Abstract
Chlamydia trachomatis is responsible for both trachoma and sexually transmitted infections causing substantial morbidity and economic cost globally. Despite this, our knowledge of its population and evolutionary genetics is limited. Here we present a detailed whole genome phylogeny from representative strains of both trachoma and lymphogranuloma venereum (LGV) biovars from temporally and geographically diverse sources. Our analysis demonstrates that predicting phylogenetic structure using the ompA gene, traditionally used to classify Chlamydia, is misleading because extensive recombination in this region masks true relationships. We show that in many instances ompA is a chimera that can be exchanged in part or whole, both within and between biovars. We also provide evidence for exchange of, and recombination within, the cryptic plasmid, another important diagnostic target. We have used our phylogenetic framework to show how genetic exchange has manifested itself in ocular, urogenital and LGV C. trachomatis strains, including the epidemic LGV serotype L2b.
INTRODUCTION
C. trachomatis is the most prevalent bacterial sexually transmitted infection (STI) world-wide, with an estimated 101.5 million new cases among adults in 20051. Additionally, ocular C. trachomatis is the leading infectious cause of blindness, with >40 million people estimated to be suffering from active disease2.
C. trachomatis comprises two biovars: the trachoma biovar includes ocular and urogenital strains characterized by localised infections of the epithelial surface of the conjunctiva or genital mucosa; strains of the LGV biovar are distinguished by their ability to spread systemically thorough the lymphatic system, causing genital ulceration and bubonic disease3. LGV is reported most frequently in Africa, South East Asia, South America and the Caribbean, while being rare in developed countries4-6. However, an ‘epidemic’ of LGV exhibiting atypical presentation and symptoms is currently in progress in Europe and North America, primarily affecting men who have sex with men (MSM).
Despite the obvious importance of C. trachomatis as a human pathogen, very little is known about the evolution of strains causing disease7. This is primarily because modern diagnosis is generally based on commercial nucleic acid amplification tests (NAATs)8,9 rather than culture, where strains would be available for further study. Most of our understanding of the diversity of circulating C. trachomatis is based on the primary surface antigen, the major outer membrane protein (MOMP) and its gene, ompA. Typing has traditionally been performed serologically using a number of antibodies against divergent epitopes within MOMP, but has more recently switched to a genotyping approach using the ompA sequence. Based on these methods, the two C. trachomatis biovars have been subdivided into 15-19 serotypes: the trachoma biovar includes ocular serotypes A to C and urogenital serotypes D to K, while the LGV biovar includes serotypes L1, L2, L3 and L2b, the serotype associated with the current LGV outbreak in Europe and North America. Although ompA genotyping provides some further differentiation within these serotypes, it provides little or no detailed information about the nature of the infecting strain or the variation in the remaining 99.88% of the genome.
Attempts have been made to reconstruct the C. trachomatis species phylogeny using ompA sequences10, 16S rRNA gene analysis11, multilocus sequence typing (MLST) approaches12-15 and whole genome sequencing16-18. However, there is still no consensus over the true evolutionary relationships between C. trachomatis strains. It is generally accepted that ompA does not reflect the phylogeny of the species14,19,20, but the lack of agreement between trees produced from other small gene sets suggests that many other regions of the C. trachomatis genome also provide conflicting phylogenetic evidence19.
Historically, horizontal gene transfer in the chlamydiae was considered unlikely due to their obligate intracellular niche and because co-infection, and therefore opportunities for recombination, are rare. Together these factors were thought to represent a barrier for effective recombination. However, it has recently become evident that not only do the chlamydiae have all the necessary recombination machinery21, they can recombine following mixed infection both in tissue culture and the human host17. It is likely that this recombination is the cause of the difficulty in resolving the relationships of C. trachomatis strains.
Recent attempts to quantify the impact of recombination by reanalysing published genomes have been limited by the small number available18. We set out to sample widely from the diversity of clinical strains, and present 36 new C. trachomatis whole genome sequences. Using these sequences and those previously published, we provide a detailed reconstruction of the evolutionary history of C. trachomatis that identifies and takes into account the effect of recombination. We show conclusively that the epidemic outbreak of L2b strains in Europe was the result of clonal expansion and transmission, and provide evidence of recombination in natural clinical strains both within and between biovars. We show the impact that recombination has had on our general understanding of C. trachomatis diversity, its implications for monitoring and epidemiological tracking of infections based on current typing techniques, and provide evidence for the first time of recent serotype switches within circulating clinical strains.
RESULTS
We sequenced 36 C. trachomatis genomes from global collections, isolated between 1959 and 2009, comprising 18 LGV, 14 urogenital and 4 ocular strains. Included are 12 L2b strains, which originate from the UK, France, Sweden, the Netherlands and Canada, and a further 6 LGV strains from South Africa and the USA, including historical L1 and L3 strains. The newly sequenced ocular strains are from Tanzania in 2000. Urogenital strains were collected in the UK (10 strains), Sweden (3) and the USA (1). To guard against potential cross contamination of strains and provide an independently verifiable set of reference genomes, a key element of our experimental design was to use live strains that were cultured individually. All strains are held as stocks except for three ocular strains that were destroyed after culture. Our analysis also includes 16 previously published genome sequences: 2 LGV (UK, USA), 3 ocular (Egypt, Tanzania and The Gambia) and 11 urogenital strains (USA, Sweden and not described), making a total of 52 strains included in this study (see Supplementary Table 1).
Whole-genome phylogeny of C. trachomatis
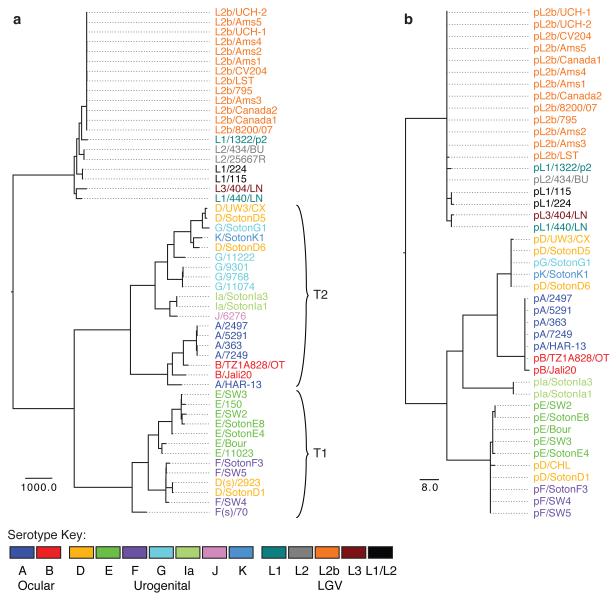
To establish whether the current understanding of C. trachomatis phylogeny is evolutionarily robust, we determined the interrelationships of our strain collection using genome-wide single nucleotide polymorphisms (SNPs). To mitigate the effect of homologous recombination between C. trachomatis strains on our phylogenetic reconstruction, we employed the method of Croucher et al.22. The resulting tree (Fig. 1a, see Fig. 1b for plasmid phylogeny, Supplementary Fig. 1 for bootstrap support values and Supplementary Fig. 2 for trees without recombination removed) provides compelling evidence that ompA serotyping (Supplementary Fig. 3a) does not reflect the evolutionary structure of C. trachomatis, with trachoma serotypes A, B, D, F, G and LGV serotype L1 all occurring in multiple distinct branches on the tree. This clearly demonstrates that exchange of the whole or part of the ompA gene is a natural phenomenon in distinct lineages of C. trachomatis. Comparison of the whole-genome phylogeny with those reconstructed using some of the available C. trachomatis MLST schemes (Supplementary Fig. 3b-d) demonstrates that multi-locus techniques based on housekeeping loci show greater congruency with our tree than the ompA phylogeny, but lack resolution.
Figure 1. Maximum likelihood reconstruction of the phylogeny of C. trachomatis with recombinations removed.
(a) C. trachomatis species phylogeny using the chromosomal sequences of 52 genomes after predicted recombinations have been removed using the method described in Croucher et al.22. Bootstrap support for nodes on the tree are shown in Supplementary Figure 1. (b) Phylogenetic reconstruction of the C. trachomatis plasmid after predicted recombinations have been removed. Strain names are colored by serotype, see key. Scale bar represents number of SNPs. Plasmid sequences were not available for all strains in a. For comparison, trees without recombination removal are shown in Supplementary Figure 2.
The whole genome tree confirms that the species is split into two distinct clades representing the trachoma and LGV biovars, separated by 4,860 SNPs. Using Chlamydia muridarum NIGG as an outgroup, the root of the tree falls on the branch between the two biovars (Supplementary Fig. 1), suggesting that this split occurred early in the evolutionary history of the species, consistent with the conclusions of previous analyses based on 16S rRNA gene sequences23.
The trachoma clade comprises two lineages (Fig. 1a T1 and T2) separated from their common ancestor by 2,374 and 2,228 SNPs, respectively. T1 is composed of clinically prevalent urogenital serotypes, while T2 contains most of the rarer urogenital serotypes24. All ocular strains form a cluster within T2, indicating that they emerged from a urogenital ancestor (Fig. 1a).
Strains within the LGV clade exhibit a far lower level of diversity than those in the trachoma lineage, illustrated by the shorter branch lengths in Figure 1a. The 13 strains from the recent L2b outbreak form a tight cluster with maximal bootstrap support (Supplementary Fig. 1). The maximum pairwise evolutionary distance between the most variant strains within the L2b serotype is just 19 SNPs. This low level of variation between the strains despite their global distribution shows conclusively that the L2b epidemic25 is a clonal outbreak that has spread throughout the world.
Our LGV collection also includes two South African strains (L1/115 and L1/224) that could not originally be classified using micro-immunofluorescence with monoclonal antibodies specific to the known MOMP types26. Sequence analysis of the ompA gene in these strains showed that the 5′ end (variable segments VS1-2) matched the L1 ompA sequence, while the 3′ end (VS3-4) matched L226. In our analysis, these strains form a clade midway between an L1+L3 clade and a group including the L2 and L2b strains. A third South African strain (L1/1322/p2), despite being genotyped as L1, is located on its own branch distinct from the other L1 strain in this study (L1/440/LN). It is evident that these three strains collected in South Africa define two new LGV lineages that are equally distinct as the accepted LGV serotype clusters. This suggests that LGV diversity is far greater than previously recognized, and that additional sampling is necessary.
Recombination is a natural phenomenon occurring widely within C. trachomatis strains
Considering the traditional view that Chlamydia do not recombine, we would expect that, given the low level of variation identified relative to the size of the genome, very few identical SNPs would have occurred multiple times independently on different branches of the tree (homoplasies). However, superimposing the SNP events on the rooted phylogenic tree using PAML27 shows that homoplasy is commonplace. Of the 17,163 sites in the genome that exhibit a SNP in at least one strain (variant sites), 4,492 (26%) are homoplasic.
Homoplasic SNPs can arise by chance, as a result of a selective pressure for a particular SNP to become fixed in a population, or by homologous recombination between divergent strains where sets of SNPs are effectively imported in one block. In cases where recombination is the cause of homoplasy, it is expected that homoplasic SNPs shared by two distant strains would be clustered along the genome sequence within the regions that have been recombined. Therefore, dense clusters of compatible homoplasies are often used as markers of recombination events. A pair of sites are incompatible if no tree can be drawn upon which both sites could be reconstructed without at least one being homoplasic28. We applied three compatibility-based recombination detection methods implemented in the PhiPack package29: maximum χ230, neighbor similarity score (NSS)31 and the pairwise homoplasy index (PHI)29 to these data. All three methods reported significant p-values (<< 0.05), indicating that compatibility is significantly higher between closely linked sites, as would be expected if recombination had occurred, but not consistent with random accumulation of homoplasies or convergent selection.
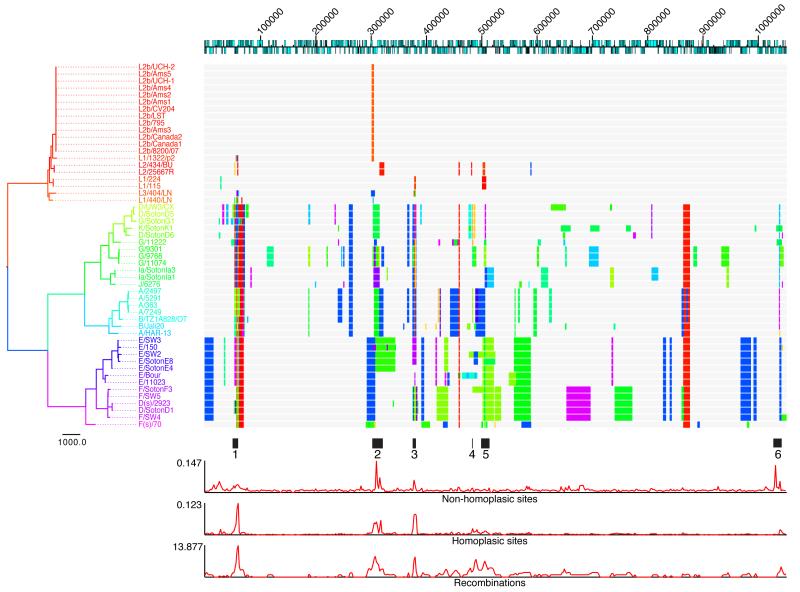
To investigate the recombination history, for each node on the tree we reconstructed the ancestral DNA sequence using PAML27, and used a scanning statistic22 to identify regions of the genome for which the node sequence was significantly more similar to a distant branch on the tree than to its direct ancestral node (see methods). Figure 2 shows a reconstruction of recombination events across the C. trachomatis tree, where the location of the recombination in the genome of the recipient branch is shown, and colors illustrate the phylogenetic position of the most likely donor. In total, 267 putative recombination blocks ranging from 3 to 50,141bp (mean = 4039bp) were reconstructed, covering 539,409bp (51%) of the aligned genome length. This provides strong evidence that recombination has occurred many times during the evolution of the species and has not only happened at a few limited ‘hotspots’ as reported in previous papers18,32, but across a large proportion of the genome. SNP density along the length of the genome is not constant, however. Some regions do show increased SNP and homoplasy density, which may be the result of diversifying selection within the species or homologous recombination importing pre-selected SNPs from outside of the species. Of particular note are six regions of the genome in which SNP density, homoplasy density and recombination density are all significantly raised (Fig. 2). These are in ompA (region 1), region 2 encoding the polymorphic outer membrane proteins, and region 3 predicted to encode four hypothetical proteins (CTL304-CTL307) and the 3′ end of hemN. Recombination density is also raised around region 4, encoding a putative membrane protein (CTL0399), region 5 spanning the plasticity zone and region 6 carrying genes predicted to encode a conserved hypothetical protein (CTL0886), a putative exported protein (CTL0887) and a putative virulence protein (mviN). Four of these regions (1-3 and 6) also show raised levels of non-homoplasic SNPs, which may be the result of further recombinations from lineages not represented on our tree.
Figure 2. Reconstruction of recombination events on the species phylogeny of C. trachomatis.
The top line represents the full chromosome structure of C. trachomatis based on L2/434/BU, with CDSs represented as blue boxes on the relevant coding strand. Numbers indicate position in the genome alignment, beginning at CTL0001 (L2/434/BU accession number AM884176). Each horizontal track represents the chromosome of a strain in the species phylogeny on the left. Blocks shown on the tracks represent the location of received homologous replacements, with their color corresponding to the color of the donor branch on the tree. Tree branches and taxon names are colored by phylogenetic distance, with more similar colors representing more closely related branches. Regions of interest along the genome are highlighted immediately below the recombination tracks. Below this are plots of the density of non-homoplasic SNP sites, homoplasic SNP sites and recombination events, based on a moving window analysis. Window size = 2000bp.
Evidence of recombination within and between biovars
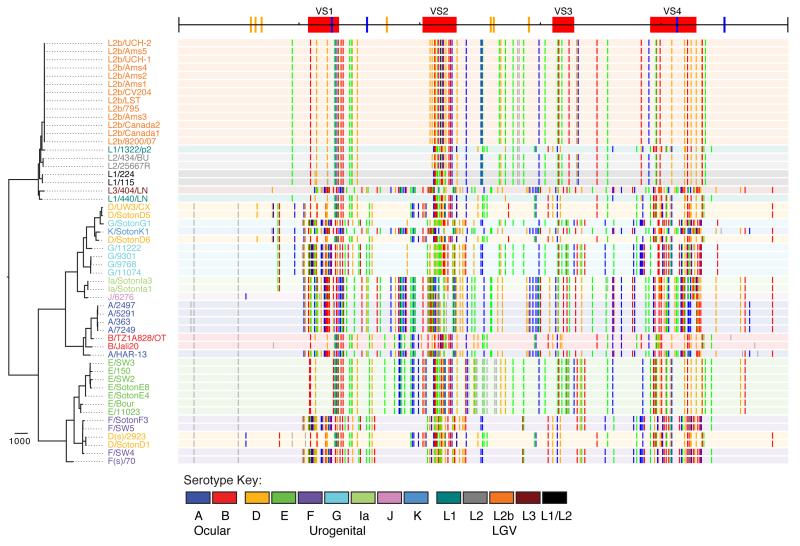
Within the LGV clade, 5% (46) of the 920 variant sites are homoplasic. Twelve recombinations were reconstructed within the clade, all of which are between different LGV serotypes (Supplementary Fig. 4) and six of which coincide with regions of the genome that were also found to recombine in other C. trachomatis lineages (Fig. 2; regions 1, 2 and 3). Three recombinations were identified as affecting ompA, and can be interpreted by visualizing the SNP distribution as a genetic ‘barcode’ (Fig. 3). A region of almost 3kb, from the 3′ end of the gene encoding the translation elongation factor, tsf, through the 3′ end of ompA to VS2 (Fig. 3) appears to have been replaced by homologous recombination between L1/440/LN and L1/1322/p2, explaining why these two strains, which are distinct on the tree, are both typed as L1. A second recombination of only 25bp in the VS2 region of ompA was identified as an exchange between the L2 and L2b clades (Fig. 3), explaining the reported serotypic results, as the two South African strains, typed as L1/L2 hybrids based on sequence (L1/115 and L1/224, Fig. 3), differ from the archetypal L2 sequence in this region. The third recombination is also located in VS2, between L1/404/LN and the L2 clade.
Figure 3. Distribution of SNPs in the ompA gene of C. trachomatis.
The top line represents the structure of the ompA gene, showing the location of variable regions (VS1-4, red blocks) and cysteine residues (conserved in blue, non-conserved in orange). On the left is the species phylogeny of C. trachomatis, with strain names colored by serotype, see key. Adjacent to each strain name is a track with a background color based on the serotype of the corresponding strain. Colored vertical lines along the tracks represent bases that differ from the ancestral sequence: grey=non-homoplasic change; colored lines represent homoplasic bases: red=A, blue=T, green=C, orange=G. The pattern of lines provide a barcode of ompA similarity between strains.
More surprisingly, there is clear evidence of recombination between the LGV and trachoma biovars, with 24 events reconstructed. 14 of these affect the ompA gene, with a particularly clear example being the similarity of the L3 ompA sequence to that of trachoma serotypes Ia, J and K (Fig. 3), which was noted when the K serotype was first recognized33. Outside ompA, the clearest inter-biovar recombination is a region of 665bp within the recD gene in which 47 SNPs have been transferred from the T2 trachoma lineage to the two African LGVs, L1/115 and L1/224 (Supplementary Fig. 5).
Within the trachoma biovar, recombination events are far more prevalent. 3,367 (26.2%) of the 15,902 variant sites exhibit homoplasy, and 162 intra-biovar recombination events were reconstructed (Fig. 2), covering 446,917bp (43%) of the length of the aligned genomes. Figure 3 highlights the extent of ompA swapping between trachoma strains. Not only have multiple serotype switches occurred, but the SNP barcodes of the ompA gene show clearly that serotypes with similar barcodes (sequences) do not necessarily fall near each other on the tree (Fig. 3). 43 recombination events were also reconstructed between ocular and urogenital branches, clearly indicating that DNA exchange occurs between these two biotypes (Fig. 2).
Evidence of recent recombination
Our analyses provide convincing evidence that recombination is an ongoing process rather than a purely historical event in the evolution of C. trachomatis. This can be clearly illustrated with two examples.
Seth-Smith et al.16 described the genome of a serotype B strain (B/TZ1A828/OT) from Kongwa, central Tanzania. In this study we have included new genomes from four serotype A strains (A/2497, A/363, A/5291 and A/7249) from Rombo, Northern Tanzania, 550km from Kongwa. Our phylogenetic reconstruction placed these new strains as the sister group to B/TZ1A828/OT rather than the reference serotype A strain, A/HAR-13. Analysis of the SNP differences between the genomes of B/TZ1A828/OT and a representative (A/2497) of the new serotype A strains (Supplementary Fig. 6a) shows that 434 of the 720 SNPs differentiating them are located in a 10kb region (4.34% divergence) around ompA (from aspC to pbpB) (Supplementary Fig. 6b), such that the ompA genes are highly divergent, yet the remaining 99% of the genome differs at only 286 nucleotide sites (0.027% divergence). The converse is true if the reference A/HAR-13 is compared with A/2497. In this comparison there are 1,242 SNP differences, with only 170 found in the same 10kb region in and around ompA, and 1,072 in the remainder of the genome (Supplementary Fig. 6a-b). Closer inspection shows that the ompA gene itself is almost identical between the genomes of A/2497 and A/HAR-13 (differing by 5 SNPs), while the ompA sequence of B/TZ1A828/OT is highly divergent (199 SNPs) (Supplementary Fig. 6c), explaining why the four new strains are serotype A. This is clear evidence for a recent recombination event where a divergent ompA gene has replaced the existing ompA sequence and changed the serotype of the circulating clone. Although it is difficult to be certain of the direction of the change: from serotype A to B or vice-versa, this example underlines the need to use genome-wide SNPs to be confident of the phylogenetic history of Chlamydia strains.
We also see evidence for a recent recombination event within the new-variant (nvCT) Swedish serotype E strain (E/SW2)34. Comparison with another Swedish serotype E strain (E/SW3) shows close similarity along most of the genome length, excepting a large number of SNPs in a 30kb region spanning CTL393 to CTL417 (Supplementary Fig. 6d). The sequence of this region in E/SW2 exactly matches the homologous region in D/UW3/CX (Supplementary Fig. 6d), indicating a recombination event in E/SW2 from a D/UW3/CX-like donor. The exchanged genes are of unknown function and phenotypic comparison of nvCT to other serotype E strains has identified no differences35.
Plasmid phylogeny and recombination
Previously16 we showed that the cryptic plasmids of a small sample of C. trachomatis strains share the same evolutionary history as their chromosomes, suggesting that the plasmid is not (or rarely) exchanged. Analyzing our much larger dataset of 43 plasmids for evidence of recombination and exchange showed a single difference in phylogenetic structure between the chromosomal and plasmid trees (Fig. 1) relating to two serotype Ia strains from Southampton, UK. In the whole genome analysis, these two strains group in the T2 cluster, while in the plasmid phylogeny they form a distinct branch between the T1 and T2 clades (Fig. 1b). Reconstructing SNP events across the plasmid tree shows that the SNPs supporting the positioning of these Ia strains are distributed along the length of the plasmid sequence (Supplementary Fig. 7): this is evidence of replacement of the Ia lineage plasmid with an equivalent plasmid, potentially from an unsampled C. trachomatis lineage. All other relationships are congruent between the plasmid and chromosome trees, suggesting that exchange of whole plasmids between distant C. trachomatis strains is indeed rare. We identified just 7 homoplasies on the plasmid tree (Supplementary Fig. 7). Applying the methods for recombination detection detailed above, we identified a single recombination event that accounted for 6 of these homoplasies (Supplementary Fig. 7). The recombination region extends from the middle of pCDS3 to the middle of pCDS5 and can be best explained by homologous recombination of this region between the ocular strains and the T1 clade. By reconstructing phylogenies of the recombination region and the rest of the plasmid it can be seen that the recombined region in the ocular strains clusters in the T1 clade rather than T2 (Supplementary Fig. 8). Alternative explanations would be recombination between the Ia plasmid and the T2 clade, or that the Ia plasmid is a chimera between an T2 plasmid and an unknown donor plasmid. We think the latter is unlikely, as the Ia plasmid does not cluster within a T2 clade in either the backbone or recombined regions.
A further interesting discovery is a previously unrecognized deletion in the plasmid of LGV strain pL3/404/LN within pCDS1 (Supplementary Fig. 7) in a different location to that of the nvCT, providing further evidence that pCDS1 is not a stable diagnostic target.
DISCUSSION
In the past, studies of the evolutionary history and diversity of C. trachomatis have almost always been based on serological classification or the sequence of a small number of genes. We have shown that these loci do not necessarily represent the true history of the species, or the true relationships between strains, and that an in-depth understanding of the population structure of the chlamydia requires the maximum resolution available: whole genome data. This resolution has allowed us to observe the extent of recombination that has occurred in the population and raises a number of issues of clinical significance.
Our analysis has confirmed that C. trachomatis comprises three distinct lineages. The species appears to have split early into LGV and urogenital clades, with the urogenital clade itself later splitting into two clades termed T1 and T2. Ocular Chlamydia is a younger lineage that, within the limits of our sampling frame, appears to have emerged only once from a urogenital ancestor within T2, although more data are needed to confirm this observation. Within the LGV biovar, the emergence of the ‘epidemic’ L2b is a clonal expansion that probably resulted from a single introduction into Europe or North America. We suggest that the lack of variation seen within the L2b genomes is indicative of relatively rapid transmission (rather than a lower rate of recombination) as exemplified by the emergence and spread of the Swedish nvCT, which evaded detection by some NAATs due to a deletion in the first coding sequence (CDS) of the plasmid. The nvCT and L2b outbreaks have shown that given a selective advantage or lack of competition, a single lineage of C. trachomatis can proliferate and spread. However, for lineages with more subtle changes occurring outside either ompA or diagnostic primer binding sites,such as drug resistance, it would be more difficult to detect such a clonal expansion using current diagnostic and molecular typing techniques. Fortunately, until now there have been only sporadic reports of antibiotic resistance in clinical strains36-39 despite the fact that it is possible to induce resistance in the laboratory40.
Although recombination in C. trachomatis was once a controversial concept it has more recently been shown to occur both in laboratory tissue culture17 and naturally between clinical strains17,20,32,41,42. We have extended these observations by demonstrating that recombination is not limited to a few ‘hotspots’ around the chromosome. Rather than true hotspots, it is likely that previous studies18,32 have simply observed higher rates of fixation of recombinations in genomic regions that are under diversifying selection pressure.
In contrast to a previous analysis18, our more comprehensive assessment of C. trachomatis diversity shows greater exchange between strains with tropism for the same site or tissue, which correlates with the majority of reports of mixed infections43-47. However, we have identified multiple examples of recombination between strains with tropisms for different tissues, and even between biovars, suggesting that there are no absolute barriers to genetic exchange. In particular, recombination between ocular and urogenital strains appears relatively frequent, again correlating with reports of both cross-site and mixed infections of ocular and urogenital serotypes47-50.
The clinical significance of our findings is extensive. We have shown clear examples in which the genetic backbone of the strain is unlinked from its serotype.
Replacement and chimaerism of ompA is most likely a process of diversification that counteracts the effect of the immune system protecting the host against immediate reinfection, and as such it is clear that the ompA gene, the main chromosomal diagnostic target used to detect C. trachomatis infections and to type strains, is a poor indicator of genetic relatedness within the species. This may explain why there are equally as many studies that have failed to draw significant association between disease severity and the nature of the infecting strain51-55 as those that have found an association44,56-59.
Multi-locus typing schemes, particularly those based on housekeeping genes under low selective pressure, more closely reflect the genome phylogeny, and may prove useful in cases where the ultimate resolution of genome-wide SNP-based techniques is not necessary. However, any scheme based on a small number of loci has the potential to be confused by recombination, so that different MLST schemes will differ in resolution and accuracy (Supplementary Fig. 3b-d). The choice of typing approach will need to be determined on a case-by-case basis depending on the resolution required to answer the question at hand.
Finally, we have shown for the first time that there has been some exchange of, and homologous recombination within, the DNA of the cryptic plasmid, providing further evidence of the potential unreliability of the plasmid for diagnostics and typing.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the core informatics, library-making and sequencing teams at the Wellcome Trust Sanger Institute. S.R.H. is grateful for the opportunity to discuss this project at the Permafrost conference. This work was funded by the Wellcome Trust grant number WT076964. B.G.S. was funded by the Wellcome Trust.
Appendix
ONLINE METHODS
Cell culture, DNA extraction and sequencing
The strains and sources of C. trachomatis used in this work are summarized in Supplementary Table 1. Cell culture and DNA extraction was performed as Seth-Smith et al.16 for all strains except A/363, A/5291 and A/7249, which were extracted from a single 24 well plate using 1N NaOH followed by neutralization with Tris. The genome of A/2497 was sequenced to a depth of 12x coverage derived from pUC18 (insert size 0.7 kb) small insert libraries using dye terminator chemistry on ABI3700 automated sequencers. End sequences from larger insert plasmid (pMAQ1 9-12 kb insert size) libraries were used as a scaffold. Sequencing, assembly, finishing and checking of this genome was as described60. The genome sequence of strain L2b/UCH-1 was improved with Illumina GAII data, 2,877,472 37bp paired end reads using iCORN61. All other genomes were sequenced using Illumina Genome Analyzer (Illumina) as summarized in Supplementary Table 1. Additionally, PCRs were performed on genomic DNA to confirm the sequences of the repetitive regions within hctB, tarp and at the plasmid origin using Platinum Pfx (Invitrogen), an annealing temperature of 60°C using primers HC2_f, HC2_r, tarp_f, tarp_r, pori_f and pori_f (see Supplementary Table 2). PCR products were sequenced using the primers above plus primers tarp_s1-tarp_s7 (see Supplementary Table 2).
Assembly and Alignment
Illumina reads were assembled using velvet v1.0.1262. Due to the small size and non-repetitive nature of the C. trachomatis genome, most assemblies produced only a small number of contigs that could be scaffolded by hand using the genome of L2/434/BU63 as a reference (acc no AM884176), with the circular genome cut at the origin immediately upstream of hemB. Manual insertion of hctB and tarp gene sequences resulted in Improved High Quality Draft Sequences64 made up of 1-11 contigs (Supplementary Table 1). Plasmid assemblies were completed by PCR across the origin to give a single contig. To double-check the assemblies and ensure than none of our cultures were mixed populations, raw data was mapped back against the assemblies using SMALT (see URLs) to check for heterozygosity. All genome and plasmid sequences have been deposited at EMBL under accession numbers given in Supplementary Table 1. Complete genomes were aligned with progressiveMauve65 using the --collinear option. The resulting alignment was manually checked and misalignments improved.
In order to allow the phylogenetic tree to be rooted, the sequence of Chlamydia muridarum Nigg (AE002160.2) was added to the alignment. Due to the divergent nature of this sequence, it was not possible to align it to the rest of the sequences with progressiveMauve. Instead, it was fragmented in silico and then mapped to the C. trachomatis alignment using SMALT.
Phylogenetic reconstruction
Due to limitations of assembly of repetitive regions using short-read data, repetitive regions of the consensus of aligned genomes were identified using REPuter66 and excluded from phylogenetic analysis. A phylogenetic reconstruction of the alignment data was carried out using RAxML v7.0.467 using a GTR model of evolution with a GAMMA correction for among site rate variation with four rate categories. To reduce the effect of recombination on the phylogeny, the iterative recombination removal method of Croucher et al.22 was employed with 100 bootstrap replicates calculated on the final tree to provide a measure of support for the relationships identified.
Ancestral sequence reconstruction and recombination detection
Ancestral sequences were reconstructed onto each node of the phylogeny using PAML27. From these ancestral sequences, SNPs were reconstructed onto branches of the tree. To identify recombination in the data we applied a moving window approach similar to that used in Croucher et al.22. Croucher et al. identified regions of high SNP density on a given branch of the Streptococcus pneumoniae PMEN1 phylogeny and hypothesized these to represent recombination events from a source outside of the PMEN1 lineage. Here the same principal was applied exclusively to homoplasic SNPs, in order to identify regions of homologous replacement within the tree. First, homoplasic SNPs were identified for each branch of the tree using the PAML reconstruction of ancestral sequences. The DNA sequences on each branch (the potential recipient) were then compared, in turn, to the corresponding bases along all other branches on the tree to identify potential donors. For all pairwise comparisons where at least three shared homoplasies were found, a moving window approach was used to identify regions where these occurred at a higher density than would be expected if they were randomly acquired. The lengths of these regions were then refined using a spatial scanning statistic as described in Croucher et al.22, which alters the length of the region until the recombination likelihood is maximized. Once the likelihood of all clusters was calculated for all recipient branches, SNPs produced by the cluster with the highest likelihood were removed and the process repeated iteratively until no significant homoplasy clusters were identified.
Footnotes
URLs: SMALT, http://www.sanger.ac.uk/resources/software/smalt/
ACCESSION NUMBERS: Short reads and assembled genomes and plasmids have been submitted to EMBL under the accession numbers listed in Supplementary Table 1.
AUTHOR CONTRIBUTIONS: S.R.H. assembled, aligned and analyzed the data and wrote the paper. I.N.C. jointly conceived the project with N.R.T. and provided samples. H.M.B.S-S. performed experiments, carried out analyses of the data and helped write the paper. L.T.C., P.M., R.J.S., M.J.H., D.M., R.W.P., D.A.L., M.U., K.P., C.B., R.B., H.J.V.d.V., S.A.M., A.W.S., C.M.B., A.S., M.C. and B.d.B. collected and cultured samples. B.G.S. and J.P. helped interpret the data and write the paper. N.R.T. conceived and ran the project and wrote the paper.
REFERENCES
- 1.WHO . Prevalence and incidence of selected sexually transmitted infections. World Health Organization; Geneva: 2011. [Google Scholar]
- 2.Mariotti SP, Pascolini D, Rose-Nussbaumer J. Trachoma: global magnitude of a preventable cause of blindness. Br J Ophthalmol. 2009;93:563–8. doi: 10.1136/bjo.2008.148494. [DOI] [PubMed] [Google Scholar]
- 3.Burgoyne RA. Lymphogranuloma venereum. Prim Care. 1990;17:153–7. [PubMed] [Google Scholar]
- 4.Behets FM, et al. Chancroid, primary syphilis, genital herpes, and lymphogranuloma venereum in Antananarivo, Madagascar. J Infect Dis. 1999;180:1382–5. doi: 10.1086/315005. [DOI] [PubMed] [Google Scholar]
- 5.Mabey D, Peeling RW. Lymphogranuloma venereum. Sex Transm Infect. 2002;78:90–2. doi: 10.1136/sti.78.2.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Viravan C, et al. A prospective clinical and bacteriologic study of inguinal buboes in Thai men. Clin Infect Dis. 1996;22:233–9. doi: 10.1093/clinids/22.2.233. [DOI] [PubMed] [Google Scholar]
- 7.Clarke IN. Evolution of Chlamydia trachomatis. Ann N Y Acad Sci. 2011;1230:E11–E18. doi: 10.1111/j.1749-6632.2011.06194.x. [DOI] [PubMed] [Google Scholar]
- 8.Gaydos CA. Nucleic acid amplification tests for gonorrhea and Chlamydia: practice and applications. Infect Dis Clin North Am. 2005;19:367–86. doi: 10.1016/j.idc.2005.03.006. ix. [DOI] [PubMed] [Google Scholar]
- 9.Fredlund H, Falk L, Jurstrand M, Unemo M. Molecular genetic methods for diagnosis and characterisation of Chlamydia trachomatis and Neisseria gonorrhoeae: impact on epidemiological surveillance and interventions. APMIS. 2004;112:771–84. doi: 10.1111/j.1600-0463.2004.apm11211-1205.x. [DOI] [PubMed] [Google Scholar]
- 10.Nunes A, Borrego MJ, Nunes B, Florindo C, Gomes JP. Evolutionary dynamics of ompA, the gene encoding the Chlamydia trachomatis key antigen. J Bacteriol. 2009;191:7182–92. doi: 10.1128/JB.00895-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pudjiatmoko, Fukushi H, Ochiai Y, Yamaguchi T, Hirai K. Phylogenetic analysis of the genus Chlamydia based on 16S rRNA gene sequences. Int J Syst Bacteriol. 1997;47:425–31. doi: 10.1099/00207713-47-2-425. [DOI] [PubMed] [Google Scholar]
- 12.Klint M, et al. High-resolution genotyping of Chlamydia trachomatis strains by multilocus sequence analysis. J Clin Microbiol. 2007;45:1410–4. doi: 10.1128/JCM.02301-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pannekoek Y, et al. Multi locus sequence typing of Chlamydiales: clonal groupings within the obligate intracellular bacteria Chlamydia trachomatis. BMC Microbiol. 2008;8:42. doi: 10.1186/1471-2180-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brunelle BW, Sensabaugh GF. The ompA gene in Chlamydia trachomatis differs in phylogeny and rate of evolution from other regions of the genome. Infect Immun. 2006;74:578–85. doi: 10.1128/IAI.74.1.578-585.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dean D, et al. Predicting phenotype and emerging strains among Chlamydia trachomatis infections. Emerg Infect Dis. 2009;15:1385–94. doi: 10.3201/eid1509.090272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seth-Smith HM, et al. Co-evolution of genomes and plasmids within Chlamydia trachomatis and the emergence in Sweden of a new variant strain. BMC Genomics. 2009;10:239. doi: 10.1186/1471-2164-10-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeffrey BM, et al. Genome sequencing of recent clinical Chlamydia trachomatis strains identifies loci associated with tissue tropism and regions of apparent recombination. Infection and Immunity. 2010;78:2544–53. doi: 10.1128/IAI.01324-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joseph SJ, Didelot X, Gandhi K, Dean D, Read TD. Interplay of recombination and selection in the genomes of Chlamydia trachomatis. Biol Direct. 2011;6:28. doi: 10.1186/1745-6150-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ikryannikova LN, Shkarupeta MM, Shitikov EA, Il’ina EN, Govorun VM. Comparative evaluation of new typing schemes for urogenital Chlamydia trachomatis isolates. FEMS Immunol Med Microbiol. 2010;59:188–96. doi: 10.1111/j.1574-695X.2010.00678.x. [DOI] [PubMed] [Google Scholar]
- 20.Millman KL, Tavare S, Dean D. Recombination in the ompA gene but not the omcB gene of Chlamydia contributes to serovar-specific differences in tissue tropism, immune surveillance, and persistence of the organism. J Bacteriol. 2001;183:5997–6008. doi: 10.1128/JB.183.20.5997-6008.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stephens RS, et al. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–9. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 22.Croucher NJ, et al. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331:430–4. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stephens RS. In: Schachter JC, Kaltenboek H, Rank K, Saikku R, Stephens S, Timms RS, Wyrick P, editors. Chlamydiae and evolution; a billion years and counting; Proceedings of the tenth International Symposium on Human Chlamydial Infections; International Chlamydia Symposium, San Francisco, CA. 2002.pp. 3–12. [Google Scholar]
- 24.Suchland RJ, Eckert LO, Hawes SE, Stamm WE. Longitudinal assessment of infecting serovars of Chlamydia trachomatis in Seattle public health clinics: 1988-1996. Sex Transm Dis. 2003;30:357–61. doi: 10.1097/00007435-200304000-00016. [DOI] [PubMed] [Google Scholar]
- 25.Nieuwenhuis RF, Ossewaarde JM, van der Meijden WI, Neumann HA. Unusual presentation of early lymphogranuloma venereum in an HIV-1 infected patient: effective treatment with 1 g azithromycin. Sex Transm Infect. 2003;79:453–5. doi: 10.1136/sti.79.6.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayes LJ, et al. Evidence for naturally occurring recombination in the gene encoding the major outer membrane protein of lymphogranuloma venereum isolates of Chlamydia trachomatis. Infect Immun. 1994;62:5659–63. doi: 10.1128/iai.62.12.5659-5663.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–6. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 28.LeQuesne W. A method of selection of characters in numerical taxonomy. Systematic Biology. 1969;18:201–205. [Google Scholar]
- 29.Bruen TC, Philippe H, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172:2665–81. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith JM. Analyzing the mosaic structure of genes. J Mol Evol. 1992;34:126–9. doi: 10.1007/BF00182389. [DOI] [PubMed] [Google Scholar]
- 31.Jakobsen IB, Easteal S. A program for calculating and displaying compatibility matrices as an aid in determining reticulate evolution in molecular sequences. Comput Appl Biosci. 1996;12:291–5. doi: 10.1093/bioinformatics/12.4.291. [DOI] [PubMed] [Google Scholar]
- 32.Gomes JP, et al. Evolution of Chlamydia trachomatis diversity occurs by widespread interstrain recombination involving hotspots. Genome Res. 2007;17:50–60. doi: 10.1101/gr.5674706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuo CC, Wang SP, Grayston JT, Alexander ER. TRIC type K, a new immunologic type of Chlamydia trachomatis. J Immunol. 1974;113:591–6. [PubMed] [Google Scholar]
- 34.Unemo M, Clarke IN. The Swedish new variant of Chlamydia trachomatis. Curr Opin Infect Dis. 2011;24:62–9. doi: 10.1097/QCO.0b013e32834204d5. [DOI] [PubMed] [Google Scholar]
- 35.Unemo M, et al. The Swedish new variant of Chlamydia trachomatis: genome sequence, morphology, cell tropism and phenotypic characterization. Microbiology. 2010;156:1394–404. doi: 10.1099/mic.0.036830-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Misyurina OY, et al. Mutations in a 23S rRNA gene of Chlamydia trachomatis associated with resistance to macrolides. Antimicrobial agents and chemotherapy. 2004;48:1347–9. doi: 10.1128/AAC.48.4.1347-1349.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones RB, Van der Pol B, Martin DH, Shepard MK. Partial characterization of Chlamydia trachomatis isolates resistant to multiple antibiotics. The Journal of infectious diseases. 1990;162:1309–15. doi: 10.1093/infdis/162.6.1309. [DOI] [PubMed] [Google Scholar]
- 38.Lefevre JC, Lepargneur JP, Guion D, Bei S. Tetracycline-resistant Chlamydia trachomatis in Toulouse, France. Pathologie-biologie. 1997;45:376–8. [PubMed] [Google Scholar]
- 39.Somani J, Bhullar VB, Workowski KA, Farshy CE, Black CM. Multiple drug-resistant Chlamydia trachomatis associated with clinical treatment failure. The Journal of infectious diseases. 2000;181:1421–7. doi: 10.1086/315372. [DOI] [PubMed] [Google Scholar]
- 40.Binet R, Maurelli AT. Fitness cost due to mutations in the 16S rRNA associated with spectinomycin resistance in Chlamydia psittaci 6BC. Antimicrobial agents and chemotherapy. 2005;49:4455–64. doi: 10.1128/AAC.49.11.4455-4464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brunham R, et al. Chlamydia trachomatis from individuals in a sexually transmitted disease core group exhibit frequent sequence variation in the major outer membrane protein (omp1) gene. J Clin Invest. 1994;94:458–63. doi: 10.1172/JCI117347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gomes JP, Bruno WJ, Borrego MJ, Dean D. Recombination in the genome of Chlamydia trachomatis involving the polymorphic membrane protein C gene relative to ompA and evidence for horizontal gene transfer. J Bacteriol. 2004;186:4295–306. doi: 10.1128/JB.186.13.4295-4306.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jurstrand M, et al. Characterization of Chlamydia trachomatis omp1 genotypes among sexually transmitted disease patients in Sweden. J Clin Microbiol. 2001;39:3915–9. doi: 10.1128/JCM.39.11.3915-3919.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Duynhoven YT, Ossewaarde JM, Derksen-Nawrocki RP, van der Meijden WI, van de Laar MJ. Chlamydia trachomatis genotypes: correlation with clinical manifestations of infection and patients’ characteristics. Clin Infect Dis. 1998;26:314–22. doi: 10.1086/516291. [DOI] [PubMed] [Google Scholar]
- 45.Moncan T, Eb F, Orfila J. Monoclonal antibodies in serovar determination of 53 Chlamydia trachomatis isolates from Amiens, France. Res Microbiol. 1990;141:695–701. doi: 10.1016/0923-2508(90)90063-v. [DOI] [PubMed] [Google Scholar]
- 46.Wagenvoort JH, Suchland RJ, Stamm WE. Serovar distribution of urogenital Chlamydia trachomatis strains in The Netherlands. Genitourin Med. 1988;64:159–61. doi: 10.1136/sti.64.3.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barnes RC, Suchland RJ, Wang SP, Kuo CC, Stamm WE. Detection of multiple serovars of Chlamydia trachomatis in genital infections. J Infect Dis. 1985;152:985–9. doi: 10.1093/infdis/152.5.985. [DOI] [PubMed] [Google Scholar]
- 48.Hanna L, Thygeson P, Jawetz E. Elementary-body virus isolated from clinical trachoma in California. Science. 1959;130:1339–40. doi: 10.1126/science.130.3385.1339. [DOI] [PubMed] [Google Scholar]
- 49.Spaargaren J, et al. Analysis of Chlamydia trachomatis serovar distribution changes in the Netherlands (1986-2002) Sex Transm Infect. 2004;80:151–2. doi: 10.1136/sti.2003.006395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dean D, Stephens RS. Identification of individual genotypes of Chlamydia trachomatis from experimentally mixed serovars and mixed infections among trachoma patients. J Clin Microbiol. 1994;32:1506–10. doi: 10.1128/jcm.32.6.1506-1510.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Machado AC, et al. Distribution of Chlamydia trachomatis genovars among youths and adults in Brazil. J Med Microbiol. 2010;60:472–6. doi: 10.1099/jmm.0.026476-0. [DOI] [PubMed] [Google Scholar]
- 52.Batteiger BE, et al. Correlation of infecting serovar and local inflammation in genital chlamydial infections. J Infect Dis. 1989;160:332–6. doi: 10.1093/infdis/160.2.332. [DOI] [PubMed] [Google Scholar]
- 53.Millman K, et al. Population-based genetic and evolutionary analysis of Chlamydia trachomatis urogenital strain variation in the United States. J Bacteriol. 2004;186:2457–65. doi: 10.1128/JB.186.8.2457-2465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Persson K, Osser S. Lack of evidence of a relationship between genital symptoms, cervicitis and salpingitis and different serovars of Chlamydia trachomatis. Eur J Clin Microbiol Infect Dis. 1993;12:195–9. doi: 10.1007/BF01967111. [DOI] [PubMed] [Google Scholar]
- 55.Lysen M, et al. Characterization of ompA genotypes by sequence analysis of DNA from all detected cases of Chlamydia trachomatis infections during 1 year of contact tracing in a Swedish County. J Clin Microbiol. 2004;42:1641–7. doi: 10.1128/JCM.42.4.1641-1647.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sturm-Ramirez K, et al. Molecular epidemiology of genital Chlamydia trachomatis infection in high-risk women in Senegal, West Africa. J Clin Microbiol. 2000;38:138–45. doi: 10.1128/jcm.38.1.138-145.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Geisler WM, Suchland RJ, Whittington WL, Stamm WE. The relationship of serovar to clinical manifestations of urogenital Chlamydia trachomatis infection. Sex Transm Dis. 2003;30:160–5. doi: 10.1097/00007435-200302000-00013. [DOI] [PubMed] [Google Scholar]
- 58.Gao X, et al. Distribution study of Chlamydia trachomatis serovars among high-risk women in China performed using PCR-restriction fragment length polymorphism genotyping. J Clin Microbiol. 2007;45:1185–9. doi: 10.1128/JCM.02076-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van de Laar MJ, et al. Differences in clinical manifestations of genital chlamydial infections related to serovars. Genitourin Med. 1996;72:261–5. [PMC free article] [PubMed] [Google Scholar]
- 60.Parkhill J, et al. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature. 2000;404:502–6. doi: 10.1038/35006655. [DOI] [PubMed] [Google Scholar]
- 61.Otto TD, Sanders M, Berriman M, Newbold C. Iterative Correction of Reference Nucleotides (iCORN) using second generation sequencing technology. Bioinformatics. 2010;26:1704–7. doi: 10.1093/bioinformatics/btq269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–9. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thomson NR, et al. Chlamydia trachomatis: genome sequence analysis of lymphogranuloma venereum isolates. Genome Res. 2008;18:161–71. doi: 10.1101/gr.7020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chain PS, et al. Genomics. Genome project standards in a new era of sequencing. Science. 2009;326:236–7. doi: 10.1126/science.1180614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kurtz S, Schleiermacher C. REPuter: fast computation of maximal repeats in complete genomes. Bioinformatics. 1999;15:426–7. doi: 10.1093/bioinformatics/15.5.426. [DOI] [PubMed] [Google Scholar]
- 67.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–90. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.