Abstract
Nuclear noncoding RNA (ncRNA) surveillance pathways play key roles in shaping the steady-state transcriptomes of eukaryotic cells. Defective and unneeded ncRNAs are primarily degraded by exoribonucleases that rely on protein cofactors to identify these RNAs. Recent studies have begun to elucidate both the mechanisms by which these cofactors recognize aberrant RNAs and the features that mark RNAs for degradation. One crucial RNA determinant is the presence of an accessible end, and the failure of aberrant RNAs to fold into compact structures and assemble with specific binding proteins likely also contributes to their recognition and subsequent degradation. To date, ncRNA surveillance has been most extensively studied in budding yeast. However, mammalian cells possess nucleases and cofactors that have no known yeast counterparts, indicating that RNA surveillance pathways may be more complex in metazoans. Importantly, there is evidence that the failure of ncRNA surveillance pathways contributes to human disease.
Keywords: noncoding RNA quality control, RNA degradation, oligoadenylation, TRAMP complex, exoribonucleases
The need for nuclear ncRNA surveillance
The use of sensitive techniques to analyze genome-wide transcription has revealed that nuclear ncRNA surveillance pathways critically influence steady-state transcriptomes [1, 2]. Experiments in which exoribonucleases have been depleted from yeast, human and plant cells have revealed the existence of enormous numbers of ncRNAs whose fate is to be rapidly degraded [1, 2]. Moreover, precursors to abundant long-lived ncRNAs, such as rRNAs, tRNAs, spliceosomal small nuclear RNAs (snRNAs) and small nucleolar RNAs (snoRNAs) account for a large fraction of eukaryotic transcripts. Defective RNA precursors can result from mutations, transcriptional errors and processing mistakes. Thus, one challenge in the nucleus is to degrade aberrant and unneeded RNAs while allowing other RNAs to undergo maturation and become functional ribonucleoproteins (RNPs).
Although ncRNAs are far more numerous than mRNAs, the nuclear quality control pathways that degrade aberrant mRNAs are better understood. These pathways monitor each step in mRNA biogenesis, including 5′ capping, splicing, 3′ end formation, polyadenylation, nuclear export and the ability to support translation of full-length proteins [3, 4]. Some ncRNAs that are destined for decay, particularly those transcribed by RNA polymerase II, resemble mature mRNAs in that they undergo nuclear export and are degraded in the cytoplasm [5–7]. However, because the biogenesis of RNA polymerase I and III transcripts [8, 9], and also some RNA polymerase II transcripts [6], differs significantly from mRNAs, recognition of these aberrant ncRNAs requires distinct mechanisms. Due to its genetic tractability, nuclear ncRNA surveillance pathways have been most extensively studied in the budding yeast Saccharomyces cerevisiae [10–12]. Excitingly, recent studies have begun to reveal an abundance of pathways by which unneeded ncRNAs are targeted for degradation in mammalian nuclei [13–19] and to provide evidence that failure to degrade ncRNAs can result in disease [19].
Lessons from budding yeast: ends matter
In budding yeast nuclei, ncRNA degradation is largely carried out by exoribonucleases, which degrade RNA from either the 5′ end (5′ to 3′ exoribonucleases) or 3′ end (3′ to 5′ exoribonucleases). One 3′ to 5′ exoribonuclease, Rrp6, functions as part of a multiprotein complex called the nuclear exosome [20, 21] (Box 1). The exosome also contains a second nuclease, Dis3/Rrp44, which is a modular protein containing both 3′ to 5′ exoribonuclease and endoribonuclease activities [20, 21]. The major 5′ to 3′ exoribonuclease involved in nuclear ncRNA surveillance is Rat1 [10]. Both the exosome nucleases and Rat1 function with partner proteins, or cofactors, that recognize aberrant RNAs and/or assist in their decay [10–12]. Because all exoribonucleases require an accessible end to initiate degradation, a critical role of some of the best-studied cofactors is to recognize and/or increase the availability of these ends [11, 12, 22, 23]. Moreover, because ncRNAs are often highly folded, these cofactors often assist exoribonucleases in degrading structured RNAs [22, 24, 25].
Box 1. Yeast and human exosomes.
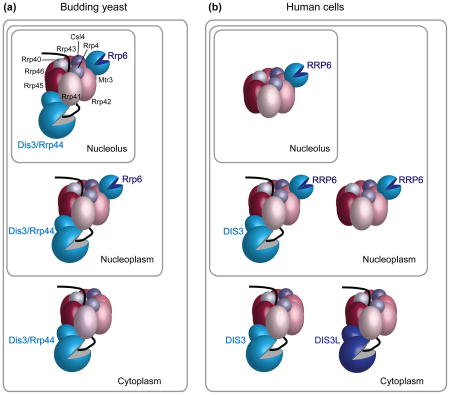
The exosome is a nuclease complex that functions in both the nucleus and cytoplasm [20, 21]. The “core exosome” consists of nine distinct subunits (Figure I). Six subunits (Rrp41, Rrp42, Rrp43, Rrp45, Rrp46 and Mtr3), which resemble bacterial RNase PH monomers but are catalytically inactive, form a hexameric ring that is topped by three RNA-binding subunits (Rrp40, Rrp4 and Csl4) [20, 21]. In yeast (a), the tenth subunit, the 3′ to 5′ exoribonuclease Dis3/Rrp44, interacts with the bottom of the channel through its N-terminal PIN domain [57]. The PIN domain harbors endonucleolytic activity [50, 74, 75], while the remainder of Dis3/Rrp44 consists of RNA-binding domains and a catalytic exoribonuclease domain. RNA threads through the central channel to reach Dis3/Rrp44 [57] and may bypass the channel to reach Rrp6. While both the cytoplasmic and nuclear exosomes contain the nine subunit core and Dis3/Rrp44, the nuclear exosome is specialized by the presence of a second 3′ to 5′ exoribonuclease, Rrp6 [20, 21]. Based on single particle electron microscopy of the Leishmania tarentolae exosome [99], Rrp6 may be located near the channel entrance.
The human exosome (b) differs from its yeast counterpart in having greater subcellular compartment-specific variation in its catalytic subunits. In place of Dis3/Rrp44, the human exosome can contain either of two related proteins, DIS3 and DIS3L [16, 100]. DIS3L appears to be exclusively cytoplasmic while DIS3 is mostly nucleoplasmic and excluded from nucleoli, but may also have a cytoplasmic pool [16, 100]. Similar to yeast, RRP6 is a specific component of the nuclear exosome; however immunofluorescence suggests a predominantly nucleolar location with some nucleoplasmic staining [21]. Thus, the cytoplasmic exosome in human cells is proposed to contain either DIS3L or DIS3 as its sole active exoribonuclease, the nucleoplasmic exosome contains DIS3 and can also contain RRP6, while the only active exoribonuclease in the nucleolar exosome may be RRP6 [16].
Figure I title: Nuclear and cytoplasmic forms of the exosome are specialized by the presence of distinct catalytic subunits.
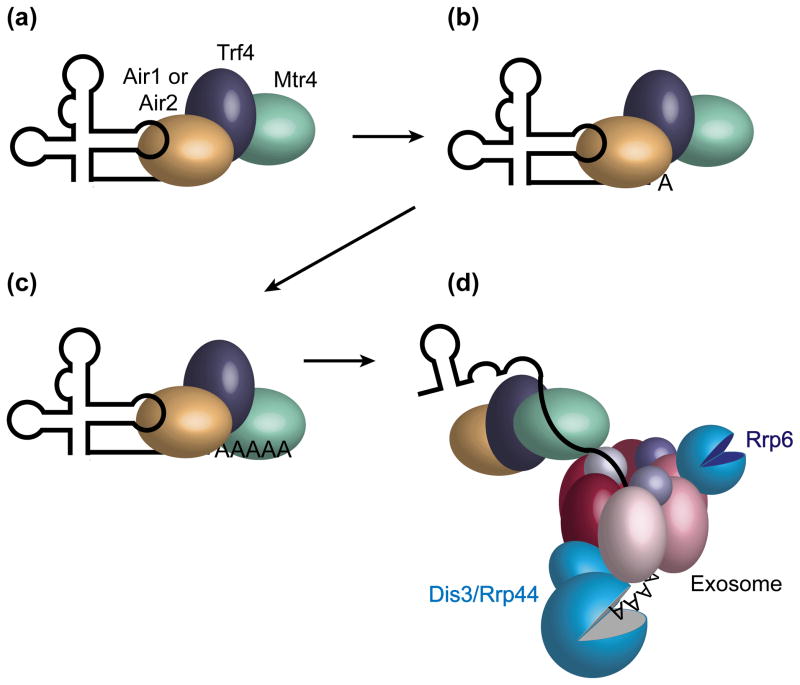
Oligo(A) addition by TRAMP
A prominent cofactor is the Trf4-Air1/2-Mtr4 polyadenylation complex (TRAMP), which targets RNAs for degradation by the nuclear exosome. Noncoding RNAs degraded by this pathway include hypomodified [26], aberrant 23S rRNA processing intermediates [24], truncated 5S rRNAs and SRP RNAs [27, 28], unspliced pre-tRNAs [28, 29], antisense RNAs [30, 31] and a large class of “cryptic unstable transcripts” (CUTs) originating from bidirectional transcription in the vicinity of RNA polymerase II promoters [32–34] (Table 1). TRAMP, which consists of the poly(A) polymerase Trf4, either of two zinc knuckle proteins, Air1 or Air2, and the RNA helicase Mtr4, adds an oligo(A) tail to RNAs destined for degradation [24, 25, 32]. In contrast to the long poly(A) tails added to mRNAs by classical poly(A) polymerases, which stabilize mRNAs from degradation [35], the tails added by TRAMP are quite short (~4–5 adenosines on average) [29, 36]. Both TRAMP and the oligo(A) tails function to recruit the exosome and enhance its activity, in part by providing an accessible 3′ end [11, 12]. TRAMP is primarily nucleoplasmic, and a closely related nucleolar complex known as TRAMP5 (in which Trf4 is replaced by its homolog Trf5), polyadenylates an overlapping set of pre-rRNAs that are aberrantly processed or present in excess [37–39].
Table 1.
Some aberrant noncoding RNAs and their nuclear surveillance pathways
| Yeast | ||||
|---|---|---|---|---|
| Noncoding RNAs | Problem | Nuclease(s) | Cofactors | References |
| pre-rRNAs | aberrant processing; failure to assemble with proteins | Exosome Rat1 |
TRAMP, TRAMP5 Rai1 |
[24, 37–39, 67] [67, 68] |
| pre-tRNAs | Absent modifications; unspliced; misfolded; decreased 3′ processing | Exosome | TRAMP, Nrd1/Nab3 | [26–29, 45, 64] |
| 5S rRNAs | truncated | Exosome | TRAMP | [27] |
| SRP RNA | truncated | Exosome | TRAMP | [28] |
| mature tRNAs | absent modifications; structurally compromised | Rat1 | Rai1 | [71] |
| CUTs | No known binding proteins | Exosome | TRAMP, Rrp47, Mpp6 | [30–32, 59, 60, 66] |
| telomeric repeat transcripts | No known binding proteins | Rat1 | Rai1 | [70] |
| Mammals | ||||
| pre-rRNAs | premature termination; aberrant processing | Exosome XRN2 |
PAPD5/ZCCHC7/MTR4 unknown |
[15, 17] [18] |
| PROMPTs | No known binding proteins | Exosome | NEXT complex | [13, 17] |
| Endogenous retroviral transcripts | mutated, divergent RNAs | Exosome XRN2 |
unknown unknown |
[83] [83] |
| dsRNAs from Alu elements | unknown | DICER1 | unknown | [19] |
Recent studies have begun to elucidate how TRAMP chooses its substrates. Trf4 lacks a recognizable RNA binding domain and requires either Air1 or Air2 to bind RNA substrates and carry out polyadenylation [25]. Both the intact TRAMP and Trf4-Air2 subcomplexes have been shown to only adenylate RNAs that have a 3′ overhang of at least 1 nucleotide [36, 40]. TRAMP also recognizes other RNA features, as a purified Trf4-Air2 subcomplex preferentially polyadenylates the misfolded forms of two tRNAs over the correctly folded versions [25]. Air1 and Air2 each contain five zinc knuckles which, in retroviral nucleocapsid proteins, interact with single-stranded RNA [41]. The C-terminal knuckle is involved in tethering Air1/2 to Trf4, while at least two other knuckles are important for substrate recognition [40, 42]. One possibility is that Trf4-Air1/2 recognizes substrates with both a 3′ overhang and single-stranded elements located within a critical distance of the end. In this scenario, the misfolded tRNAs that are preferentially polyadenylated may have more single-stranded regions due to secondary or tertiary structure alterations.
In vivo, where many TRAMP substrates are 3′ extended precursors, TRAMP competes with at least some RNA-binding proteins and processing enzymes for access to 3′ ends [28, 43–45]. An RNA-binding protein, the La protein Lhp1, binds the UUUOH at the 3′ end of all newly synthesized RNA polymerase III transcripts and some snRNA and snoRNA processing intermediates [46, 47]. Lhp1 protects these ends from exoribonucleases [46, 47] and also from TRAMP, as TRAMP-mediated degradation of hypomodified pre-tRNAMeti increases in lhp1Δ strains [44] and decreases upon LHP1 overexpression [43]. TRAMP also competes with the nonexosomal exoribonuclease Rex1, which matures the 3′ trailers of both 5S rRNA and certain pre-tRNAs [28, 45, 48]. In rex1Δ strains, some RNAs that are normally Rex1 substrates undergo increased TRAMP-mediated polyadenylation [45].
As noted, the TRAMP-added tails are much shorter than those added by the classical poly(A) polymerase to mRNAs. In vivo, TRAMP-added tails are usually less than 10 nt and can be as short as 1–2 nt [29, 36, 49, 50]. The short tails eliminate the Lhp1 binding site and preclude binding by the poly(A) binding protein Pab1, which requires 12 adenosines for high affinity binding [51], ensuring the 3′ ends of TRAMP substrates remain available to exonucleases. Mtr4 is important for modulating the length of TRAMP-added tails, because in the presence of Mtr4, addition of more than 4 adenosines by recombinant Trf4p/Air2p is disfavored [36]. Notably, hyperadenylated TRAMP substrates accumulate when Mtr4 is mutated or depleted from yeast cells [37, 52], indicating that Mtr4 also modulates tail-length and/or contributes to exosome recruitment in vivo.
Based on these data, we propose a model in which Trf4-Air1/2 carries out the initial target recognition by adenylating RNAs containing protein-free 3′ extensions and nearby single-stranded elements (Figure 1). Once the first adenosine is added, Mtr4 modulates the rate of subsequent adenylation [36], favoring formation of short tails. Mtr4 may serve as a three-way bridge between the substrate, TRAMP and the exosome, as Mtr4 binds both the substrate and the Trf4-Air2 subcomplex [53, 54] and has a conserved surface that may interact with the exosome [55]. Because degradation of structured RNAs could require multiple rounds of polyadenylation, Mtr4 may function throughout the decay process. Additionally, Mtr4 likely unwinds structured RNAs through its helicase activity. In support of this role, Mtr4 carries out ATP-dependent unwinding of 3′ overhang-containing RNA duplexes in vitro [52, 56] and a single-stranded tail of 31–33 nt is needed for an RNA to span the exosome central channel and reach the active site of the Dis3/Rrp44 exoribonuclease [57] (Box 1).
Figure 1. Model for TRAMP function in yeast.
Trf4 and an Air protein (Air1 or Air2) recognize RNAs that contain a protein-free 3′ extension (a) and begins oligoadenylation (b). As soon as a single 3′ adenosine is added, Mtr4 modulates the activity of the polymerase to favor production of RNAs with short A tails [36] (c). Binding by Mtr4 to the oligo(A) tail may help recruit the exosome to the RNA/TRAMP complex (d). Mtr4, through its helicase activity, unwinds the structured RNA. Although only Dis3/Rrp44 is shown degrading the substrate, Rrp6 also participates in degradation.
Nrd1/Nab3 recruits TRAMP and the exosome to some RNAs
Although TRAMP recognition of many ncRNAs likely occurs after transcription, TRAMP and the exosome can be recruited co-transcriptionally to RNA polymerase II transcripts such as CUTs [30, 31, 58]. In contrast to pre-mRNAs, where termination is usually coupled to cleavage by the 3′ end processing machinery, termination of CUTs involves binding of the Nrd1/Nab3 heterodimer and the putative helicase Sen1 to sequences in the nascent RNA [59, 60]. Nrd1 binds the Ser5-phosphorylated C-terminal domain of the RNA polymerase II large subunit [61] and co-purifies with TRAMP and the exosome [58], suggesting it acts as an adaptor to recruit TRAMP to the transcript. After the generation of a free 3′ end by release of the nascent transcript and/or endonucleolytic cleavage, CUTs undergo oligoadenylation and 3′ trimming by the TRAMP/exosome complex [32, 60].
Interestingly, the biogenesis of S. cerevisiae snoRNAs resembles that of CUTs in that, following termination of pre-snoRNAs by the Nrd1/Nab3/Sen1 pathway, TRAMP and the exosome carry out 3′ end trimming [49, 62, 63]. How do TRAMP and the exosome distinguish mature snoRNAs from RNAs that should be degraded? In the case of snoRNAs, assembly with snoRNP core proteins likely acts as a roadblock to continued degradation [49]. For CUTs, the lack of specific binding proteins may allow complete decay [3].
Although Nrd1/Nab3/Sen1-mediated recruitment of the TRAMP/exosome complex was assumed to be restricted to RNA polymerase II transcripts, Nrd1/Nab3 may also recruit TRAMP to defective RNA polymerase III RNAs after their transcription is complete [29, 64]. Recently, using in vivo RNA-protein crosslinking, Nrd1 and Nab3 were found to associate with fragments derived from 5S rRNA, tRNAs and ribonuclease P (RNase P) RNA [29, 64]. Many of the fragments contained oligo(A) tails consistent with TRAMP addition, and also contained 5′ leader, 3′ trailer or intron sequences (in the case of pre-tRNAs), indicating they were derived from precursors [29, 64]. In support of a role for Nrd1/Nab3 in targeting defective pre-tRNAs for decay, unspliced pre-tRNAs accumulate when Nrd1 or Nab1 is depleted [29]. However, because an RNA polymerase II subunit crosslinks to similar RNAs, some of the Nrd1/Nab3-bound RNAs could originate from aberrant initiation by RNA polymerase II [64].
Additional nuclear exosome cofactors
The proteins Rrp47 and Mpp6 also assist substrate recognition by exosome exoribonucleases [12], although the mechanisms by which they function are less well understood. In vitro, Rrp47 binds structured RNAs and interacts directly with Rrp6 [65]. In yeast lacking Rrp47, many Rrp6 substrates accumulate, including CUTs [12, 60, 66]. CUTs also accumulate in mpp6Δ strains; however, Mpp6 preferentially binds polyuridine in vitro [66]. Consistent with this functional redundancy, strains lacking both Rrp47 and Mpp6 are inviable [66]. As strains lacking both Rrp6 and Mpp6 are also inviable, one possibility is that Rrp47 is a specific cofactor for Rrp6, while Mpp6 assists RNA recognition by the Dis3/Rrp44 nuclease [66].
Other nucleases
A second prominent nuclear ncRNA surveillance pathway involves the 5′ to 3′ exoribonuclease Rat1. Substrates of this pathway include aberrantly processed pre-rRNAs [67, 68], antisense RNAs [69], transcripts from telomeric repeats [70], structurally compromised mature tRNAs [71], and the ncRNAs that remain attached to RNA polymerase II following cleavage of pre-mRNAs by the 3′ end processing endonuclease [72] (Table 1). Notably, both the aberrant pre-rRNAs and telomeric transcripts contain long poly(A) tails that are added by the classical poly(A) polymerase [70, 73], which may indicate that 3′ decay is blocked by bound poly(A) binding proteins. Rat1 functions with the cofactor Rai1, which stabilizes Rat1 activity and assists in degrading structured RNAs [22]. Consistent with a role in recognizing 5′ termini and generating accessible ends, Rai1 converts the 5′ triphosphate that is the initial 5′ end of all transcripts to a 5′ monophosphate [22] and can cleave unmethylated caps from newly synthesized mRNAs [23]. Although only mRNAs have been shown to be Rai1 substrates [23], the ability of Rai1 to remove 5′ triphosphates and unmethylated caps could contribute to the degradation of telomeric transcripts and also allow Rat1 to degrade both CUTs and aberrant RNA polymerase III transcripts when the exosome pathway is inactive. In support of functional overlap between the Rat1 and exosome pathways, levels of polyadenylated rRNAs and telomeric-repeat RNAs increase when both Rat1 and components of the TRAMP/exosome pathway are inactivated [67, 70].
Endoribonucleases also participate in nuclear ncRNA surveillance. The endonucleolytic activity of Dis3/Rrp44 contributes to the degradation of some RNAs, including CUTs and rRNA spacers, possibly by generating more ends for the exoribonuclease activities of Dis3/Rrp44 and Rrp6 [50, 74, 75]. Additionally, numerous ncRNAs, including CUTs and antisense RNAs, accumulate in yeast harboring a mutation in, or depleted for, the endoribonuclease RNase P [76, 77]. Although the accumulation of these ncRNAs has not been demonstrated to be due to failure of RNase P to cleave the transcripts (rather than an indirect effect), these data raise the possibility that, in addition to its established function in pre-tRNA maturation [78], RNase P contributes to ncRNA surveillance.
Impacts on cell function
Noncoding RNA surveillance pathways both impact gene expression and maintain genome integrity. Some antisense RNAs mediate transcriptional silencing of their sense loci through the recruitment of histone-modifying enzymes [79]. By modulating the antisense RNA levels, ncRNA surveillance pathways affect the degree to which the sense loci are silenced [69, 80, 81]. Additionally, by degrading rDNA-derived CUTs, TRAMP and the exosome help prevent recombination-induced loss of rDNA repeats [30, 31]. Rat1, by degrading antisense transcripts from telomeres [70], prevents hybridization of these RNAs to the transcribed DNA strand. In rat1-1 mutant strains, these “R-loops” impede telomere replication, resulting in telomere shortening [70]. Moreover, by degrading the ncRNA that remains attached to RNA polymerase II following cleavage of nascent pre-mRNAs by the 3′ end processing machinery, Rat1 contributes to termination by this polymerase [63].
Although aberrant pre-tRNAs and rRNAs are prominent TRAMP targets, the consequences of their accumulation in budding yeast are unknown. However, in the fission yeast Schizosaccharomyces pombe that (unlike S. cerevisiae) has a functional RNA interference (RNAi) pathway, TRAMP-mediated degradation prevents entry of excess rRNA precursors into the RNAi pathway [82]. In S. pombe lacking the Trf4 ortholog Cid14, the levels of centromeric siRNAs decrease, due to competition between pre-rRNAs and centromeric transcripts for the siRNA-generating machinery [82].
Nuclear ncRNA surveillance in mammalian cells
Because the pervasive transcription of mammalian genomes produces numerous short-lived ncRNAs [1, 2], ncRNA surveillance pathways likely play major roles in shaping mammalian transcriptomes. As in budding yeast, exoribonucleases are prominent components of the identified pathways. Experiments in which exosome proteins were depleted from human and mouse cells have revealed that the exosome degrades excised rRNA spacers [14], prematurely terminated pre-rRNAs [15], endogenous retroviral transcripts [83] and short-lived RNAs called PROMPTs (promoter upstream transcripts) [13, 16] that resemble yeast CUTs in being 3′ adenylated transcripts derived from RNA polymerase II initiation in the vicinity of promoters [84]. Similar depletion experiments have revealed that the Rat1 ortholog XRN2 degrades prematurely terminated pre-rRNAs, aberrant pre-rRNAs, excised pre-rRNA spacers [18], and the ncRNAs that remain attached to RNA polymerase II following cleavage of nascent pre-mRNAs [85] (Table 1).
Although mammalian cells contain potential orthologs of most yeast exoribonuclease cofactors [10, 11], the extent to which these proteins function similarly is still unclear. In particular, TRAMP-mediated adenylation has not yet been shown to contribute strongly to ncRNA surveillance in mammalian cells. Moreover, there is already evidence that mammalian cells possess a wider range of exosome cofactors and ncRNA surveillance pathways than are present in budding yeast.
Evidence for a TRAMP-mediated decay pathway
Mammalian cells contain Trf4, Air1/2 and Mtr4 orthologs that can assemble into complexes resembling yeast TRAMP. For example, when human cells overexpressing epitope-tagged Trf4 orthologs are used in immunoprecipitation experiments, the Air1/2 ortholog ZCCHC7 is detected in the immunoprecipitates [17, 42]. Similarly, the Mtr4 ortholog MTR4 and exosome proteins are present in immunoprecipitates from cells overexpressing tagged ZCCHC7 [17]. Consistent with observations that Trf4 and Mtr4 have reduced half-lives in air1 mutant yeast [42], the level of ZCCHC7 decreases when either the Trf4 ortholog PAPD5 or MTR4 is depleted with siRNAs [42]. However, a stoichiometric complex consisting of either of the two human Trf4 orthologs (PAPD5 and PAPD7) with ZCCHC7 and MTR4 has not been reported.
Importantly, components of the putative human TRAMP contribute to adenylation of short-lived ncRNAs. Some prematurely terminated pre-rRNAs contain A tails that require PAPD5 and ZCCHC7 for their formation [15, 17] and the A tails of PROMPTs are likely also added by PAPD5 [84]. When MTR4 is depleted using siRNAs, the levels of the adenylated pre-rRNAs and PROMPTs increase [17]. Thus, as in yeast [36, 37], MTR4 may modulate the tail lengths and/or association of these RNAs with the exosome.
Surprisingly, unlike yeast Trf4, PAPD5 binds RNA and is catalytically active by itself [86]. RNA binding requires amino acids in the PAPD5 C-terminus that, although absent from Trf4, are conserved in other vertebrate orthologs [86]. PAPD5 can also discriminate on its own between native initiator yeast tRNAMet, which is correctly folded, and the unmodified tRNA that has an altered structure [86]. Notably, while ZCCHC7 is nucleolar [17, 42], PAPD5 is both nucleolar and nucleoplasmic [17, 86]. Together, these data suggest that PAPD5 may function in the nucleolus as part of TRAMP and have an independent role in ncRNA surveillance in the nucleoplasm.
The nuclear exosome targeting (NEXT) complex
In addition to its probable role in a TRAMP-like complex, MTR4 associates with the putative RNA binding protein RBM7 and the Zn-knuckle protein ZCCHC8 to form another exosome cofactor, the Nuclear EXosome Targeting (NEXT) complex [17]. Remarkably, depletion of any NEXT component results in stabilization of PROMPTs, while depletion of the TRAMP orthologs PAPD5 and ZCCHC7 has no effect [17]. Thus, although PAPD5 likely carries out adenylation of PROMPTs [84], their degradation requires the NEXT complex. In support of a role for MTR4 in two spatially distinct complexes, MTR4-GFP fusions are both nucleoplasmic and nucleolar, while the other NEXT complex members (RBM7 and ZCCHC8) are confined to the nucleoplasm [17]. Interestingly, the nucleoplasmic and nucleolar forms of the human exosome also contain different sets of exoribonucleases [16] (Box 1). Specifically, while the nucleoplasmic exosome contains the Dis3/Rrp44 and Rrp6 orthologs DIS3 and RRP6, the nucleolar exosome may contain only RRP6 [16] (Box 1). Consistent with a role for the nucleoplasmic exosome in PROMPT degradation, depletion of both DIS3 and RRP6 is required to detect strong accumulation of these RNAs [16, 17]. Together, these data suggest that the NEXT complex is a nucleoplasm-restricted exosome cofactor, while the PAPD5/ZCCHC7/MTR4 complex targets defective RNAs for exosome-mediated degradation in nucleoli. Whether the segregation of specific exoribonucleases and cofactors to different subnuclear compartments is functionally related to the types of substrates encountered in these compartments remains to be addressed.
Evidence for additional pathways
Recently, the miRNA processing endonuclease DICER1 was reported to cleave double-stranded RNAs derived from transcription of Alu repeats [19]. Reduced DICER1 levels results in accumulation of Alu RNAs and degeneration of retinal pigment epithelial (RPE) cells in patients with age-related maculodegeneration and in genetically engineered mice [19]. Consistent with a role in nuclear surveillance, DICER1 is nuclear and cytoplasmic in human RPE cells and DICER1 depletion results in increased Alu RNAs in both compartments [19].
Notably, the aberrant pre-tRNAs, truncated 5S and SRP RNAs, and 3′ extended snRNAs and snoRNAs that accumulate in yeast lacking components of the TRAMP/exosome pathways [24, 27, 28, 32, 62] have not been detected in mammalian cells when siRNAs are used to deplete TRAMP or exosome orthologs. One possibility is that these RNAs can be degraded by either the exosome or the 5′ to 3′ decay pathway. Indeed, efficient degradation of prematurely terminated rRNAs in mammalian cells requires both XRN2 and the nuclear exosome [18]. An alternative but not exclusive possibility is that as yet uncharacterized surveillance pathways contribute to the surveillance of some of these ncRNAs. This may be especially likely for pre-snRNAs and pre-snoRNAs, since the biogenesis of these human ncRNAs differs in multiple respects from their budding yeast orthologs [87].
Moreover, although a large fraction of both the mouse and human genomes consists of retrotransposon-derived sequences and satellite elements [88], little is known as to how most transcripts from these repeats are degraded. Some endogenous retroviral sequences are degraded by the nuclear exosome and XRN2 [83], but the cofactors involved and the extent to which other surveillance pathways contribute are unknown. Increased transcription of satellites and retrotransposon-derived sequences occurs following many types of environmental stress [89, 90] and some satellite RNAs are elevated ~100-fold in a variety of cancers [91]. Although the increased levels of satellite transcripts in tumors are likely due to de-repression of heterochromatic loci [91], ncRNA surveillance pathways could contribute to maintaining low levels of these RNAs in normal cells by degrading RNAs that escape transcriptional silencing.
There are already several candidates for components of additional surveillance pathways. One possibility is the second Trf4 ortholog, PAPD7, whose function is currently unknown. Human cells also contain at least five other members of this family of noncanonical poly(A) polymerases, several of which add oligo(U) tails to specific cytoplasmic mRNAs and pre-miRNAs, resulting in degradation of the tailed RNAs [92]. Since some family members are nuclear [92], these polymerases could potentially contribute to nuclear ncRNA surveillance. Some tRNAs with unstable acceptor stems have an extra CCA added to their 3′ ends by the CCA-adding enzyme, which is proposed to target them for decay [93]. Another protein that could be involved in ncRNA surveillance is the Ro 60 kDa autoantigen, a ring shaped protein that binds misfolded ncRNAs in some vertebrate nuclei [94]. In support of a role as a 3′ to 5′ exoribonuclease cofactor, a prokaryotic Ro ortholog functions with 3′ to 5′ exoribonucleases to mature 23S pre-rRNA during heat stress and to degrade rRNAs during starvation [94].
“Ending” remarks
Both the extent of the ncRNA transcriptome and the nuclear mechanisms that recognize and degrade aberrant ncRNAs are only beginning to be understood. One obvious goal is the generation of a high-resolution structure of TRAMP in complex with both an RNA substrate and the exosome. Analyses of the NEXT complex should reveal whether this complex, like TRAMP, recognizes accessible 3′ ends and whether additional features of aberrant RNAs contribute to their recognition. The application of in vivo RNA-protein crosslinking technology [95, 96] and next-generation sequencing techniques [97] should reveal the diversity of ncRNAs subject to surveillance by the identified pathways in mammalian nuclei. The report that failure to degrade Alu RNAs contributes to age-related macular degeneration [19] will likely be followed by more examples where the malfunction of ncRNA surveillance contributes to human disease. Excitingly, it has been proposed that excess repeat-derived ncRNAs, derived from increased transcription or a failure of degradation, could contribute to the initiation of the rheumatic disease systemic lupus erythematosus [98]. Elucidating both the full complement of nuclear ncRNA surveillance pathways in human cells and their RNA targets will be invaluable for determining the extent to which these pathways contribute to maintaining cellular homeostasis and preventing disease.
Acknowledgments
We apologize to anyone whose work was omitted due to space constraints. Our work on ncRNA surveillance is supported by National Institutes of Health grants R01GM048410 and R01GM073863.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jacquier A. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat Rev Genet. 2009;10:833–844. doi: 10.1038/nrg2683. [DOI] [PubMed] [Google Scholar]
- 2.Rougemaille M, Libri D. Control of cryptic transcription in eukaryotes. Adv Exp Med Biol. 2011;702:122–131. doi: 10.1007/978-1-4419-7841-7_10. [DOI] [PubMed] [Google Scholar]
- 3.Schmid M, Jensen TH. Nuclear quality control of RNA polymerase II transcripts. Wiley Interdiscip Rev RNA. 2010;1:474–485. doi: 10.1002/wrna.24. [DOI] [PubMed] [Google Scholar]
- 4.van Hoof A, Wagner EJ. A brief survey of mRNA surveillance. Trends Biochem Sci. 2011;36:585–592. doi: 10.1016/j.tibs.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thompson DM, Parker R. Cytoplasmic decay of intergenic transcripts in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:92–101. doi: 10.1128/MCB.01023-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marquardt S, et al. Distinct RNA degradation pathways and 3′ extensions of yeast non-coding RNA species. Transcription. 2011;2:145–154. doi: 10.4161/trns.2.3.16298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Dijk EL, et al. XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature. 2011;475:114–117. doi: 10.1038/nature10118. [DOI] [PubMed] [Google Scholar]
- 8.Dieci G, et al. The expanding RNA polymerase III transcriptome. Trends Genet. 2007;23:614–622. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 9.Henras AK, et al. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 11.Anderson JT, Wang X. Nuclear RNA surveillance: no sign of substrates tailing off. Crit Rev Biochem Mol Biol. 2009;44:16–24. doi: 10.1080/10409230802640218. [DOI] [PubMed] [Google Scholar]
- 12.Butler JS, Mitchell P. Rrp6, Rrp47 and cofactors of the nuclear exosome. Adv Exp Med Biol. 2011;702:91–104. doi: 10.1007/978-1-4419-7841-7_8. [DOI] [PubMed] [Google Scholar]
- 13.Preker P, et al. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- 14.Kent T, et al. The 5′ external transcribed spacer in mouse ribosomal RNA contains two cleavage sites. RNA. 2009;15:14–20. doi: 10.1261/rna.1384709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shcherbik N, et al. Polyadenylation and degradation of incomplete RNA polymerase I transcripts in mammalian cells. EMBO Rep. 2010;11:106–111. doi: 10.1038/embor.2009.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomecki R, et al. The human core exosome interacts with differentially localized processive RNases: hDIS3 and hDIS3L. EMBO J. 2010;29:2342–2357. doi: 10.1038/emboj.2010.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lubas M, et al. Interaction profiling identifies the human nuclear exosome targeting complex. Mol Cell. 2011;43:624–637. doi: 10.1016/j.molcel.2011.06.028. [DOI] [PubMed] [Google Scholar]
- 18.Wang M, Pestov DG. 5′-end surveillance by Xrn2 acts as a shared mechanism for mammalian pre-rRNA maturation and decay. Nucleic Acids Res. 2011;39:1811–1822. doi: 10.1093/nar/gkq1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaneko H, et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 2011;471:325–330. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Januszyk K, Lima CD. Structural components and architectures of RNA exosomes. Adv Exp Med Biol. 2011;702:9–28. doi: 10.1007/978-1-4419-7841-7_2. [DOI] [PubMed] [Google Scholar]
- 21.Chlebowski A, et al. Catalytic properties of the eukaryotic exosome. Adv Exp Med Biol. 2011;702:63–78. doi: 10.1007/978-1-4419-7841-7_6. [DOI] [PubMed] [Google Scholar]
- 22.Xiang S, et al. Structure and function of the 5′-->3′ exoribonuclease Rat1 and its activating partner Rai1. Nature. 2009;458:784–788. doi: 10.1038/nature07731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiao X, et al. Identification of a quality-control mechanism for mRNA 5′-end capping. Nature. 2010;467:608–611. doi: 10.1038/nature09338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.LaCava J, et al. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 25.Vanacova S, et al. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kadaba S, et al. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in. S cerevisiae Genes Dev. 2004;18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kadaba S, et al. Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA. 2006;12:508–521. doi: 10.1261/rna.2305406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Copela LA, et al. Competition between the Rex1 exonuclease and the La protein affects both Trf4p-mediated RNA quality control and pre-tRNA maturation. RNA. 2008;14:1214–1227. doi: 10.1261/rna.1050408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wlotzka W, et al. The nuclear RNA polymerase II surveillance system targets polymerase III transcripts. EMBO J. 2011;30:1790–1803. doi: 10.1038/emboj.2011.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Houseley J, et al. Trf4 targets ncRNAs from telomeric and rDNA spacer regions and functions in rDNA copy number control. EMBO J. 2007;26:4996–5006. doi: 10.1038/sj.emboj.7601921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vasiljeva L, et al. Transcription termination and RNA degradation contribute to silencing of RNA polymerase II transcription within heterochromatin. Mol Cell. 2008;29:313–323. doi: 10.1016/j.molcel.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 32.Wyers F, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 33.Neil H, et al. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- 34.Xu Z, et al. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin G, Keller W. RNA-specific ribonucleotidyl transferases. RNA. 2007;13:1834–1849. doi: 10.1261/rna.652807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jia H, et al. The RNA helicase Mtr4p modulates polyadenylation in the TRAMP complex. Cell. 2011;145:890–901. doi: 10.1016/j.cell.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Houseley J, Tollervey D. Yeast Trf5p is a nuclear poly(A) polymerase. EMBO Rep. 2006;7:205–211. doi: 10.1038/sj.embor.7400612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dez C, et al. Roles of the HEAT repeat proteins Utp10 and Utp20 in 40S ribosome maturation. RNA. 2007;13:1516–1527. doi: 10.1261/rna.609807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wery M, et al. The nuclear poly(A) polymerase and exosome cofactor Trf5 is recruited cotranscriptionally to nucleolar surveillance. RNA. 2009;15:406–419. doi: 10.1261/rna.1402709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hamill S, et al. Structure and function of the polymerase core of TRAMP, a RNA surveillance complex. Proc Natl Acad Sci USA. 2010;107:15045–15050. doi: 10.1073/pnas.1003505107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.D’Souza V, Summers MF. Structural basis for packaging the dimeric genome of Moloney murine leukaemia virus. Nature. 2004;431:586–590. doi: 10.1038/nature02944. [DOI] [PubMed] [Google Scholar]
- 42.Fasken MB, et al. Air1 zinc knuckles 4 and 5 and a conserved IWRXY motif are critical for the function and integrity of the Trf4/5-Air1/2-Mtr4 polyadenylation (TRAMP) RNA quality control complex. J Biol Chem. 2011;286:37429–37445. doi: 10.1074/jbc.M111.271494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anderson J, et al. The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev. 1998;12:3650–3662. doi: 10.1101/gad.12.23.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Calvo O, et al. GCD14p, a repressor of GCN4 translation, cooperates with Gcd10p and Lhp1p in the maturation of initiator methionyl-tRNA in. Saccharomyces cerevisiae Mol Cell Biol. 1999;19:4167–4181. doi: 10.1128/mcb.19.6.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ozanick SG, et al. Rex1p deficiency leads to accumulation of precursor initiator tRNAMet and polyadenylation of substrate RNAs in Saccharomyces cerevisiae. Nucleic Acids Res. 2009;37:298–308. doi: 10.1093/nar/gkn925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolin SL, Cedervall T. The La protein. Ann Rev Biochem. 2002;71:375–402. doi: 10.1146/annurev.biochem.71.090501.150003. [DOI] [PubMed] [Google Scholar]
- 47.Bayfield MA, et al. Conserved and divergent features of the structure and function of La and La-related proteins (LARPs) Biochim Biophys Acta. 2010;1799:365–378. doi: 10.1016/j.bbagrm.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Hoof A, et al. Three conserved members of the RNase D family have unique and overlapping functions in the processing of 5S, 5.8S, U4, U5, RNase MRP and RNase P RNAs in yeast. EMBO J. 2000;19:1357–1365. doi: 10.1093/emboj/19.6.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grzechnik P, Kufel J. Polyadenylation linked to transcription termination directs the processing of snoRNA precursors in yeast. Mol Cell. 2008;32:247–258. doi: 10.1016/j.molcel.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lebreton A, et al. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature. 2008;456:993–996. doi: 10.1038/nature07480. [DOI] [PubMed] [Google Scholar]
- 51.Sachs AB, et al. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol Cell Biol. 1987;7:3268–3276. doi: 10.1128/mcb.7.9.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang X, et al. Degradation of hypomodified tRNA(iMet) in vivo involves RNA-dependent ATPase activity of the DExH helicase Mtr4p. RNA. 2008;14:107–116. doi: 10.1261/rna.808608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bernstein J, et al. Unique properties of the Mtr4p-poly(A) complex suggest a role in substrate targeting. Biochemistry. 2010;49:10357–10370. doi: 10.1021/bi101518x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weir JR, et al. Structural analysis reveals the characteristic features of Mtr4, a DExH helicase involved in nuclear RNA processing and surveillance. Proc Natl Acad Sci USA. 2010;107:12139–12144. doi: 10.1073/pnas.1004953107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jackson RN, et al. The crystal structure of Mtr4 reveals a novel arch domain required for rRNA processing. EMBO J. 2010;29:2205–2216. doi: 10.1038/emboj.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bernstein J, et al. Characterization of the essential activities of Saccharomyces cerevisiae Mtr4p, a 3′–>5′ helicase partner of the nuclear exosome. J Biol Chem. 2008;283:4930–4942. doi: 10.1074/jbc.M706677200. [DOI] [PubMed] [Google Scholar]
- 57.Bonneau F, et al. The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell. 2009;139:547–559. doi: 10.1016/j.cell.2009.08.042. [DOI] [PubMed] [Google Scholar]
- 58.Vasiljeva L, Buratowski S. Nrd1 interacts with the nuclear exosome for 3′ processing of RNA polymerase II transcripts. Mol Cell. 2006;21:239–248. doi: 10.1016/j.molcel.2005.11.028. [DOI] [PubMed] [Google Scholar]
- 59.Thiebaut M, et al. Transcription termination and nuclear degradation of cryptic unstable transcripts: a role for the nrd1-nab3 pathway in genome surveillance. Mol Cell. 2006;23:853–864. doi: 10.1016/j.molcel.2006.07.029. [DOI] [PubMed] [Google Scholar]
- 60.Arigo JT, et al. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol Cell. 2006;23:841–851. doi: 10.1016/j.molcel.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 61.Vasiljeva L, et al. The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat Struct Mol Biol. 2008;15:795–804. doi: 10.1038/nsmb.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Egecioglu DE, et al. Contributions of Trf4p- and Trf5p-dependent polyadenylation to the processing and degradative functions of the yeast nuclear exosome. RNA. 2006;12:26–32. doi: 10.1261/rna.2207206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kuehner JN, et al. Unravelling the means to an end: RNA polymerase II transcription termination. Nat Rev Mol Cell Biol. 2011;12:283–294. doi: 10.1038/nrm3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jamonnak N, et al. Yeast Nrd1, Nab3, and Sen1 transcriptome-wide binding maps suggest multiple roles in post-transcriptional RNA processing. RNA. 2011;17:2011–2025. doi: 10.1261/rna.2840711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stead JA, et al. The PMC2NT domain of the catalytic exosome subunit Rrp6p provides the interface for binding with its cofactor Rrp47p, a nucleic acid-binding protein. Nucleic Acids Res. 2007;35:5556–5567. doi: 10.1093/nar/gkm614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Milligan L, et al. A yeast exosome cofactor, Mpp6, functions in RNA surveillance and in the degradation of noncoding RNA transcripts. Mol Cell Biol. 2008;28:5446–5457. doi: 10.1128/MCB.00463-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fang F, et al. Rat1p and Rai1p function with the nuclear exosome in the processing and degradation of rRNA precursors. RNA. 2005;11:1571–1578. doi: 10.1261/rna.2900205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sahasranaman A, et al. Assembly of Saccharomyces cerevisiae 60S ribosomal subunits: role of factors required for 27S pre-rRNA processing. EMBO J. 2011;30:4020–4032. doi: 10.1038/emboj.2011.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Geisler S, et al. Decapping of Long Noncoding RNAs Regulates Inducible Genes. Mol Cell. 2012;45:279–291. doi: 10.1016/j.molcel.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Luke B, et al. The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol Cell. 2008;32:465–477. doi: 10.1016/j.molcel.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 71.Whipple JM, et al. The yeast rapid tRNA decay pathway primarily monitors the structural integrity of the acceptor and T-stems of mature tRNA. Genes Dev. 2011;25:1173–1184. doi: 10.1101/gad.2050711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim M, et al. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- 73.Kuai L, et al. Polyadenylation of rRNA in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2004;101:8581–8586. doi: 10.1073/pnas.0402888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schaeffer D, et al. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat Struct Mol Biol. 2009;16:56–62. doi: 10.1038/nsmb.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schneider C, et al. The N-terminal PIN domain of the exosome subunit Rrp44 harbors endonuclease activity and tethers Rrp44 to the yeast core exosome. Nucleic Acids Res. 2009;37:1127–1140. doi: 10.1093/nar/gkn1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Samanta MP, et al. Global identification of noncoding RNAs in Saccharomyces cerevisiae by modulating an essential RNA processing pathway. Proc Natl Acad Sci USA. 2006;103:4192–4197. doi: 10.1073/pnas.0507669103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Marvin MC, et al. Accumulation of noncoding RNA due to an RNase P defect in Saccharomyces cerevisiae. RNA. 2011;17:1441–1450. doi: 10.1261/rna.2737511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marvin MC, Engelke DR. Broadening the mission of an RNA enzyme. J Cell Biochem. 2009;108:1244–1251. doi: 10.1002/jcb.22367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Camblong J, et al. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell. 2007;131:706–717. doi: 10.1016/j.cell.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 81.Houseley J, et al. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol Cell. 2008;32:685–695. doi: 10.1016/j.molcel.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Buhler M, et al. TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nat Struct Mol Biol. 2008;15:1015–1023. doi: 10.1038/nsmb.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kammler S, et al. The RNA exosome component hRrp6 is a target for 5-fluorouracil in human cells. Mol Cancer Res. 2008;6:990–995. doi: 10.1158/1541-7786.MCR-07-2217. [DOI] [PubMed] [Google Scholar]
- 84.Preker P, et al. PROMoter uPstream Transcripts share characteristics with mRNAs and are produced upstream of all three major types of mammalian promoters. Nucleic Acids Res. 2011;39:7179–7193. doi: 10.1093/nar/gkr370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.West S, et al. Human 5′ --> 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432:522–525. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- 86.Rammelt C, et al. PAPD5, a noncanonical poly(A) polymerase with an unusual RNA-binding motif. RNA. 2011;17:1737–1746. doi: 10.1261/rna.2787011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Matera AG, et al. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 88.Waterston RH, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 89.Rudin CM, Thompson CB. Transcriptional activation of short interspersed elements by DNA-damaging agents. Genes Chromosomes Cancer. 2001;30:64–71. [PubMed] [Google Scholar]
- 90.Valgardsdottir R, et al. Transcription of Satellite III non-coding RNAs is a general stress response in human cells. Nucleic Acids Res. 2008;36:423–434. doi: 10.1093/nar/gkm1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ting DT, et al. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science. 2011;331:593–596. doi: 10.1126/science.1200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schmidt MJ, Norbury CJ. Polyadenylation and beyond: emerging roles for noncanonical poly(A) polymerases. WIREs RNA. 2010;1:142–151. doi: 10.1002/wrna.16. [DOI] [PubMed] [Google Scholar]
- 93.Wilusz JE, et al. tRNAs marked with CCACCA are targeted for degradation. Science. 2011;334:817–821. doi: 10.1126/science.1213671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sim S, Wolin SL. Emerging roles for the Ro 60 kDa autoantigen in noncoding RNA metabolism. WIREs RNA. 2011;2:686–689. doi: 10.1002/wrna.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Darnell RB. HITS-CLIP: panoramic views of protein-RNA regulation in living cells. Wiley Interdiscip Rev RNA. 2010;1:266–286. doi: 10.1002/wrna.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ascano M, et al. Identification of RNA-protein interaction networks using PAR-CLIP. Wiley Interdiscip Rev RNA. 2011;3 doi: 10.1002/wrna.1103. epublished ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang Z, et al. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Crow MK. Long interspersed nuclear elements (LINE-1): potential triggers of systemic autoimmune disease. Autoimmunity. 2010;43:7–16. doi: 10.3109/08916930903374865. [DOI] [PubMed] [Google Scholar]
- 99.Cristodero M, et al. The Leishmania tarentolae exosome: purification and structural analysis by electron microscopy. Mol Biochem Parasitol. 2008;159:24–29. doi: 10.1016/j.molbiopara.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 100.Staals RH, et al. Dis3-like 1: a novel exoribonuclease associated with the human exosome. EMBO J. 2010;29:2358–2367. doi: 10.1038/emboj.2010.122. [DOI] [PMC free article] [PubMed] [Google Scholar]