Abstract
Percent mammographic breast density (PMD) is a strong heritable risk factor for breast cancer. However, the pathways through which this risk is mediated are still unclear. To explore whether PMD and breast cancer have a shared genetic basis, we identified genetic variants most strongly associated with PMD in a published meta-analysis of five genome-wide association studies (GWAS) and used these to construct risk scores for 3628 breast cancer cases and 5190 controls from the UK2 GWAS of breast cancer. The signed per-allele effect estimates of SNPs were multiplied with the respective allele counts in the individual and summed over all SNPs to derive the risk score for an individual. These scores were included as the exposure variable in a logistic regression model with breast cancer case-control status as the outcome. This analysis was repeated using ten different cut-offs for the most significant density SNPs (1-10% representing 5,222-50,899 SNPs). Permutation analysis was also performed across all 10 cut-offs. The association between risk score and breast cancer was significant for all cut-offs from 3-10% of top density SNPs, being most significant for the 6% (2-sided P=0.002) to 10% (P=0.001) cut-offs (overall permutation P=0.003). Women in the top 10% of the risk score distribution had a 31% increased risk of breast cancer [OR= 1.31 (95%CI 1.08-1.59)] compared to women in the bottom 10%. Together, our results demonstrate that PMD and breast cancer have a shared genetic basis that is mediated through a large number of common variants.
Keywords: breast cancer, mammographic density, SNPs, polygenic, Mendelian Randomisation
INTRODUCTION
Percent mammographic breast density (PMD) is defined as the proportion of a mammographic image occupied by radiodense tissue (largely stromal and epithelial tissues, appearing as white regions on the mammogram) as opposed to nondense, fatty tissue (the darker regions of the image). PMD is one of the strongest known risk-factors for breast cancer. Women with dense tissue in more than 75% of the breast have been shown to be at a four-five fold increased risk of breast cancer when compared to women who have mostly fatty breasts (1). The pathways through which the strong association between mammographic density and breast cancer is mediated are still unclear. Breast cancer has an estimated familial relative risk to first degree relatives of approximately two (2), and the heritability estimates for PMD range between 63 and 67% (3), hence it is plausible that these two traits share a common genetic basis.
A recent meta-analysis of five genome-wide association studies (GWAS) identified variants within intron 4 of the ZNF365 gene that are associated with PMD, adjusted for age and body mass index (BMI) (4). One of the ZNF365 variants, rs10995190, had previously been shown in a GWAS to be associated with breast cancer in the same direction as the corresponding PMD association (5). The meta-analysis also examined associations between 22 known breast cancer susceptibility loci and percent density. Two breast cancer SNPs, rs2046210 (ESR1, P=0.005) and rs3817198 (LSP1, P=0.04) were found to be associated with percent density in the same direction as determined by the corresponding breast cancer associations i.e the same allele was associated with higher density and with higher breast cancer risk (4). These findings suggest the existence of common genetic factors affecting density and breast cancer. Few, if any, other SNPs have been robustly confirmed as being associated with breast density (6).
Several recent studies have shown the potential value of using a score based on a combination of thousands of individual SNPs to predict disease status, and hence provide insights into the underlying genetic model of inheritance for that disease e.g. (7,8). To further examine evidence for shared genetic basis between density and breast cancer, we identified a set of SNPs that showed the strongest evidence of association in the combined GWAS of breast density and examined their association with breast cancer risk in a separate GWAS, to determine whether the SNPs in combination were associated with breast cancer risk.
METHODS
Breast cancer case-control study population
The present analysis was based on genotype data from the UK2 GWAS, as previously described in (5). The cases were recruited from 23 clinical genetics centres in the UK through the Familial Breast Cancer Study (FBCS) and from oncology clinics in the UK through the Prospective study of Outcomes in Sporadic versus Hereditary (POSH) breast cancer study. Cases were preferentially selected to have at least two affected first- or second-degree relatives, and had been screened to exclude BRCA1 or BRCA2 mutations. The controls used in this study were genotyped as part of the Wellcome Trust Case Control Consortium (WTCCC) study and were drawn from the 1958 Birth Cohort (a population-based study in the United Kingdom of individuals born in one week in 1958) and from the UK National Blood Service (9). After quality control exclusions, the analysis was based on data from 3628 cases and 5190 controls. The constituent studies were all approved by their appropriate ethics committees.
Breast cancer case-control genotyping
Genotypes for cases were generated using a custom Illumina Infinium 670k array and controls were genotyped using an Illumina Infinium 1.2M array at the Wellcome Trust Sanger Institute. Genotypes were called using the Illuminus algorithm; we utilised genotypes with a posterior probability >0.95. Quality controls steps are as described in (5). Briefly, the analyses were based on individuals with a call rate of >97%. We eliminated cryptic duplicates and first degree relative pairs based on identity-by-state (IBS) probabilities, and individuals with non-European ancestry based on comparison with genotypes in Hapmap phase 2. We also excluded SNPs with a call rate less than 95% if their minor allele frequency (MAF) was >5%, SNPs with a call rate of <99% if the MAF was ≤ 5%, SNPs with MAF <1% and SNPs whose frequencies departed from HWE at P < 0.00001 in controls or P<10−12 in cases. Cluster plots were inspected manually for the top SNPs as part of the UK2 GWAS. After exclusions, data were available for 531,158 SNPs that were successfully genotyped in 3,628 breast cancer cases and 5,190 controls (5)
The SNP data were used to impute genotypes for ~2.6 million SNPs in HapMap 2 CEU (release 22; build 36) using the program MACH (10). Chromosome X was imputed using HapMap 2 CEU, release 21 (build 35). Only SNPs for which the imputation r2 was greater than 0.3 were included in the analyses, where r2 is the estimated squared correlation between imputed and true genotypes.
Mammographic density measurements and genotyping
Associations between SNPs and density were based on the meta-analysis of five GWAS (totaling 4,887 individuals) previously reported by the MODE consortium (4). The five studies varied in their proportions of premenopausal women (0-46%) and in their proportions of women using hormone replacement therapy at the time of their mammogram (9-46%). All studies estimated density using the CUMULUS program (11). Percent density (dense area expressed as a proportion of total breast area), adjusted for age, BMI and population stratification (using principal components analysis), was taken as the phenotype. All women were of self-described European descent and the majority (89%) were post-menopausal at the time of mammogram. Each study used multivariate adjustments on square-root or natural logarithm transformed percent density before using appropriate regression methods to generate estimates of per-allele effects. Imputation was then used to generate statistics for ~2.6M SNPs, based on Hapmap2 CEU as a reference. One of the studies selected samples based on the upper and lower extremes of the density distribution, treated density as a binary trait and estimated per-allele odds ratios using logistic regression, while the remaining studies used samples unselected for density and analyzed density as a continuous trait using linear regression. Since the effect size estimates for the different studies were not directly comparable, the meta-analysis was performed by deriving signed Z-scores for each SNP for each study, and deriving a test statistic by summing the Z-scores weighted by the square-root of the sample size (4).
Written informed consent was given by all participants. The individual studies were approved by The Committee on the Use of Human Subjects in Research at Brigham and Women’s Hospital; The Norfolk and Norwich Hospital Ethics Committee; the Ethical Review Board at Karolinska Institutet; the Mayo Clinic Institutional Review Board and the University Health Network, Toronto, respectively.
The Sisters in Breast Screening study (SIBS)
The SIBS study of mammographic breast density was not included in the MODE meta-analysis of five GWAS (although it was included as a replication set for the most significant SNP in that study (4)), and so it was possible to test the association of the polygenic risk score with percent breast density using this set of women. The SIBS study is described in (12). Briefly, families were identified through the National Health Service breast screening program in the United Kingdom. Eligibility was restricted to families in which two or more female blood relatives (sisters, half sisters, first cousins, or aunt-niece) had had mammographic screening. The study was approved by the local research ethical committee. Study recruitment commenced in October 2002. The current analysis was limited to families whose data including mammographic density measurements were completed by July 2007.
For each participant, all available mammograms were retrieved from the local screening unit and were digitized. The mammograms were scanned using the Array 2905 Laser Film Digitizer and the program DICOM ScanPro Plus Version 1.3E (Array Corp), with 50-μm pixel resolution and 12-bit digitization, and an absorbance of 4.7. For each individual, we aimed to collect the earliest and most recently available mammograms. Mammographic density was measured using the CUMULUS program (11). Mammograms were analyzed in a random order and the reader was blinded to the sequence of the mammograms and to the visual density evaluation that had been done by another reader.
This analysis was based on 1145 women from 563 families who had been genotyped as part of an ongoing genomewide association study. Samples were genotyped by Illuminia (San Diego) using the Illumina HumanCytoSNP-12 platform. Of 1160 genotyped women, 6 were excluded on the basis of non-European ethnicity, and the twin with the lower call rate was excluded for each of the 9 pairs of monozygotic twins. The quality control steps were as for the UK2 GWAS; after exclusions genotypes were available for 184,838 SNPs. Genotype imputation was performed in the same way as for the UK2 GWAS.
Statistical Methods
All analyses were done within R and utilized the GenABEL package (13) implemented in R. A list of the most significant 10% of imputed SNPs (236,090 SNPs) from the density meta-analysis was intersected with the complete list of SNPs genotyped in the UK2 GWAS (531,158 SNPs). The 50,899 SNPs found on both lists were used in the analyses. The same exercise was performed for other cut-offs of density SNPs between 1% and 10%. As a sensitivity analysis we repeated the analyses of the 1% to 5% cut-offs using the intersection of the imputed density SNPs with the imputed SNPs in the UK2 GWAS.
To derive an overall test statistic, we assumed that each SNP associated with density would confer an approximately proportional effect on breast cancer risk (as would be expected if density were an intermediate phenotype). Specifically, we assumed that the per-allele log-relative risk of breast cancer conferred by each SNP would be (approximately) proportional to the per-allele mean density difference. The aim was to create a more informative score than would be obtained by simply summing the number of unweighted risk alleles. We further assumed that the combined effect of SNPs was additive for density and log-additive for breast cancer risk (as has been observed for susceptibility SNPs identified to date).
Under this model, the log-odds of breast cancer for individual i would be related to the density SNPs using a polygenic risk score (PRS) of the form:
where βk is the per-allele change in mean density for SNP k and Gik is the genotype for SNP k in individual i (0,1 or 2) in the breast cancer GWAS. However, because the meta-analysis was done by combining P-values, the effect sizes were only available as signed Z scores (Zk) rather than parameter estimates (βk). Although the above test statistic could be based on the Z scores rather than the βk, this would be inefficient because for a given P-value, a rarer SNP (lower MAF) would have had a higher effect estimate compared to a more common SNP (higher MAF), and hence a higher predicted relative risk of breast cancer.
For a standard linear regression analysis of density against SNP k, the standard error of βk is given approximately by:
where n is the sample size, qk and 1-qk are the allele frequencies and σ is the (residual) trait variance in the density study. For simplicity, we approximated the allele frequencies qk from the density studies by the allele frequencies pk in the breast cancer study. Hence:
so that for a given sample size, as here, βk is proportional to . (The ranked density SNPs were based on a meta-analysis of imputations carried out by each group to the same reference panel, and so we would expect n to be the same for all SNPs, except in the case of SNPs that were poorly imputed in one or more studies).
The polygenic risk score that we used for each individual was therefore of the form:
In computing the score, missing genotypes were replaced with the value 0, equivalent to assuming that the individual has a common homozygote genotype at that SNP (individuals with ≥3% of genotypes missing had been excluded).
In practice, only a subset of the SNPs will be associated with density (the remainder being false positives) and the effect sizes will often be overestimated. Nevertheless, provided that some of the associated SNPs are also associated with risk, the composite score, PRSi, should be associated with breast cancer risk. To test this hypothesis, PRS was included as the exposure variable in a logistic regression model with breast cancer case-control status as the outcome. This analysis was repeated for different cut-offs of the top density SNPs (1-10%). For the sensitivity analysis using imputed SNPs in the breast cancer GWAS (rather than just the observed SNPs) the genotypes Gik were replaced by the estimated allele doses.
To evaluate the probability of obtaining a significant association at any cut-off by chance, a permutation P-value was obtained by randomly permuting the case-control status 5000 times and repeating the association tests with the PRS calculated for each cut-off. The number of permutations for which the P-value was less than or equal to the smallest actual P-value were counted to estimate an empirical P-value. The use of a permutation approach should make the test robust to the non-independence of SNPs within the same linkage disequilibrium block.
To quantify the predictive value of the risk score, we calculated odds ratios by percentiles of PRS, using logistic regression.
To assess the association of the PRS with breast density we intersected the SNPs used to calculate the PRS in the case-control study (50,899 SNPs) with the list of SNPs successfully imputed in the SIBS study. Thus the PRS for the SIBS women were calculated using 49,912 SNPs. Percent densities for up to four mammograms per woman (both breasts and two time points) were adjusted for age at mammogram and BMI, and the mean over the four values (or over all available images, if less than four) was used. Linear regression was performed to assess the association between the PRS and adjusted percent density, allowing for the non-independence of relatives.
Statistical analyses were performed using R and Stata version 10.0. All P-values are 2-sided.
RESULTS
This study examined the association between breast cancer risk in 3,628 cases and 5,190 controls and a polygenic risk score based on a woman’s genotypes at the top ~5000 to ~50,000 ranked SNPs from a genome-wide meta-analysis of mammographic breast density. The PRS based on the top10% of density SNPs ranged between −5274 and 6982 (mean = 355, sd=1594). The mean 10% PRS was 421 (sd=1582) among the cases and 310 (sd=1600) in the controls (t-test P=0.0012). A 1-point increase in the PRS was associated with an increase in percent breast density of 6.9×10−4 percentage points (P=0.014) in the SIBS study.
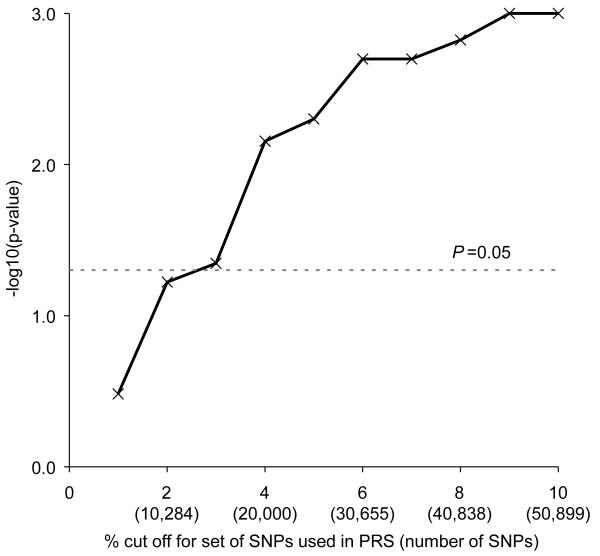
The significance of the association between PRS and breast cancer risk for different cut-offs of significant density SNPs are shown in figure 1. The PRS based on the top 1% of density SNPs (5,222 SNPs) was not significantly associated with breast cancer (P = 0.33) but the association was significant at the 5% level for all cut-offs from 3% to 10%, being most significant for cut-offs between 6 and 10% (P = 0.002 for the 6% cut-off; P = 0.001 for the 10% cut-off) (Figure 1). For example, an increase of 1000 PRS points (for the 10% cut-off) was associated with a 4.5% increase in breast cancer risk (95% CI = 1.7%-7.3%). Permutation analysis done across all 10 cut-off points (5000 permutations) gave an overall empirical P-value of 0.003.
Figure 1.
Significance of the mammographic breast density polygenic risk score (PRS) for the prediction of breast cancer risk, according to percent cut-off of SNPs used in the PRS. Significance levels were obtained using unconditional logistic regression.
The analysis was also repeated using the imputed genotype dosages for the top 1-5% of density SNPs. The results followed a similar pattern as seen with genotyped SNPs. The score based on the top 5% of imputed SNPs was significantly associated with breast cancer risk at P=0.01 (Supplementary Fig 1). Having established that the use of imputed SNPs did not change the broad pattern of the results we did not extend the imputed analysis beyond the top 5% of SNPs for computational reasons.
We next categorised individuals by their risk score based on the top 10% of genotyped density SNPs (50,899 SNPs). We chose to use the scores based on the 10% cut-off for convenience, although we note that the 10% scores are only slightly better than those obtained using the top 6% of SNPs. Women in the lowest decile of the PRS distribution were at a significantly lower risk of breast cancer than women with higher scores, with women in the top decile of PRS having an estimated OR=1.31 (95% CI 1.08-1.58) compared to the bottom decile (table 1). There does not appear to be a steady gradient in risk, with women between the 10th-25th percentiles, the 25th-75th percentiles and the 75th-95th percentiles of the PRS distribution all having an approximately 19% higher risk than the lowest 10% of women. The group of SIBS women with the highest adjusted breast density were actually those between the 75th-90th percentiles of PRS distribution (P=0.014 versus the lowest decile), but the standard deviations are large, given the relatively small numbers (table 1).
Table 1.
Breast cancer odds ratios estimated for individuals within different percentiles of the polygenic risk score calculated using the top 10% (50,899) of SNPs associated with percent mammographic density.
| UK2 study | SIBS study | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| Percentiles of the PRS |
No. casesa |
No. controlsa |
Odd Ratio (95% CI) | P-value | No. womenb | Mean PMDc (SD) |
| <10% | 306 | 519 | Baseline comparison group | 115 | −1.78 (11.8) | |
|
| ||||||
| 10-25% | 547 | 779 | 1.19 (1.00 – 1.42) | 0.055 | 172 | −1.42 (13.0) |
|
| ||||||
| 25-75% | 1830 | 2595 | 1.20 (1.03 – 1.39) | 0.022 | 572 | −0.094 (12.7) |
|
| ||||||
| 75-90% | 544 | 778 | 1.19 (0.99 – 1.42) | 0.062 | 172 | 2.01 (13.8) |
|
| ||||||
| >90% | 401 | 519 | 1.31 (1.08 – 1.59) | 0.0058 | 114 | 0.95 (15.2) |
percentiles taken from the PRS distribution in UK2 controls
percentiles taken from the distribution of the PRS in the SIBS study (values were not exactly comparable to those in the UK2 study because of the slightly different set of SNPs for which data were available).
Mean Percent Mammographic Density, adjusted for age at mammogram and body mass index (standard deviation)
DISCUSSION
In this study, we tested the hypothesis that common genetic factors contribute to the association between mammographic density and breast cancer. We calculated a polygenic risk score based on the signed Z-scores for the set of SNPs showing the strongest association with density in a meta-analysis of five breast density GWAS, and examined whether this risk score was predictive of breast cancer status. We found that a risk score based on the top 10% of SNPs was significantly associated with breast cancer risk, while the risk score based on the top 1% was not. Consistent with this result women whose SNP profile is such as to give them a risk score in the top decile of the PRS distribution had a 31% increased risk of breast cancer compared to women in the lowest decile (P=0.0058), although the effect on breast cancer risk appears only to be detectable at either extreme of the polygenic allele distribution.
The aim was not to identify the specific SNPs that are associated with both traits; we note that we observed only two SNPs that were associated with both PMD and breast cancer with P-values <10−4, one of which has been previously described (rs10995190) (4,5) and the other SNP (rs10509168) is also in the ZNF365 gene (the two are not strongly correlated, r2=0.13). Our results suggest that more such SNPs should be reliably identifiable through larger meta-analyses of GWAS. Exclusion of SNPs rs10995190 and rs10509168 in ZNF365 and all correlated SNPs (r2>0.2), made no material difference to the results (data not shown).
This study is the first to provide direct evidence from genotyping that mammographic breast density has a polygenic mode of inheritance, in line with other quantitative human traits, such as height and BMI (14). Using a Mendelian Randomisation approach (15) with the PRS as the instrumental variable, we predicted that a 1000-point increase in the PRS would be associated with a ~1.4% increase in breast cancer risk (95% CI = 0.041%-4.7%), based on our observed effect of the PRS on breast density and assuming a 1% increase in density is associated with a 2% increase in breast cancer risk (16,17). This prediction is slightly smaller than the observed OR of 1.045 per 1000 point increase in PRS (95% CI=1.017-1.073, P=0.001). This supports the hypothesis that percent breast density is causally related to breast cancer (if the observed effect had instead been non-significant we would have concluded that the frequently observed association between density and breast cancer was a consequence of residual confounding). As the observed effect was slightly larger than that predicted it might be the case that, in addition to their effect on density, these SNPs act pleiotropically to also increase breast cancer risk via other pathways, or that Cumulus-estimated percent mammographic density does not fully capture the most relevant aspects of breast composition, and thus the PRS underestimates the true effects of these SNPs. The MODE meta-analysis included density results from two breast cancer case-control studies (hence 1324 of the 4877 women in MODE were breast cancer patients), which may have resulted in some confounding by breast cancer status, which in turn could have inflated the observed effect of the density PRS on breast cancer risk. We were not able to re-analyse the meta-GWAS adjusting for case-control status, but the original report noted that for the most significant SNP (rs10995190) neither adjusting for case-control status nor excluding the cases changed the significance, suggesting that confounding was not present, at least for this SNP (4). We should also bear in mind that the set of cases used in this study were preferentially selected for an early age at breast cancer diagnosis and/or a positive family history of breast cancer, both of which may have upwardly biased the magnitude of the observed association (5).
One might have anticipated that the PRS based on the top 1% of density SNPs would show the most significant association with breast cancer risk. In fact, however, the significance of the association increased as further SNPs were added to the score, until about the 6% cut-off, beyond which there was little improvement (a pattern broadly similar to that seen in the Multiple Sclerosis polygenic score study (18).) This suggests the existence of large numbers of SNPs associated with both traits, and that these are not limited to those variants showing the strongest associations with density. The flattening out of the graph beyond the 6% cut-off may indicate a lack of power to detect variants with smaller shared effects, rather than an absence of such variants. Although one of the strengths of this study is the large number of samples in the density meta-GWAS from which the SNP rankings were obtained, the lower significance seen for the top 1% of SNPs may nevertheless point to a lack of power in the density study, resulting in imprecision in the ranking; an ongoing meta-analysis incorporating additional individual density studies may improve the precision with which SNPs are ranked. It should also be noted that this study only considered SNPs associated with percent mammographic density. It is possible that the absolute volume of dense tissue in the breast is more relevant for breast cancer risk, and hence that a polygenic risk score based on SNPs associated with absolute dense area (as a hypothesised better surrogate for volume) would show stronger evidence of a shared genetic basis with breast cancer risk.
To the best of our knowledge this is the first such study to use SNPs ranked on the basis of their association with a quantitative trait to predict risk of a related disease. Other groups have used similar approaches to obtain disease-risk information from SNPs that are not individually significant, with mixed results. For example, using WTCCC data Evans et al (8) found that disease-specific polygenic scores gave only limited discrimination between cases and controls of the same disease for bipolar disorder, coronary heart disease, hypertension, Crohn’s disease, rheumatoid arthritis and type II diabetes, and that the discriminatory ability of the type I diabetes score was markedly reduced after the exclusion of SNPs in and around the MHC (a region known to be associated with type I diabetes). However, separate studies of Multiple Sclerosis and schizophrenia both found significant associations between disease risk and scores based on multiple SNPs, with the schizophrenia-derived polygenic score also predicting the risk of bipolar disease, but not of the six non-psychiatric diseases in the WTCCC study described above (7,18). Our results are in contrast to those of Machiela et al (19) who, using a related approach, found no association between an unweighted polygenic risk score constructed using ranked SNPs from a breast cancer GWAS with breast cancer risk, after excluding 13 established breast cancer SNPs. Witte and Hoffmann also found that a similar polygenic model did not significantly predict breast cancer status (20). This presumably reflects the low power of both studies due to small training and testing sets (<1200 cases and controls in total for each) and demonstrates the need for large studies when considering variants with small effects. The power of our study was additionally boosted by the enrichment of the set of breast cancer cases for a positive family history of the disease.
Theoretically, the association we observed could result from population stratification, if there were strata with higher density and higher breast cancer risk. However, this is unlikely since the GWAS for density were conducted in several populations in which there was little evidence of population stratification. Moreover, the individual risk scores were computed using many, largely unlinked tag-SNPs that are highly unlikely to be related to population structure. In the present analysis, for individuals with missing genotype data, the missing genotypes were replaced with the value 0, which is equivalent to assuming that the individual has a common homozygote genotype at that SNP. An alternative approach would have been to substitute the missing genotype count with the mean count of risk alleles for the respective SNP. However, this should not make any material difference as only individuals with at least 97% call rates were included in the breast cancer GWAS and the call rates were not significantly different among cases and controls (5). A recent study reported no significant difference in their results with or without substitution of missing genotypes (21).
This analysis demonstrates directly that a suitably chosen set of SNPs that do not reach genome-wide significance levels individually for one trait can nevertheless predict a genetically related trait when used in combination. The method we applied here could be used to look for shared genetic influences between other risk factors and breast cancer, or indeed between any pair of correlated heritable traits.
The recent identification of common SNPs associated with breast cancer risk indicate that susceptibility to breast cancer is at least partly polygenic (22). Our results suggest that a much larger number of common variants are involved, and, moreover that there is at least some overlap between the set of genetic variants involved in breast density and those involved in breast cancer risk. This observation in turn confirms that the well-established correlation between the two traits is genuine, and not the result of uncorrected confounding. It is not possible at this stage to determine whether the observed effect is attributable to variants which increase cancer risk via the creation of a dense breast, or to variants which act pleiotropically to affect both traits, or to a mixture of both types of variants. The identification of the relevant variants should help to elucidate the biological mechanisms underlying breast density and the reasons for its association with breast cancer.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge the role of Tina Audley in co-ordinating the SIBS study.
Funding:
MODE breast density GWAS: This study was supported by Public Health Service Grants CA131332, CA087969, CA049449 from the National Cancer Institute, National Institutes of Health, Department of Health and Human Services. The NHS breast cancer cases and controls were genotyped with support from the National Cancer Institute’s Cancer Markers of Susceptibility (CGEMS) initiative. Data evaluation of mammograms and analysis of the EPIC-Norfolk study was supported by Cancer Research UK. The SASBAC study was supported by Märit & Hans Rausing’s Initiative against Breast Cancer, National Institute of Health, Susan Komen Foundation and Agency for Science, Technology and Research of Singapore (A*STAR). Genotyping in the TORONTO/MELBOURNE subjects was supported by the Campbell Family Institute for Breast Cancer Research. Support was also provided by the Ontario Ministry of Health and Long Term Care.
UK2 breast cancer GWAS: This work was supported by the Wellcome Trust and by Cancer Research UK. C.T. is funded by a Medical Research Council Clinical Research Fellowship. The samples were collected and screened for BRCA mutations through funding from Cancer Research UK; US Military Acquisition (ACQ) Activity, Era of Hope Award (W81XWH-05-1-0204) and the Institute of Cancer Research (UK). This study makes use of data generated by the Wellcome Trust Case Control Consortium (WTCCC) 2. A full list of the investigators who contributed to the generation of the data is available from the WTCCC website. We acknowledge use of DNA from the British 1958 Birth Cohort collection, funded by the Medical Research Council grant G0000934 and the Wellcome Trust grant 068545/Z/02. Funding for this project was provided by the Wellcome Trust under award 085475.
JSV is funded by The Cambridge Commonwealth Trust and a Cambridge Overseas Research Scholarship. D.F.E. is a Principal Research Fellow of Cancer Research UK
References
- 1.McCormack VA, dos Santos Silva I. Breast density and parenchymal patterns as markers of breast cancer risk: a meta-analysis. Cancer Epidemiol Biomarkers Prev. 2006;15:1159–69. doi: 10.1158/1055-9965.EPI-06-0034. [DOI] [PubMed] [Google Scholar]
- 2.Collaborative Group on Hormonal Risk Factors in Breast Cancer Familial breast cancer: collaborative reanalysis of individual data from 52 epidemiological studies including 58,209 women with breast cancer and 101,986 women without the disease. Lancet. 2001;358:1389–99. doi: 10.1016/S0140-6736(01)06524-2. [DOI] [PubMed] [Google Scholar]
- 3.Boyd NF, Dite GS, Stone J, Gunasekara A, English DR, McCredie MR, et al. Heritability of mammographic density, a risk factor for breast cancer. N Engl J Med. 2002;347:886–94. doi: 10.1056/NEJMoa013390. [DOI] [PubMed] [Google Scholar]
- 4.Lindstrom S, Vachon CM, Li J, Varghese J, Thompson D, Warren R, et al. Common variants in ZNF365 are associated with both mammographic density and breast cancer risk. Nat Genet. 2011;43:185–87. doi: 10.1038/ng.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turnbull C, Ahmed S, Morrison J, Pernet D, Renwick A, Maranian M, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42:504–7. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelemen LE, Sellers TA, Vachon CM. Can genes for mammographic density inform cancer aetiology? Nat Rev Cancer. 2008;8:812–23. doi: 10.1038/nrc2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–52. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans DM, Visscher PM, Wray NR. Harnessing the information contained within genome-wide association studies to improve individual prediction of complex disease risk. Hum Mol Genet. 2009;18:3525–31. doi: 10.1093/hmg/ddp295. [DOI] [PubMed] [Google Scholar]
- 9.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Byng JW, Boyd NF, Fishell E, Jong RA, Yaffe MJ. The quantitative analysis of mammographic densities. Phys Med Biol. 1994;39:1629–38. doi: 10.1088/0031-9155/39/10/008. [DOI] [PubMed] [Google Scholar]
- 12.Kataoka M, Antoniou A, Warren R, Leyland J, Brown J, Audley T, et al. Genetic models for the familial aggregation of mammographic breast density. Cancer Epidemiol Biomarkers Prev. 2009;18:1277–84. doi: 10.1158/1055-9965.EPI-08-0568. [DOI] [PubMed] [Google Scholar]
- 13.Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–96. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 14.Yang J, Manolio TA, Pasquale LR, Boerwinkle E, Caporaso N, Cunningham JM, et al. Genome partitioning of genetic variation for complex traits using common SNPs. Nat Genet. 2011;43:519–25. doi: 10.1038/ng.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheehan NA, Didelez V, Burton PR, Tobin MD. Mendelian randomisation and causal inference in observational epidemiology. PLoS Med. 2008;5:e177. doi: 10.1371/journal.pmed.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyd NF, Byng JW, Jong RA, Fishell EK, Little LE, Miller AB, et al. Quantitative classification of mammographic densities and breast cancer risk: results from the Canadian National Breast Screening Study. J Natl Cancer Inst. 1995;87:670–675. doi: 10.1093/jnci/87.9.670. [DOI] [PubMed] [Google Scholar]
- 17.Mitchell G, Antoniou AC, Warren R, Peock S, Brown J, Davies R, et al. Mammographic density and breast cancer risk in BRCA1 and BRCA2 mutation carriers. Cancer Res. 2006;66:1866–72. doi: 10.1158/0008-5472.CAN-05-3368. [DOI] [PubMed] [Google Scholar]
- 18.Bush WS, Sawcer SJ, de Jager PL, Oksenberg JR, McCauley JL, Pericak-Vance MA, et al. Evidence for polygenic susceptibility to multiple sclerosis--the shape of things to come. Am J Hum Genet. 2010;86:621–25. doi: 10.1016/j.ajhg.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Machiela MJ, Chen CY, Chen C, Chanock SJ, Hunter DJ, Kraft P. Evaluation of polygenic risk scores for predicting breast and prostate cancer risk. Genet Epidemiol. 2011 doi: 10.1002/gepi.20600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Witte JS, Hoffmann TJ. Polygenic modeling of genome-wide association studies: an application to prostate and breast cancer. OMICS. 2011;15:393–98. doi: 10.1089/omi.2010.0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li S, Zhao JH, Luan J, Ekelund U, Luben RN, Khaw KT, et al. Physical activity attenuates the genetic predisposition to obesity in 20,000 men and women from EPIC-Norfolk prospective population study. PLoS Med. 2010;7 doi: 10.1371/journal.pmed.1000332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pharoah PD, Antoniou A, Bobrow M, Zimmern RL, Easton DF, Ponder BA. Polygenic susceptibility to breast cancer and implications for prevention. Nat Genet. 2002;31:33–36. doi: 10.1038/ng853. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.