Abstract
Purpose
PTEN deletions in prostate cancer are associated with tumor aggression and poor outcome. Recent studies have implicated PTEN as a determinant of homologous-recombination (HR) through defective RAD51 function. Similar to BRCA1/2-defective tumor cells, PTEN-null prostate and other cancer cells have been reported to be sensitive to PARP inhibitors (PARPi). To date, no direct comparison between PTEN and RAD51 expression in primary prostate tumors has been reported.
Experimental Design
Prostate cancer cell lines and xenografts with known PTEN status (22RV1-PTEN+/+; DU145-PTEN+/−; PC3-PTEN−/−) and H1299 and HCT116 cancer cells were used to evaluate how PTEN loss affects RAD51 expression and PARPi sensitivity. Primary prostate cancers with known PTEN status were analyzed for RAD51 expression.
Results
PTEN status is not associated with reduced RAD51 mRNA or protein expression in primary prostate cancers. Decreased PTEN expression did not reduce RAD51 expression or clonogenic survival following PARPi amongst prostate cancer cells that vary in TP53 and PTEN. PARPi sensitivity instead associated with a defect in MRE11 expression. PTEN-deficient cells had only mild PARPi sensitivity and no loss of HR or RAD51 recruitment. Clonogenic cell survival following a series of DNA-damaging agents was variable: PTEN-deficient cells were sensitive to ionizing radiation, mitomycin-C, UV, H2O2 and methyl-methanesulfonate; but not to cisplatin, camptothecin, or paclitaxel.
Conclusions
These data suggest that the relationship between PTEN status and survival following DNA damage is indirect and complex. It is unlikely that PTEN status will be a direct biomarker for HR status or PARPi response in prostate cancer clinical trials.
Keywords: PTEN, radiotherapy, chemotherapy, PARP, prostate cancer, RAD51
Introduction
The PTEN gene encodes a dual specificity lipid/protein phosphatase which antagonizes the activation of the phosphatidylinositol-3′-OH-kinase (PI3K)/AKT pathway. Mono- and bi-alleleic losses of the PTEN gene has been implicated in prostate cancer progression and inferior clinical outcome (1-6). In a number of models, the PTEN protein mediates its anti-tumorigenic effects via PI3K/AKT-dependent and -independent pathways (7) and PTEN-independent AKT signaling has been implicated in the MRE11-ATM DNA damage response (DDR) (8). Murine embryonic fibroblasts (MEFs) lacking PTEN were recently observed to have high levels of genomic instability and increased endogenous DNA double strand breaks (DSB) associated with a reduction in the expression of RAD51 (a key gene involved in homologous recombination (HR) repair of DSBs). Restoration of PTEN in PTEN−/− MEFs restored RAD51 expression in a manner independent of its phosphatase activity (6). However, subsequent reports in human tumor cell lines have shown conflicting data as to whether PTEN loss is associated with a reduced expression of RAD51 (9, 10). To date, no information exists as to whether PTEN gene status determines RAD51 expression in primary prostate cancers in vivo.
A defined link between PTEN status and HR function in prostate and other human tumors would be important as it would support the treatment for PTEN-null tumors using agents targeted against defects in DNA repair. An example of this approach is the use of inhibitors of poly(ADP ribose) polymerase (PARP) as single agents in germline BRCA1/2-defective ovarian, breast and prostate cancers that are HR-defective (11). The PARP1 and 2 proteins are required for repair of DNA single strand breaks (SSB) and in cells treated with small molecule inhibitors of PARP (PARPi), unrepaired SSBs at replication forks are converted into DSBs which require HR-mediated repair to offset cell lethality. Tumor cells lacking HR function (e.g. deficient in BRCA1 or BRCA2 expression) are exquisitely sensitive to PARPi due the inability to repair replication-associated DSBs; this results in ‘synthetic cell lethality’ (12-15). The results of clinical trials using PARPi in germline BRCA1- and BRCA2-deficient tumors are promising, but not perfect. It is now recognized that biomarkers that predict functional losses in DNA repair activity, in addition to mutations in DNA repair genes, will be required to more accurately predict clinical PARPi efficacy (11, 16, 17).
As a potentially important biomarker of DNA repair status, recent reports have suggested that tumor cells that lack PTEN have a marked reduction in RAD51-dependent HR and are therefore sensitive to PARPi in vitro and in vivo (18-20). This suggests that many sporadic tumors could be amenable to PARPi-specific treatments or other agents that are highly toxic to HR-deficient tumor cells such as mitomycin C (MMC), cis-platinum (cDDP) and ionizing radiation (IR) (21-23). Novel trials utilizing PARPi in prostate and other cancers could therefore stratify patients on the basis of intact or abrogated function of the HR, FA, DDR (MRE11-ATM) and now, PTEN pathways (24-26). Based on a recent prostate cancer-specific report, they may also be stratified by the presence or absence of aberrant signaling associated with a TMPRSS2:ERG fusion (27, 28). BRCA2-deficient prostate cancers are particularly aggressive and the use of PARPi in their treatment could help with overcoming castration-resistance (29). Similarly, as PTEN loss and TMPRSS2:ERG fusions are common events in high-grade and castrate-resistant prostate cancers (2), the additional use of PARPi in these tumors would be an important new therapeutic option (27, 28).
We previously reported that prostate cancer cells were defective in SSB, DSB, and BER gene expression and selected functional repair endpoints when compared to normal prostate epithelium or stromal cells (30). We therefore evaluated whether PTEN loss in human prostate cancer cells is associated with loss of RAD51 expression and HR and leads to altered clonogenic sensitivity. The current report represents, to our knowledge, the first systematic study of the relationship between PTEN status and RAD51 expression in primary prostate cancers and cell lines.
Materials and Methods
Cell Culture
H1299 human lung carcinoma cells were cultured in αMEM supplemented with 10mM HEPES. Prostate cancer cell lines with varying TP53 status (31) included DU145 (mutant TP53), 22RV-1 (wild-type TP53), and PC3(null TP53); these were cultured as previously reported (30). These cell lines are all negative for the TMPRSS2:ERG fusion (32). HCT116-PTEN+/+ and -PTEN−/− cells (a generous gift from Dr. Todd Waldman, Georgetown University, Washington DC) and U2OS cells (American Type Culture Collection, Manassas, VA) were cultured in McCoy’s 5A media. Ataxia telangiectasia-like disorder (ATLD; MRE11 deficient) fibroblasts were cultured as previously reported (8). All cells media were supplemented with 10% fetal calf serum (FCS), penicillin, streptomycin, and were grown at 5% CO2, 21% O2, 37°C. The origin and correct status of all cell lines used for this study was confirmed by STR (short tandem repeat) DNA analyses as previously described (8, 33).
siRNA Transfections and Homologous Recombination Assay
Cells were seeded for 18h in 6-well plates such that their density on the day of transfection was ~35%. Cells were transfected with siRNA duplexes to RAD51 (0.25 nM), PTEN (1 nM), or control siRNA using Lipofectamine 2000 (Invitrogen; Carlsbad, CA), according to the manufacturer’s instructions. HR-dependent DNA DSB repair was assessed using the DR-GFP/ISce-I assay, as previously described (34).
Western blot analysis
Cells were lysed and subjected to Western blot analysis as previously reported (34). Primary antibodies were as follows: rabbit anti-RAD51 (Santa Cruz Biotechnologies, Santa Cruz, CA; 1:1000), rabbit anti-PTEN (Cell Signaling Technologies, Danvers, MA; 1:1000), rabbit anti-phospho-AKT (S473) (Cell Signaling, 1:1000), rabbit anti-Actin (Sigma-Aldrich, St. Louis, MO; 1:10,000). Membranes were washed three times in TBS containing 0.01% Tween-20 (TBS-T), and then incubated with IRDye 800 Donkey anti-Rabbit or IRDye 700 Donkey anti-Mouse (LiCor Biosciences), at room temperature in the dark for 1h. Blots were scanned on a LiCor Odyssey.
Clonogenic, Proliferation, and Cell Cycle Assays
Cells were seeded in 6-well plates (two dilutions, in triplicate, per 6-well plate), treated as indicated, and then returned to 37°C, 5% CO2 for the duration of the experiment. Once colonies of >50 cells were observed, the cells were stained with methylene blue for 1h, washed, and then allowed to dry overnight at 37°C. Colonies were counted manually and survival calculated as previously described (34).
ATLD fibroblasts (and isogenic fibroblasts expressing either wtMRE11 or an endonuclease-deficient mtMRE11; see (8) for details) were counted manually (using Trypan Blue exclusion to ensure seeding of viable cells) and then seeded in triplicate at a density of ~6500 cells per cm2 in 6-well plates in the presence of PARP inhibitor (KU-0059436, Olaparib; PARPi) at a final concentration of 1 μM, or in the presence of DMSO as a vehicle control. Cells were harvested by trypsinization at 1, 2, 4, and 8 days post-seeding, and cell number was assessed by manual counting using a haemocytometer. For cell cycle analyses, cells were stained with propidium iodide and analysed using a FACS Aria flow cytometer, as previously described (35).
PARP Activity Assay
Cells growing in log phase were pre-treated for 1h with 2.5 μM PARPi (or DMSO as control). Cells were lysed in 1X PARP Lysis Buffer from the Universal Chemiluminescent PARP Assay Kit (Trevigen Inc, Gaithersburg, MD). Protein concentrations were determined and PARP activity was assessed using 30 μg of total protein per experimental group, according to the manufacturer’s instructions. Activated DNA was omitted from some reactions in order to obtain a measure of basal PARP activity in treated vs. untreated cells.
Primary Prostate Cancer and Xenograft Studies
A tissue microarray (TMA) was constructed using donor cores from 142 radical prostatectomy (RP) specimens with usual acinar-type prostate carcinoma obtained at University Health Network (UHN) between 2001 and 2002. The RP slides from all 142 cases were reviewed in order to identify prostate cancer foci that would suitable for sampling. Where possible, the TMA was constructed using up to six 0.6 mm donor cores from each RP specimen. In cases of multi focal and bilateral carcinoma, we attempted to include 3 donor cores from the largest foci in each lobe of the prostate. Distinct tumor foci were defined as those separated by a distance of ≥ 3mm in a single donor block or ≥ 4 mm in adjacent donor blocks (above or below). Where possible, we also specifically sampled areas with different Gleason patterns within each focus of tumor. The resulting TMA comprised 733 donor cores distributed in three paraffin blocks. Standard 5 μm H&E sections from each TMA block were scanned using an Aperio ScanScope CS (Aperio, Vista, CA). The digital slides were reviewed to confirm the presence of representative tumor in each donor core and, when applicable, to annotate specific Gleason patterns in each core. Standard clinical follow-up data, representing 7-9 years of follow-up, were compiled for each case represented on the TMA using a UHN RP clinical database. Prostate cancer cell line xenografts and human prostate cancer tissue microarrays were stained with rabbit anti-RAD51 (Santa Cruz Biotechnologies, Santa Cruz, CA) or rabbit monoclonal anti-phospho-AKT (Ser473; Cell Signaling), as previously described (33). In selected studies, the hypoxic biomarker EF5 was injected into tumor-bearing animals prior to sacrifice as previously described (Chan et al.; Can Res; 2010). RAD51 IHC signal was quantified using a custom algorithm that considers both IHC signal intensity and signal distribution, normalized for total area. Results are presented as arbitrary relative units.
Fluoresence In Situ Hybridization (FISH)
Three color interphase FISH (for PTEN and TMPRSS2:ERG) was applied to formalin-fixed paraffin-embedded (FFPE) prostate cancer TMAs, as previously described (2). Analyses were done using an epi-fluorescence microscope (Zeiss Axio Imager, Gò’ttingen, Germany) equipped with a triple bandpass filter set (DAPI/Green/Orange), dual bandpass filter set (Green/Orange) and single bandpass fitlers (DAPI,Green, Red & Orange) (Chroma, Bellows Falls, VT). Image capture was done using a digital ProgRes MF video camera (Jenoptik, Germany) and the fluorescence image acquisition software ISIS (MetaSystems, Germany). PTEN genomic losses and TMPRSS2:ERG fusions were evaluated for each probe by counting signals in 100 non-overlapped, intact interphase nuclei per tumor tissue. Nuclei with signals from each fluorochrome and complete DAPI nuclear staining were randomly selected for scoring from tumor regions.
RNA Isolation, Reverse Transcription PCR, and Real-Time Quantitative PCR
Total RNA was isolated using the RNeasy isolation kit (Qiagen). Sample RNA or human reference RNA (Stratagene) was treated with DNase I (Roche Diagnostics). Reverse transcription PCR (RT-PCR) was performed using the TaqMan Reverse Transcription kit (Applied Biosystems). Quantitative real-time PCR analysis of RAD51 was performed as previously described (34).
UV Laser Microirradiation and Immunofluoresence Microscopy
UV laser microirradiation (UVLM) was performed as previously described (8). Briefly, cells to be subjected to UVLM were seeded on round #1.5 coverslips, pre-treated with 5-bromo-2′-deoxyuridine (BrdU) for 24h, and then transferred to a humidified, temperature-controlled live cell chamber for irradiation. Cells were fixed in paraformaldehyde 30min post-irradiation, and double-stained using indicated antibodies. Nuclei were visualized by staining with 4′,6-amidino-2-phenylindole (DAPI).
Stained cells were imaged as previously described (8), using an Olympus Spinning Disk Confocal microscope with a 100x Olympus objective lens, producing 0.16 × 0.16 μm pixels. Z-stacks were obtained at a resolution of 0.25 μm per slice, yielding 0.16 × 0.16 × 0.25 μm voxels. Images were subjected to 3D deconvolution, as previously described (8).
Gene Expression Analyses
Gene expression analyses in 54 primary castrate-resistant prostate cancers were conducted as previously reported (36), using Complementary DNA-mediated Annealing Selection and Ligation (DASL; Illumina, San Diego, CA).
Statistical Analyses
Quantitative data is shown as the mean +/− one standard error of the mean (SEM) for at least 3 independent experiments. Comparative statistics utilized the paired Student’s T-test, Mann-Whitney test, or two-way Analysis of Variance (ANOVA) with Tukey post-hoc tests to evaluate differences between experimental groups. p-values of <0.05 were considered statistically significant.
Results
Lack of PTEN expression or TMPRSS2:ERG fusion Does Not Alter RAD51 Expression in Prostate Cancer Cell Lines or Primary Human Prostate Tumours
Several recent reports have demonstrated that loss of PTEN is often associated with down-regulation of RAD51 (7, 18, 19), a key regulator of HR-mediated DNA DSB repair. Since PTEN loss is common in human prostate cancer, we evaluated whether PTEN status is associated with differential RAD51 expression in prostate cancer cells. To that end, we first assessed PTEN and RAD51 protein levels in whole-cell lysates of 22RV1 (PTEN +/+), DU145 (PTEN +/−), and PC3 (PTEN −/−) prostate cancer cells that also vary in TP53 status (31) as functional TP53 has been linked to basal and IR-induced RAD51 expression (37). Table 1 summarizes the various DNA repair and checkpoint defects that characterize the cell lines used in this study. 22RV-1 and DU145 cells expressed PTEN, while PTEN was not detectable in PC3 cells consistent with their reported PTEN nullizygous status (Figure 1A, left panel). In contrast, all of the cell lines expressed similar levels of RAD51, despite their disparate PTEN genotypes. Similar effects were observed in xenografts derived from these cell lines; whereas PTEN loss was associated with increased phospho-AKT immunostaining in these xenografts, there was no differential RAD51 expression in vivo amongst this xenograft panel (Figure 1A, right panel).
Table 1. Known Genetic Aberrations in Cell Lines Used in this Study.
| 22RV1 | DU145 | PC3 | HCT116 | H1299 | |
|---|---|---|---|---|---|
| p53 Genotype | WT | MT | Null | WT | Null |
| IR-induced p21WAF1 |
Yes | No | No | Yes | No |
| IR-Induced G1/S Checkpoint |
Minimal | No | No | Intact | No |
| PTEN | WT | Heterozygous | Null | WT | WT |
| Known DNA Repair Gene Defects |
MMR (MSH2) | MMR (MLH1, PMS2, MSH2) |
None | MMR (MLH1) | |
|
Other
reported mutations (COSMIC[1]) |
PIK3CA | CDKN2A, RB1, STK11, |
PIK3CA, CDKN2A, CTNNB1, KRAS, |
NRAS |
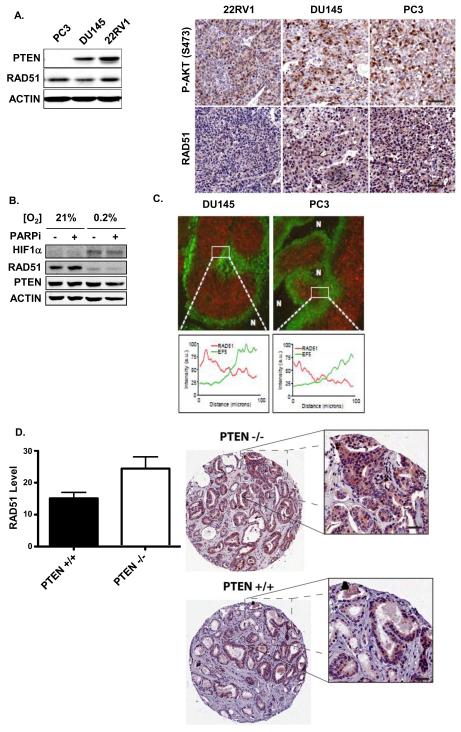
Figure 1. Effects of PTEN loss on RAD51 expression in Prostate Cancer Cells and Primary Human Prostate Tumours (A; left panel).
PTEN and RAD51 protein content was measured by Western blot of whole-cell lysates of DU145, PC3, and 22RV1 prostate cancer cells. Actin served as a loading control. (right panel) Xenografts of prostate cancer cell lines were produced as described in Materials and Methods. Sections were stained for phospho-AKT (S473) or RAD51 by immunohistochemistry. (B) DU145 cells were grown in either 21% or 0.2% O2 for 72h, and HIF1α, PTEN and RAD51 protein contents were assessed in whole-cell lysates. Actin served as a loading control. (C) (upper panel) Xenografts of prostate cancer cell lines were produced as described in Materials and Methods. Tumors were excised and stained for RAD51 by immunohistochemistry. (lower panel) Line plots of RAD51 and EF5 staining intensity within the indicated region of interest in prostate cancer xenografts. (D) A human prostate tumor tissue microarray (TMA) was produced and PTEN status confirmed by Fluoresence In-Situ Hybridization, as detailed in Materials and Methods. The TMA was stained for RAD51 by immunohistochemistry. Staining intensity was quantified in PTEN+/+ and PTEN−/− tumors (29 of each genotype) using ImageScope software. * − p<0.05
Little data are available on RAD51 expression in vivo in relation to PTEN status in which the microenvironment can alter protein expression. As we had previously shown that hypoxia can down-regulate RAD51 expression (33), we first determined the potential effects of hypoxia on RAD51 expression with PTEN-null or PTEN-intact status (34). As a positive control, culturing DU145 cells under hypoxia in 0.2% O2 for 72h markedly down-regulated RAD51, as previously reported (34, 38); however there was no concomitant effect of this hypoxic gassing on PTEN expression (Figure 1B). We also observed that RAD51 staining was inversely correlated with hypoxia (as demarcated by EF5 staining) in the tested xenografts, independent of their PTEN status (Figure 1C, lower panel). Pre-treatment with a PARP inhibitor (PARPi) did not affect either RAD51 or PTEN levels.
While PTEN loss has been linked to suppression of RAD51 expression in vitro, to our knowledge no data exist directly comparing the expression of RAD51 in primary human cancers with differential PTEN status. To that end, we stained twenty-nine PTEN+/+ and twenty-nine PTEN−/− human prostate tumors from a tissue microarray of radical prostatectomy specimens that had been evaluated for PTEN status by fluorescence in situ hybridization (FISH; Supplemental Figure 1A). Quantification of the IHC signal intensity and distribution revealed that PTEN loss was not associated with diminished RAD51 expression (Figure 1D). Indeed, we observed a significant increase in RAD51 expression in PTEN−/− tumors, relative to PTEN+/+ tumors (p<0.01). RAD51 expression was also unrelated to TMPRSS2:ERG fusion status (Supplemental Figure 1B). We also compared mRNA expression of PTEN and RAD51 using a public database from a prostate cancer cohort of 218 primary localized and metastatic tumors and xenografts following array CGH and RNA expression studies(5). In this cohort, we found no correlation between reduced PTEN and reduced RAD51 expression (Supplemental Figure 2); indeed, in no single case was PTEN loss associated with reduced RAD51 expression. Finally, in a cohort of castrate-resistant prostate cancer (CRPC), the expression of genes thought to be synthetically lethal to PARPi (RAD51, ATM, PRKDC, BRCA1, BRCA2, MRE11, CDK6, MSH2) was not correlated to PTEN or TMPRSS2:ERG status (Supplemental Figure 1C).
Effects of PTEN loss on Sensitivity to DNA Damaging Agents in Prostate Cancer Cells
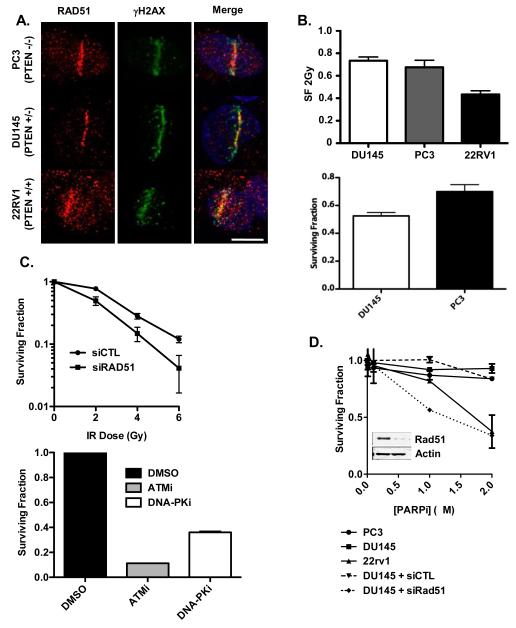
Previous reports have suggested that PTEN loss causes a defect in HR-mediated DNA DSB repair by attenuating RAD51 gene expression and recruitment to DNA DSB (9, 18). To test whether PTEN status is associated with diminished RAD51 recruitment to DSB in prostate cancer cells, we employed UV laser micro-irradiation (UVLM), which produces a sub-nuclear track of DNA DSBs that can be visualized by indirect immunofluoresence against γH2AX, and to which RAD51 is recruited (39). RAD51 was present at UVLM-induced DSB in 22RV1, DU145 and PC3 cells, demonstrating that altered PTEN status does not correlate with RAD51 recruitment to DSB in prostate cancer cells (Figure 2A).
Figure 2. Effects of PTEN loss on Sensitivity to DNA Damaging Agents in Prostate Cancer Cells.
(A) DU145, PC3, and 22RV1 prostate cancer cells were subjected to UV laser microirradiation, as indicated in Materals and Methods. Cells were stained for RAD51 and γH2AX by indirect immunofluoresence. (B; upper panel) DU145, PC3, and 22RV1 cells were treated with 2Gy of IR (or sham irradiated) and seeded for clonogenic survival assay. Surviving fraction was calculated as detailed in Materials and Methods. (lower panel) DU145 and PC3 cells were treated with 0.5 μg/ml mitomycin C (MMC) for 1h, and then seeded for clonogenic survival assay. (C; upper panel) DU145 cells were transfected with siRNA to RAD51 (or control siRNA) and treated with the indicated dose of ionizing radiation or (lower panel) pre-treated with ATMi (10 μM), or DNA-PKcsi (4 μM), and then irradiated with 2Gy of IR (or sham irradiated), and then seeded for clonogenic survival assay. (D) Clonogenic survival assays in DU145, PC3, and 22RV1 prostate cancer cells treated with PARPi. DU145 were also transfected with siRNA to RAD51 (or control siRNA) as a positive control for PARPi sensitivity. Inset shows siRNA-mediated down-regulation of RAD51 in DU145 cells. (D) 22RV1, DU145, and PC3 cells were pretreated with DMSO or PARPi (2.5 μM; 1h) and then subjected to in vitro PARP assay in the absence (upper panel) or presence (lower panel) of activated DNA.
If PTEN loss produces a defect in HR-mediated DSB repair, it may be possible to treat patients whose tumours have mono- or bi-allelic loss of PTEN with agents that preferentially kill HR-defective cells, such as ionizing radiation (IR) or mitomycin C (MMC) (34). However, PTEN status did not correlate with sensitivity to either agent (Figure 2B). By contrast, experimental down-regulation of RAD51 or treatment with ATM or DNA-PKcs inhibitors markedly sensitized cells to IR (Figure 2C).
A recent report demonstrated that loss of PTEN is associated with sensitivity to PARPi (18), as would be predicted if PTEN loss does, in fact, produce a defect in HR. However, we did not observe a correlation between PARPi sensitivity and PTEN status in prostate cancer cells, although experimental down-regulation of RAD51 markedly sensitized DU145 cells to PARPi (Figure 2D). Indeed, we found that PTEN wild-type 22RV1 cells were the most sensitive of the prostate cancer cell lines to PARPi.
Gottipati and colleagues recently demonstrated that cells with defects in HR-mediated DSB repair have elevated PARP activity and are sensitive to PARPi (24). Since PTEN status has been linked to alterations in HR, and since we observed differential PARPi sensitivity in prostate cancer cells, we next evaluated basal PARP activity in these cell lines in the absence or presence of PARPi; the assay was also performed with and without activated DNA. Absolute basal PARP activity was highest in 22RV1 cells (Supplemental Figure 3). Of note, PTEN-null PC3 cells did not show elevated basal PARP activity relative to 22RV1 or DU145 cells. In all cell lines, basal PARP activity was markedly attenuated by PARPi.
PTEN knock-out or down-regulation does not alter RAD51 expression
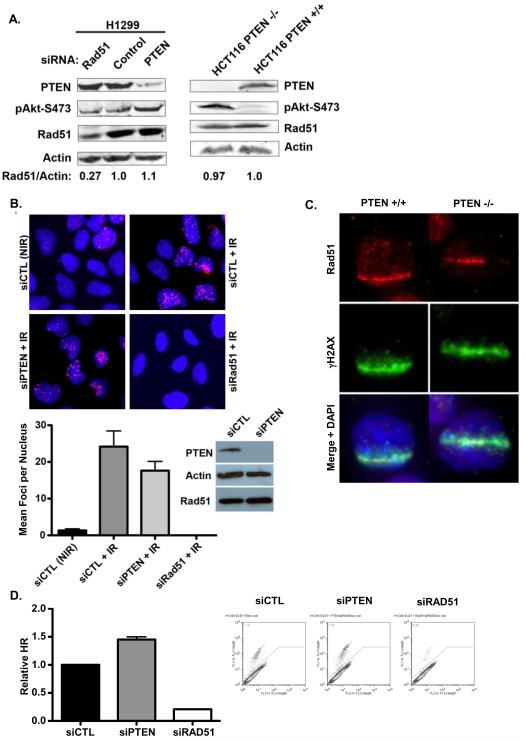
To further evaluate PTEN as a biomarker of chemosensitivity, null-TP53 H1299 cells were transfected with siRNA to RAD51 or PTEN, and expression of these proteins were evaluated by Western blotting. siRNA to RAD51 or PTEN effectively down-regulated the respective protein, and down-regulation of PTEN increased phospho-AKT content, demonstrating the functional loss of PTEN (Figure 3A, left panel). However, PTEN down-regulation did not affect RAD51 expression, consistent with the results shown in Figure 1 in prostate cancer cells. Similarly, we observed no change in RAD51 expression in HCT116 colon cancer cells in which the PTEN gene has been knocked out (HCT116-PTEN−/−), relative to the parental cells (HCT116-PTEN+/+), despite an increase in phospho-AKT content (indicative of loss of PTEN function; Figure 3A, right panel). Indeed, we observed no down-regulation of RAD51 in two independent clones of HCT116-PTEN−/− (data not shown). Moreover, we did not observe any decrease in RAD51 mRNA content in either H1299 cells transfected with PTEN siRNA or HCT116-PTEN−/− cells, relative to the respective control (Supplemental Figure 4A).
Figure 3. PTEN knock-out or down-regulation does not alter RAD51 expression (A; left panel).
H1299 cells were transfected with siRNA to RAD51 or PTEN (or control siRNA) and RAD51, PTEN, and phospho-AKT contents were assessed by Western blot. Actin was used as a loading control. (right panel) PTEN, phospho-AKT, and RAD51 contents in whole-cell lysates of PTEN knockout HCT116 cells (or wild-type control cells) (B) U2OS cells were transfected with siRNA to PTEN or control siRNA and then treated with 4Gy of IR and fixed 4h later. Down-regulation of PTEN was confirmed by Western blot. RAD51 nuclear foci were detected by immunofluoresence and siRNA to RAD51 was used to confirm the specificity of the staining. DNA was stained with DAPI (blue) (C) HCT116-PTEN+/+ or -PTEN−/− cells were subjected to UV laser microirradiation and stained for γH2AX and RAD51 by indirect immunofluoresence. DNA was stained with DAPI (blue). (D) H1299 were transfected with siRNA to RAD51 or PTEN (or control siRNA) for 48h, and HR-mediated DSB repair was assayed using the DR-GFP reporter assay, as detailed in Materials and Methods. * − p<0.05, ** − p<0.01
Shen and colleagues previously demonstrated that PTEN promotes RAD51 expression in MEFs by promoting the binding of E2F1 to the RAD51 promoter (7). To examine whether PTEN down-regulation or knockout has a similar effect in human cancer cells, we performed chromatin immunoprecipitation (ChIP) in H1299 cells transfected with siRNA to RAD51 and in HCT116-PTEN−/− cells, using an antibody against E2F1 and primers directed against the RAD51 promoter. E2F1 was bound to the RAD51 promoter in H1299 cells, and this was not affected by down-regulation of PTEN (Supplemental Figure 4B). Similarly, knockout of PTEN in HCT116 cells did not markedly affect E2F1 binding to the RAD51 promoter (Supplemental Figure 4B).
Mendes-Pereira and colleagues previously demonstrated that PTEN loss reduced the formation of DSB-induced RAD51 foci (18), although previous reports have shown that RAD51 foci are induced normally in cells lacking PTEN (40). To assess whether down-regulation of PTEN affects recruitment of RAD51 to DSB, we transfected U2OS osteosarcoma cells with siRNA to PTEN or RAD51, and treated these cells with ionizing radiation (IR; 4Gy), and then evaluated RAD51 recruitment to IR-induced DSB by immunofluoresence. As shown in Figure 3B, and similar to the effects shown above in H1299 and HCT116 cells, down-regulation of PTEN in U2OS cells did not affect RAD51 protein levels. While IR induced the formation of RAD51 nuclear foci, and while this was completely abrogated by down-regulation of RAD51, down-regulation of PTEN did not significantly affect the formation of RAD51 foci in response to IR (Figure 3B).
To further investigate whether PTEN affects RAD51 recruitment to DSB, HCT116-PTEN+/+ and HCT116-PTEN−/− cells were subjected to UVLM, to monitor RAD51 recruitment to sub-nuclear DSB. As shown in Figure 3C, RAD51 accumulation occurred in HCT116 cells lacking PTEN as well as in the parental cells.
To directly evaluate the effects of PTEN on HR-mediated DSB repair, we transfected H1299 cells expressing the DR-GFP plasmid HR reporter system (34) with siRNA to RAD51 or PTEN, and then induced DNA DSB by expression of I-SceI. Down-regulation of RAD51 produced an ~90% reduction in HR-mediated repair, without a reduction in S- and G2/M-phase cell cycle distribution (Figure 3D and Supplemental Figure 4C). In contrast, down-regulation of PTEN did not produce an HR defect, and instead produced a significant increase in HR-mediated repair in this reporter system.
Disruption of PTEN Produces a Complex Pattern of Sensitivities to DNA Damaging Agents
While our data do not support the hypothesis that PTEN status affects RAD51 expression or recruitment to DSB, or HR-mediated DSB repair, PTEN loss could still be associated with increased sensitivity to PARPi or other DNA damaging agents. If true, this would suggest that tumors with loss of PTEN may be amenable to treatment with novel agents, similar to a strategy in BRCA1/2 breast and ovarian cancer (11, 18, 25). However, in experimental isogenic models of PTEN loss, knock-down or knockout of PTEN did not increase basal PARP activity (Supplemental Figure 4D).
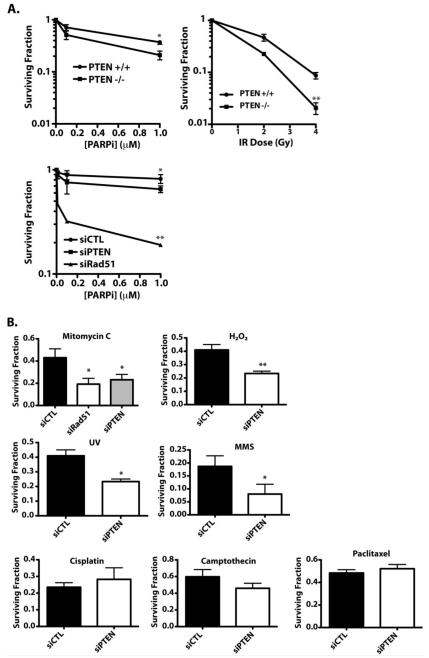
Consistent with the results of Mendes-Pereira and colleagues, PTEN knockout in HCT116 cells produced a small, but significant sensitization to PARPi (Figure 4A, upper panel, left). Similarly, transfection of PTEN siRNA in H1299 cells sensitized the cells to PARPi, albeit to a much lesser extent than the dramatic sensitization induced by down-regulation of RAD51 (Figure 4A, lower panel, left).
Figure 4. Disruption of PTEN Produces a Complex Pattern of Sensitivities to DNA Damaging Agents.
(A) HCT116-PTEN+/+ and -PTEN−/− cells (upper panels) or H1299 cells transfected with siRNA to PTEN or RAD51 (or control siRNA) (lower panel) were treated with 0-1μM PARPi or 0-6 Gy IR and cell survival was assessed by clonogenic assay. (B) H1299 cells were transfected with indicated siRNA and then treated with one of the following agents: mitomycin C (0.5 μg/ml, 1h), H2O2 (50 μM, 4h), UV (10 J/m2), MMS (0.5 mM, 4h), cisplatin (0.5 μg/ml, 24h), camptothecin (10 nM, 24h), or paclitaxel (10 nM, 4h), and cell survival was assessed by clonogenic assay.
These PTEN−/− cells have previously been shown to be sensitive to IR, relative to the parental cells (41). To validate this phenotype, we performed clonogenic survival assays using HCT116-PTEN+/+ and -PTEN−/− cells following irradiation with 0-6Gy of IR. As shown in Figure 4A (upper panel, right), HCT116 cells lacking PTEN were sensitive to IR, relative to the parental cells, consistent with the original experiments by Lee and colleagues (41). We also observed that PTEN down-regulation sensitized H1299 cells to UV irradiation and to MMC, H2O2, and methylmethanesulfonate (MMS), but did not affect sensitivity to cisplatin, camptothecin, or taxol (Figure 4B). Underscoring the heterogeneity associated with PTEN status, we found that PTEN knock-down or knockout produced a highly variable pattern of mRNA expression of genes involved in the DNA damage response (Supplemental Figure 5 and Supplemental Table 1), and did not produce a gene ‘signature’ that could be used as a surrogate marker for either PTEN loss or sensitivity to specific DNA damaging agents.
Disruption of MRE11 Sensitizes Cells to PARP Inhibition
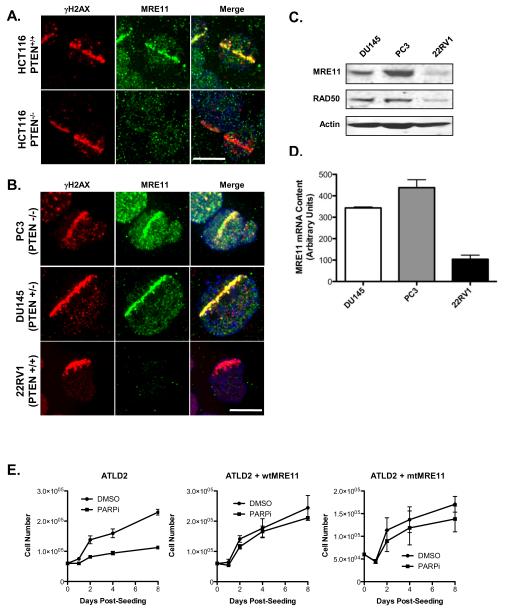
Previous reports have suggested that PARP1 is required for recruitment of MRE11 to DSB, and that PARP1 interacts with MRE11 to facilitate replication fork restart following DNA replication blocks (26, 42). These data suggest that MRE11 may be an important component of PARP1 biology. Moreover, a recent report demonstrated that PTEN knock-out HCT116 cells have a defect in MRE11 recruitment to DSB (43), consistent with our own findings (Figure 5A). For these reasons, we assessed whether MRE11 recruitment to DNA DSB is associated with altered PARPi sensitivity. We subjected 22RV1, DU145, or PC-3 cells to UVLM and monitored MRE11 and γH2AX by immunofluoresence. While MRE11 was recruited to DSB in DU145 and PC-3 cells, we did not observe any recruitment of MRE11 in 22RV-1 cells (Figure 5B), which were also the most PARPi-sensitive of the prostate cancer cells lines we evaluated (Figure 2). We confirmed that MRE11 is down-regulated in 22RV1 cells at the protein and mRNA levels (Figure 5C and 5D).
Figure 5. MRE11 Deficiency Increases Sensitivity to PARP Inhibition.
(A) HCT116-PTEN+/+ and −/− cells were subjected to UV laser microirradiation, and stained for γH2AX (red) and MRE11 (green). DNA was stained with DAPI (blue). (B) PC3, DU145, and 22RV1 cells were subjected to UVLM as in panel (A). (C) Whole-cell lysates of DU145, PC3, and 22RV1 cells were analysed by Western blot using antibodies against MRE11, RAD50, or actin. (D) MRE11 mRNA levels were analysed by quantitative RT-PCR. (E) MRE11-deficient ATLD2 fibroblasts were treated with 1 μM PARPi (or DMSO) and cell proliferation was assessed over 8 days. ATLD2 fibroblasts expressing wild-type MRE11 (ATLD2-wtMRE11) or endonuclease-deficient MRE11 (ATLD2-mtMRE11) were also analysed.
To evaluate whether MRE11 can specifically influence PARPi toxicity, we treated MRE11-deficient ATLD fibroblasts (ATLD2) with PARPi, and assessed cell proliferation over 8 days as clonogenic survival experiments were not possible on these cells due to poor plating efficiency. As a control, we performed the same experiment in ATLD fibroblasts expressing either wild-type MRE11 (ATLD2-wtMRE11) or endonuclease-deficient MRE11 (ATLD2-mtMRE11). PARPi significantly attenuated proliferation in ATLD2 fibroblasts, relative to DMSO treatment, an effect that was completely abrogated by reconstitution of either wild-type or mutant MRE11 (Figure 5E).
Discussion
Mono- and bi-allelic deletions of the PTEN gene are amongst the most frequently observed molecular aberrations in human cancer. While the vast majority of data surrounding the tumour suppressor role of PTEN pertain to its ability to antagonize the oncogenic PI3K/AKT pathway, recent reports have suggested that disruption of the PTEN gene is associated with reduction in the expression of the homologous recombination factor RAD51 (7, 18, 19). This suggested that PTEN-mediated tumorigenesis may, in part, be secondary to RAD51-associated genomic instability (7, 18). These findings are potentially of immense clinical importance because of the high frequency of PTEN loss in human tumors, and because they suggest that PTEN loss could potentially be used a biomarker for tumors that may be amenable to treatment with agents that preferentially target HR-deficient cells, including PARPi, IR, MMC or cisplatinum. Recent studies have suggested that PTEN deletion is a fairly late event during prostate cancer development (5), suggesting that genomic alterations in these tumours may occur in a PTEN-independent manner. It is likely that PTEN loss promotes cell cycle progression late in the development of prostate cancer, likely through AKT-dependent inhibition of CHK1, as previously reported (44). This may contribute to overall genomic instability, since CHK1 inhibition has been associated with the induction of DNA DSB (45).
PTEN loss was originally associated with down-regulation of RAD51 by Shen and colleagues, who suggested that PTEN promotes tumourigenesis by suppressing E2F1-dependent RAD51 gene transcription, leading to suppressed HR and genomic instability. Two subsequent reports showed that loss of PTEN is associated with RAD51 down-regulation and PARPi sensitivity (18, 19). However, several other studies are not in agreement with these findings. In particular, Gupta and colleagues (10) failed to observe an association between PTEN loss and reduced RAD51 expression, using the same cells as both Shen and Mendes-Pereira. Moreover, our laboratory previously reported that expression of RAD51 mRNA and protein in two PTEN-null prostate cancer cell lines (PC-3 and LNCaP) were not reduced, relative to PTEN+/− DU145 cells, and were in fact significantly elevated relative to normal prostate epithelial cells (30). Similar IR-induced up-regulation of RAD51 was observed in xenografts derived from PC3 and DU145 cells. These data are consistent with those shown here in Figure 1, and suggest that RAD51 expression is independent of PTEN status. In addition, independent studies from both Gospodinov and Wakasugi found that RAD51 is expressed in PC3 cells, Cummings and colleagues demonstrated the presence of cisplatin-induced RAD51 foci in PC3 cells (40, 46, 47), and Russell found normal RAD51 expression in PTEN-null U251 glioblastoma cells (48). In the current study, we observed no correlation between PTEN status and RAD51 expression or biological function in prostate cancer cell lines or xenografts. Likewise, our isogenic experimental models of PTEN loss (using siRNA or gene knockout) revealed no correlation between PTEN and RAD51 expression. These results contrast with those reported by both Shen and Mendes-Pereira and colleagues (7, 41). McEllin and colleagues (9) likewise showed no difference in RAD51 mRNA or protein expression in PTEN-null astrocytes vs. wild-type, but did observe a significant decrease in mRNA expression of the RAD51 paralogs RAD51B, C, and D, reduced HR-mediated repair, and sensitivity to MNNG, camptothecin (CPT), and a PARP inhibitor. They furthermore observed that PTEN-null astrocytes were resistant to ionizing radiation, though it is not immediately clear how this can be reconciled with an HR-deficient phenotype, which would be expected to increase radiosensitivity, as we observed in Figure 2C. In contrast, we failed to observe CPT or PARPi sensitivity in our models of PTEN loss. As such, the data presented herein are not consistent with those of Shen, Mendes-Pereira or McEllin (7, 9, 18), with respect to the effect of PTEN loss on RAD51 expression and/or HR capacity, but are consistent with both Gupta and colleagues (10) and with the broader previous literature that has failed to show a correlation between PTEN loss and RAD51 expression. It is possible that the observed disparity results from a complex interaction between PTEN and various DNA damage response and repair pathways, which may be cell and/or tissue-type specific. Nevertheless, it is clear that PTEN status cannot be used universally as a biomarker of either RAD51 expression, HR capacity, or PARPi sensitivity.
In agreement with this, we observed that PTEN status de novo is not a biomarker of RAD51 expression in patients specimens based on FISH and protein expression in 48 patients with localized disease and RNA expression in 218 patients from a recently-published prostate cancer outcomes cohort (5). RAD51 expression (as well as that of other genes previously implicated in the response to PARPi) was also not correlated with PTEN or TMPRSS2:ERG status in a cohort of castrate-resistant prostate cancers. This represents, to our knowledge, the first detailed examination of the expression of RAD51 in primary human prostate cancers with disparate PTEN status. Taken together, these data strongly argue against an association of PTEN or TMPRSS2:ERG status and RAD51 expression in primary prostate cancers.
Previous studies have evaluated the effects of PTEN on sensitivity to numerous agents, with mixed results. For instance, Kao and colleagues demonstrated that inducible reconstitution of PTEN in PTEN-null U251 cells attenuated DNA DSB repair and sensitized the cells to IR (49), suggesting that PTEN promotes radiosensitivity. By contrast, we observed that depletion of PTEN in PTEN+/+ H1299 or HCT116 cells resulted in increased sensitivity to IR, consistent with data previously reported by Lee (41). One possible explanation for these discrepant results is that cancer cells that have developed within the context of a PTEN-null genotype may function differently than cells in which PTEN has been removed experimentally, perhaps due to secondary genetic aberrations that develop due to the absence of PTEN during tumourigenesis. Our data lend support to the hypothesis that experimental down-regulation of PTEN does not phenocopy PTEN loss that occurs within the context of tumourigenesis. If this is indeed the case, it would have important ramifications for the study of PTEN biology, since it would imply that experimental models of PTEN deficiency need to be chosen very carefully in order to properly relate the findings to the potential clinical use of PTEN loss as a biomarker to stratify patients to disparate therapies.
Consistent with this hypothesis, sensitivity to IR, MMC, or PARPi in prostate cancer cells was not correlated with PTEN status, while experimental down-regulation of PTEN resulted in sensitization to all three agents. Similarly, PTEN knockout in HCT116 cells attenuated MRE11 accumulation at DNA DSB, consistent with a recent report (43), while PTEN-null PC3 cells showed no MRE11 defect. These results strongly suggest that the phenotype of prostate cancer and other cells that lack PTEN is complex and cell-specific. Moreover, these results suggest that the clinical utility of PARPi in PTEN-null tumours is likely to be highly context-specific, and suggest that further studies are needed to more completely elucidate the complex relationship between PTEN status and sensitivity to PARPi and other agents, and to identify other genes and pathways that, when altered in tumours, may confer sensitivity to PARPi.
To that end, MRE11 recruitment to DSBs was correlated to PARPi sensitivity in both prostate cancer cells and in HCT116 cells, while experimental down-regulation of MRE11 sensitized cells to PARPi. PARP1 has recently been shown to interact with MRE11 (42), while MRE11 is required for activation of the ATM kinase in response to DSB, suggesting that the sensitivity of cells with suppressed MRE11 may be secondary to a diminished ATM response, given the previously reported synthetic lethality between loss or inhibition of PARP and ATM (50). Our finding that restoration of either wild-type or endonuclease-deficient MRE11, both of which restore MRE11-dependent ATM activation (8), supports this hypothesis and the use of MRE11 function as a potential biomarker of PARPi sensitivity. Indeed, we have recently shown that AKT can bind to DSBs following MRE11-ATM activation in a PTEN-independent manner (8). As such, combining biomarkers that reflect MRE11, ATM, BRCA1/2 and AKT status in tumors may provide additional utility to individualize patient treatment strategies based on defective germline or somatic DNA repair in tumors.
Taken together, our data suggest that in prostate cancer, PTEN status is not a biomarker of RAD51 expression or biological activity, or of sensitivity to PARPi, and further suggest that PTEN loss promotes a complex pattern of sensitivity to DNA damaging agents. These findings suggest that further studies will be required to identify novel cellular and molecular biomarkers that predict clinical response to PARPi (e.g. MRE11).
Supplementary Material
Translational Statement.
Biomarkers that reflect functional DNA repair deficiencies are urgently needed to personalize cancer treatment using synthetic lethality. PARP inhibitors (PARPi) preferentially kill tumor cells with defective homologous recombination (HR) and/or DNA damage response (DDR) during the repair of DNA single-strand and double-strand breaks. The PTEN gene is lost in many prostate cancers and PTEN status has been reported to confer sensitivity to PARPi via RAD51 downregulation, which raises the possibility of individualizing treatment of PTEN-null tumours with PARPi. Using pre-clinical prostate cancer models and primary prostate cancer tissues, we show that PTEN loss does not predict HR function nor RAD51 expression as the basis for profound sensitivity to irradiation, chemotherapy or PARPi. Instead, other DDR genes, including MRE11, may confer PARPi sensitivity. PTEN status alone is unlikely to be a direct and predictive biomarker of HR or RAD51 function in prostate cancer radiotherapy, chemotherapy, or PARPi clinical trials.
Acknowledgements and Grant Support
We thank Mark O’Connor, Brad Wouters, Vuk Stambolic, and Gaetano Zafarana for helpful comments and discussion. KU-0059436 was a gift of AstraZeneca. These studies were supported by operating grants from the Terry Fox Foundation-Canadian Cancer Society Project Program, Prostate Cancer Canada, Canadian Cancer Society Research Institute and a Canadian Foundation for Innovation grant to the STTARR Innovation Facility. This research was funded in part by the Ontario Ministry of Health and Long Term Care. The views expressed do not necessarily reflect those of the Ontario Ministry of Health and Long Term Care to RGB. RGB is a Canadian Cancer Society Research Scientist. MF was funded by a Terry Fox Foundation Post-PhD Fellowship.
REFERENCES
- 1.McCall P, Witton CJ, Grimsley S, Nielsen KV, Edwards J. Is PTEN loss associated with clinical outcome measures in human prostate cancer? Br J Cancer. 2008;99:1296–301. doi: 10.1038/sj.bjc.6604680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoshimoto M, Joshua AM, Cunha IW, Coudry RA, Fonseca FP, Ludkovski O, et al. Absence of TMPRSS2:ERG fusions and PTEN losses in prostate cancer is associated with a favorable outcome. Mod Pathol. 2008;21:1451–60. doi: 10.1038/modpathol.2008.96. [DOI] [PubMed] [Google Scholar]
- 3.Bedolla R, Prihoda TJ, Kreisberg JI, Malik SN, Krishnegowda NK, Troyer DA, et al. Determining risk of biochemical recurrence in prostate cancer by immunohistochemical detection of PTEN expression and Akt activation. Clin Cancer Res. 2007;13:3860–7. doi: 10.1158/1078-0432.CCR-07-0091. [DOI] [PubMed] [Google Scholar]
- 4.Yoshimoto M, Cunha IW, Coudry RA, Fonseca FP, Torres CH, Soares FA, et al. FISH analysis of 107 prostate cancers shows that PTEN genomic deletion is associated with poor clinical outcome. Br J Cancer. 2007;97:678–85. doi: 10.1038/sj.bjc.6603924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen M, Pratt CP, Zeeman ME, Schultz N, Taylor BS, O’Neill A, et al. Identification of PHLPP1 as a Tumor Suppressor Reveals the Role of Feedback Activation in PTEN-Mutant Prostate Cancer Progression. Cancer Cell. 2011;20:173–86. doi: 10.1016/j.ccr.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen WH, Balajee AS, Wang J, Wu H, Eng C, Pandolfi PP, et al. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell. 2007;128:157–70. doi: 10.1016/j.cell.2006.11.042. [DOI] [PubMed] [Google Scholar]
- 8.Fraser M, Harding SM, Zhao H, Coackley C, Durocher D, Bristow RG. MRE11 promotes AKT phosphorylation in direct response to DNA double-strand breaks. Cell Cycle. 2011;10 doi: 10.4161/cc.10.13.16305. [DOI] [PubMed] [Google Scholar]
- 9.McEllin B, Camacho CV, Mukherjee B, Hahm B, Tomimatsu N, Bachoo RM, et al. PTEN loss compromises homologous recombination repair in astrocytes: implications for glioblastoma therapy with temozolomide or poly(ADP-ribose) polymerase inhibitors. Cancer Res. 70:5457–64. doi: 10.1158/0008-5472.CAN-09-4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta A, Yang Q, Pandita RK, Hunt CR, Xiang T, Misri S, et al. Cell cycle checkpoint defects contribute to genomic instability in PTEN deficient cells independent of DNA DSB repair. Cell Cycle. 2009;8:2198–210. doi: 10.4161/cc.8.14.8947. [DOI] [PubMed] [Google Scholar]
- 11.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–34. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 12.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–7. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 13.Helleday T, Bryant HE, Schultz N. Poly(ADP-ribose) polymerase (PARP-1) in homologous recombination and as a target for cancer therapy. Cell Cycle. 2005;4:1176–8. doi: 10.4161/cc.4.9.2031. [DOI] [PubMed] [Google Scholar]
- 14.Lord CJ, McDonald S, Swift S, Turner NC, Ashworth A. A high-throughput RNA interference screen for DNA repair determinants of PARP inhibitor sensitivity. DNA Repair (Amst) 2008;7:2010–9. doi: 10.1016/j.dnarep.2008.08.014. [DOI] [PubMed] [Google Scholar]
- 15.Turner NC, Lord CJ, Iorns E, Brough R, Swift S, Elliott R, et al. A synthetic lethal siRNA screen identifying genes mediating sensitivity to a PARP inhibitor. EMBO J. 2008;27:1368–77. doi: 10.1038/emboj.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dent RA, Bristow RG. In situ DNA repair assays as guides to personalized breast cancer chemotherapeutics: ready for prime time? J Clin Oncol. 2011;29:2130–2. doi: 10.1200/JCO.2010.34.1990. [DOI] [PubMed] [Google Scholar]
- 17.Guha M. PARP inhibitors stumble in breast cancer. Nat Biotechnol. 2011;29:373–4. doi: 10.1038/nbt0511-373. [DOI] [PubMed] [Google Scholar]
- 18.Mendes-Pereira AM, Martin SA, Brough R, McCarthy A, Taylor JR, Kim JS, et al. Synthetic lethal targeting of PTEN mutant cells with PARP inhibitors. EMBO Mol Med. 2009;1:315–22. doi: 10.1002/emmm.200900041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dedes KJ, Wetterskog D, Mendes-Pereira AM, Natrajan R, Lambros MB, Geyer FC, et al. PTEN deficiency in endometrioid endometrial adenocarcinomas predicts sensitivity to PARP inhibitors. Sci Transl Med. 2010;2:53ra75. doi: 10.1126/scitranslmed.3001538. [DOI] [PubMed] [Google Scholar]
- 20.Forster MD, Dedes KJ, Sandhu S, Frentzas S, Kristeleit R, Ashworth A, et al. Treatment with olaparib in a patient with PTEN-deficient endometrioid endometrial cancer. Nat Rev Clin Oncol. 2011;8:302–6. doi: 10.1038/nrclinonc.2011.42. [DOI] [PubMed] [Google Scholar]
- 21.Vesprini D, Narod SA, Trachtenberg J, Crook J, Jalali F, Preiner J, et al. The therapeutic ratio is preserved for radiotherapy or cisplatin treatment in BRCA2-mutated prostate cancers. Can Urol Assoc J. 2011;5:E31–5. doi: 10.5489/cuaj.10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chalmers AJ, Lakshman M, Chan N, Bristow RG. Poly(ADP-ribose) polymerase inhibition as a model for synthetic lethality in developing radiation oncology targets. Semin Radiat Oncol. 2010;20:274–81. doi: 10.1016/j.semradonc.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 23.Liu SK, Coackley C, Krause M, Jalali F, Chan N, Bristow RG. A novel poly(ADP-ribose) polymerase inhibitor, ABT-888, radiosensitizes malignant human cell lines under hypoxia. Radiother Oncol. 2008;88:258–68. doi: 10.1016/j.radonc.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 24.Gottipati P, Vischioni B, Schultz N, Solomons J, Bryant HE, Djureinovic T, et al. Poly(ADP-ribose) polymerase is hyperactivated in homologous recombination-defective cells. Cancer Res. 70:5389–98. doi: 10.1158/0008-5472.CAN-09-4716. [DOI] [PubMed] [Google Scholar]
- 25.Fong PC, Yap TA, Boss DS, Carden CP, Mergui-Roelvink M, Gourley C, et al. Poly(ADP)-Ribose Polymerase Inhibition: Frequent Durable Responses in BRCA Carrier Ovarian Cancer Correlating With Platinum-Free Interval. J Clin Oncol. doi: 10.1200/JCO.2009.26.9589. [DOI] [PubMed] [Google Scholar]
- 26.Dedes KJ, Wilkerson PM, Wetterskog D, Weigelt B, Ashworth A, Reis-Filho JS. Synthetic lethality of PARP inhibition in cancers lacking BRCA1 and BRCA2 mutations. Cell Cycle. 2011;10:1192–9. doi: 10.4161/cc.10.8.15273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brenner JC, Ateeq B, Li Y, Yocum AK, Cao Q, Asangani IA, et al. Mechanistic Rationale for Inhibition of Poly(ADP-Ribose) Polymerase in ETS Gene Fusion-Positive Prostate Cancer. Cancer Cell. 2011;19:664–78. doi: 10.1016/j.ccr.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sebastian de Bono J, Sandhu S, Attard G. Beyond hormone therapy for prostate cancer with PARP inhibitors. Cancer Cell. 2011;19:573–4. doi: 10.1016/j.ccr.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 29.Narod SA, Neuhausen S, Vichodez G, Armel S, Lynch HT, Ghadirian P, et al. Rapid progression of prostate cancer in men with a BRCA2 mutation. Br J Cancer. 2008;99:371–4. doi: 10.1038/sj.bjc.6604453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fan R, Kumaravel TS, Jalali F, Marrano P, Squire JA, Bristow RG. Defective DNA strand break repair after DNA damage in prostate cancer cells: implications for genetic instability and prostate cancer progression. Cancer Res. 2004;64:8526–33. doi: 10.1158/0008-5472.CAN-04-1601. [DOI] [PubMed] [Google Scholar]
- 31.Meng AX, Jalali F, Cuddihy A, Chan N, Bindra RS, Glazer PM, et al. Hypoxia down-regulates DNA double strand break repair gene expression in prostate cancer cells. Radiother Oncol. 2005;76:168–76. doi: 10.1016/j.radonc.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 32.Mertz KD, Setlur SR, Dhanasekaran SM, Demichelis F, Perner S, Tomlins S, et al. Molecular characterization of TMPRSS2-ERG gene fusion in the NCI-H660 prostate cancer cell line: a new perspective for an old model. Neoplasia. 2007;9:200–6. doi: 10.1593/neo.07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan N, Pires IM, Bencokova Z, Coackley C, Luoto KR, Bhogal N, et al. Contextual synthetic lethality of cancer cell kill based on the tumor microenvironment. Cancer Res. 2010;70:8045–54. doi: 10.1158/0008-5472.CAN-10-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan N, Koritzinsky M, Zhao H, Bindra R, Glazer PM, Powell S, et al. Chronic hypoxia decreases synthesis of homologous recombination proteins to offset chemoresistance and radioresistance. Cancer Res. 2008;68:605–14. doi: 10.1158/0008-5472.CAN-07-5472. [DOI] [PubMed] [Google Scholar]
- 35.Choudhury A, Zhao H, Jalali F, Al Rashid S, Ran J, Supiot S, et al. Targeting homologous recombination using imatinib results in enhanced tumor cell chemosensitivity and radiosensitivity. Mol Cancer Ther. 2009;8:203–13. doi: 10.1158/1535-7163.MCT-08-0959. [DOI] [PubMed] [Google Scholar]
- 36.Rickman DS, Chen YB, Banerjee S, Pan Y, Yu J, Vuong T, et al. ERG cooperates with androgen receptor in regulating trefoil factor 3 in prostate cancer disease progression. Neoplasia. 2010;12:1031–40. doi: 10.1593/neo.10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lazaro-Trueba I, Arias C, Silva A. Double bolt regulation of Rad51 by p53: a role for transcriptional repression. Cell Cycle. 2006;5:1062–5. doi: 10.4161/cc.5.10.2764. [DOI] [PubMed] [Google Scholar]
- 38.Bindra RS, Schaffer PJ, Meng A, Woo J, Maseide K, Roth ME, et al. Down-regulation of Rad51 and decreased homologous recombination in hypoxic cancer cells. Mol Cell Biol. 2004;24:8504–18. doi: 10.1128/MCB.24.19.8504-8518.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan MB, Bartek J, et al. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J Cell Biol. 2006;173:195–206. doi: 10.1083/jcb.200510130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gospodinov A, Tsaneva I, Anachkova B. RAD51 foci formation in response to DNA damage is modulated by TIP49. Int J Biochem Cell Biol. 2009;41:925–33. doi: 10.1016/j.biocel.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 41.Lee C, Kim JS, Waldman T. PTEN gene targeting reveals a radiation-induced size checkpoint in human cancer cells. Cancer Res. 2004;64:6906–14. doi: 10.1158/0008-5472.CAN-04-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bryant HE, Petermann E, Schultz N, Jemth AS, Loseva O, Issaeva N, et al. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 2009;28:2601–15. doi: 10.1038/emboj.2009.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu N, Hegarat N, Black EJ, Scott MT, Hochegger H, Gillespie DA. Akt/PKB suppresses DNA damage processing and checkpoint activation in late G2. J Cell Biol. 2010;190:297–305. doi: 10.1083/jcb.201003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Puc J, Keniry M, Li HS, Pandita TK, Choudhury AD, Memeo L, et al. Lack of PTEN sequesters CHK1 and initiates genetic instability. Cancer Cell. 2005;7:193–204. doi: 10.1016/j.ccr.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 45.Puc J, Parsons R. PTEN loss inhibits CHK1 to cause double stranded-DNA breaks in cells. Cell Cycle. 2005;4:927–9. doi: 10.4161/cc.4.7.1795. [DOI] [PubMed] [Google Scholar]
- 46.Cummings M, Higginbottom K, McGurk CJ, Wong OG, Koberle B, Oliver RT, et al. XPA versus ERCC1 as chemosensitising agents to cisplatin and mitomycin C in prostate cancer cells: role of ERCC1 in homologous recombination repair. Biochem Pharmacol. 2006;72:166–75. doi: 10.1016/j.bcp.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 47.Wakasugi T, Izumi H, Uchiumi T, Suzuki H, Arao T, Nishio K, et al. ZNF143 interacts with p73 and is involved in cisplatin resistance through the transcriptional regulation of DNA repair genes. Oncogene. 2007;26:5194–203. doi: 10.1038/sj.onc.1210326. [DOI] [PubMed] [Google Scholar]
- 48.Russell JS, Brady K, Burgan WE, Cerra MA, Oswald KA, Camphausen K, et al. Gleevec-mediated inhibition of Rad51 expression and enhancement of tumor cell radiosensitivity. Cancer Res. 2003;63:7377–83. [PubMed] [Google Scholar]
- 49.Kao GD, Jiang Z, Fernandes AM, Gupta AK, Maity A. Inhibition of phosphatidylinositol-3-OH kinase/Akt signaling impairs DNA repair in glioblastoma cells following ionizing radiation. J Biol Chem. 2007;282:21206–12. doi: 10.1074/jbc.M703042200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bryant HE, Helleday T. Inhibition of poly (ADP-ribose) polymerase activates ATM which is required for subsequent homologous recombination repair. Nucleic Acids Res. 2006;34:1685–91. doi: 10.1093/nar/gkl108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.