Abstract
Repetitive DNA sequences constitute 30% of the human genome, and are often sites of genomic rearrangement. Recently, it has been found that several constitutional translocations, especially those that involve chromosome 22, take place utilizing palindromic sequences on 22q11 and on the partner chromosome. Analysis of translocation junction fragments shows that the breakpoints of such palindrome-mediated translocations are localized at the center of palindromic AT-rich repeats (PATRRs). The presence of PATRRs at the breakpoints, indicates a palindrome-mediated mechanism involved in the generation of these constitutional translocations. Identification of these PATRR-mediated translocations suggests a universal pathway for gross chromosomal rearrangement in the human genome. De novo occurrences of PATRR-mediated translocations can be detected by PCR in normal sperm samples but not somatic cells. Polymorphisms of various PATRRs influence their propensity for adopting a secondary structure, which in turn affects de novo translocation frequency. We propose that the PATRRs form an unstable secondary structure, which leads to double-strand breaks at the center of the PATRR. The double-strand breaks appear to be followed by a non-homologous end-joining repair pathway, ultimately leading to the translocations. This review considers recent findings concerning the mechanism of meiosis-specific, PATRR-mediated translocations.
The genomic structure of 22q11 mediates rearrangements
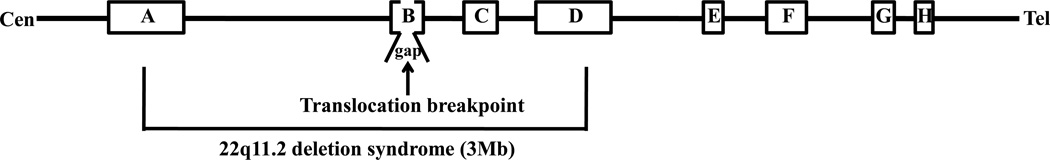
It was previously thought that most genomic rearrangements were formed randomly, but more recent data indicates that is not the case. The 22q11 region is a hotspot for nonrandom chromosomal rearrangements. Deletions, duplications and translocations at 22q11 occur in greater than 1/3000–4000 livebirths [1]. Rearrangements of 22q11 include deletions or duplications associated with congenital developmental defects. The 22q11 deletion syndrome includes DiGeorge, velocardiofacial and conotruncal anomaly face syndromes while the duplications include Cat Eye syndrome [2]. Most interstitial deletions and duplications within 22q11 are attributed to the presence of a specific genomic structure, chromosome-specific low copy repeats (LCRs). The 22q11 region harbors eight large paralogous LCRs [3–5] (LCR-A through LCR-H, Figure 1). Unequal crossovers between DNA segments of similar sequence contained within the LCRs, or non-allelic homologous recombination (NAHR), can produce such chromosomal duplications and deletions [6].
Figure 1. The region of 22q11 containing LCR-A–LCR-H is shown.
A schematic diagram of proximal chromosome 22 indicating the position of the low copy repeats (LCRs) in 22q11.2. Boxes indicate the position of the LCRs. The bracket is indicates the recurrent 3Mb deletion of chromosome 22q11.2. The translocation breakpoint region on chromosome 22 is located within one of the remaining unclonable gap region from human genome project.
Balanced translocations with 22q11 breakpoints also represent non-random genomic rearrangements in this region. A number of constitutional translocations involving 22q11 have been reported, including the recurrent t(11;22)(q23;q11.2), t(17;22)(q11.2;q11.2), and t(8;22)(24.1;q11.2), as well as several non-recurrent rearrangements such as a t(4;22)(q35.1;q11.2), and a t(1;22)(p21.1;q11.2) (Figure 2A). Balanced t(11;22) carriers have no clinical symptoms, but 3:1 meiotic malsegregation of the small der(22) chromosome results in progeny with the supernumerary-der(22)t(11;22) syndrome or Emanuel syndrome (MIM#609029) [7,8*,9–12]. Similar to the t(11;22), phenotypically normal t(8;22) carriers segregate their rearranged chromosomes 3:1 to produce offspring with unbalanced karyotypes [13,14*]. The birth of such chromosomally unbalanced offspring usually results in the testing and detection of their phenotypically normal translocation carrier parent. In contrast, the two reported balanced t(17;22)s are associated with neurofibromatosis type 1 in which disruption of the NF1 gene is thought to be the primary cause of the phenotype (MIM#162200) [15,16].
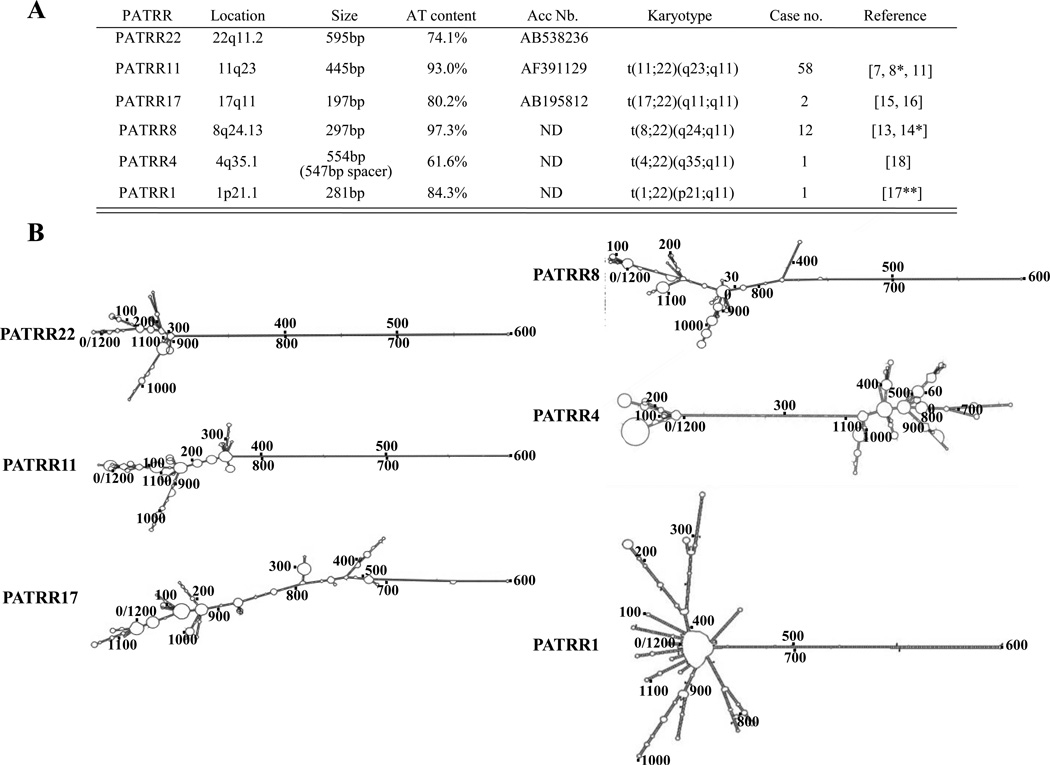
Figure 2. Characterization of PATRR sequences and their secondary structure.
(A) PATRRs and their mediated translocations are listed. Deduced palindromic sequences of PATRR8, 4 and 1 are formed from translocation junction fragments. (B) Reconstructed translocation breakpoint regions for each translocation analyzed by M-fold to determine potential secondary structures [27]
(http://mfold.rna.albany.edu/?q=mfold/DNA-Folding-Form). Breakpoint regions of all PATRRs are located around 600nt.
The molecular etiology of these translocations has been revealed by studies of the genomic structure of breakpoint junction fragments [17**,18,19]. Most constitutional translocations involving 22q11 share the same 22q11.2 LCR breakpoint located within LCR-B (Figure 1). These chromosome 22 translocation breakpoints are localized at the center of an AT-rich palindromic sequence. The breakpoints of the partner chromosomes (11q23, 8q24.1, 17q11.2, 1p21.1, and 4q35.1) also lie at the center of a palindrome sequence, whose length spans several hundred base pairs. Thus, a palindrome-mediated mechanism has been invoked for the generation of these constitutional translocations [8*,9,13,14*,16,17**,18]. Therefore, we suggest palindrome-mediated translocation as one of the universal pathways for human chromosomal rearrangements. The AT-rich palindromic sequences located at these translocation breakpoints have been designated as palindromic AT-rich repeats (PATRRs).
Identification of PATRR sequences at the breakpoints of palindrome-mediated translocations
Detailed analysis of the genomic configuration of the chromosome 11 and 22 breakpoint regions was initially quite difficult because the palindromic sequence is highly unstable, representing a hotspot for deletion and recombination in bacteria, yeast, and mammals [20–22,23**]. To overcome these difficulties, we established a permissive PCR strategy, sequencing by RNA polymerase and cloning in recombination-deficient E. coli cells to accomplish PATRR genotyping [24]. Using these methods, we were able to elucidate three translocation-associated breakpoint regions where we identified highly AT-rich sequence and palindromic structures among the PATRR11, 17, and 22 (Figure 2A). For the other translocations, we could not analyze the entire PATRR directly. Therefore, we designed primer pairs for the breakpoint region on each of the relevant chromosomes and amplified each of the derivative chromosomes using translocation breakpoint-specific PCR. Thus, the breakpoint regions at 8q24.1, 1p21.1, and 4q35.1 were inferred based on reconstructed junction fragments derived from both derivative chromosomes. Inferred sequence of the normal homologous demonstrated palindromic sequence at both 22q11 and at each partner breakpoint region. All partner breakpoints were AT-rich, except 4q35.1 (Figure 2A).
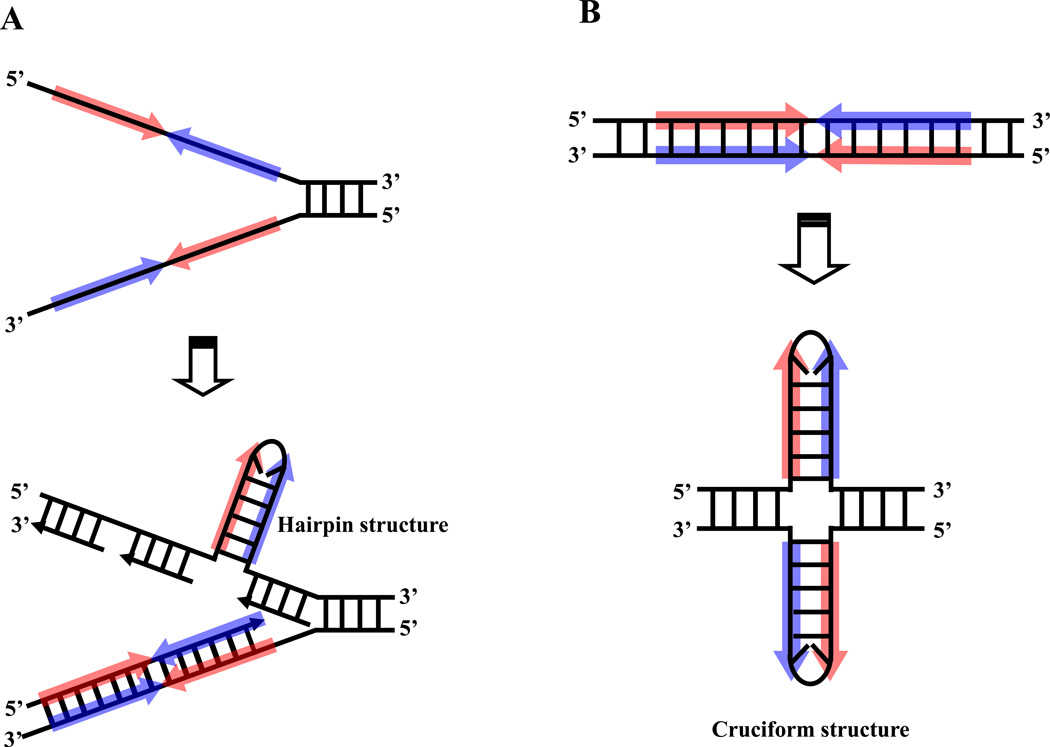
Palindromic sequence, which contains two head-to-head consensus motifs adjacent to one another, is theoretically capable of forming a single-stranded hairpin structure or a double-stranded cruciform structure (Figure 3A, B). It is said that palindrome instability is primarily mediated by its propensity for secondary structure formation [23**,25,26]. A sequence analysis package was used to investigate the potential for secondary structure to form. M-fold (http://mfold.rna.albany.edu/?q=mfold/DNA-Folding-Form) was used to examine 1200nt of sequence at the site of each translocation breakpoint including the PATRR region [17**,27] (Figure 2B). All are capable of forming a symmetrical single-stranded secondary structure with the exception of the palindrome on chromosome 4. Furthermore, we have directly observed cruciform extrusions from the PATRR-plasmid using atomic force microscopy (AFM) [28]. It is likely that formation of such unusual secondary structures would induce genomic instability leading to translocations. Although there are numerous palindromic sequences in the human genome [29], only a few translocation-mediating palindrome sequences have been identified to date. Generally, translocations are considered largely random events. However specific sequence or secondary structure features are probably responsible for the existence of breakage prone sites with translocation susceptibility [30,31] and we speculate that PATRRs possess specific features that make them susceptible as translocation targets.
Figure 3. Possible involvement of DNA secondary structure.
Palindromic sequence possesses the potential for forming an unusual secondary structure by intrastrand base pairing in single stranded DNA. (A) Palindromic DNA may form a hairpin structure during DNA replication. (B) Palindromic DNA can form a double-stranded cruciform structure. Palindromic regions are depicted blue arrows. The red arrows represent the complement of the sequence indicated by blue arrows.
Detection of de novo t(11;22)s in the sperm of normal males by translocation-specific PCR
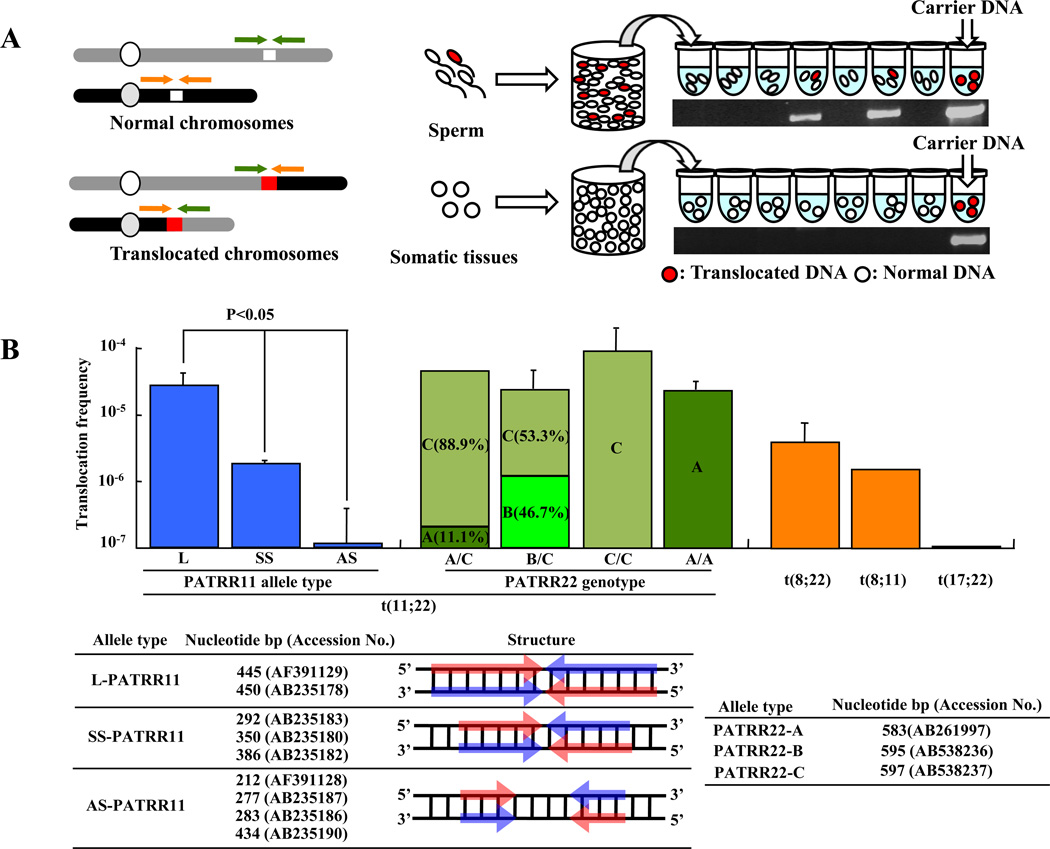
Because t(11;22) translocations have a tightly confined recurrent breakpoint region, we hypothesized that de novo translocations might be detectable in sperm from normal males by PCR [8*,10]. Utilizing the information derived from translocation carriers, we established t(11;22) translocation-specific PCR methodology to assess translocation prevalence in sperm from normal individuals [32**] (Figure 4A). When we amplified multiple aliquots of sperm DNA, translocation-specific PCR products were detected in a substantial number of reactions. On the other hand, PCR products were never found in DNA derived from a variety of somatic tissues. The frequency of de novo translocation events was calculated based on the presence of positive PCR reactions. In our initial analysis, we examined four randomly selected healthy male volunteers, and the estimated frequency of the translocation was approximately 1 × 10−5 in all subjects. The presence of translocation product in sperm from all subjects was surprising and higher than expected [32**]. Subsequently, other de novo PATRR-mediated translocations were successfully detected in sperm [14*].
Figure 4. Detection of de novo translocations by PCR of sperm DNA.
(A) Diagram of the strategy used for estimation of translocation frequency by PCR. The green and orange arrowheads indicate the location of each individual PCR primer. White chromosomal regions indicate PATRR at the breakpoint; red chromosomal regions indicate the presence of a translocation. Combining the forward and reverse primers from different chromosomes allows the detection of the translocated product. Genomic DNA was isolated from sperm samples. Translocation-specific PCR was performed using multiple aliquots of template DNA. The translocation frequency was calculated using the equation, q = 1 - (1 - p)1/n; with n = number of haploid genomes per aliquot, p = the probability that an aliquot sustained a translocation [32]. The gel images show representative PCR results derived from sperm and lymphoblast DNA samples. The positive control is translocation carrier DNA. (B) PATRR polymorphisms affect the de novo t(11;22) translocation frequencies. In the histogram the vertical axis indicates the de novo translocation frequency for different alleles of PATRR11 (allele type L, SS, AS) and for different genotypes of PATRR22 (allele type A, B, C) in sperm. Also shown is frequency of the t(8;22), t(8;11) and t(17;22). Nucleotide size and accession number of PATRR11 and PATRR22 allele types are listed. Arrows indicate each arm of the palindromic sequences. Red and Blue arrows indicate complementary strands.
PATRR polymorphisms affect the de novo translocation propensity of several PATRRs
Previously, we demonstrated that PATRR11 manifests size polymorphisms as a result of deletions within the PATRR, and that this polymorphism influences the frequency of de novo t(11;22)s in sperm [33**]. Long symmetrical PATRR11s (L-PATRR11, 442-450bp) produce de novo translocations in approximately 10−5 gametes. The translocation frequencies of symmetrical short alleles (SS-PATRR11, 292–386bp) are approximately 10-fold lower than L-PATRR11s, while asymmetrical short alleles (AS-PATRR11, 212-434bp) seldom produce de novo translocation products (Figure 4B). The size and symmetry of the PATRRs also reflect their propensity for secondary structure formation, which seems a feature likely to affect translocation frequency, as observed by mobility shift assays and direct visualization by atomic force microscopy [34,35]. However, in contrast to PATRR11 variation, PATRR22 structure has slight difference among alleles. Variation of PATRR22 does not appear to have as great an effect on the frequency of de novo t(11;22)s (Figure 4B). Nonetheless, comparison of the allelic origin of the translocations has demonstrated some allelic bias [36].
De novo t(8;22) translocations are also detected in sperm samples (Figure 4B). The translocation frequency is similar to the t(11;22) frequency derived from a SS-PATRR11 [14*]. Even though there have been no constitutional t(8;11)s reported in the literature, de novo t(8;11)s are detected in sperm. The identification of a palindrome-mediated translocation that does not involve 22q11 provides evidence that palindrome-mediated translocation represents a general rearrangement mechanism. There were two previous reports of a familial constitutional t(3;8) segregating in association with renal cell carcinoma [37,38]. The breakpoints of these rearrangements have now been identified in the same PATRR8 and a PATRR-like sequence on chromosome 3 (B.S.E unpublished data). This recent finding adds further support to the hypothesis that PATRR-mediated translocations represent a universal pathway for gross chromosomal rearrangement.
Although there have been two unrelated t(17;22) carriers reported [15,16,24] (Figure 4B), de novo t(17;22)s have never been observed in sperm samples. Thus, it appears that PATRR17 is less susceptible to double-strand breaks than are PATRR8, 11 or 22. The number of reported cases of recurrent rearrangements involving these sites appears to correlate with translocation frequency observed in sperm samples. Thus, it is possible that additional t(17;22)s might be detected in sperm if a far greater number of genome equivalents were studied.
Chromosomal instability mediated by unusual DNA structures-several scenarios
As mentioned previously, de novo t(11;22)s can only be detected in sperm and not in other somatic tissues [32**]. Further, all de novo t(11;22)s examined have been determined to be paternal in origin [39]. From these results, we hypothesize that PATRRs form secondary structures, which in turn induce translocations during gametogenesis, especially spermatogenesis. The timing and mechanisms of secondary structure and translocation formation in male germ cells are potentially threefold 1) prior to meiosis, 2) during meiosis, and 3) after meiosis. We will discuss these scenarios in turn. In general, such genomic instability prior to meiosis has been explained invoking a replication-dependent mechanism. PATRRs should be capable of forming hairpin structures within long single-stranded regions of DNA on the lagging–strand during DNA replication [23**,40–43>] (Figure 3A). If the translocation takes place during meiosis, the four-way junction of the cruciform structure is analogous to the Holliday junction in homologous genetic recombination [23**]. If the translocation takes place after meiosis during the spermatid stage, chromatin remodeling leads to the accumulation of negatively supercoiled DNA, from which cruciform structures are likely to extrude [44–46] (Figure 3B). These unusual DNA conformations could potentially act as targets for a structure-specific nuclease and contribute to palindrome-mediated translocations. To explore these possibilities, several experimental approaches have been undertaken.
Chromosomal abnormalities in humans often demonstrate a gender bias with respect to parental origin [47]. Structural abnormalities predominantly occur in the paternal germline [48]. This is due to a greater number of cell divisions occurring during spermatogenesis. If translocations take place by a replication error, one would expect a positive relationship between paternal age and de novo translocations [49*]. However the de novo t(11;22) frequency does not appear to increase with increasing paternal age [50]. This data suggests that translocations do not occur via a pre-meiotic replication-dependent mechanism. Further, despite the obvious AT richness at the breakpoint, no significant homology is apparent between PATRR11 and PATRR22, suggesting that t(11;22) events result from double-strand break repair occurring by a non-homologous end joining mechanism [8*,51]. In contrast to translocation formation, deletions within the PATRRs appear to utilize much more extensive microhomology [5,51]. Furthermore, inhibition of DNA replication induces deletions but not translocations [52]. These results imply that the mechanism for the generation of a palindrome-mediated rearrangement differs between deletions and translocations.
Some recurrent constitutional translocations take place via NAHR, during genetic recombination [53*,54–56]. For PATRRs, the potential cruciform conformation is analogous to a Holliday junction and potentially could act as a substrate for the Holliday junction resolvase [23**]. However such an enzyme has not been identified in mammals [57*]. Further, t(11;22) translocation breakpoints do not coincide with recombination hotspots [58] and the t(11;22) translocation frequency is neither facilitated nor altered by PRDM9 [59]. Thus, it appears that meiotic recombination pathways may not be involved in the mechanism of the translocation.
Regarding a post-meiotic hypothesis, it has previously been shown that triplet-repeat expansion is limited to the post-meiotic, haploid cell and therefore does not involve mitotic replication or recombination [60]. However for these translocations, some unusual de novo der(22)t(11;22)s have been reported. A patient with supernumerary der(22) syndrome, was born to normal parents as the result of a de novo translocation followed by aberrant adjacent-I segregation in meiosis [61]. This contradicts a post meiotic origin. Also, we previously studied another unusual carrier, whose karyotype is mosaic 46,XX/46,XX, t(11;22) suggesting that the translocation might have taken place early in embryogenesis as a mitotic event [62]. Thus, each of these potential scenarios has some contradictions.
Other factors may influence PATRR-mediated translocations. It has been shown that the spatial organization of chromosomes is non-random, and is a contributing factor in the formation of specific somatic translocations [63–65]. Perhaps the spatial proximity of chromosome 11 and 22 in meiosis play a role in generating the translocation. To test this hypothesis, the position of the 11q23 and 22q11 breakpoint regions were examined by FISH analysis during male and female meiosis. The proximity between 11q23 and 22q11 is closer than that of a control region and 22q11, suggesting that spatial proximity during meiosis might play a role in the generation of recurrent translocations [58*]. Thus, using various methodologies and approaches, attempts have been made to clarify the mechanism behind PATRR-mediated translocations. Nonetheless, despite numerous speculations and experiments, the mechanism of PATRR-mediated translocations remains elusive. Thus additional studies will be required to determine the enzymatic pathway(s) and the timing involved in formation of these translocations in gametogenesis.
Acknowledgements
The authors wish to thank Molly B. Sheridan, April M. Hacker and Colleen P. Franconi for suggestions. These studies were supported by Award Number R01CA039926 from the National Cancer Institute (B.S.E.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. The studies were also supported by funds from the Charles E.H. Upham Chair (B.S.E.). One of the authors, (T.K.), was supported by the JSPS Postdoctoral Fellowships for Research Abroad.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This review is dedicated in memory of E. Stephen Emanuel, M.D. His unerring support for intellectual curiosity about and interest in this topic will not be forgotten.
References
- 1.Burn J, Goodship J. Developmental genetics of the heart. Curr Opin Genet Dev. 1996;6:322–325. doi: 10.1016/s0959-437x(96)80009-8. [DOI] [PubMed] [Google Scholar]
- 2.Emanuel BS. Molecular mechanisms and diagnosis of chromosome 22q11.2 rearrangements. Dev Disabil Res Rev. 2008;14:11–18. doi: 10.1002/ddrr.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edelmann L, Pandita RK, Spiteri E, Funke B, Goldberg R, Palanisamy N, Chaganti RS, Magenis E, Shprintzen RJ, Morrow BE. A common molecular basis for rearrangement disorders on chromosome 22q11. Hum Mol Genet. 1999;8:1157–1167. doi: 10.1093/hmg/8.7.1157. [DOI] [PubMed] [Google Scholar]
- 4.Shaikh TH, Kurahashi H, Saitta SC, O'Hare AM, Hu P, Roe BA, Driscoll DA, McDonald-McGinn DM, Zackai EH, Budarf ML, et al. Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: genomic organization and deletion endpoint analysis. Hum Mol Genet. 2000;9:489–501. doi: 10.1093/hmg/9.4.489. [DOI] [PubMed] [Google Scholar]
- 5.Shaikh TH, O'Connor RJ, Pierpont ME, McGrath J, Hacker AM, Nimmakayalu M, Geiger E, Emanuel BS, Saitta SC. Low copy repeats mediate distal chromosome 22q11.2 deletions: sequence analysis predicts breakpoint mechanisms. Genome Res. 2007;17:482–491. doi: 10.1101/gr.5986507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- 7.Edelmann L, Spiteri E, Koren K, Pulijaal V, Bialer MG, Shanske A, Goldberg R, Morrow BE. AT-rich palindromes mediate the constitutional t(11;22) translocation. Am J Hum Genet. 2001;68:1–13. doi: 10.1086/316952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kurahashi H, Emanuel BS. Long AT-rich palindromes and the constitutional t(11;22) breakpoint. Hum Mol Genet. 2001;10:2605–2617. doi: 10.1093/hmg/10.23.2605. In this study, junction fragments of 40 unrelated t(11;22)s were isolated by PCR and analyzed. All breakpoints are nearly identical and are located at the center of palindromic sequence on 11 and 22
- 9.Kurahashi H, Shaikh TH, Hu P, Roe BA, Emanuel BS, Budarf ML. Regions of genomic instability on 22q11 and 11q23 as the etiology for the recurrent constitutional t(11;22) Hum Mol Genet. 2000;9:1665–1670. doi: 10.1093/hmg/9.11.1665. [DOI] [PubMed] [Google Scholar]
- 10.Shaikh TH, Budarf ML, Celle L, Zackai EH, Emanuel BS. Clustered 11q23 and 22q11 breakpoints and 3:1 meiotic malsegregation in multiple unrelated t(11;22) families. Am J Hum Genet. 1999;65:1595–1607. doi: 10.1086/302666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tapia-Paez I, Kost-Alimova M, Hu P, Roe BA, Blennow E, Fedorova L, Imreh S, Dumanski JP. The position of t(11;22)(q23;q11) constitutional translocation breakpoint is conserved among its carriers. Hum Genet. 2001;109:167–177. doi: 10.1007/s004390100560. [DOI] [PubMed] [Google Scholar]
- 12.Zackai EH, Emanuel BS. Site-specific reciprocal translocation, t(11;22) (q23;q11), in several unrelated families with 3:1 meiotic disjunction. Am J Med Genet. 1980;7:507–521. doi: 10.1002/ajmg.1320070412. [DOI] [PubMed] [Google Scholar]
- 13.Gotter AL, Nimmakayalu MA, Jalali GR, Hacker AM, Vorstman J, Conforto Duffy D, Medne L, Emanuel BS. A palindrome-driven complex rearrangement of 22q11.2 and 8q24.1 elucidated using novel technologies. Genome Res. 2007;17:470–481. doi: 10.1101/gr.6130907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sheridan MB, Kato T, Haldeman-Englert C, Jalali GR, Milunsky JM, Zou Y, Klaes R, Gimelli G, Gimelli S, Gemmill RM, et al. A palindrome-mediated recurrent translocation with 3:1 meiotic nondisjunction: the t(8;22)(q24.13;q11.21) Am J Hum Genet. 2010;87:209–218. doi: 10.1016/j.ajhg.2010.07.002. In this study a new recurrent palindrome-mediated translocation is characterized. De novo t(8;22)s and t(8;11)s are detected in normal sperm. This study suggests that PATRR-mediated translocations represent a universal pathway for gross chromosomal rearrangement in the human genome
- 15.Kehrer-Sawatzki H, Haussler J, Krone W, Bode H, Jenne DE, Mehnert KU, Tummers U, Assum G. The second case of a t(17;22) in a family with neurofibromatosis type 1: sequence analysis of the breakpoint regions. Hum Genet. 1997;99:237–247. doi: 10.1007/s004390050346. [DOI] [PubMed] [Google Scholar]
- 16.Kurahashi H, Shaikh T, Takata M, Toda T, Emanuel BS. The constitutional t(17;22): another translocation mediated by palindromic AT-rich repeats. Am J Hum Genet. 2003;72:733–738. doi: 10.1086/368062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gotter AL, Shaikh TH, Budarf ML, Rhodes CH, Emanuel BS. A palindrome-mediated mechanism distinguishes translocations involving LCR-B of chromosome 22q11.2. Hum Mol Genet. 2004;13:103–115. doi: 10.1093/hmg/ddh004. In this study, M-fold analysis of several palindrome-mediated translocations demonstrates that breakpoint sequences have the potential to form unstable hairpin or cruciform structures.
- 18.Nimmakayalu MA, Gotter AL, Shaikh TH, Emanuel BS. A novel sequence-based approach to localize translocation breakpoints identifies the molecular basis of a t(4;22) Hum Mol Genet. 2003;12:2817–2825. doi: 10.1093/hmg/ddg301. [DOI] [PubMed] [Google Scholar]
- 19.Kurahashi H, Inagaki H, Hosoba E, Kato T, Ohye T, Kogo H, Emanuel BS. Molecular cloning of a translocation breakpoint hotspot in 22q11. Genome Res. 2007;17:461–469. doi: 10.1101/gr.5769507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akgun E, Zahn J, Baumes S, Brown G, Liang F, Romanienko PJ, Lewis S, Jasin M. Palindrome resolution and recombination in the mammalian germ line. Mol Cell Biol. 1997;17:5559–5570. doi: 10.1128/mcb.17.9.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collick A, Drew J, Penberth J, Bois P, Luckett J, Scaerou F, Jeffreys A, Reik W. Instability of long inverted repeats within mouse transgenes. EMBO J. 1996;15:1163–1171. [PMC free article] [PubMed] [Google Scholar]
- 22.Gordenin DA, Lobachev KS, Degtyareva NP, Malkova AL, Perkins E, Resnick MA. Inverted DNA repeats: a source of eukaryotic genomic instability. Mol Cell Biol. 1993;13:5315–5322. doi: 10.1128/mcb.13.9.5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Leach DR. Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. Bioessays. 1994;16:893–900. doi: 10.1002/bies.950161207. Here it is shown that long palindromes form hairpin or cruciform structures. In E coli , palindrome-mediated mutagenesis occurs by strand-slippage during replication with a preference for deletions on the lagging strand.
- 24.Inagaki H, Ohye T, Kogo H, Yamada K, Kowa H, Shaikh TH, Emanuel BS, Kurahashi H. Palindromic AT-rich repeat in the NF1 gene is hypervariable in humans and evolutionarily conserved in primates. Hum Mutat. 2005;26:332–342. doi: 10.1002/humu.20228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nag DK, Kurst A. A 140-bp-long palindromic sequence induces double-strand breaks during meiosis in the yeast Saccharomyces cerevisiae. Genetics. 1997;146:835–847. doi: 10.1093/genetics/146.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith GR. Meeting DNA palindromes head-to-head. Genes Dev. 2008;22:2612–2620. doi: 10.1101/gad.1724708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurahashi H, Inagaki H, Yamada K, Ohye T, Taniguchi M, Emanuel BS, Toda T. Cruciform DNA structure underlies the etiology for palindrome-mediated human chromosomal translocations. J Biol Chem. 2004;279:35377–35383. doi: 10.1074/jbc.M400354200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu L, Jia H, Droge P, Li J. The human genome-wide distribution of DNA palindromes. Funct Integr. 2007;7:221–227. doi: 10.1007/s10142-007-0047-6. [DOI] [PubMed] [Google Scholar]
- 30.Kurahashi H, Inagaki H, Ohye T, Kogo H, Kato T, Emanuel BS. Palindrome-mediated chromosomal translocations in humans. DNA Repair (Amst) 2006;5:1136–1145. doi: 10.1016/j.dnarep.2006.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raghavan SC, Swanson PC, Wu X, Hsieh CL, Lieber MR. A non-B-DNA structure at the Bcl-2 major breakpoint region is cleaved by the RAG complex. Nature. 2004;428:88–93. doi: 10.1038/nature02355. [DOI] [PubMed] [Google Scholar]
- 32. Kurahashi H, Emanuel BS. Unexpectedly high rate of de novo constitutional t(11;22) translocations in sperm from normal males. Nat Genet. 2001;29:139–140. doi: 10.1038/ng1001-139. In this study, translocation-specific PCR was used to detect de novo t(11;22)s in normal sperm. PCR products are detected in sperm, but not somatic DNA samples, indicating that the t(11;22) arises during gametogenesis
- 33. Kato T, Inagaki H, Yamada K, Kogo H, Ohye T, Kowa H, Nagaoka K, Taniguchi M, Emanuel BS, Kurahashi H. Genetic variation affects de novo translocation frequency. Science. 2006;311:971. doi: 10.1126/science.1121452. This study shows, for the first time, that size polymorphisms of the palindrome at the chromosome 11 breakpoint affect translocation frequency in sperm.
- 34.Kato T, Inagaki H, Tong M, Kogo H, Ohye T, Yamada K, Tsutsumi M, Emanuel BS, Kurahashi H. DNA secondary structure is influenced by genetic variation and alters susceptibility to de novo translocation. Mol Cytogenet. 2011;4:18. doi: 10.1186/1755-8166-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kogo H, Inagaki H, Ohye T, Kato T, Emanuel BS, Kurahashi H. Cruciform extrusion propensity of human translocation-mediating palindromic AT-rich repeats. Nucleic Acids Res. 2007;35:1198–1208. doi: 10.1093/nar/gkm036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tong M, Kato T, Yamada K, Inagaki H, Kogo H, Ohye T, Tsutsumi M, Wang J, Emanuel BS, Kurahashi H. Polymorphisms of the 22q11.2 breakpoint region influence the frequency of de novo constitutional t(11;22)s in sperm. Hum Mol Genet. 2010;19:2630–2637. doi: 10.1093/hmg/ddq150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cohen AJ, Li FP, Berg S, Marchetto DJ, Tsai S, Jacobs SC, Brown RS. Hereditary renal-cell carcinoma associated with a chromosomal translocation. N Engl J Med. 1979;301:592–595. doi: 10.1056/NEJM197909133011107. [DOI] [PubMed] [Google Scholar]
- 38.Poland KS, Azim M, Folsom M, Goldfarb R, Naeem R, Korch C, Drabkin HA, Gemmill RM, Plon SE. A constitutional balanced t(3;8)(p14;q24.1) translocation results in disruption of the TRC8 gene and predisposition to clear cell renal cell carcinoma. Genes Chromosomes Cancer. 2007;46:805–812. doi: 10.1002/gcc.20466. [DOI] [PubMed] [Google Scholar]
- 39.Ohye T, Inagaki H, Kogo H, Tsutsumi M, Kato T, Tong M, Macville MV, Medne L, Zackai EH, Emanuel BS, et al. Paternal origin of the de novo constitutional t(11;22)(q23;q11) Eur J Hum Genet. 2010;18:783–787. doi: 10.1038/ejhg.2010.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trinh TQ, Sinden RR. Preferential DNA secondary structure mutagenesis in the lagging strand of replication in E. coli. Nature. 1991;352:544–547. doi: 10.1038/352544a0. [DOI] [PubMed] [Google Scholar]
- 41.Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 42.Mirkin SM, Smirnova EV. Positioned to expand. Nat Genet. 2002;31:5–6. doi: 10.1038/ng0502-5. [DOI] [PubMed] [Google Scholar]
- 43.Zhang H, Freudenreich CH. An AT-rich sequence in human common fragile site FRA16D causes fork stalling and chromosome breakage in S. cerevisiae. Mol Cell. 2007;27:367–379. doi: 10.1016/j.molcel.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Braun RE. Packaging paternal chromosomes with protamine. Nat Genet. 2001;28:10–12. doi: 10.1038/ng0501-10. [DOI] [PubMed] [Google Scholar]
- 45.Ward WS. Function of sperm chromatin structural elements in fertilization and development. Mol Hum Reprod. 2010;16:30–36. doi: 10.1093/molehr/gap080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ward WS. Regulating DNA supercoiling: sperm points the way. Biol Reprod. 2011;84:841–843. doi: 10.1095/biolreprod.111.090951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2001;2:280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- 48.Buwe A, Guttenbach M, Schmid M. Effect of paternal age on the frequency of cytogenetic abnormalities in human spermatozoa. Cytogenet Genome Res. 2005;111:213–228. doi: 10.1159/000086892. [DOI] [PubMed] [Google Scholar]
- 49. Crow JF. The origins, patterns and implications of human spontaneous mutation. Nat Rev Genet. 2000;1:40–47. doi: 10.1038/35049558. Here the author reviews the evidence that the germline mutation rate in males versus females is higher as the result of more numerous germ-cell divisions.
- 50.Kato T, Yamada K, Inagaki H, Kogo H, Ohye T, Emanuel BS, Kurahashi H. Age has no effect on de novo constitutional t(11;22) translocation frequency in sperm. Fertil Steril. 2007;88:1446–1448. doi: 10.1016/j.fertnstert.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kato T, Inagaki H, Kogo H, Ohye T, Yamada K, Emanuel BS, Kurahashi H. Two different forms of palindrome resolution in the human genome: deletion or translocation. Hum Mol Genet. 2008;17:1184–1191. doi: 10.1093/hmg/ddn008. [DOI] [PubMed] [Google Scholar]
- 52.Kurahashi H, Inagaki H, Kato T, Hosoba E, Kogo H, Ohye T, Tsutsumi M, Bolor H, Tong M, Emanuel BS. Impaired DNA replication prompts deletions within palindromic sequences, but does not induce translocations in human cells. Hum Mol Genet. 2009;18:3397–3406. doi: 10.1093/hmg/ddp279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bandyopadhyay R, Heller A, Knox-DuBois C, McCaskill C, Berend SA, Page SL, Shaffer LG. Parental origin and timing of de novo Robertsonian translocation formation. Am J Hum Genet. 2002;71:1456–1462. doi: 10.1086/344662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Giglio S, Calvari V, Gregato G, Gimelli G, Camanini S, Giorda R, Ragusa A, Guerneri S, Selicorni A, Stumm M, et al. Heterozygous submicroscopic inversions involving olfactory receptor-gene clusters mediate the recurrent t(4;8)(p16;p23) translocation. Am J Hum Genet. 2002;71:276–285. doi: 10.1086/341610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maas NM, Van Vooren S, Hannes F, Van Buggenhout G, Mysliwiec M, Moreau Y, Fagan K, Midro A, Engiz O, Balci S, et al. The t(4;8) is mediated by homologous recombination between olfactory receptor gene clusters, but other 4p16 translocations occur at random. Genet Couns. 2007;18:357–365. [PubMed] [Google Scholar]
- 56.Ou Z, Stankiewicz P, Xia Z, Breman AM, Dawson B, Wiszniewska J, Szafranski P, Cooper ML, Rao M, Shao L, et al. Observation and prediction of recurrent human translocations mediated by NAHR between nonhomologous chromosomes. Genome Res. 2011;21:33–46. doi: 10.1101/gr.111609.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lobachev KS, Gordenin DA, Resnick MA. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell. 2002;108:183–193. doi: 10.1016/s0092-8674(02)00614-1. [DOI] [PubMed] [Google Scholar]
- 58. Ashley T, Gaeth AP, Inagaki H, Seftel A, Cohen MM, Anderson LK, Kurahashi H, Emanuel BS. Meiotic recombination and spatial proximity in the etiology of the recurrent t(11;22) Am J Hum Genet. 2006;79:524–538. doi: 10.1086/507652. This study demonstrates that physical proximity between 11q23 and 22q11 in meiosis plays an important role in the generation of the constitutional t(11;22).
- 59.Berg IL, Neumann R, Lam KW, Sarbajna S, Odenthal-Hesse L, May CA, Jeffreys AJ. PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat Genet. 2010;42:859–863. doi: 10.1038/ng.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kovtun IV, McMurray CT. Trinucleotide expansion in haploid germ cells by gap repair. Nat Genet. 2001;27:407–411. doi: 10.1038/86906. [DOI] [PubMed] [Google Scholar]
- 61.Dawson AJ, Mears AJ, Chudley AE, Bech-Hansen T, McDermid H. DeR22)t(11;22) resulting from a paternal de novo translocation, adjacent 1 segregation, and maternal heterodisomy of chromosome 22. J Med Genet. 1996;33:952–956. doi: 10.1136/jmg.33.11.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kurahashi H, Shaikh TH, Zackai EH, Celle L, Driscoll DA, Budarf ML, Emanuel BS. Tightly clustered 11q23 and 22q11 breakpoints permit PCR-based detection of the recurrent constitutional t(11;22) Am J Hum Genet. 2000;67:763–768. doi: 10.1086/303054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Misteli T. Spatial positioning: a new dimension in genome function. Cell. 2004;119:153–156. doi: 10.1016/j.cell.2004.09.035. [DOI] [PubMed] [Google Scholar]
- 64.Parada LA, Sotiriou S, Misteli T. Spatial genome organization. Exp Cell Res. 2004;296:64–70. doi: 10.1016/j.yexcr.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 65.Roix JJ, McQueen PG, Munson PJ, Parada LA, Misteli T. Spatial proximity of translocation-prone gene loci in human lymphomas. Nat Genet. 2003;34:287–291. doi: 10.1038/ng1177. [DOI] [PubMed] [Google Scholar]