Abstract
Intracellular proteins are degraded by a number of proteases including the ubiquitin-proteasome pathway (UPP). Impairments in the UPP occur during the aging of a variety of tissues, although little is known in regards to age-related alterations to the UPP during the aging of adipose tissue. The UPP is known to be involved in regulating the differentiation of a variety of cell types, although the potential changes in UPP during adipose differentiation have not been fully elucidated. Simultaneously elucidating how the UPP is altered in aging adipose tissue and adipocyte differentiation, and determining the effects of proteasome inhibition on adipocyte homeostasis and differentiation, are critical issues to elucidate experimentally. Adipogenesis continues throughout the life of adipose tissue, with continual differentiation of pre-adipocytes essential to maintaining tissue function during aging, while UPP alterations in mature adipocytes is likely to directly modulate adipose function during aging. In the current study we demonstrate that aging induces alterations in the activity and expression of principal components of the UPP. Additionally, we show that multiple changes in the UPP occur during the differentiation of 3T3-L1 cells into adipocytes. In vitro data link observed UPP alterations to increased levels of oxidative stress and altered adipose biology relevant to both aging and differentiation. Taken together, these data demonstrate that changes in the UPP occur in response to adipose aging and adipogenesis, and strongly suggest that proteasome inhibition is sufficient to decrease adipose differentiation, as well as increase oxidative stress in mature adipocytes, both of which likely promote deleterious effects on adipose aging.
Keywords: adipocyte, aging, insulin resistance, obesity, proteolysis, ubiquitin, pathology
Introduction
Proteasome is a large multisubunit complex, responsible for majority of intracellular proteolysis, degrading proteins in an ATP-dependent pathway as well as an ATP-independent pathway (1–2). The activity of the proteasome is mediated by the coordinated functions of the 20S (ATP-independent) and 26S (ATP-dependent) proteasome complexes (1,3–7), with the activity of these two complexes measured by taking advantage of the differences in molecular weight, and the ability of complex specific co-activators to stimulate proteasome activities. The ubiquitin-proteasome pathway (UPP) has a well established role in a number of physiological and pathological processes (8–10). Numerous studies have linked proteasome-dependent proteolysis to the degradation of oxidized proteins (6, 11–13), identifying the proteasome as a modulator of oxidative stress in a variety of settings.
The UPP is present in all cells of the body, and plays a vital role in regulating cellular homeostasis (14–17), with proteasome inhibition known to be inhibited in a number of tissues with aging (8,18–19). The biological importance of these findings is highlighted by reports that proteasome inhibition is sufficient to mimic multiple aspects of aging (8,19–20). In addition to aging, the proteasome is known to be important in the process of cellular differentiation (21–24).
Adipose tissue is known to undergo significant changes during aging including changes in distribution (25–26), function (27–29), and differentiation (25, 30). Because of the increasing levels of adiposity in Western societies around the world, and the acceleration in the number of elderly around the world, there is an increasing urgency for understanding the interactions between adiposity and aging. This is highlighted by the numerous studies demonstrating the links between adiposity and the development of metabolic disease and a host of morbidities. Currently, little is known in regards to whether aging alters the UPP in adipose tissue, or whether the UPP is altered during the differentiation of adipocytes. In the present study we demonstrate that the UPP undergoes dramatic changes in response to both aging and adipose differentiation, with proteasome inhibition inhibiting adipocyte differentiation and promoting oxidative stress in mature adipocytes. The importance of these findings to the biology and pathophysiology of adipose tissue aging is discussed.
Materials and Methods
Materials
The substrates Boc-Leu-Arg-Arg-AMC (PGPH-L activity) and Suc-Leu-Leu-Val-Tyr-AMC (ChT-L activity) were purchased from Bachem America’s Inc., (Torrance, CA). The substrate Boc-Leu-Ser-Thr-Arg-7-Amido-4-methylcoumarin (T-L activity) was purchased from Sigma Aldrich Company (St Louis, MO). The antibodies used in this study were antibodies against β-actin (SC-47778) and ubiquitin (SC -8017) (Santa Cruz, CA, USA), Alpha 7 or C8 (Clone MCP72) subunit 20S proteasome-mouse mAb, 19S regulator ATPase subunit Rpt3 (S6b,TBP7) -mouse mAb, Subunit β5 for 20S proteasome-Rbt, 11S regulator subunit PA28 alpha rabbit polyclonal antibody (BIOMOL International, Plymouth Meeting, PA, USA). Antibodies against PPARγ and c/EBPβ were purchased from Cell Signaling Technology, Incorporated (Danvers, MA). MG132 was purchased from EMD chemicals (Gibbstown, NJ). All the HRP conjugated secondary antibodies were purchased from Vector Laboratories, USA. All electrophoresis and immunoblot reagents were purchased from Bio-Rad Laboratories (Hercules, CA, USA). Dulbecco's modified Eagle's medium containing high glucose and glutamine (DMEM, Cat. No: 10-017-CV), Fetal bovine serum (FBS-SH30071), calf serum (SH3011803), trypsin (Cat No: SH 30236.02) and antibiotics penicillin G/streptomycin (16-700-49) were purchased from Fisher Scientifics (Pittsburgh, PA). All other items and chemicals including Insulin (I1882), 3-isobutyl-1-methylxanthine (IBMX-I5879), dexamethasone (D4902), Oil Red O stain and MTT were purchased from Sigma-Aldrich Corp (St. Louis, MO, USA).
Animal tissues
All animal experiments were approved by the IACUC of Pennington Biomedical Research Center. Animals at 2 month, 12 month and 22 month of age were used in the present study. Animals were male and Ad Libitum chow fed C57Bl/6 mice obtained from the contract colony of the National Institute on Aging maintained by Charles River Laboratories. Animals were fasted overnight and euthanatized by isoflurane anesthesia followed by rapid decapitation. Tissues were manually dissected and frozen at −80°C until further use.
Adipocyte differentiation
Murine 3T3-L1 preadipocytes purchased from the American Type Culture Collection (Cat. No: CL-173, Manassas, VA) were cultured in DMEM high glucose containing 10% calf serum and antibiotics (100 units/ml penicillin G and 100 µg/ml streptomycin). The medium was changed every 48 h. To obtain fully differentiated adipocytes, the 3T3-L1 preadipocytes were plated and grown in 6 well plates to 1 day post confluence and induced to differentiate by changing the medium to DMEM containing 10% FBS and 0.5 mM IBMX, 1 µM dexamethasone and 1.7 µM insulin (MDI) as previously described with some modifications (31,32). After 48 hours, medium was replaced with DMEM high glucose supplemented with 10% FBS, pen/strep and 0.425 µM insulin. Thereafter medium was replaced every 2 days with DMEM high glucose medium containing 10% FBS. Cells were used at 3, 6, 9 and 12 days after induction of differentiation for experiments. The extent of differentiation for adipocyte cultures in the present study is indicated by days, which refers to days post induction with MDI medium.
To see the effect of MG 132 on lipid accumulation in 3T3L1 adipocytes, 3T3L1 fibroblast differentiation in to adipocytes was induced as described above with the addition of indicated amounts of MG132 to the medium for 7 days. Fresh media with vehicle or MG132 was administered daily. The lipid accumulation in the 7th day cells after induction of differentiation were observed by staining the cells with Oil Red O stain.
Oil Red O staining
Oil Red O staining was performed to visualize the lipid accumulation in differentiating adipocytes following treatment with proteasome inhibitor, MG132 for 15hours, as described previously (31) with some modifications. Cells were gently washed with PBS and fixed in 10% of formalin for 1 hour. 0.5% Oil Red O stock solution was prepared in 99% Isopropanol and then the working solution was made by mixing 4 parts of Oil Red O stock solution to 1 part of water. After 1 hour incubation of cells in formalin, 60% isopropanol was added to each well and incubated for 5 minutes. After 5 minutes, 60% isopropanol was removed and then the cells were incubated with filtered Oil Red O working solution for one hour at room temperature. After that, cells were washed gently with distilled water to clear the background and analyzed under microscope.
Analysis of proteasome activities
Assays for proteasome activity were performed as described previously by our laboratory (33) using the fluorogenic substrates for Post Glutamyl Peptidase like (PGPH-L), Chymotrypsin like (ChT-L), and Trypsin like (T-L) activities. The reaction was conducted in 250 µl of activity assay buffer in a 96 well black assay plate with clear bottom containing 50 µg/ml of protein lysate, 20mM Tris HCl, pH 7.8, 1 mM EDTA, 0.5 mM DTT and 5mM MgCl2, 2mM ATP and 50 µM of the corresponding substrate. The reaction mixture was incubated for 1 hour at 37°C and the fluorescence of the released AMC product was measured in Molecular Devices plate reader at an emission wavelength of 355 nm and an excitation wavelength of 460 nm. The background fluorescence values obtained by incubating the lysates with MG132 were subtracted from activity values as described previously by our laboratory (33, 34). The proteasome activity per mg of the protein per hour was calculated from the fluorescence values and all subsequent data expressed as percent control values.
Western blot analysis
The protein samples were analyzed by SDS-PAGE and immunoblotted with specified antibodies as described previously by our laboratory (33).
Analysis of oxidized proteins
4-Hydroxynonenal (HNE) modified proteins in cell or tissue lysates were analyzed by western blot analysis. Antibody against HNE and the following working protocol was kindly provided by Dr Luke Szweda (Oklahoma Medical Research Foundation). Briefly, the proteins were transferred on to the PVDF membrane and the membrane was incubated in freshly made solution containing 250 mM sodium borohydride in 100 mM MOPS, pH 8.0 for 15 minutes, which was done to chemically reduce the adduct for antibody recognition. After 15 minutes of incubation the membrane was washed for 3 times with water and another 3 times with PBS, with 5 minutes used for each washing step. The membrane was blocked in 5% milk for 1 hour and then incubated overnight with 1:2000 dilution of HNE antibody in 5% milk. The membrane was developed using ECL reagent. Protein carbonyl levels were analyzed using Oxyblot kit (Millipore) as described by the manufacturer. Briefly, cell or tissue lysates were derivatized with DNP and then the derivatized products were detected by the Western blot analysis as described by the manufacturer.
Results
Analysis of 20S and 26S proteasome activity in aging adipose
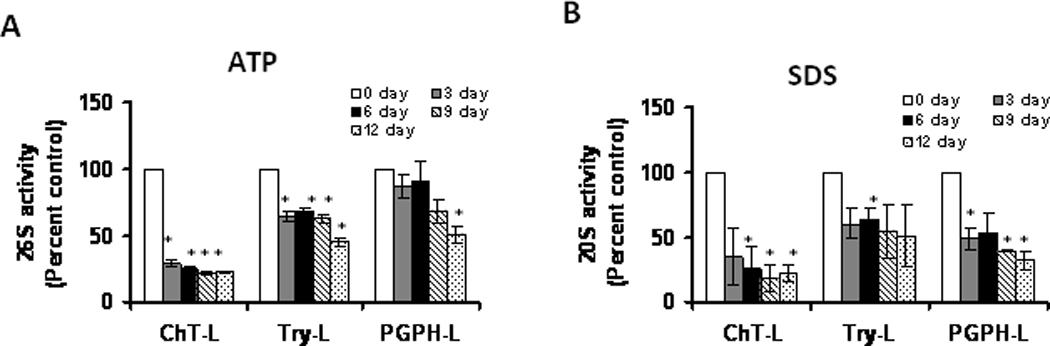
In our first set of studies we sought to determine how the relative levels of 20S and 26S proteasome activity are altered during aging. These studies used adipose tissue collected from 2-, 12- and 22-month old male C57Bl/6 mice. In these studies we observed that the 3 major proteolytic activities of the 26S proteasome decreased during adipose aging (Figure 1A). Similarly, the 20S activities of the proteasome were decreased during adipose aging (Figure 1B). Interestingly, Post Glutamyl Peptidase like (PGPH-L) activities of the 20S and 26S proteasome were the least affected by aging (Figure 1), with chymotrypsin-like (ChT-L) activity of the 20S proteasome the most inhibited during adipose aging. These data indicate that there are gross impairments in the 20S and 26S proteasome complexes in adipose tissue with aging.
Figure 1. Proteasome activity decreases with age in adipose tissue.
Adipose tissue lysates were assayed for (A) 26S proteasome (+ATP) and for (B) 20S proteasome (+0.02% SDS) activity as described in methods section. Results represent the mean of 5 sets of experiments (n =5), where each animal represents an n =1, conducted under identical experimental conditions. 2M: 2 month ; 12M: 12 month ;22M: 22 month; AL: Ad Libitum. *P=0.05 when compared with 2M AL.
Effects of aging on proteasome subunit expression
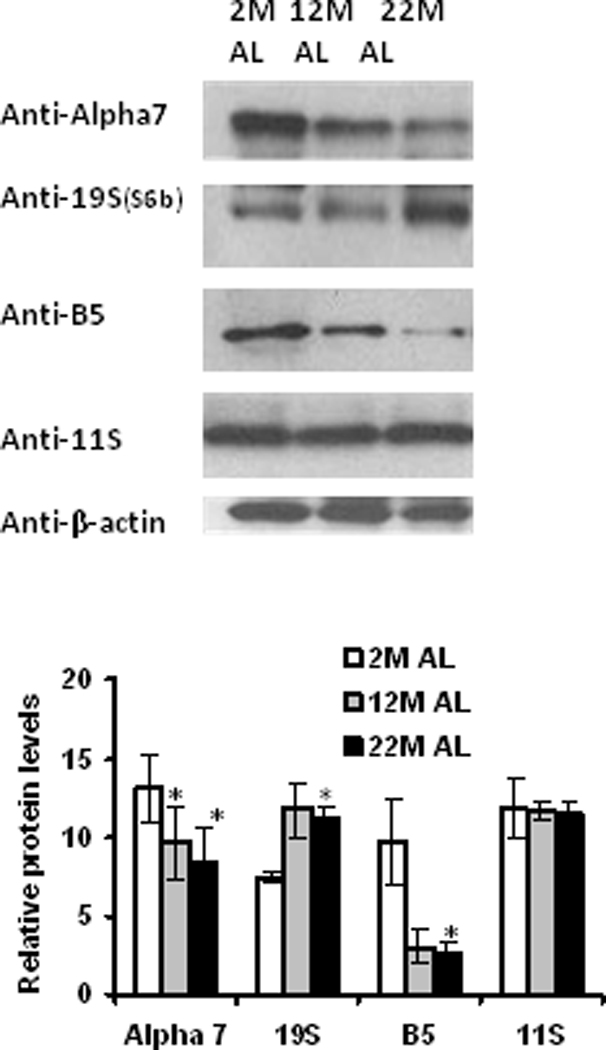
Aging significantly decreased alpha-7 and beta-5 proteasome subunit expression in fat adipose tissue (Figure 2). These data suggest that both non-catalytic (alpha) and catalytic (beta) subunits of the core proteolytic complex exhibit decreased expression during aging. Aging increased the levels of 19S subunit expression in adipose tissue (Figure 2). No significant changes in 11S subunits of the proteasome complex were observed during aging (Figure 2). These data are consistent with 26S cap-associated proteins of the proteasome being differentially impacted by aging in adipose tissue.
Figure 2. Proteasome subunit expression is altered with aging in adipose tissue.
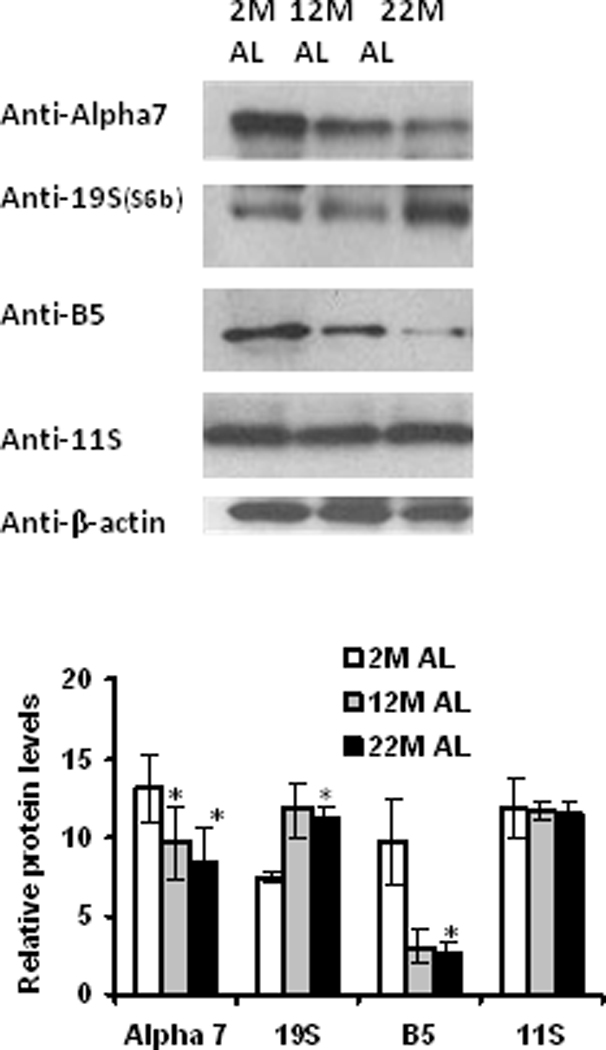
Adipose tissue lysates were analyzed for the proteasome subunit expression using Western blot analysis. Results represent the mean of 5 sets (n =5) of experiments, where each animal represents an n =1, conducted under identical experimental conditions. 2M: 2 month; 12M: 12 month; 22M: 22 month; AL: Ad Libitum. *P=0.05 respectively, when compared with 2M AL.
Effects of aging on levels of ubiquitinated and oxidized proteins
In order to understand the influence of aging on ubiquitinated protein levels, we analyzed the levels of ubiquitinated proteins in the same tissues used for measurements of proteasome activity and expression. In these studies we surprisingly observed that even though proteasome activity was decreased with aging, the levels of ubiquitinated protein did not significantly increase in response to aging (Figure 3A). Studies then identified that these same samples exhibited a significant age-related increase in both oxidized proteins (Figure 3B) as well as 4-hydroxynonenal (HNE) conjugated proteins (Figure 3C). These data are consistent with the presence of increased oxidative stress in adipose tissue during aging.
Figure 3. Changes in ubiquitinated and oxidized protein levels with aging in adipose tissue.
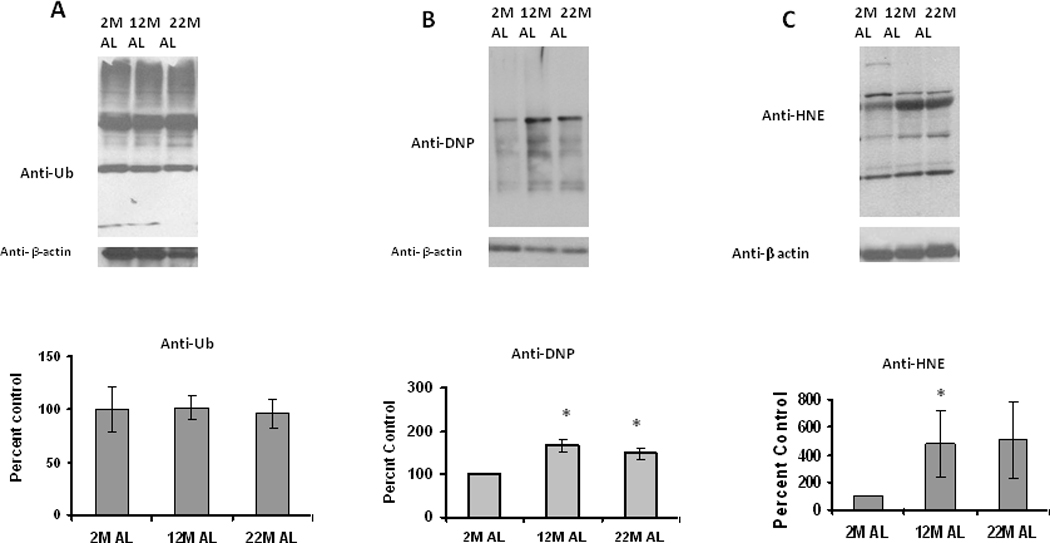
Adipose tissue lysates were analyzed for (A) ubiquitinated, (B) oxidized and (C) HNE (4-hydroxy-2-nonenal) modified proteins, using Western blot analysis. Results represent the mean of 5 sets (n=5) of experiments, where each animal represents an n =1, conducted under identical experimental conditions. 2M: 2 month; 12M: 12 month; 22M: 22month; AL: Ad Libitum. *P=0.05 when compared with 2M AL.
Changes in proteasome activities during adipose differentiation
Pre-adipocytes within adipose tissue continually differentiate, and this development of new adipocytes is critical to maintain adipose tissue homeostasis (28,29). Understanding the changes in proteasome activity that occur during adipocyte differentiation are important to elucidate, since they could contribute to development of proteasome inhibition during aging. In this experimentation we sought to elucidate how individual 20S and 26S proteasome activities were altered during adipose differentiation. In these studies we observed that the levels of chymotrypsin (Ch-L), trypsin (T-L) and Post Glutamyl Peptidase like (PGPH-L) activities of the 26S proteasome were decreased during adipocyte differentiation (Figure 4A). ChT-L activity of the 26S proteasome was the most effected, undergoing significant decreases starting at day 3 differentiation (Figure 4A). In contrast to the sustained inhibition of 26S ChT-L and T-L activity, the activity of 26S PGPH-L was not significantly inhibited until day 12 of differentiation (Figure 4A). Initial decreases in 20S T-L activity were transient and not significantly different than control values by day 9 of adipocytes differentiation. The 20S ChT-L and PGPH-L activities were both inhibited at day 9 and 12 of adipocyte differentiation (Figure 4B). These data indicate that while not identical to aging, differentiation of cells into adipocytes is associated with gross impairments in multiple proteasome activities.
Figure 4. Proteasome activity changes during adipocyte differentiation.
Lysates of 3T3L1 adipocytes following 3, 6, 9 and 12 day of differentiation were assayed for (A) 26S proteasome (+ATP) and for (B) 20S proteasome (+0.02% SDS) as described in methods section. Results represent the mean of 5 sets (n=5) of experiments), where each animal represents an n =1, conducted under identical experimental conditions. *P=0.05 when compared with 3T3 L1 preadipocytes, which was taken as percent control in the represented graphs.
Changes in proteasome subunit expression during adipose differentiation
Similar to aging, the levels of beta-5 expression were decreased with increasing differentiation (Figure 5). The alpha-7 subunit, in contrast to aging, did not undergo any significant decline in expression (Figure 5). No significant or consistent change was observed with the proteasome cap protein S6b (Figure 5), even though the levels of the cap protein 11S significantly increased with adipose differentiation (Figure 5A). These data identify the complex nature of proteasome subunit changes which accompany adipocyte differentiation.
Figure 5. Changes in proteasome subunit protein expression during adipocyte differentiation.
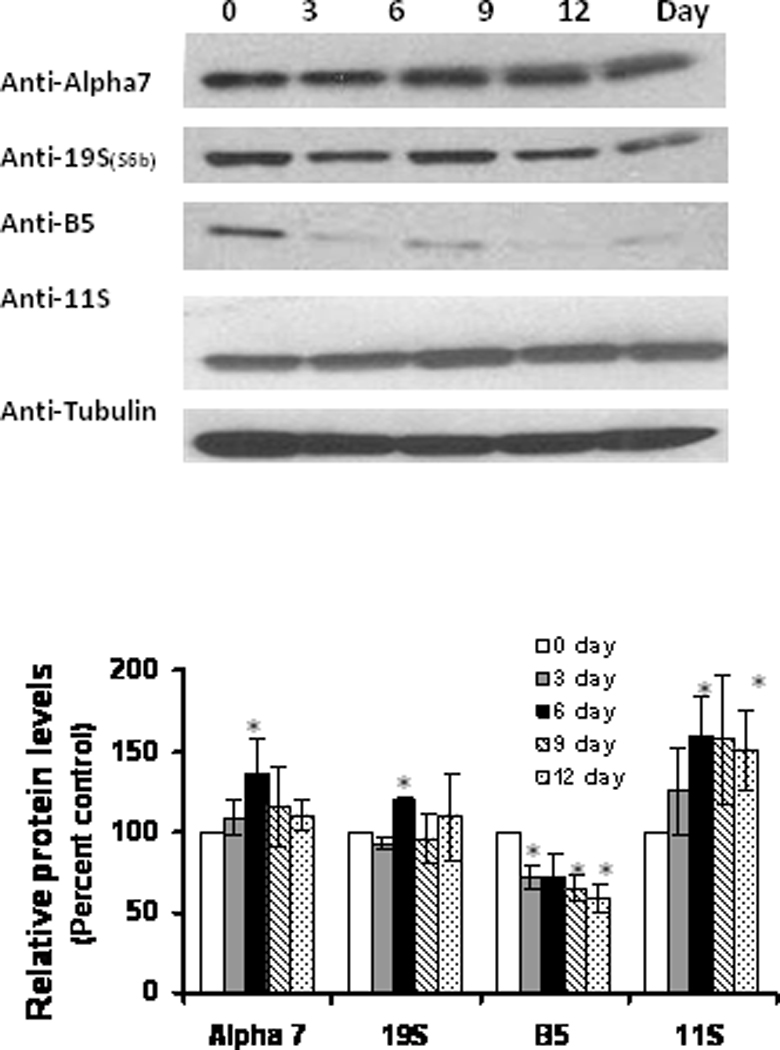
Lysates of 3 T3L1 adipocytes following 3, 6, 9 and 12 day of differentiation were analyzed for proteasome subunit protein levels, using western blot analysis. Results represent the mean of 5 sets (n=5) of experiments, where each cell culture dish represents an n =1, conducted under identical experimental conditions. *P=0.05 when compared with 3T3 L1 preadipocytes, which was taken as percent control in the represented graphs.
Effects of differentiation on levels of ubiquitinated and oxidized proteins
Differentiation did not significantly alter the levels of ubiquitinated protein (Figure 6A) in adipose cells, similar to what was observed during aging. Differentiation of adipocytes produced a significant and transient increase in the amount of oxidized proteins (Figure 6B), while at the same time inducing a sustained increase in HNE-modified proteins (Figure 6C), consistent with the presence of oxidative stress during differentiation.
Figure 6. Changes in ubiquitinated and oxidized protein levels during adipocyte differentiation.
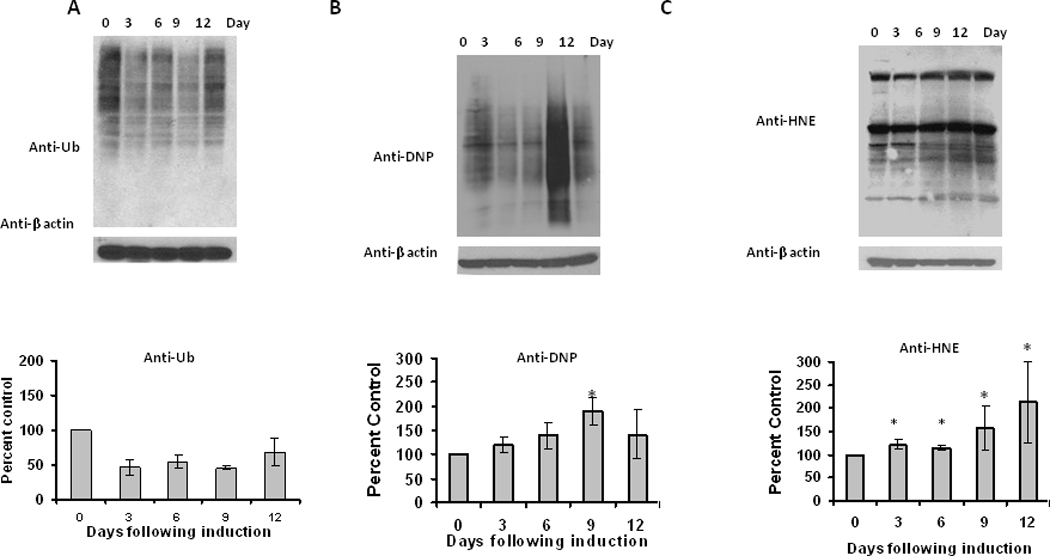
Lysates of 3 T3L1 adipocytes were analyzed for (A) ubiquitinated, (B) oxidized and (C) HNE (4-hydroxy-2-nonenal) modified proteins, using Western blot analysis. Results represent the mean of 5 sets (n=5) of experiments, where each cell culture dish represents an n =1, conducted under identical experimental conditions. *P=0.05 when compared with 3T3 L1 preadipocytes.
Effect of acute proteasome inhibition on mature differentiated adipocytes
Next we analyzed the acute effects of the proteasome inhibitor MG132 on survival of mature differentiated adipocytes. Oil Red O staining with adipocytes following MG132 treatment showed a decrease in the lipid content with increasing amounts of MG132 (Figure 7). Following administration of proteasome inhibitors there was also a dose-dependent increase in ubiquitinated, oxidized, and HNE-modified proteins (Figure 8). These data are consistent with proteasome inhibition in mature adipocytes being sufficient to promote decreased lipid content, and increased oxidative stress.
Figure 7. Effect of acute proteasome inhibition on lipid accumulation of differentiated 3T3-L1 adipocytes.
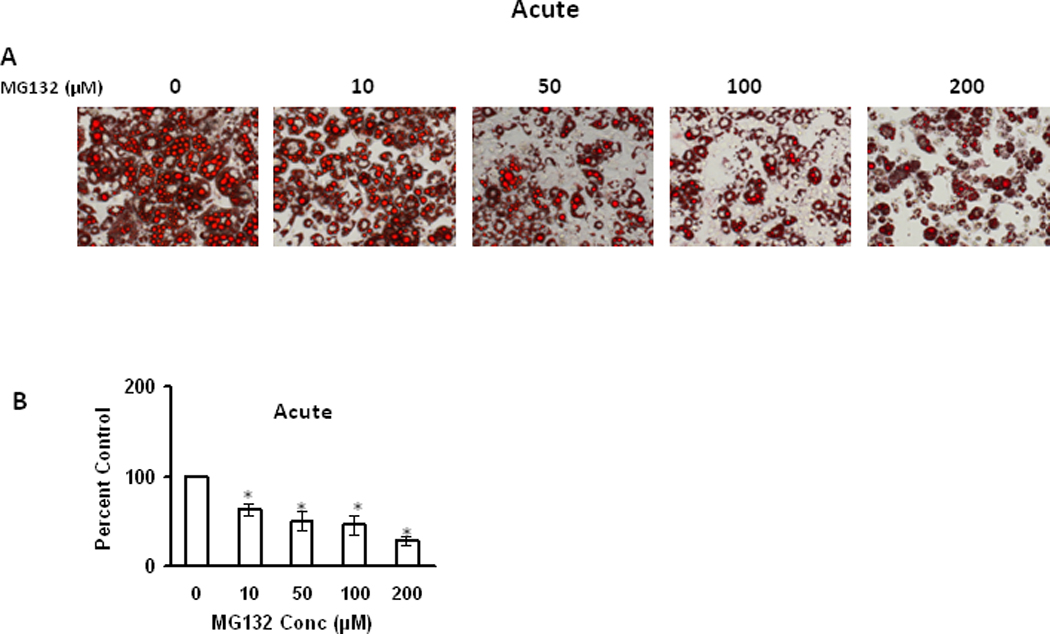
9 day (post induction) differentiated adipocytes were treated with different concentrations of MG132 for 15 hours and then analyzed by Oil Red O staining to visualize the effect of MG132 on lipid accumulation (A) and Oil red O stained lipid amount was quantified by measuring the absorbance at 570nm after extraction with ethanol (B). Results represent the mean of 5 sets (n=5) of experiments, where each culture dish represents an n =1, conducted under identical experimental conditions. *P=0.05 when compared with untreated 3T3 L1 adipocytes, which was taken as percent control in the represented graphs.
Figure 8. Effect of proteasome inhibition on ubiquitinated and oxidized protein levels in differentiated 3T3-L1 adipocytes.
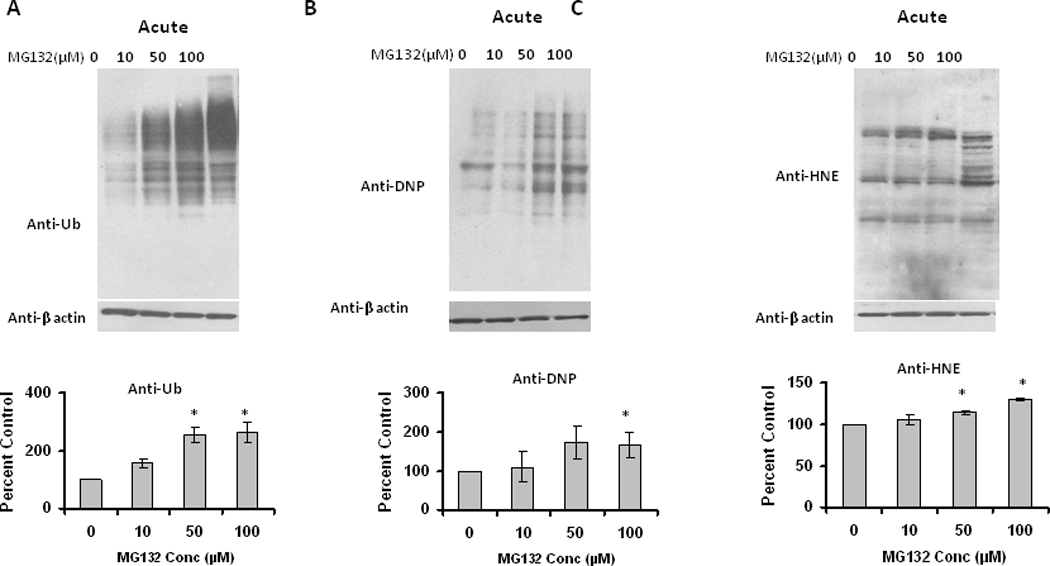
9 day (post induction) differentiated adipocytes were treated with different amounts of MG132 for 15 hrs. Cell lysates were analyzed for (A) ubiquitinated, (B) oxidized and (C) HNE (4-hydroxy-2-nonenal) modified proteins, using Western blotting analysis. Results represent the mean of 5 sets (n=5) of experiments, where each culture dish represents an n =1, conducted under identical experimental conditions. *P=0.05 when compared with untreated 3T3 L1 adipocytes, which was taken as percent control in the represented graphs.
Effect of chronic proteasome inhibition on differentiating immature adipocytes
While the previous studies identify that proteasome inhibition occurs in response to aging and differentiation of adipocytes/adipose tissue, and identify proteasome inhibition being sufficient to promote oxidative stress, they do not provide insight as to the potential for proteasome inhibition altering adipocyte differentiation. Because continual differentiation of pre-adipocytes within adipose tissue is essential to maintaining homeostasis, it is possible that stressors promoting inhibition of basal proteasome activity in pre-adipocytes in adipose tissue, may inhibit adipocyte differentiation during aging. Therefore, we next sought to elucidate if proteasome inhibitors had any effect on differentiating adipocytes prior to their maturation into adipocytes. Chronic treatment with low level proteasome inhibitors resulted in dramatic decreases in Oil Red O staining (Figure 9), consistent with decreased lipid content. In contrast to acute treatment in mature adipocytes, chronic administration of proteasome inhibitors to differentiating cells resulted in a low level transient increase in ubiquitinated, oxidized, and HNE modified proteins (Figure 10). Surprisingly, this same treatment significantly decreased the levels of key adipogenesis transcription factors PPARγ and C/EBPβ (Figure 10). These data are consistent with chronic proteasome inhibition in differentiating adipocytes having similar effects as acute treatment in mature adipocytes in regards to reduced lipid content, which is accompanied by a reduced level of adipogenic transcription factors.
Figure 9. Effect of chronic proteasome inhibition on lipid accumulation of differentiating 3T3-L1 preadipocytes.
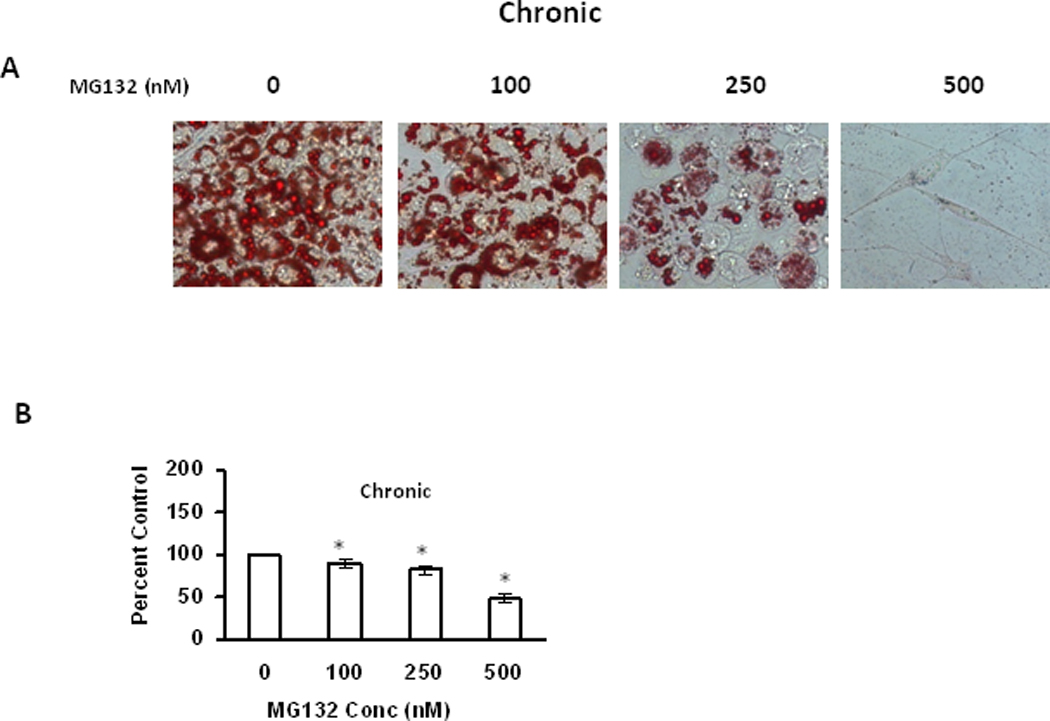
3T3-L1 preadipocytes were induced for adipocyte differentiation as described in methods with the addition of indicated amounts of MG132 to the medium for 7 days. On day 7,differentiated adipocytes were analyzed by Oil Red O staining to visualize the effect of MG132 on lipid accumulation (A) and Oil Red O stained lipid amount was quantified by measuring the absorbance at 570nm after extraction with ethanol (B). Results represent the mean of 5 sets (n=5) of experiments, where each culture dish represents an n =1, conducted under identical experimental conditions. *P=0.05 when compared with untreated 7 day 3T3 L1 adipocytes.
Figure 10. Effect of chronic proteasome inhibition on ubiquitinated, oxidized, and adipogenic protein expression in 3T3-L1 preadipocytes.
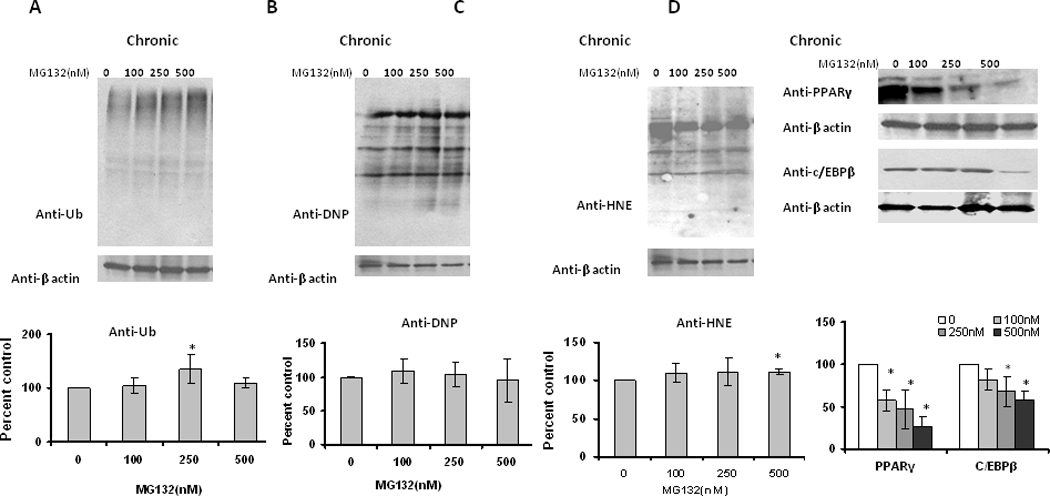
3T3-L1 preadipocytes were induced for adipocyte differentiation as described in methods with the addition of indicated amounts of MG132. Fresh media with MG132 was added daily to the cultures. On day 7, differentiated adipocytes for (A) ubiquitinated, (B) oxidized, (C) HNE (4-hydroxy-2-nonenal) modified proteins, and (D) PPARγ & C/EBPβ levels. Results represent the mean of 5 sets (n=5) of experiments, where each culture dish represents an n =1, conducted under identical experimental conditions. *P=0.05 when compared with untreated 7 day 3T3 L1 adipocytes.
Discussion
The current study is the first to show that the activity and expression of the proteasome is altered within aging adipose tissue. The fact that both 26S and 20S activities of the proteasome are inhibited suggests that there are gross perturbations in the functionality of the proteasome occur and thereby alter the function of the UPP. Interestingly, this decrease in proteasome activity occurs even though we observed the basal levels of proteasome activity to be lower in adipose tissue as compared to tissues such as the liver (data not shown). It is likely that the impairments in proteasome activity contribute to age-related changes in adipose function and adipose differentiation, consistent with numerous other studies of the UPS and aging in non-adipose tissue. For example, it has been shown that proteasome activity and proteasome subunit expression alters with age in variety of tissues including liver, brain, heart, spleen and spinal cord (8, 33–35). Numerous studies in non-adipose tissue have demonstrated that inhibition of the proteasome is sufficient to induce cytoxicity, oxidative stress, and pathology. These data raise possibility of decreased UPP function triggering pathology in aging adipose tissue. Interestingly, the UPP is believed to influence the development of obesity and insulin signaling in type 2 diabetes (36–40) and regulate the degradation of proteins essential to adipose function. Insulin receptor protein (IRS1) involved in insulin signaling, and is known to be inactivated by degradation through the UPP (41–43). Taken together, these data suggest that age-related impairments in UPP function may be particularly important to the genesis of age-related insulin resistance in adipose tissue, as well as other features of adipocyte pathogenesis during aging (44–46). Additionally, our studies indicate a role for proteasome inhibition for reducing adipogenesis (reduced lipid content and key adipogenic transcription factors) in differentiating cells. These data are consistent with proteasome inhibition promoting significant and deleterious changes to aging adipose depots due to effects on both mature as well as differentiating adipose cells.
Based on our in vitro proteasome inhibitor experiments it appears that proteasome inhibition may directly contribute to increased oxidative stress in mature adipose cells during aging. Increases in oxidized proteins are increasingly linked to adipose aging and obesity (47,48). Studies have linked oxidative stress in the adipocytes to insulin resistance and metabolic dysfunction, with the mechanism of such pathophysiology remaining largely unknown (49–51). Currently it is not known how such increases in oxidative stress occur within adipose tissue. Our data indicates that proteasome inhibition may be a central mechanism by which deleterious increases in both oxidized and HNE-modified proteins become elevated in adipose tissue. Recently we have shown that aging also increases the levels of hypoxia in adipose tissue (52), with hypoxia known to promote impairments in proteasome function (53). Alternatively, the presence of readily oxidized lipid within adipocytes may act as a potential modulator of UPP activity. Studies from our laboratory and numerous others (19, 20, 54–56) have demonstrated lipid peroxidation products such as HNE are potent inhibitors of the UPP. Taken together, these data open the potential for a feed forward cascade of increased levels of oxidized and HNE-modified proteins and proteasome impairment occurring with adipose aging and obesity.
Proteasome activity has been shown to be at its highest level during the early stages of differentiation, in human adipose derived stem cells (45), decreasing in activity as the stem cells become differentiated. In the present study we differentiated a cell line into adipocytes, using one of the most common models of adipose differentiation, and observed a decline in proteasome activity with increasing differentiation. Alterations in the UPP during adipose differentiation almost certainly are a part of organized patterns of protein regulation, which is required for adipose differentiation. Identifying those UPP substrates which are key to successful adipose differentiation is important not only advancing our understanding of adipose aging and adipose differentiation, but will also likely provide clues for designing therapeutic strategies for the prevention of adipose tissue expansion in clinical disorders including obesity, dyslipidemia, and type 2 diabetes. Of particular interest is the determination of how proteasome inhibition decreases the levels of both PPARγ and C/EBPβ, which are both key regulators of adipogenesis. It is almost certain that this down regulation of pro-adipogenic factors is due to the complicated changes in the proteome which occurs in response to proteasome inhibition, which likely results in the modulation of both inhibitors as well as inducers of PPARγ and C/EBPβ expression.
Because the continual differentiation of pre-adipocytes into mature adipocytes is essential to maintaining adipose homeostasis (38,39), the inhibition of basal proteasome function in pre-adipocytes may be another important way whereby age-related proteasome inhibition deleteriously alters adipose function. In this scenario stressors during aging result in a chronic loss of differentiating pre-adipocytes within the adipose tissue. In this model, the effects of proteasome inhibition on mature adipocytes within the aging tissue would be magnified, due to a lack of new mature adipocytes occurring replacing and/or diluting the effects of aging on older adipocytes.
Interestingly, even though proteasome activity is decreased with aging and differentiation, we did not see the expected increase in ubiquitinated protein levels. While the basis for this is not known, we anticipate that it may be mediated in part to a decrease in protein synthesis following proteasome inhibition. In studies from our laboratory and others, it has been established that impairments in proteasome function result in rapid and reversible impairments in protein synthesis (18, 57, 58), which means there are fewer proteins available for ubiquitination.
Our studies for the first time link changes in proteasome expression and function to increases in oxidative stress during adipose aging and adipose differentiation. Our studies indicate that whether occurring in mature adipocytes or differentiating adipocytes, proteasome inhibition is sufficient to induce oxidative stress and profound changes in adipose biology. Given the unprecedented increase in both aging and obesity in modern society, these data highlight the importance for age-related impairments in proteasome activity within adipose tissue being a potential mechanism for the continued rise in metabolic disease. Developing interventions which prevent or delay proteasome impairment in adipose tissue, as interventions which delay the downstream effects of proteasome inhibition on adipocyte pathogenesis, will likely be a very important avenue for the treatment and prevention of metabolic disease in the elderly.
Highlights.
Aging impairs proteasome function in adipose tissue. Proteasome inhibition is sufficient to modify adipocyte homeostasis. Proteasome inhibition may contribute to adipose aging.
Acknowledgements
This work was generously supported by grants from the NIA (AG029885 and AG025771) to J.N.K.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ciechanover A, Orian A, Schwartz AL. The ubiquitin-mediated proteolytic pathway: mode of action and clinical implications. J. Cell. Biochem. Suppl. 2000;34:40–51. doi: 10.1002/(sici)1097-4644(2000)77:34+<40::aid-jcb9>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 2.Pickart CM. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 3.Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 4.Dahlmann B, Kuehn L, Reinauer H. Studies on the activation by ATP of the 26 S proteasome complex from rat skeletal muscle. Biochem J. 1995;309:195–202. doi: 10.1042/bj3090195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hilt W, Wolf DH. Proteasomes: destruction as a programme. Trends Biochem Sci. 1996;21:96–102. [PubMed] [Google Scholar]
- 6.Grune T, Reinheckel T, Davies KJ. Degradation of oxidized proteins in K562 human hematopoietic cells by proteasome. J Biol Chem. 1996;271:15504–15509. doi: 10.1074/jbc.271.26.15504. [DOI] [PubMed] [Google Scholar]
- 7.Jung T, Catalgol B, Grune T. The proteasomal system. Mol Aspects Med. 2009;30:191–296. doi: 10.1016/j.mam.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 8.Keller JN, Huang FF, Markesbery WR. Decreased levels of proteasome activity and proteasome expression in aging spinal cord. Neuroscience. 2000;98:149–156. doi: 10.1016/s0306-4522(00)00067-1. [DOI] [PubMed] [Google Scholar]
- 9.de Vrij FM, Fischer DF, van Leeuwen FW, Hol EM. Protein quality control in Alzheimer's disease by the ubiquitin proteasome system. Prog Neurobiol. 2004;74:249–270. doi: 10.1016/j.pneurobio.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 10.Hung CC, Davison EJ, Robinson PA, Ardley HC. The aggravating role of the ubiquitin-proteasome system in neurodegenerative disease. Biochem Soc Trans. 2006;34:743–745. doi: 10.1042/BST0340743. [DOI] [PubMed] [Google Scholar]
- 11.Poppek D, Grune T. Proteasomal defense of oxidative protein modifications. Antioxid Redox Signal. 2006;8:173–184. doi: 10.1089/ars.2006.8.173. [DOI] [PubMed] [Google Scholar]
- 12.Jung T, Grune T. The proteasome and its role in the degradation of oxidized proteins. IUBMB Life. 2008;60:743–752. doi: 10.1002/iub.114. [DOI] [PubMed] [Google Scholar]
- 13.Shringarpure R, Grune T, Mehlhase J, Davies KJ. Ubiquitin conjugation is not required for the degradation of oxidized proteins by proteasome. J Biol Chem. 2003;278:311–318. doi: 10.1074/jbc.M206279200. [DOI] [PubMed] [Google Scholar]
- 14.Morimoto RI, Cuervo AM. Protein homeostasis and aging: taking care of proteins from the cradle to the grave. J Gerontol A Biol Sci Med Sci. 2009;64:167–170. doi: 10.1093/gerona/gln071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calabrese V, Cornelius C, Mancuso C, Lentile R, Stella AM, Butterfield DA. Redox homeostasis and cellular stress response in aging and neurodegeneration. Methods Mol Biol. 2010;610:285–308. doi: 10.1007/978-1-60327-029-8_17. [DOI] [PubMed] [Google Scholar]
- 16.Koga H, Kaushik S, Cuervo AM. Protein homeostasis and aging: The importance of exquisite quality control. Ageing Res Rev. 2011;10:205–215. doi: 10.1016/j.arr.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friguet B, Bulteau AL, Chondrogianni N, Conconi M, Petropoulos I. Protein degradation by the proteasome and its implications in aging. Ann N Y Acad Sci. 2000;908:143–154. doi: 10.1111/j.1749-6632.2000.tb06643.x. [DOI] [PubMed] [Google Scholar]
- 18.Ding Q, Dimayuga E, Markesbery WR, Keller JN. Proteasome inhibition induces reversible impairments in protein synthesis. FASEB J. 2006;20:1055–1063. doi: 10.1096/fj.05-5495com. [DOI] [PubMed] [Google Scholar]
- 19.Keller JN, Hanni KB, Markesbery WR. Possible involvement of proteasome inhibition in aging: implications for oxidative stress. Mech Ageing Dev. 2000;113:61–70. doi: 10.1016/s0047-6374(99)00101-3. [DOI] [PubMed] [Google Scholar]
- 20.Keller JN, Huang FF, Dimayuga ER, Maragos WF. Dopamine induces proteasome inhibition in neural PC12 cell line. Free Radic Biol Med. 2000;29:1037–1042. doi: 10.1016/s0891-5849(00)00412-3. [DOI] [PubMed] [Google Scholar]
- 21.Mugita N, Honda Y, Nakamura H, Fujiwara T, Tanaka K, Omura S, Shimbara N, Ogawa M, Saya H, Nakao M. The involvement of proteasome in myogenic differentiation of murine myocytes and human rhabdomyosarcoma cells. Int J Mol Med. 1999;3:127–137. [PubMed] [Google Scholar]
- 22.Guo W, Shang F, Liu Q, Urim L, Zhang M, Taylor A. Ubiquitin-proteasome pathway function is required for lens cell proliferation and differentiation. Invest Ophthalmol Vis Sci. 2006;47:2569–2575. doi: 10.1167/iovs.05-0261. [DOI] [PubMed] [Google Scholar]
- 23.Ichihara A, Tanaka K. Roles of proteasomes in cell growth. Mol Biol Rep. 1995;21:49–52. doi: 10.1007/BF00990970. [DOI] [PubMed] [Google Scholar]
- 24.Tuoc TC, Stoykova A. Roles of the ubiquitin-proteosome system in neurogenesis. Cell Cycle. 2010;15:3174–3180. doi: 10.4161/cc.9.16.12551. [DOI] [PubMed] [Google Scholar]
- 25.Kuk JL, Saunders TJ, Davidson LE, Ross R. Age-related changes in total and regional fat distribution. Ageing Res Rev. 2009;8:339–348. doi: 10.1016/j.arr.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 26.Ohkawa S, Odamaki M, Ikegaya N, Hibi I, Miyaji K, Kumagai H. Association of age with muscle mass, fat mass and fat distribution in non-diabetic haemodialysis patients. Nephrol Dial Transplant. 2005;20:945–951. doi: 10.1093/ndt/gfh643. [DOI] [PubMed] [Google Scholar]
- 27.Huffman DM, Barzilai N. Contribution of adipose tissue to health span and longevity. Interdiscip Top Gerontol. 2010;37:1–19. doi: 10.1159/000319991. [DOI] [PubMed] [Google Scholar]
- 28.Kirkland JL, Tchkonia T, Pirtskhalava T, Han J, Karagiannides I. Adipogenesis and aging: does aging make fat go MAD? Exp Gerontol. 2002;37:757–767. doi: 10.1016/s0531-5565(02)00014-1. [DOI] [PubMed] [Google Scholar]
- 29.Cartwright MJ, Tchkonia T, Kirkland JL. Aging in adipocytes: potential impact of inherent, depot-specific mechanisms. Exp Gerontol. 2007;42:463–547. doi: 10.1016/j.exger.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.St-Onge MP. Relationship between body composition changes and changes in physical function and metabolic risk factors in aging. Curr Opin Clin Nutr Metab Care. 2005;8:523–528. [PubMed] [Google Scholar]
- 31.Kilroy G, Burk DH, Floyd ZE. High efficiency lipid-based siRNA transfection of adipocytes in suspension. PLoS One. 2009;11:e6940. doi: 10.1371/journal.pone.0006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Floyd ZE, Stephens JM. Interferon-gamma-mediated activation and ubiquitin-proteasome-dependent degradation of PPARgamma in adipocytes. J Biol Chem. 2002;277:4062–4068. doi: 10.1074/jbc.M108473200. [DOI] [PubMed] [Google Scholar]
- 33.Dasuri K, Zhang L, Ebenezer P, Liu Y, Fernandez-Kim SO, Keller JN. Aging and dietary restriction alter proteasome biogenesis and composition in the brain and liver. Mech Ageing Dev. 2009;130:777–783. doi: 10.1016/j.mad.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li F, Zhang L, Craddock J, Bruce-Keller AJ, Dasuri K, Nguyen A, Keller JN. Aging and dietary restriction effects on ubiquitination, sumoylation, and the proteasome in the heart. Mech Ageing Dev. 2008;129:515–521. doi: 10.1016/j.mad.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang L, Li F, Dimayuga E, Craddock J, Keller JN. Effects of aging and dietary restriction on ubiquitination, sumoylation, and the proteasome in the spleen. FEBS Lett. 2007;581:5543–5547. doi: 10.1016/j.febslet.2007.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rome S, Meugnier E, Vidal H. The ubiquitin-proteasome pathway is a new partner for the control of insulin signaling. Curr Opin Clin Nutr Metab Care. 2004;7:249–254. doi: 10.1097/00075197-200405000-00002. [DOI] [PubMed] [Google Scholar]
- 37.Balasubramanyam M, Sampathkumar R, Mohan V. Is insulin signaling molecules misguided in diabetes for ubiquitin-proteasome mediated degradation? Mol Cell Biochem. 2005;275:117–125. doi: 10.1007/s11010-005-1083-y. [DOI] [PubMed] [Google Scholar]
- 38.Chang TL, Chang CJ, Lee WY, Lin MN, Huang YW, Fan K. The roles of ubiquitin and 26S proteasome in human obesity. Metabolism. 2009;58:1643–1648. doi: 10.1016/j.metabol.2009.05.020. [DOI] [PubMed] [Google Scholar]
- 39.Wang J, Maldonado MA. The ubiquitin-proteasome system and its role in inflammatory and autoimmune diseases. Cell Mol Immunol. 2006;3:255–261. [PubMed] [Google Scholar]
- 40.Sun XJ, Goldberg JL, Qiao LY, Mitchell JJ. Insulin-induced insulin receptor substrate-1 degradation is mediated by the proteasome degradation pathway. Diabetes. 1999;48:1359–1364. doi: 10.2337/diabetes.48.7.1359. [DOI] [PubMed] [Google Scholar]
- 41.Zhande R, Mitchell JJ, Wu J, Sun XJ. Molecular mechanism of insulin-induced degradation of insulin receptor substrate 1. Mol Cell Biol. 2002;22:1016–1026. doi: 10.1128/MCB.22.4.1016-1026.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee AV, Gooch JL, Oesterreich S, Guler RL, Yee D. Insulin-like growth factor I-induced degradation of insulin receptor substrate 1 is mediated by the 26S proteasome and blocked by phosphatidylinositol 3'-kinase inhibition. Mol Cell Biol. 2000;20:1489–1496. doi: 10.1128/mcb.20.5.1489-1496.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haruta T, Uno T, Kawahara J, Takano A, Egawa K, Sharma PM, Olefsky JM, Kobayashi MA. Rapamycin-sensitive pathway down-regulates insulin signaling via phosphorylation and proteasomal degradation of insulin receptor substrate-1. Mol Endocrinol. 2000;14:783–794. doi: 10.1210/mend.14.6.0446. [DOI] [PubMed] [Google Scholar]
- 44.Sakamoto K, Sato Y, Shinka T, Sei M, Nomura I, Umeno M, Ewis AA, Nakahori Y. Proteasome subunits mRNA expressions correlate with male BMI: implications for a role in obesity. Obesity. 2009;17:1044–1049. doi: 10.1038/oby.2008.612. [DOI] [PubMed] [Google Scholar]
- 45.Sakamoto K, Sato Y, Sei M, Ewis AA, Nakahori Y. Proteasome activity correlates with male BMI and contributes to the differentiation of adipocyte in hADSC. Endocrine. 2010;37:274–279. doi: 10.1007/s12020-009-9298-4. [DOI] [PubMed] [Google Scholar]
- 46.De Barros S, Zakaroff-Girard A, Lafontan M, Galitzky J, Bourlier V. Inhibition of human preadipocyte proteasomal activity by HIV protease inhibitors or specific inhibitor lactacystin leads to a defect in adipogenesis, which involves matrix metalloproteinase-9. J Pharmacol Exp Ther. 2007;320:291–299. doi: 10.1124/jpet.106.111849. [DOI] [PubMed] [Google Scholar]
- 47.Findeisen HM, Pearson KJ, Gizard F, Zhao Y, Qing H, Jones KL, Cohn D, Heywood EB, de Cabo R, Bruemmer D. Oxidative stress accumulates in adipose tissue during aging and inhibits adipogenesis. PLoS One. 2011;6:e18532. doi: 10.1371/journal.pone.0018532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frohnert BI, Sinaiko AR, Serrot FJ, Foncea RE, Moran A, Ikramuddin S, Choudry U, Bernlohr DA. Increased Adipose Protein Carbonylation in Human Obesity. Obesity (Silver Spring) 2011 doi: 10.1038/oby.2011.115. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rudich A, Tirosh A, Potashnik R, Hemi R, Kanety H, Bashan N. Prolonged oxidative stress impairs insulin-induced GLUT4 translocation in 3T3-L1 adipocytes. Diabetes. 1998;47:1562–1569. doi: 10.2337/diabetes.47.10.1562. [DOI] [PubMed] [Google Scholar]
- 50.Tirosh A, Potashnik R, Bashan N, Rudich A. Oxidative stress disrupts insulin-induced cellular redistribution of insulin receptor substrate-1 and phosphatidylinositol 3-kinase in 3T3-L1 adipocytes. A putative cellular mechanism for impaired protein kinase B activation and GLUT4 translocation. J Biol Chem. 1999;274:10595–10602. doi: 10.1074/jbc.274.15.10595. [DOI] [PubMed] [Google Scholar]
- 51.Urakawa H, Katsuki A, Sumida Y, Gabazza EC, Murashima S, Morioka K, Maruyama N, Kitagawa N, Tanaka T, Hori Y, Nakatani K, Yano Y, Adachi Y. Oxidative stress is associated with adiposity and insulin resistance in men. J Clin Endocrinol Metab. 2003;88:4673–4676. doi: 10.1210/jc.2003-030202. [DOI] [PubMed] [Google Scholar]
- 52.Zhang L, Ebenezer PJ, Dasuri K, Fernandez-Kim SO, Francis J, Mariappan N, Gao Z, Ye J, Bruce-Keller A, Keller JN. Aging is associated with hypoxia and oxidative stress in adipose tissue: Implications for adipose function. Am J Physiol Endocrinol Metab. 2011 doi: 10.1152/ajpendo.00059.2011. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Keller JN, Huang FF, Zhu H, Yu J, Ho YS, Kindy TS. Oxidative stress-associated impairment of proteasome activity during ischemia-reperfusion injury. J Cereb Blood Flow Metab. 2000;20:1467–1473. doi: 10.1097/00004647-200010000-00008. [DOI] [PubMed] [Google Scholar]
- 54.Friguet B, Szweda LI. Inhibition of the multicatalytic proteinase (proteasome) by 4-hydroxy-2-nonenal cross-linked protein. FEBS Lett. 1997;405:21–25. doi: 10.1016/s0014-5793(97)00148-8. [DOI] [PubMed] [Google Scholar]
- 55.Okada K, Wangpoengtrakul C, Osawa T, Toyokuni S, Tanaka K, Uchida K. 4-Hydroxy-2-nonenal-mediated impairment of intracellular proteolysis during oxidative stress. Identification of proteasomes as target molecules. J Biol Chem. 1999;274:23787–23793. doi: 10.1074/jbc.274.34.23787. [DOI] [PubMed] [Google Scholar]
- 56.Dasuri K, Nguyen A, Zhang L, Fernandez-Kim OS, Bruce-Keller AJ, Blalock BA, Cabo RD, Keller JN. Comparison of rat liver and brain proteasomes for oxidative stress-induced inactivation: Influence of ageing and dietary restriction. Free Radic Res. 2009;43:28–36. doi: 10.1080/10715760802534812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dasuri K, Ebenezer PJ, Zhang L, Fernandez-Kim SO, Uranga RM, Gavilán E, Di Blasio A, Keller JN. Selective vulnerability of neurons to acute toxicity after proteasome inhibitor treatment: implications for oxidative stress and insolubility of newly synthesized proteins. Free Radic Biol Med. 2010;49:1290–1297. doi: 10.1016/j.freeradbiomed.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mytilineou C, McNaught KS, Shashidharan P, Yabut J, Baptiste RJ, Parnandi A, Olanow CW. Inhibition of proteasome activity sensitizes dopamine neurons to protein alterations and oxidative stress. J Neural Transm. 2004;111:1237–1251. doi: 10.1007/s00702-004-0167-2. [DOI] [PubMed] [Google Scholar]


