Abstract
Background
Genome-wide association (GWA) studies have identified several susceptibility loci for metabolic syndrome (MetS) component traits, but have had variable success in identifying susceptibility loci to the syndrome as an entity. We conducted a GWA study on MetS and its component traits in four Finnish cohorts consisting of 2637 MetS cases and 7927 controls, both free of diabetes, and followed the top loci in an independent sample with transcriptome and NMR-based metabonomics data. Furthermore, we tested for loci associated with multiple MetS component traits using factor analysis and built a genetic risk score for MetS.
Methods and Results
A previously known lipid locus, APOA1/C3/A4/A5 gene cluster region (SNP rs964184), was associated with MetS in all four study samples (P=7.23×10−9 in meta-analysis). The association was further supported by serum metabolite analysis, where rs964184 associated with various VLDL, TG, and HDL metabolites (P=0.024-1.88×10−5). Twenty-two previously identified susceptibility loci for individual MetS component traits were replicated in our GWA and factor analysis. Most of these associated with lipid phenotypes and none with two or more uncorrelated MetS components. A genetic risk score, calculated as the number of alleles in loci associated with individual MetS traits, was strongly associated with MetS status.
Conclusions
Our findings suggest that genes from lipid metabolism pathways have the key role in the genetic background of MetS. We found little evidence for pleiotropy linking dyslipidemia and obesity to the other MetS component traits such as hypertension and glucose intolerance.
Keywords: metabolic syndrome, risk factors, genome-wide association study, meta-analysis, lipids
Introduction
Metabolic syndrome (MetS) is characterized by the clustering of several risk factors, which increase the risk of cardiovascular disease (CVD) and type 2 diabetes mellitus (T2DM). 1, 2 Definition of MetS is debated 3, 4 and the individual MetS components usually include dyslipidemia, hypertension, insulin resistance, and anthropometric measures. To date, linkage, candidate gene, and more recently, genome-wide association (GWA) studies 5 have identified several susceptibility regions and genes for individual MetS traits. These established associations to common and rare variants typically affect only a small part of the broad spectrum of MetS traits and explain only a fraction of the heritability that is estimated to be up to 90% for some of the traits and ~30% for MetS (reviewed in 6-8). Studies on families have nevertheless suggested that undetected genetic loci may mediate clustering of MetS components, 9-11 although some studies have failed to find unifying factors. 12, 13 More recently, the common genetic background hypothesis has been supported by emerging evidence of GWA loci where variants associate with more than one metabolic phenotype. 14-16
Several candidate gene studies have analyzed MetS as a dichotomous trait using one of the available case-control definitions, and successfully identified genes from specific pathways that are known to be relevant to MetS component phenotypes (reviewed in 6-8). In these, association with overall MetS status is typically explained by a strong association with one or two of the MetS components. The relatively few GWA studies, which have analyzed MetS status, have had variable success in identifying susceptibility loci. 5, 17, 18 The current evidence of MetS as an entity is still mostly based on clinical data on the component traits, and not on biological data connecting MetS susceptibility genes into pathways relevant to the broad spectrum of MetS components.
We searched for MetS susceptibility loci and evidence of a common genetic background for MetS by testing four types of associations: 1) individual SNPs and MetS (defined by the International Diabetes Federation, IDF 1), 2) SNPs and MetS component traits, 3) SNPs and MetS factors that combine evidence across component traits, and 4) MetS diagnosis and genetic risk score (GRS) derived from loci associated with one or more component traits of MetS.
Methods
Each of the DNA donors in our study has given individual consent to the use of their DNA sample in scientific research. The studies have been approved by the relevant research ethics boards.
Study subjects
Four Finnish population cohorts were included in the study sample (Table 1). We used IDF definition of metabolic syndrome 1 to identify cases and controls (Table 1, Table S1, Table S2, and Figure S1). In all, our study included 11616 subjects from which 23% fulfilled criteria for MetS cases (N=2637) and 68% for controls (N=7927). Our replication study samples in the GRS analysis consisted of 906 non-diabetic men from the Metabolic Syndrome in Men (METSIM) study population 19 and a subset (N=731) of male and female participants from the FINRISK 2007 study. 20
Table 1.
Characteristics of the study sample.
| H2000* | HBCS | YFS | NFBC1966 | |
|---|---|---|---|---|
| N (passed genotyping QC) |
2124 | 1676 | 2443 | 5373 |
| Men (%) | 49 | 43 | 46 | 48 |
| MetS† case (%) | 48 (men 49, women 47) |
38 (men 41, women 35) |
18 (men 22, women 14) |
10 (men 14, women 7) |
| Individuals on hypolipidemic medication (%) |
6 | 18 | 2 | 0.1 |
| Individuals on antihypertensive medication (%) |
21 | 36 | 6 | 0.2 |
| Individuals on hypoglycaemic medication (%) |
0.2 | 7 | 1 | 0.5 |
|
|
||||
| Mean ± SD | ||||
|
|
||||
| Age (y) | 51 ± 11 | 61 ± 3 | 38 ± 5 | 31 ± ~0.3 |
| Waist circumference (cm) |
94.0 ± 12.6 (men 98.7 ± 10.7, women 89.5 ± 12.7) |
95.3 ± 13.2 (men 100.8 ± 11.3, women 91.1 ± 13.0) |
88.5 ± 13.5 (men 94.4 ± 12.0, women 83.5 ± 12.6) |
83.8 ± 12.1 (men 88.9 ± 9.9, women 78.8 ± 12.0) |
| Triglycerides (mmol/l) | 1.63 ± 0.98 | 1.53 ± 1.11 | 1.39 ± 0.92 | 1.20 ± 0.73 |
| HDL cholesterol (mmol/l) |
1.33 ± 0.37 (men 1.21 ± 0.33, women 1.44 ± 0.37) |
1.61 ± 0.44 (men 1.45 ± 0.38, women 1.73 ± 0.44) |
1.34 ± 0.33 (men 1.21 ± 0.29, women 1.44 ± 0.33) |
1.56 ± 0.38 (men 1.41 ± 0.33, women 1.70 ± 0.38) |
| Systolic blood pressure (mmHg) |
134.3 ± 19.7 | 145.6 ± 20.3 | 120.7 ± 14.2 | 124.8 ± 13.5 |
| Diastolic blood pressure (mmHg) |
83.5 ± 10.8 | 89.0 ± 10.4 | 75.8 ± 11.3 | 77.4 ± 11.5 |
| HOMA-IR | 2.07 ± 1.62 | 2.88 ± 2.77 | 2.35 ± 3.96 | 2.02 ± 2.07 |
| Glucose (mmol/l) | 5.36 ± 0.49 | 5.85 ± 1.37 | 5.34 ± 0.92 | 5.06 ± 0.72 |
The H2000 study sample has an approximately equal number of cases and controls due to sample selection criteria (Supplementary Methods).
Metabolic syndrome defined by the IDF criteria (Table S1). From the 11616 genotyped subjects 23% qualified as MetS cases (N=2637) and 68% as controls (N=7927). The characteristics are presented separately for men and women for those MetS components (waist, HDL) that have sex-specific cut-off thresholds in the IDF MetS definition. Individuals receiving hypolipidemic, antihypertensive, or hypoglycaemic medications are included in the calculations. H2000, Health 2000; HBCS, Helsinki Birth Cohort Study; YFS, Cardiovascular Risk in Young Finns Study; NFBC1966, Northern Finland Birth Cohort 1966, HOMA-IR; Homeostatic Model Assessment Insulin Resistance.
Genotyping
Studies used Illumina genotyping arrays for genome wide genotyping: Illumina Infinium HD Human610-Quad BeadChip in Health 2000 (H2000), Illumina HumanCNV370-Duo BeadChip in the Northern Finland Birth Cohort 1966 (NFBC1966), and Illumina Human670K custom BeadChip in the Cardiovascular Risk in Young Finns Study (YFS) and the Helsinki Birth Cohort Study (HBCS). To increase the amount of genotypic data in our analysis, we imputed SNP genotype data to HapMap3 resolution with a HMM-based imputation program MACH 1.0.16 21, 22 (Supplementary Methods).
GWA analyses
Using directly measured and imputed genotypes in logistic or linear regression analysis, we tested for association between SNPs and MetS (and quantitative MetS component traits) in each study separately using PLINK v. 1.07 23 for directly genotyped SNPs and ProbABEL v. 0.1-6 24 for imputed SNPs, and then combined the results in a genome-wide fixed effects meta-analysis across all samples (2637 cases and 7927 controls after excluding individuals with diabetes) with Metabel v. 0.03, applying genomic control correction to each GWAS data before pooling. 25 We computed (based on β-estimates and SE) I2 statistics as measures of heterogeneity between studies. We adjusted the analyses for age (except in NFBC1966) and gender, and used additive model for the SNP effect. In MetS component trait analyses, individuals taking hypolipidemic, antihypertensive, or hypoglycaemic medication were not included in the relevant trait analyses. Details and power calculations in Supplementary Methods.
Copy Number Polymorphism tag-SNP analysis
We extended our genetic analysis from single nucleotides to structural variation by assessing the association between previously identified genomic copy number polymorphisms (CNPs) and MetS. For this, we had directly genotyped or imputed genotypes for 620 SNPs that were in linkage disequilibrium with CNPs and could be used as proxies for the CNP alleles of 649 CNPs in our data. All GWA analyses for these CNP tag-SNPs were done as part of the main SNP GWA analysis (Supplementary Methods section).
SNP associations with gene expression and serum metabolites
Expression Quantitative Trait Locus (eQTL) and serum metabolite data were available from an independent study sample of 518 Finns. A total of 135 metabolic measures were obtained from nuclear magnetic resonance (NMR) spectroscopy data of both native serum and lipid extracts. (Supplementary Methods).
Risk score analysis
We selected for the GRS analysis the SNP with strongest statistical evidence (lowest P-value) of an association with any MetS component trait from each of the 22 genome-wide significant loci (Table S3). We then formed and evaluated the GRS using two independent cohorts by calculating the average number of risk increasing alleles of each variant for each study individual and divided our study samples into quartiles based on the GRS distribution, separately for each study sample. We compared the risk of MetS in these quartiles in an age and gender adjusted logistic regression model in R, 26 using the lowest quartile as a reference group (Supplementary Methods). Albeit for example for cardiovascular diseases it would be relatively easy to compare the effect of GRS and conventional risk factors for an association with a disease, a difficulty in the case of MetS arises from the definition of the syndrome; the definition itself already includes many conventional risk factors such as dyslipidemia, central obesity, hypertension, and insulin resistance. For this reason, we assessed the value of our GRS for the risk of MetS in a simple model adjusted for age and gender only.
Factor analysis
Factor analysis has been applied to MetS in several studies with the aim to identify unifying entities (factors) for the same quantitative MetS component traits that were analyzed individually (Supplementary Methods). We estimated factor loadings (Table S4) based on correlations of individual MetS components and created factor scores as linear combinations based on three factor solutions: 1) Lipids/waist circumference, 2) Lipids/waist circumference/HOMA-IR, and 3) Systolic and diastolic blood pressure (SBP, DBP). Factor scores were analyzed as quantitative traits similarly to the individual MetS component traits (Supplementary Methods). Individuals taking hypolipidemic, antihypertensive, or hypoglycaemic medication were not included in the relevant factor analyses.
Results
Metabolic syndrome, case-control analysis
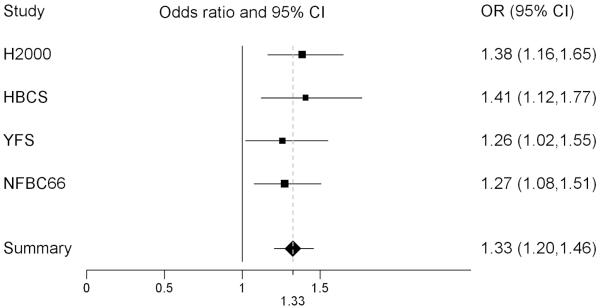
Our study sample of 11616 non-diabetic Finnish individuals comprised four GWA scans (2637 cases and 7927 controls by IDF criteria of MetS)(Table 1). We conducted the four GWA analyses and their meta-analysis using 1257079 directly genotyped or imputed SNPs. In meta-analysis of the four data sets, the minor allele of SNP rs964184, on chromosome 11q23.3 in a known lipid locus, apolipoprotein APOA1/C3/A4/A5 gene cluster region, associated with increased risk for MetS (P=7.23×10−9) (Figures S2 and S3). The associations were to the same direction in all four study samples (Figure 1) with an overall effect size OR of 1.33 (95% CI 1.20-1.46). Five SNPs that associated with MetS in a recent GWA study5 were not replicated on a genome-wide significance level in our data, although a meta-analysis combining both studies found these SNPs highly significant (Table S5).
Figure 1.
Forest plot of the MetS case-control association result for SNP rs964184. The plot shows odds ratios (OR) and 95% confidence intervals (CI) separately for the four study samples and for their meta-analysis (in the Summary row). A separate analysis of the ‘older’ and ‘younger’ study samples resulted in very similar odds ratios and overlapping 95% confidence intervals: HBCS+H2000 1.40 (1.22-1.60) and YFS+NFBC 1.27 (1.11-1.44). A meta-analysis of the three older cohorts, excluding the youngest and largest cohort NFBC, resulted in an odds ratio of 1.35 (1.20-1.52). The OR is on the x-axis of the plot and the lines represent the confidence intervals. H2000, Health 2000; HBCS, Helsinki Birth Cohort Study; YFS, Cardiovascular Risk in Young Finns Study; NFBC1966, Northern Finland Birth Cohort 1966.
In parallel to the complete GWA scan, we analyzed a subset of the SNPs (N=620) as CNP tag-SNPs, to test for an association between MetS and structural variation in the genome. Pooling results from all four data sets failed to identify significant associations between MetS and genomic loci.
Factor analysis and gene loci with multiple phenotype associations
Our factor analysis sought to identify unifying factors for the individual MetS traits based on their correlation, and resulted in three separate MetS component trait combinations; 1) TG/HDL/waist circumference (F1), 2) TG/HDL/waist circumference/HOMA-IR (F2), and 3) systolic/diastolic blood pressure (F3) factors. While SNP associations for the blood pressure factor were absent, several SNPs from four established lipid gene loci in literature associated with the TG/HDL/waist -factor at the genome-wide significance level; apolipoprotein B (APOB) locus, APOA1/C3/A4/A5 gene cluster region, lipoprotein lipase (LPL) locus, and cholesteryl ester transfer protein (CETP) locus (Figure 2, Figure S4). Analyzing the TG-HDL-waist-HOMA factor generated only suggestive associations (top hit P= 3.19×10−07) for loci that included CAMK2D, LPL, the YPEL4/CLP1/ZDHHC5/MED19/TMX2/CTNND1 gene region, APOA1/C3/A4/A5 gene cluster region, CETP, RNF157, and ETFB.
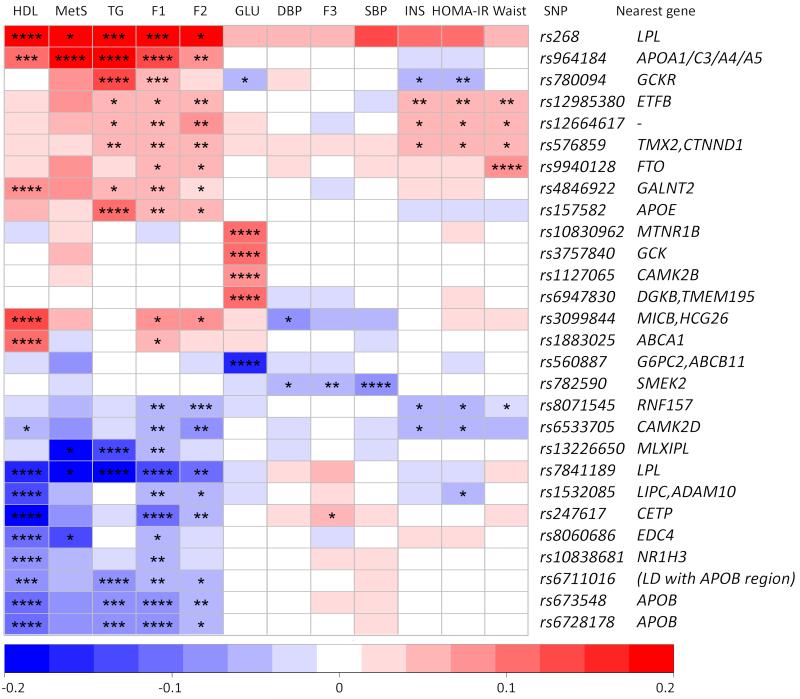
Figure 2.
SNP associations across MetS phenotypes. The plot shows SNP-phenotype associations for the twenty-two top hit SNPs from the MetS component GWA analysis (Figure S6 and Table S3), four top hit SNPs from the TG/HDL/waist circumference –factor analysis, and five SNPs with a suggestive association in the TG/HDL/waist circumference/HOMA-IR –factor analysis (overlapping SNPs excluded). The colours correspond to beta effect size values for the minor allele of the SNP (values above 0.2 have been recoded to 0.2 and values below −0.2 to −0.2 and positive values refer to a risk effect and negative values to a protective effect). Psig-values for the tests are shown as: * P < 0.01, ** P < 1×10−4, *** P < 1×10−6, **** P < 5×10−8. HDL, high-density lipoprotein; MetS, metabolic syndrome case-control status; TG, triglycerides; F1, TG/HDL/waist circumference -factor; F2, TG/HDL/waist circumference/HOMA-IR -factor; GLU, glucose; DBP, diastolic blood pressure; F3, SBP/DBP blood pressure factor; SBP, systolic blood pressure; INS, insulin; HOMA_IR, Homeostatic Model Assessment Insulin Resistance.
Some of the GWA top hit SNPs associated with more than one MetS related phenotype (Figure 2). Several of the established lipid loci SNPs associated with HDL, TG, MetS, and the factors F1 and F2 (containing lipids, waist, and insulin resistance (HOMA-IR)) from the factor analysis. A few loci also showed suggestive association with waist circumference, HOMA-IR, and the factors F1 and F2. The glucose and SBP associated GWA SNPs did not show association with other phenotypes. These SNP associations with phenotypes were in concordance with correlations between those phenotypes in our data (Supplementary Results and Figure S5). An exception to this association and correlation pattern was observed for the GCKR gene locus SNP rs780094, which had a strong positive association with TG and the TG/HDL/waist circumference -factor, but at the same time suggestive negative associations with glucose and HOMA-IR.
Dissecting the underlying mechanism of the associations with MetS
We examined the underlying factors of our MetS association by studying the association between the relevant SNP rs964184 and the component traits that were used to define the case-control status. Rs964184 associated with TG (P=2.59×10−31) and HDL (P=5.83×10−8), but not with waist circumference (P=0.52), systolic BP (P=0.68), diastolic BP (P=0.98), or glucose (P=0.73). When we adjusted the case-control analysis for TG and/or HDL, the association of rs964184 disappeared (P=0.41 with TG and HDL in the model, P=0.62 with TG in the model) or was reduced (P=0.0017 with HDL in the model). This suggested that the main driving force behind the case-control association signal of rs964184 was its association with lipid components of MetS.
We proceeded to analyze metabolome and transcriptome data from an independent Finnish study sample of 518 individuals. The association analysis of rs964184 and 135 metabolites supported the conclusion that lipid components were behind the MetS association; rs964184 associated with several metabolites, most significantly with all 30 of the very low density lipoprotein (VLDL) particles (P=0.024-1.88×10−5, Bonferroni corrected p-value cut-off for significance Psig=3.70×10-4 ). Other significant and suggestive associations included, unsurprisingly, TG metabolites (details in the Supplementary Results). We also examined metabolome associations for the five MetS SNPs proposed by the STAMPEED Consortium 5 and confirmed by our additional meta-analysis (Table S5). All SNPs except rs295 associated with several lipid related metabolites, including VLDL and IDL particles (P=0.029-9.30×10−05) and apolipoprotein B (P=0.012-2.25×10−06). None of these five MetS SNPs associated with glucose or glycoproteins (P>0.14).
Our leukocyte-derived gene expression data analysis failed to identify any cis- or trans-regulated genes whose expression was significantly affected by the rs964184 alleles, possibly due to differential lipid gene expression in leucocytes and other more relevant tissues such as fat or liver, and a relatively small number of samples analyzed (Supplementary Results).
Metabolic syndrome, risk score analysis
We extended our GWA analysis to the individual components of the MetS phenotype: waist circumference, triglyceride (TG) and high-density lipoprotein (HDL) levels, SBP, DBP, and fasting plasma glucose. We observed SNP associations with HDL, TG, and glucose concentrations at the genome-wide significance level in several previously identified gene loci (Figure S6). We then proceeded to test how GRS, combined with age and gender, would help to identify individuals with a high risk of developing MetS. We created a GRS using the twenty-two risk variants that were associated with the five IDF MetS component traits at a genome-wide significance level in our study (Figure S6) and assigned study subjects to GRS quartiles based on the number of risk alleles they carried (Table S6 and Figure S7). The risk of MetS increased linearly across quartiles in the combined data of the four cohorts (P for linear trend = 6.91×10−11). Odds ratio for the risk of MetS in the highest GRS quartile compared to the lowest GRS quartile was 1.55 (95% CI 1.35-1.77) (Table S6). We replicated our GRS analysis in a meta-analysis of two independent study cohorts: an independent study sample of 906 non-diabetic men (METSIM study 19) and 611 study subjects from the FINRISK 2007 study 20. The OR for the risk of MetS in the highest GRS quartile compared to the lowest quartile was 1.35 (P = 0.045) (GRS/Q2: OR 1.17, 95% CI 0.88-1.56; GRS/Q3: OR 1.33, 95% CI 0.98-1.80; GRS/Q4: OR 1.35, 95% CI 1.01-1.82).
We conducted another GRS analysis using the five MetS SNPs reported by the STAMPEED Consortium 5 (Table S5). The GRS comprising twenty-two risk variants for MetS component traits (GRS1) associated more strongly with MetS than the GRS comprising five risk variants for MetS (GRS2); ORs for GRS quartile 4 were 1.45 (GRS1/Q4, 95% CI 1.23-1.69, P=6.00×10−6) and 1.32 (GRS2/Q4, 95% CI 1.10-1.58, P=2.62×10−3) in the study sample H2000/HBCS/YFS, and 1.35 (GRS1/Q4, 95% CI 1.01-1.82, P=0.045) and 1.17 (GRS2/Q4, 95% CI 0.85-1.62, P=0.33) in the replication study sample METSIM/FINRISK2007.
Discussion
Our GWA study of metabolic syndrome identified an association between MetS diagnosis and a previously known lipid locus APOA1/C3/A4/A5 gene cluster region on chromosome 11. 5, 16, 27, 28 The risk effect of SNP rs964184 in our study was consistent across our cohorts which have very different age profiles. Our GWA and serum metabolite data suggested that the SNP effect was mainly on the lipid component of MetS, which was also recently suggested in a metabolite analysis of another cohort of European ancestry. 29 This lipid-driven association is somewhat surprising considering that our data included 650 individuals who met the MetS case criteria only because of high glucose level and hypertension, compared to only 169 individuals who were identified as cases solely due to abnormal TG and HDL levels. The five SNPs recently reported to associate with MetS by the STAMPEED Consortium 5 also associated with several lipid metabolites, and not with glucose, in our metabolite analysis.
Identification of novel genetic loci harboring genes that simultaneously affect several MetS component traits would provide important clues to the biological background of MetS as a whole and not merely a sum of the component traits, and may aid in genetic risk prediction of MetS. Our factor analysis and GWA SNP results identified many gene loci associated with the lipid components of MetS and MetS phenotype factors that included waist and HOMA-IR but were strongly based on HDL and TG. Glucose and blood pressure SNP associations seemed to be somewhat separate from these lipid-driven associations. The GCKR gene (glucokinase [hexokinase 4] regulator) region was an interesting exception; its minor allele (A/T) associated with higher TG levels and lower HOMA-IR. This result is supported by previous observations; SNP rs780094 T-allele has been associated with increased TG levels, 30, 31 and the major allele C with higher glucose and insulin levels and HOMA-IR. 15, 30, 31 Although our relatively small metabolite data supported only the TG association of rs780094, it is likely that the gene has an effect, seemingly an opposite one, on both TG and insulin traits. This opposing effect may contribute to the lack of association between GCKR and MetS status in our data. On the whole, our factor analysis supported the GWA SNP results, indicating that there was little evidence for connections between the blood pressure and lipid/waist, and glucose components of MetS.
Our genetic risk score analysis suggested that combining genetic association information across the MetS component traits can help in identifying individuals with a higher genetic susceptibility for developing MetS. Using information from the several individual MetS component trait associations to build a GRS seemed to give better results than building a GRS using a smaller number of loci of MetS case-control association from a previous study. 5 Extensive research is, however, needed before any GRS can be utilized in clinical practice.
The exclusion of diabetic individuals can be seen both as a strength and a limitation of this study. Since people with diagnosed diabetes are usually treated with several drugs and/or insulin, we reduce the variation among cases by excluding the individuals with a diagnosis of diabetes. At the same time these excluded individuals may have a genetic predisposition to insulin resistance which limits our analysis of MetS component traits of glucose and HOMA-IR. In our tests, additional novel SNP associations with MetS were not observed after including diabetic individuals in the analysis. Another possible limitation in our study is the sample size (N=11616), when compared to some of the current GWA studies that are using even larger number of subjects (>100000). In all probability, however, not all previously undetected susceptibility loci are uncovered merely by increasing the sample size. Here, we aimed to eliminate some of the obscuring factors in gene identification by utilizing a culturally homogenous and genetically unique population. 32, 33 We had four independent study samples enabling comparison of the results between data sets, and meta-analysis. In addition, a moderate portion of our sample is rather young, in terms of a late-onset syndrome such as MetS and its component traits 34 and thus individuals diagnosed with MetS may have a stronger genetic component and less environmental factors masking the effect of genes, which may aid in unearthing the genetic susceptibility loci. Due to our homogenous population any generalization of the results has to be made cautiously.
As a whole, our GWAS, factor analysis, and serum metabolome analysis revealed a strong lipid gene contribution to the observed MetS associations and showed little evidence for pleiotropy linking dyslipidemia and obesity to the other types of metabolic abnormalities: glucose intolerance and hypertension. We thus conclude that there are probably no gene variants predisposing to the metabolic syndrome by affecting multiple components of its definition that have a clinically relevant effect size. Our findings suggest that the genetic predisposition to MetS is likely to result from relatively small effects of multiple genes predisposing to dyslipidemia and abdominal obesity, possibly complemented by low frequency and rare variation, and/or epigenetic modifications. From a clinical point of view it thus may make more sense to study the individual component traits instead of MetS as an entity, and to direct efforts especially to the lipid components that seemed to play the key role in our study.
Supplementary Material
Metabolic syndrome (MetS) is characterized by the clustering of several risk factors, such as dyslipidemia, hypertension, insulin resistance, and anthropometric measures. To date, genetic studies have identified several susceptibility regions in the genome for MetS component traits, but have had variable success in identifying susceptibility loci to the syndrome as an entity. We searched for MetS susceptibility loci by conducting a genome-wide association (GWA) study on MetS and its component traits in >10000 individuals. Furthermore, we searched for evidence of a common genetic background for MetS by investigating the top GWA loci with transcriptome and NMR-based metabonomics data and testing for loci associated with multiple MetS component traits using factor analysis. We also examined whether a genetic risk score derived from loci associated with one or more component traits of MetS associates with the MetS diagnosis. Importantly, we saw a strong lipid gene contribution to the observed MetS associations. There was little evidence for pleiotropy linking dyslipidemia and obesity to the other types of metabolic abnormalities: glucose intolerance and hypertension. We conclude that there are probably no common gene variants predisposing to the metabolic syndrome by affecting multiple components of its definition that have a clinically relevant effect size. Accordingly, our findings suggest that in the prevention and treatment of metabolic syndrome the main emphasis should be put on lifestyle factors.
Acknowledgments
The authors would like to thank the many colleagues who contributed to collection and phenotypic characterization of the clinical samples, and DNA extraction and genotyping of the data, especially Eija Hämäläinen, Minttu Sauramo, Outi Törnwall, Päivi Laiho, and the staff from the Genotyping Facilities at the Wellcome Trust Sanger Institute, and also Professor Paula Rantakallio (launch of NFBC1966 and 1986). They would also like to acknowledge those who agreed to participate in these studies.
Funding Sources:
This research was supported through funds from the European Community’s Seventh Framework Programme (FP7/2007-2013), BioSHaRE Consortium, grant agreement 261433, Academy of Finland, Orion-Farmos Research Foundation, Finnish Foundation for Cardiovascular Research, Sigrid Jusélius Foundation, Diabetes Research Foundation, Finnish Foundation for Pediatric Research, Novo Nordisk Foundation, Jenny and Antti Wihuri Foundation, Biotechnology and Biological Research Council, Wellcome Trust, Finnish Diabetes Research Society, Samfundet Folkhälsan, Juho Vainio Foundation, Finska Läkaresällskapet, Päivikki and Sakari Sohlberg Foundation, Signe and Ane Gyllenberg Foundation, Yrjö Jahnsson Foundation, Social Insurance Institution of Finland, University Hospital Medical funds to Tampere, and Turku University Hospitals, Emil Aaltonen Foundation, University Hospital Oulu, Biocenter, University of Oulu, NHLBI, ENGAGE, Medical Research Council, and Biocentrum Helsinki. See Online Data Supplement for details.
Footnotes
Conflict of Interest Disclosures: Dr Salomaa has received lecture fees from Roche Diagnostics. Dr Collins has ownership interests (mutual funds only).
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Alberti KG, Zimmet P, Shaw J. The metabolic syndrome--a new worldwide definition. Lancet. 2005;366:1059–1062. doi: 10.1016/S0140-6736(05)67402-8. [DOI] [PubMed] [Google Scholar]
- 2.Eckel RH, Grundy SM, Zimmet PZ. The metabolic syndrome. Lancet. 2005;365:1415–1428. doi: 10.1016/S0140-6736(05)66378-7. [DOI] [PubMed] [Google Scholar]
- 3.Kahn R. Metabolic syndrome: Is it a syndrome? Does it matter? Circulation. 2007;115:1806–1810. doi: 10.1161/CIRCULATIONAHA.106.658336. discussion 1811. [DOI] [PubMed] [Google Scholar]
- 4.Reaven GM. The metabolic syndrome: Time to get off the merry-go-round? J Intern Med. 2011;269:127–136. doi: 10.1111/j.1365-2796.2010.02325.x. [DOI] [PubMed] [Google Scholar]
- 5.Kraja AT, Vaidya D, Pankow JS, Goodarzi MO, Assimes TL, Kullo IJ, et al. A bivariate genome-wide approach to metabolic syndrome: Stampeed consortium. Diabetes. 2011;60:1329–1339. doi: 10.2337/db10-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joy T, Lahiry P, Pollex RL, Hegele RA. Genetics of metabolic syndrome. Curr Diab Rep. 2008;8:141–148. doi: 10.1007/s11892-008-0025-y. [DOI] [PubMed] [Google Scholar]
- 7.Monda KL, North KE, Hunt SC, Rao DC, Province MA, Kraja AT. The genetics of obesity and the metabolic syndrome. Endocr Metab Immune Disord Drug Targets. 2010;10:86–108. doi: 10.2174/187153010791213100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teran-Garcia M, Bouchard C. Genetics of the metabolic syndrome. Appl Physiol Nutr Metab. 2007;32:89–114. doi: 10.1139/h06-102. [DOI] [PubMed] [Google Scholar]
- 9.Carmelli D, Cardon LR, Fabsitz R. Clustering of hypertension, diabetes, and obesity in adult male twins: Same genes or same environments? Am J Hum Genet. 1994;55:566–573. [PMC free article] [PubMed] [Google Scholar]
- 10.Liese AD, Mayer-Davis EJ, Tyroler HA, Davis CE, Keil U, Schmidt MI, et al. Familial components of the multiple metabolic syndrome: The aric study. Diabetologia. 1997;40:963–970. doi: 10.1007/s001250050775. [DOI] [PubMed] [Google Scholar]
- 11.Hong Y, Rice T, Gagnon J, Despres JP, Nadeau A, Perusse L, et al. Familial clustering of insulin and abdominal visceral fat: The heritage family study. J Clin Endocrinol Metab. 1998;83:4239–4245. doi: 10.1210/jcem.83.12.5312. [DOI] [PubMed] [Google Scholar]
- 12.Benyamin B, Sorensen TI, Schousboe K, Fenger M, Visscher PM, Kyvik KO. Are there common genetic and environmental factors behind the endophenotypes associated with the metabolic syndrome? Diabetologia. 2007;50:1880–1888. doi: 10.1007/s00125-007-0758-1. [DOI] [PubMed] [Google Scholar]
- 13.Sjogren M, Lyssenko V, Jonsson A, Berglund G, Nilsson P, Groop L, et al. The search for putative unifying genetic factors for components of the metabolic syndrome. Diabetologia. 2008;51:2242–2251. doi: 10.1007/s00125-008-1151-4. [DOI] [PubMed] [Google Scholar]
- 14.Chambers JC, Elliott P, Zabaneh D, Zhang W, Li Y, Froguel P, et al. Common genetic variation near mc4r is associated with waist circumference and insulin resistance. Nat Genet. 2008;40:716–718. doi: 10.1038/ng.156. [DOI] [PubMed] [Google Scholar]
- 15.Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, Schadt EE, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zabaneh D, Balding DJ. A genome-wide association study of the metabolic syndrome in indian asian men. PLoS One. 2010;5:e11961. doi: 10.1371/journal.pone.0011961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park YM, Province MA, Gao X, Feitosa M, Wu J, Ma D, et al. Longitudinal trends in the association of metabolic syndrome with 550 k single-nucleotide polymorphisms in the framingham heart study. BMC Proc. 2009;3:S116. doi: 10.1186/1753-6561-3-s7-s116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stancčáková A, Kuulasmaa T, Paananen J, Jackson AU, Bonnycastle LL, Collins FS, et al. Association of 18 confirmed susceptibility loci for type 2 diabetes with indices of insulin release, proinsulin conversion, and insulin sensitivity in 5 327 non-diabetic finnish men. Diabetes. 2009;58:2129–2136. doi: 10.2337/db09-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vartiainen E, Laatikainen T, Peltonen M, Juolevi A, Männisto S, Sundvall J, et al. Thirty-five-year trends in cardiovascular risk factors in finland. Int J Epidemiol. 2010;39:504–518. doi: 10.1093/ije/dyp330. [DOI] [PubMed] [Google Scholar]
- 21.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. Mach: Using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. Plink: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aulchenko YS, Struchalin MV, van Duijn CM. Probabel package for genome-wide association analysis of imputed data. BMC Bioinformatics. 2010;11:134. doi: 10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. Genabel: An r library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 26.R Development Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2009. http://www.R-project.org. [Google Scholar]
- 27.Miller M, Rhyne J, Chen H, Beach V, Ericson R, Luthra K, et al. Apoc3 promoter polymorphisms c-482t and t-455c are associated with the metabolic syndrome. Arch Med Res. 2007;38:444–451. doi: 10.1016/j.arcmed.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamada Y, Kato K, Hibino T, Yokoi K, Matsuo H, Segawa T, et al. Prediction of genetic risk for metabolic syndrome. Atherosclerosis. 2007;191:298–304. doi: 10.1016/j.atherosclerosis.2006.05.035. [DOI] [PubMed] [Google Scholar]
- 29.Illig T, Gieger C, Zhai G, Romisch-Margl W, Wang-Sattler R, Prehn C, et al. A genome-wide perspective of genetic variation in human metabolism. Nat Genet. 2010;42:137–141. doi: 10.1038/ng.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sparso T, Andersen G, Nielsen T, Burgdorf KS, Gjesing AP, Nielsen AL, et al. The gckr rs780094 polymorphism is associated with elevated fasting serum triacylglycerol, reduced fasting and ogtt-related insulinaemia, and reduced risk of type 2 diabetes. Diabetologia. 2008;51:70–75. doi: 10.1007/s00125-007-0865-z. [DOI] [PubMed] [Google Scholar]
- 31.Orho-Melander M, Melander O, Guiducci C, Perez-Martinez P, Corella D, Roos C, et al. Common missense variant in the glucokinase regulatory protein gene is associated with increased plasma triglyceride and c-reactive protein but lower fasting glucose concentrations. Diabetes. 2008;57:3112–3121. doi: 10.2337/db08-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nelis M, Esko T, Magi R, Zimprich F, Zimprich A, Toncheva D, et al. Genetic structure of europeans: A view from the north-east. PLoS One. 2009;4:e5472. doi: 10.1371/journal.pone.0005472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Novembre J, Johnson T, Bryc K, Kutalik Z, Boyko AR, Auton A, et al. Genes mirror geography within europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hildrum B, Mykletun A, Hole T, Midthjell K, Dahl AA. Age-specific prevalence of the metabolic syndrome defined by the international diabetes federation and the national cholesterol education program: The norwegian hunt 2 study. BMC Public Health. 2007;7:220. doi: 10.1186/1471-2458-7-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.