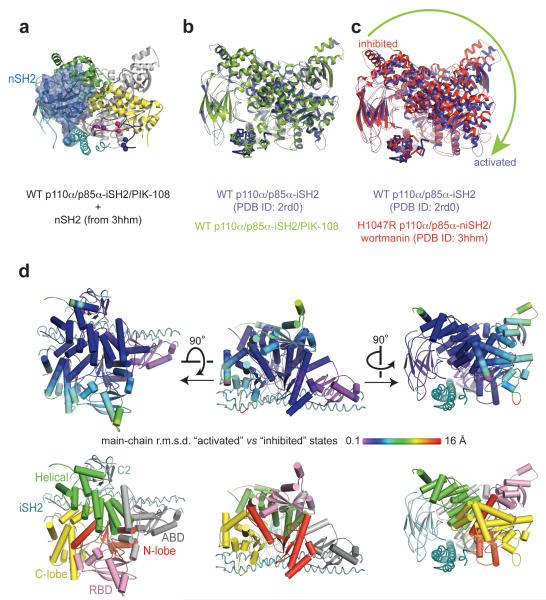
The p85α nSH2 does not contact the lipid binding elements in the kinase C-lobe (Figure 4a) but appears to control their access to membrane (Figure 3f-j). This suggests the activation mechanism by nSH2 to be allosteric. By performing structural alignments on the iSH2 of p110α/p85α structures, we observe a global conformational difference between those lacking the inhibitory contacts with nSH2 (as represented by the WT p110α/p85α-iSH2 structures) and those with the nSH2 interactions (as represented by the H1047R p110α/p85α-niSH2 structures) (Figure 4b,c). Viewing down the iSH2 coiled coil axis from the nSH2 side, the p110α subunit rotates clockwise from the state with an ordered nSH2 to the state lacking the nSH2 (Figure 4c). The ABD and the C2, which are anchored on the iSH2, form the pivot points of the movement (Figure 4d). The movement appears to be transmitted from the helical domain to the RBD and the kinase C-lobe, which exhibit the greatest degree of displacement (Figure 4d). This global conformational change suggests how nSH2 disengagement from p110α could allosterically cause local conformational change in the kinase C-lobe that might impact on activity. This is noteworthy considering that our WT p110α/p85α-iSH2/PIK-108 structure and the apo WT p110α/p85α-iSH2 structure experience very different crystal packing environments but superimpose well (Figure 4b).
Figure 4.
Global conformational change in p110α. (a) Spatial relationship between the nSH2 inhibitory contact site and the lipid binding elements in the kinase C-lobe. (b,c) Pairwise superposition of the p110α/p85α structures was performed by aligning the main-chain atoms of p85α iSH2 domain. Structures without p85α nSH2 represent the ‘activated’ form, the p110α/p85α-niSH2 structure represents the ‘inhibited’ form. The arrow indicates the direction of p110α domain movement around the p85α iSH2 coiled coil axis. (d) Representation of the r.m.s.d. calculated between main-chain atoms of the superposed p110α subunits shown in (c), as projected on the coordinates of 2rd0. To assist domain recognition, the lower panel shows the enzyme in the same orientation but coloured by domains.
Activation of all class IA PI3Ks by phosphopeptide increases lipid binding
Activation of p110β/p85α-nicSH2 and p110δ/p85α-nicSH2 complexes by phosphopeptide also induces lipid binding (Figure 5a). This is in agreement with the recent report for p110δ in a complex with full-length p85α (Burke et al., 2011). The relative contributions of electrostatic and hydrophobic components of binding differ among the three p110 isoforms, probably reflecting their sequence variability in the key lipid binding sites (Figure 1). In the primary sequences, the equivalent of H1047 in p110α is a Leu in p110β and p110δ. For PS/PtdIns(4,5)P2-containing liposomes, the total lipid binding level of activated H1047L p110α is a few fold higher than those of activated p110β and p110δ (Figure 5b). These results indicate that there is more than this hotspot position that differentiates the lipid binding capabilities among the three p110 isoforms.
Figure 5.
Lipid binding activities of class IA PI3-kinases. (a) Comparison of the three isoforms (protein concentration=1 μM). (b) Comparison of total lipid binding of the three isoforms with the p110α H1047L mutant (protein concentration=0.5 μM). The data represent the mean ± s.d. of two independent experiments.
Discussion
We have directly demonstrated that phosphopeptide-induced lipid binding underlies the activation mechanism for WT p110α/p85α. For most cancer-linked mutants of p110α/p85α, both basal and phosphopeptide-induced membrane binding is enhanced. For WT p110α, the primary lipid binding sites reside in the kinase domain, comprising the polybasic activation loop and hydrophobic elements in the C-terminal tail. We have identified activating cancer-linked mutants in the kinase C-terminal tail (H1047L, H1047R, G1049R) as well as mutants affecting the C2-iSH2 interaction (C420R and p85α-N564D) that enhance intrinsic lipid binding. Our binding data and structural analysis support the dual roles of C420R and N564D in disrupting C2-iSH2 contact and enhancing lipid binding. It is likely that this occurs via direct interactions between the C2 and/or the iSH2 domains with membranes. This additional membrane-binding site could both enhance total lipid binding and influence the choice of the membrane used by the WT enzyme or by particular p110α/p85α mutants in a cellular context.
Our data indicate that the inhibitory contact formed between the nSH2 and p110α C2/helical/kinase domains suppresses the basal kinase activity by inhibiting membrane binding. This is consistent with previous biochemical results for enzyme activity (Yu et al., 1998), in that the p85α nSH2, but not the cSH2, plays a principal role in regulating the activity of p110α. Mechanistically, we postulate that relieving the nSH2 inhibitory contact from p110α could result in a concerted conformational change propagating from the helical domain to the kinase C-lobe, as suggested by our structural comparison of activated and inhibited forms of p110α/p85α (Figure 4). This might trigger the activation loop and the C-terminal region to adopt lipid-binding competent conformations. The polybasic activation loop both facilitates long range electrostatic attraction to anionic lipids, and positions the PtdIns(4,5)P2 headgroup to the bound ATP for phosphoryl transfer. Since the ATP binding pocket is recessed in the kinase domain, partial membrane penetration by the C-terminal region would facilitate approach of the ATP γ-phosphate to the inositol ring of PtdIns(4,5)P2 (Figure 6).
Figure 6.
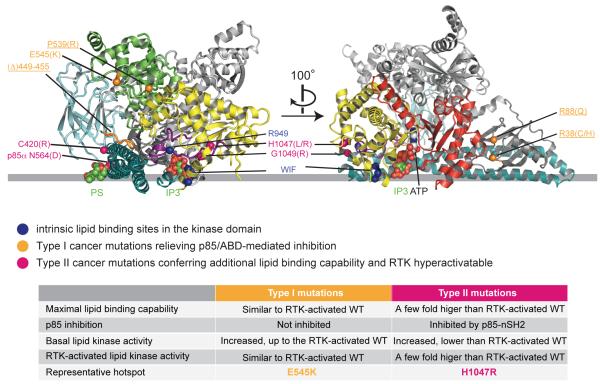
Summary and modelling of membrane binding. The grey line approximates the putative membrane surface and is drawn collinear with the phosphate in the PS and P1 in the Ins(1,4,5)P3 (IP3). Positions of PS, IP3 and ATP are derived from the following structures: 1dsy (Verdaguer et al., 1999), 1w2c (Gonzalez et al., 2004) and 1e8x (Walker et al., 1999). Details of how PS, IP3 and ATP were docked, and modelling of the kinase C-terminal helices are illustrated in Supplementary Figure S4. The Cα atoms of the labelled residues are represented in spheres. For clarity, the kinase domain N-lobe is colored red in the right panel only, and the helical domain is green on the left panel only. Type I activating cancer-linked mutations that are shown or predicted to relieve the p85α/ABD-mediated inhibition are highlighted in orange (the mutations that we predict would be type I, but have not been tested are underlined). The type II mutants, which are RTK hyperactivatable, are highlighted in pink. Note that docking of PS is solely for the purpose of approximating the lipid-binding site in the p110α C2 domain, and does not imply that this domain binds PS specifically.
We have demonstrated that binding of activated p110/p85α to lipid membrane has both an electrostatic and a hydrophobic component. Basic residues in the activation loop contribute to the electrostatic component. The C-terminal tail is clearly important for membrane interaction, but its contribution is complex. SPR measurements show that mutating the hydrophobic WIF motif to “AAA” abolishes measureable electrostatic interactions with anionic lipids. Conversely, the activated H1047L, H1047R and G1049R mutants increase both hydrophobic and electrostatic interactions with lipids. These observations attest to the importance of cooperation between the electrostatic and the hydrophobic membrane-binding elements in p110α/p85α, as seen for other membrane-binding proteins (Cho and Stahelin, 2005). Our data also elucidate the influence of lipid composition on lipid kinase activities for both WT and mutant p110/p85 complexes (Mandelker et al., 2009).
Our structure reveals an alternative, small molecule binding site in the kinase C-lobe. Current inhibitors for class I PI3K are all ATP-competitive compounds (Marone et al., 2008). This induced binding pocket might be exploited to develop a novel class of inhibitors that are not ATP competitive but could interfere with membrane binding. Although we could not detect any inhibition of lipid binding with PIK-108 (up to 50 μM for the CD_E545K mutant, assayed in the same manner described in Figure 3i,j, data not shown), it might be that compounds with enhanced affinity for this site could act as PI3K inhibitors. An example of a membrane-binding inhibitor that is in clinical trials is Perifosine, an alkylphospholipid that targets the PH domain of AKT (Gills and Dennis, 2009).
It appears that the p110α and p85α mutations can be classified as two basic types: (I) those that relieve p85-mediated inhibition and (II) those that lead to additional lipid binding capability (Figure 6). Type I mutations up-regulate kinase activity by unmasking the intrinsic lipid-binding potential of WT p110α/p85α. These mutants have elevated basal activities and binding of RTK phosphopeptides does not significantly enhance their activities beyond that of activated WT enzyme. We have shown that the hotspot mutant E545K belongs to this class, in accordance with previous work (Miled et al., 2007; Carson et al., 2008). Several other activating mutations that are distant from the putative membrane interface, such as R38C/H, R88Q, P539R, and deletion of residues 449-455 in the C2β7-β8 loop (Ikenoue et al., 2005; Oda et al., 2008; Rudd et al., 2011), could also belong to this class (Figure 6). Type II mutations are still subjected to p85α nSH2-mediated inhibition. We and others have shown that H1047R belongs to this type, which can be significantly activated by a phosphorylated RTK peptide (this study) or the adaptor protein IRS (Carson et al., 2008). Although basal activities of type II mutants are higher than that of WT enzyme, their maximal lipid binding and kinase activities achieved by RTK phosphopeptide binding greatly exceed those of the activated WT enzyme (3-7 fold with our experimental setup).
Our in vitro results imply that the biological outputs of the type II mutants can be greatly augmented by growth factor/RTK signalling. This notion is supported by reports that compared the E545K and H1047R mutants in cells. Human cancer cell lines bearing helical domain mutations showed lower AKT phosphorylation level and AKT-dependent signalling than cell lines bearing kinase domain mutations coupled with either HER2 amplification or EGFR hyperactivation (Vasudevan et al., 2009). H1047R, but not E545K, was found to enhance HER2-mediated transformation of breast cancer cells via autocrine proliferative stimulation (Chakrabarty et al., 2010). Therefore, cancers with the type II mutations might be better treated by co-inhibition of p110α and RTKs, because these mutants strongly depend on RTK stimulation to reach a critical threshold of membrane binding/activity necessary for transformation. For E545K to achieve a similar level of transforming activity as H1047R in chicken embryonic fibroblasts, it required additionally Ras binding (Zhao and Vogt, 2008). It is likely that Ras could increase recruitment of p110α to lipid membrane. Cancers with type I mutations such as E545K might be better treated by co-inhibition of the p110α and the Ras pathway, because these mutants could be more dependent on “topping-up” by Ras to achieve the membrane binding threshold necessary for transformation. Type I and type II mutants could also confer distinct functional significance during different stages of cancer biology. For example, the E545K mutant was found to have a stronger metastatic phenotype than the H1047R mutant in tumor xenograft models using isogenic, human breast cancer cells (Pang et al., 2009). The ability to induce tumour formation thus far has been demonstrated only for the H1047R mutant, but not yet for the E545K mutant, in engineered mouse models of cancers (Engelman et al., 2008; Adams et al., 2011; Meyer et al., 2011).
Both growth factor independence and growth factor signalling manipulation are traits that enable cancer cells to sustain proliferative signalling (Witsch et al., 2010; Hanahan and Weinberg, 2011). Investigating the influence of upstream RTK signalling could elucidate the yet unclear roles of the different PI3K mutations in cancer initiation, progression and maintenance, and in identifying suitable biomarkers for cancer therapy targeting the PI3K pathway (Engelman, 2009) .
Materials and methods
Plasmids
Details of expression constructs for p110α, p110β, p110δ (all mouse), p85α (mouse, except for the ‘ni’ fragment which is human), and Grp1-PH (human) are provided in Supplementary materials and methods. Domain boundaries of p85α fragments are shown in Figure 1.
Protein expression and purification
All p110/p85 complexes were co-expressed in Sf9 insect cells. The N-terminal His6-tag on p110α was kept for proteins used for crystallization but was cleaved off for those used for functional analyses. GST-Grp1-PH was expressed in E coli. Details of expression and purification are described in Supplementary materials and methods.
Crystallization and structure determination
The complex of His6-(TEV)-tagged mouse p110α with human p85α-niSH2 was co-crystallized with PIK-108. The crystals diffracted to 3.5 Å, and belong to the spacegroup I222 with one heterodimer per asymmetric unit. The human apo WT p110α/p85α-iSH2 structure ((Huang et al., 2007), PDB ID: 2rd0) was used as the search model for molecular replacement. Details of crystallization and structure determination are described in Supplementary materials and methods. Crystallographic statistics are presented in Supplementary Table S1. All structural graphics were prepared with PyMOL (www.pymol.org).
Lipid kinase activity assay
Liposomes comprising PC/PE/PS/PtdIns(4,5)P2/cholesterol were in mol% ratios of either 38:25:20:2:15 or 38:35:10:2:15. PtdIns(3,4,5)P3 production was measured by a modified fluorescence polarization assay (Echelon Biosciences) originally developed for use with diC8-PtdIns(4,5)P2 as a substrate (Drees et al., 2003). Reactions were carried out at room temperature (25 °C) in 384-well microtitre plates. A 10 μl reaction consisted of 50 μM PtdIns(4,5)P2 in 2.5 mM liposomes, p110/p85 (500 nM for non-activated WT p110α/p85α-nicSH2, 5 nM for the rest), 100 μM ATP, 1 mM MgCl2, and with or without 1 μM pY2-peptide. The reactions were quenched with 5 μl buffer containing 20 mM Hepes (pH 7.5), 150 mM NaCl, 20 mM EDTA, and 400 nM GST-Grp1-PH, followed by addition of 5 μl of 40 nM TAMRA-Ins(1,3,4,5)P4 in HNT buffer. The plates were read in a PHERAstar spectrofluorometer (BMG Labtech, Germany) using the FP/540-20/590-20/590-20 optical module. Standard curves were performed with diC8-PtdIns(3,4,5)P3 with or without pY2-peptide. Only a minor difference due to presence of pY2-peptide was observed under our assay condition, which is different from that published previously (Carson et al., 2008).
SPR measurement of lipid binding
Liposome binding experiments were performed at 25 °C on a BIAcore2000, using the L1 sensor chip (GE Healthcare). HNT was used as the running buffer. Liposomes comprising PC/PE/PS/PtdIns(4,5)P2/cholesterol were in mol% ratios of either 60:25:0:0:15 (Fc1), 50:25:10:0:15 (Fc2), 48:25:10:2:15 (Fc3) or 38:25:20:2:15 (Fc4). Liposomes (0.1 – 0.2 mM) were injected onto each flow-cell at 5 μl/min flow rate to achieve ~3400 RU immobilization level. This was followed by two injections of 0.2 M Na2CO3 to remove loosely bound liposomes. Chicken egg white albumin (0.2 mg/ml) was injected over the flow-cells to mask any residual hydrophobic surface not covered by the liposomes, followed by multiple injections of 0.2 M Na2CO3 to stabilize the baseline. Steady-state binding was measured by injecting p110/p85 complexes (concentration at either 500 nM or 1 μM) at 2 μl/min flow rate for 5 min. Each measurement was followed by injections of 0.2 M Na2CO3 to regenerate the liposome surface. Independent experiments were carried out with freshly immobilized liposomes after stripping the sensor surface sequentially with 0.1 N NaOH, isopropanol:0.1 N NaOH (2:3) and 6 M guanidinium HCl.
Supplementary Material
Acknowledgements
We are grateful to Kevan Shokat and co-workers (UCSF, USA), including Beth Apsel, Benjamin Houseman, Eli Zunder, Morri Feldman and Zachary Knight, for their generous gift of PI3K inhibitors, which were crucial for crystallization screening. We would like to thank the beamline scientists and members of staff at the ESRF beamlines ID14-1, ID14-2, ID14-4, ID23-1, ID23-2, and ID29 (Grenoble, France) and Diamond Light Source beamlines I02 and I03 (United Kingdom). We also thank past and present members of the Williams’s group, particularly Simon Miller, for assistance with remote crystal data collection, Olga Perisic for providing baculovirus of mouse p110β, Alexey Murzin for his structural insight of the WIF motif conformation, Andrew Leslie and Nicolas Soler for providing scripts to create Figure 4d. This work was supported by the Medical Research Council, UK.
Footnotes
Conflict of interest The authors declare that there are no competing financial interests in relation to the work described.
References
- Adams JR, Xu K, Liu JC, Agamez NM, Loch AJ, Wong RG, et al. Cooperation between Pik3ca and p53 mutations in mouse mammary tumor formation. Cancer Res. 2011;71:2706–2717. doi: 10.1158/0008-5472.CAN-10-0738. [DOI] [PubMed] [Google Scholar]
- Backer JM, Myers MGJ, Shoelson SE, Chin DJ, Sun XJ, Miralpeix M, et al. Phosphatidylinositol 3′-kinase is activated by association with IRS-1 during insulin stimulation. EMBO J. 1992;11:3469–3479. doi: 10.1002/j.1460-2075.1992.tb05426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayascas JR. PDK1: the major transducer of PI 3-kinase actions. Curr Top Microbiol Immunol. 2010;346:9–29. doi: 10.1007/82_2010_43. [DOI] [PubMed] [Google Scholar]
- Berndt A, Miller S, Williams O, Le DD, Houseman BT, Pacold JI, et al. The p110 delta structure: mechanisms for selectivity and potency of new PI(3)K inhibitors. Nat Chem Biol. 2010;6:117–124. doi: 10.1038/nchembio.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke JE, Vadas O, Berndt A, Finnegan T, Perisic O, Williams RL. Dynamics of the phosphoinositide 3-kinase p110δ interaction with p85α and membranes reveals aspects of regulation distinct from p110α. Structure. 2011 doi: 10.1016/j.str.2011.06.003. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter CL, Auger KR, Chanudhuri M, Yoakim M, Schaffhausen B, Shoelson S, et al. Phosphoinositide 3-kinase is activated by phosphopeptides that bind to the SH2 domains of the 85-kDa subunit. J Biol Chem. 1993;268:9478–9483. [PubMed] [Google Scholar]
- Carracedo A, Pandolfi PP. The PTEN-PI3K pathway: of feedbacks and cross-talks. Oncogene. 2008;27:5527–5541. doi: 10.1038/onc.2008.247. [DOI] [PubMed] [Google Scholar]
- Carson JD, Van Aller G, Lehr R, Sinnamon RH, Kirkpatrick RB, Auger KR, et al. Effects of oncogenic p110alpha subunit mutations on the lipid kinase activity of phosphoinositide 3-kinase. Biochem J. 2008;409:519–524. doi: 10.1042/BJ20070681. [DOI] [PubMed] [Google Scholar]
- Chakrabarty A, Rexer BN, Wang SE, Cook RS, Engelman JA, Arteaga CL. H1047R phosphatidylinositol 3-kinase mutant enhances HER2-mediated transformation by heregulin production and activation of HER3. Oncogene. 2010;29:5193–5203. doi: 10.1038/onc.2010.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalhoub N, Baker SJ. PTEN and the PI3-kinase pathway in cancer. Annu Rev Pathol. 2009;4:127–150. doi: 10.1146/annurev.pathol.4.110807.092311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W, Stahelin RV. Membrane-protein interactions in cell signaling and membrane trafficking. Annu Rev Biophys Biomol Struct. 2005;34:119–151. doi: 10.1146/annurev.biophys.33.110502.133337. [DOI] [PubMed] [Google Scholar]
- Drees BE, Weipert A, Hudson H, Ferguson CG, Chakravarty L, Prestwich GD. Competitive fluorescence polarization assays for the detection of phosphoinositide kinase and phosphatase activity. Comb Chem High Throughput Screen. 2003;6:321–330. doi: 10.2174/138620703106298572. [DOI] [PubMed] [Google Scholar]
- Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14:1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke TF. PI3K/Akt: getting it right matters. Oncogene. 2008;27:6473–6488. doi: 10.1038/onc.2008.313. [DOI] [PubMed] [Google Scholar]
- Gills JJ, Dennis PA. Perifosine: update on a novel Akt inhibitor. Curr Oncol Rep. 2009;11:102–110. doi: 10.1007/s11912-009-0016-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez B, Schell MJ, Letcher AJ, Veprintsev DB, Irvine RF, Williams RL. Structure of a human inositol 1,4,5-trisphosphate 3-kinase: substrate binding reveals why it is not a phosphoinositide 3-kinase. Mol Cell. 2004;15:689–701. doi: 10.1016/j.molcel.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Guerrero-Valero M, Ferrer-Orta C, Querol-Audi J, Marin-Vicente C, Fita I, Gomez-Fernandez JC, et al. Structural and mechanistic insights into the association of PKCalpha-C2 domain to PtdIns(4,5)P2. Proc Natl Acad Sci U S A. 2009;106:6603–6607. doi: 10.1073/pnas.0813099106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gymnopoulos M, Elsliger MA, Vogt PK. Rare cancer-specific mutations in PIK3CA show gain of function. Proc Natl Acad Sci U S A. 2007;104:5569–5574. doi: 10.1073/pnas.0701005104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Huang CH, Mandelker D, Schmidt-Kittler O, Samuels Y, Velculescu VE, Kinzler KW, et al. The structure of a human p110alpha/p85alpha complex elucidates the effects of oncogenic PI3Kalpha mutations. Science. 2007;318:1744–1748. doi: 10.1126/science.1150799. [DOI] [PubMed] [Google Scholar]
- Ikenoue T, Kanai F, Hikiba Y, Obata T, Tanaka Y, Imamura J, et al. Functional analysis of PIK3CA gene mutations in human colorectal cancer. Cancer Res. 2005;65:4562–4567. doi: 10.1158/0008-5472.CAN-04-4114. [DOI] [PubMed] [Google Scholar]
- Jaiswal BS, Janakiraman V, Kljavin NM, Chaudhuri S, Stern HM, Wang W, et al. Somatic mutations in p85alpha promote tumorigenesis through class IA PI3K activation. Cancer Cell. 2009;16:463–474. doi: 10.1016/j.ccr.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- Mandelker D, Gabelli SB, Schmidt-Kittler O, Zhu J, Cheong I, Huang CH, et al. A frequent kinase domain mutation that changes the interaction between PI3Kalpha and the membrane. Proc Natl Acad Sci U S A. 2009;106:16996–17001. doi: 10.1073/pnas.0908444106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marone R, Cmiljanovic V, Giese B, Wymann MP. Targeting phosphoinositide 3-kinase: moving towards therapy. Biochim Biophys Acta. 2008;1784:159–185. doi: 10.1016/j.bbapap.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Meyer DS, Brinkhaus H, Muller U, Muller M, Cardiff RD, Bentires-Alj M. Luminal Expression of PIK3CA Mutant H1047R in the Mammary Gland Induces Heterogeneous Tumors. Cancer Res. 2011;71:4344–4351. doi: 10.1158/0008-5472.CAN-10-3827. [DOI] [PubMed] [Google Scholar]
- Miled N, Yan Y, Hon WC, Perisic O, Zvelebil M, Inbar Y, et al. Mechanism of two classes of cancer mutations in the phosphoinositide 3-kinase catalytic subunit. Science. 2007;317:239–242. doi: 10.1126/science.1135394. [DOI] [PubMed] [Google Scholar]
- Miller S, Tavshanjian B, Oleksy A, Perisic O, Houseman BT, Shokat KM, et al. Shaping development of autophagy inhibitors with the structure of the lipid kinase Vps34. Science. 2010;327:1638–1642. doi: 10.1126/science.1184429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan K, Lemmon MA. Determining selectivity of phosphoinositide-binding domains. Methods. 2006;39:122–133. doi: 10.1016/j.ymeth.2006.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda K, Okada J, Timmerman L, Rodriguez-Viciana P, Stokoe D, Shoji K, et al. PIK3CA cooperates with other phosphatidylinositol 3′-kinase pathway mutations to effect oncogenic transformation. Cancer Res. 2008;68:8127–8136. doi: 10.1158/0008-5472.CAN-08-0755. [DOI] [PubMed] [Google Scholar]
- Pang H, Flinn R, Patsialou A, Wyckoff J, Roussos ET, Wu H, et al. Differential enhancement of breast cancer cell motility and metastasis by helical and kinase domain mutations of class IA phosphoinositide 3-kinase. Cancer Res. 2009;69:8868–8876. doi: 10.1158/0008-5472.CAN-09-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perisic O, Paterson HF, Mosedale G, Lara-Gonzalez S, Williams RL. Mapping the phospholipid-binding surface and translocation determinants of the C2 domain from cytosolic phospholipase A2. J Biol Chem. 1999;274:14979–14987. doi: 10.1074/jbc.274.21.14979. [DOI] [PubMed] [Google Scholar]
- Pirola L, Zvelebil MJ, Bulgarelli-Leva G, Van Obberghen E, Waterfield MD, Wymann MP. Activation loop sequences confer substrate specificity to phosphoinositide 3-kinase a (PI3Ka) J Biol Chem. 2001;276:21544–21554. doi: 10.1074/jbc.M011330200. [DOI] [PubMed] [Google Scholar]
- Rudd ML, Price JC, Fogoros S, Godwin AK, Sgroi DC, Merino MJ, et al. A unique spectrum of somatic PIK3CA (p110alpha) mutations within primary endometrial carcinomas. Clin Cancer Res. 2011;17:1331–1340. doi: 10.1158/1078-0432.CCR-10-0540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels Y, Diaz LAJ, Schmidt-Kittler O, Cummins JM, Delong L, Cheong I, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561–573. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- Stahelin RV, Rafter JD, Das S, Cho W. The molecular basis of differential subcellular localization of C2 domains of protein kinase C-alpha and group IVa cytosolic phospholipase A2. J Biol Chem. 2003;278:12452–12460. doi: 10.1074/jbc.M212864200. [DOI] [PubMed] [Google Scholar]
- Vasudevan KM, Barbie DA, Davies MA, Rabinovsky R, McNear CJ, Kim JJ, et al. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer Cell. 2009;16:21–32. doi: 10.1016/j.ccr.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdaguer N, Corbalan-Garcia S, Ochoa WF, Fita I, Gomez-Fernandez JC. Ca(2+) bridges the C2 membrane-binding domain of protein kinase Calpha directly to phosphatidylserine. EMBO J. 1999;18:6329–6338. doi: 10.1093/emboj/18.22.6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker EH, Perisic O, Ried C, Stephens L, Williams RL. Structural insights into phosphoinositide 3-kinase catalysis and signalling. Nature. 1999;402:313–320. doi: 10.1038/46319. [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witsch E, Sela M, Yarden Y. Roles for growth factors in cancer progression. Physiology (Bethesda) 2010;25:85–101. doi: 10.1152/physiol.00045.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Shekar SC, Flinn RJ, El-Sibai M, Jaiswal BS, Sen KI, et al. Regulation of Class IA PI 3-kinases: C2 domain-iSH2 domain contacts inhibit p85/p110alpha and are disrupted in oncogenic p85 mutants. Proc Natl Acad Sci U S A. 2009;106:20258–20263. doi: 10.1073/pnas.0902369106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Wjasow C, Backer JM. Regulation of the p85/p110alpha phosphatidylinositol 3′-kinase. Distinct roles for the n-terminal and c-terminal SH2 domains. J Biol Chem. 1998;273:30199–30203. doi: 10.1074/jbc.273.46.30199. [DOI] [PubMed] [Google Scholar]
- Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Liu G, Dziubinski M, Yang Z, Ethier SP, Wu G. Comprehensive analysis of oncogenic effects of PIK3CA mutations in human mammary epithelial cells. Breast Cancer Res Treat. 2008;112:217–227. doi: 10.1007/s10549-007-9847-6. [DOI] [PubMed] [Google Scholar]
- Zhang X, Vadas O, Perisic O, Anderson KE, Clark J, Hawkins PT, et al. Structure of lipid kinase p110beta/p85beta elucidates an unusual SH2-domain-mediated inhibitory mechanism. Mol Cell. 2011;41:567–578. doi: 10.1016/j.molcel.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao JJ, Liu Z, Wang L, Shin E, Loda MF, Roberts TM. The oncogenic properties of mutant p110alpha and p110beta phosphatidylinositol 3-kinases in human mammary epithelial cells. Proc Natl Acad Sci U S A. 2005;102:18443–18448. doi: 10.1073/pnas.0508988102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Vogt PK. Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc Natl Acad Sci U S A. 2008;105:2652–2657. doi: 10.1073/pnas.0712169105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.