Abstract
Endonuclease VIII-like 3 (Neil3) is one of the five DNA glycosylases found in mammals that recognize and remove oxidized bases, and initiate the Base Excision Repair (BER) pathway. Previous attempts to express and purify the mouse and human orthologs of Neil3 in their active form have not been successful. Here we report the construction of bicistronic expression vectors for expressing in Escherichia coli the full-length mouse Neil3 (MmuNeil3), its glycosylase domain (MmuNeil3Δ324), as well as the glycosylase domain of human Neil3 (NEIL3Δ324). The purified Neil3 proteins are all active, and NEIL3Δ324 exhibits similar glycosylase/lyase activity as MmuNeil3Δ324 on both single-stranded and double-stranded substrates containing thymine glycol (Tg), spiroiminodihydantoin (Sp) or an abasic site (AP). We show that N-terminal initiator methionine processing is critical for the activity of both mouse and human Neil3 proteins. Co-expressing an E. coli methionine aminopeptidase (EcoMap) Y168A variant with MmuNeil3, MmuNeil3Δ324 and NEIL3Δ324 improves the N-terminal methionine processing and increases the percentage of active Neil3 proteins in the preparation. The purified Neil3 proteins are suitable for biochemical, structural and functional studies.
Keywords: DNA glycosylases, Endonuclease VIII-Like 3 (Neil3), Bicistronic vector, Escherichia coli methionine aminopeptidase (EcoMap)
Introduction
Cellular DNA is constantly being challenged by oxidative stress from both endogenous and exogenous sources, such as metabolism, inflammation, ionizing radiation, and various chemical reagents [1]. To protect cells from oxidative DNA damage and mutagenesis, organisms possess multiple DNA glycosylases to recognize and remove the damaged bases and to initiate the Base Excision Repair (BER) pathway. In mammals, five DNA glycosylases have thus far been identified that are specific for oxidized bases: endonuclease III (Nth1), 8-oxoguanine-DNA-glycosylase (Ogg1), endonuclease VIII-like 1 (Neil1), endonuclease VIII-like 2 (Neil2), and endonuclease VIII-like 3 (Neil3) (reviewed in [2–4]). These oxidative DNA glycosylases are bifunctional, with a glycosylase activity that cleaves the N-glycosylic bond releasing the base lesion from the sugar backbone, and a lyase activity that cleaves the DNA backbone at the apurinic or apyrimidinic (AP) site that results from glycosylase action [5]. Our knowledge of the substrate specificity, catalytic mechanisms and structural features of the oxidative DNA glycosylases has been facilitated by the ability to overexpress and purify large quantities of these proteins. However, expressing and purifying active Neil3 proteins has been a daunting problem that has limited the enzymatic, structural and functional characterization of the Neil3 proteins.
Several expression systems have been utilized to express and purify the mouse and human orthologs of Neil3 (MmuNeil3 and NEIL3, respectively) [6–10]. Most of these methods, however, failed to produce purified Neil3 proteins with both glycosylase and lyase activities. A reticulocyte lysate system was used to express the full-length MmuNeil3 and NEIL3 in vitro, but no glycosylase or lyase activity was detected in the purified proteins [7, 9]. A baculovirus system was also used to express full-length NEIL3; however, the expression level was too low to allow purification of the protein [10]. Moreover, over-expression of NEIL3 in mammalian cells caused extreme cytotoxicity to the cells [7]. Using Escherichia coli as the host to express recombinant full-length NEIL3 resulted in only partial purification without any detectable glycosylase activity [6, 7]. Because of the difficulties in producing the full-length MmuNeil3 and NEIL3 proteins, several truncated versions containing the glycosylase domain of MmuNeil3 or NEIL3 have been expressed and purified in E. coli [6–8]. One of these, a C-terminal His-tagged NEIL3 (containing residues 1–290) showed weak lyase activity on a single-stranded substrate, but no glycosylase activity [6]. In 2010 we published the characterization of the glycosylase/lyase activities of both a full-length and a truncated version of mouse Neil3 [11] based on the purification protocol described here.
For many reasons, the E. coli expression system is desirable for expressing heterologous genes. However, a common and sometimes major problem is the retention of an initiator methionine residue that must be cleaved in order to produce active recombinant proteins [12–15]. For instance, using E. coli as a host, the purified pro-inflammatory cytokine RANTES was completely inactive with the retention of the N-terminal methionine [14]. Similarly, if expressed in E. coli, the lack of removal of the N-terminal methionine of onconase, a cytotoxic ribonuclease with antitumor properties, resulted in little catalytic activity and reduced thermostability [13]. The problem of no or incomplete N-terminal initiator methionine processing has limited the application of the E. coli expression system to produce active heterogeneous proteins.
In E. coli, the removal of the initiator N-formylmethionine involves two-step enzymatic processing, first by peptide deformylase to release the formyl group, followed by methionine aminopeptidase (EcoMap) to cleave the N-terminal methionine (reviewed in [16]). While the de-formylation step is tightly coupled to the translation process, the cleavage of the N-terminal methionine by EcoMap is determined by the penultimate residue of the protein being translated [17, 18]. It is generally accepted that cleavage occurs if the side chain is small, as is the case for Ala, Cys, Pro, Ser, Thr, and Val [17, 19]. However, previous studies have shown that a valine at the second position results in partial or incomplete processing of the heterologous proteins expressed in E. coli [18]. In addition, the antepenultimate residue, the amino acid at the third position, also affects the efficiency of methionine processing. In particular, a glutamic acid has been shown to further decrease the processing efficiency [18]. According to these results, Neil3 proteins are predicted to have incomplete cleavage of the N-terminal methionine if expressed in E. coli because of their N-terminal penultimate valine and antepenultimate glutamic acid residues.
Importantly, cleavage of the N-terminal initiator methionine is required for the catalytic activity of Neil3. Based on structure and sequence homology, Neil3 proteins are members of the Fpg/Nei family [20, 21], which is named after two bacterial proteins, formamidopyrimidine-DNA glycosylase (Fpg) [22–25] and endonuclease VIII (Nei) [26–28]. Most Fpg/Nei family members, including NEIL1 and NEIL2, utilize the secondary amine of the penultimate proline residue to form a transient imine intermediate (Schiff base) for catalysis (reviewed in [29]). Upon characterizing the mouse ortholog of Neil3, we confirmed that the primary amine of the penultimate valine residue serves the same function [11]. Therefore, the correct processing of the N-terminal methionine is required in order to produce active Neil3 proteins.
Co-expression of the E. coli methionine aminopeptidase (EcoMap) with the target gene is one strategy that has been used to remove the N-terminal methionine from recombinant proteins in vivo. Successful examples include co-expressing EcoMap with synthetic human α- and β-globin to produce native, unmodified hemoglobin A [15], co-expression of EcoMap with recombinant Methanothermus fervidus histone A [30], and co-expression of EcoMap with the Arf family of GTPases to increase acylation and homogeneity [31]. Recently, an engineered EcoMap (EcoMapY168A) has been shown to improve N-terminal processing of recombinant proteins with valine at the penultimate position compared to the wild-type EcoMap [32, 33]. These methods paved the way for us to develop a protocol for producing active Neil3 proteins with their native N-terminus using an E. coli expression system.
In the present report, the protocols for over-expressing and purifying active Neil3 proteins, including the full-length MmuNeil3 and the glycosylase domain of both MmuNeil3 and NEIL3 (MmuNeil3Δ324 and NEIL3Δ324, respectively), are described. E. coli bicistronic expression vectors, constructed in our laboratory, that contain an ORF6 leader sequence [34] were used to increase translation efficiency and EcoMapY168A was co-expressed with the Neil3 proteins under the control of separate T7/lac promoters. Whether co-expression of EcoMapY168A facilitates the N-terminal methionine processing and enhances the percentage of the active Neil3 proteins in the preparation was examined. We also demonstrate that NEIL3Δ324 and MmuNeil3Δ324 possess similar glycosylase/lyase activities. Our method of expression and purification of the Neil3 proteins has led to the successful characterization of the MmuNeil3 proteins [11], and paved the way for further biochemical, structural and functional studies.
Materials and methods
Materials
The full-length cDNA clone of Homo sapiens NEIL3 (MGC-39163) was purchased from RIKEN (Japan) and the full-length cDNA clone of Mus musculus Neil3 (MmuNeil3, MGC-36916) was purchased from American Type Culture Collection (Manassas, Virginia). The primers were chemically synthesized by Midland Certified Reagent Company, Inc. (Midland, TX). Restriction enzymes were purchased from New England Biolabs (NEB, Beverly, MA). Cloned Pfu DNA polymerase was purchased from Stratagene (Cedar Creek, TX). Expression vectors pET30a and pETDuet1 were purchased from Novagen (Madison, WI). E. coli endonuclease III (EcoNth) and E. coli endonuclease VIII (EcoNei) were from our laboratory stocks and were purified as previously described [35].
Construction of bicistronic expression vectors
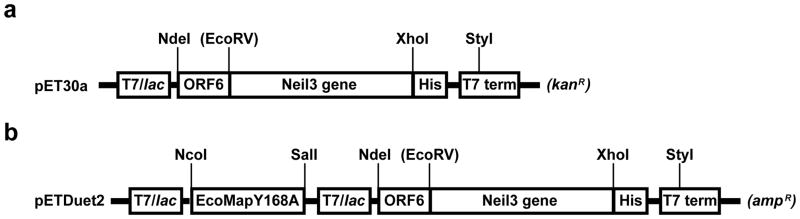
Details of the construction of the pET30a-ORF6 vector and the sequence of the ORF6 leader sequence have been previously described [34]. To create the pETDuet2- T7EcoMapY168A-ORF6 vector, we first amplified the EcoMapY168A gene from pBS-Map(Y168A)-cGSTM1 plasmid (kindly provided by Dr. Ming F. Tam, Institute of Molecular Biology, Academia Sinica, [32]) using the following primers: 5′catgccatggc tatctcaatcaagacccc3′ and 5′acgcgtcgacttattcgtcgtgcgagattatcgc3′. The PCR product was digested and ligated into the NcoI-SalI-cut pETDuet1 plasmid, resulting in the pETDuet1-T7EcoMap plasmid. We then introduced the fragment coding the ORF6 leader sequence and the hexa-histidine tag from the pET30a-ORF6 vector (between the NdeI site and the StyI site in the T7 terminator) into the pETDuet1-T7EcoMap plasmid, resulting in the pETDuet2-T7EcoMap-ORF6 vector, which allows expression of recombinant proteins containing a C-terminal hexa-histidine tag (Fig. 1). All plasmids were amplified in E. coli TOP10 cells (Invitrogen, Carlsbad, CA) and plasmid DNA was purified using Wizard Plus Midiprep DNA Purification System (Promega, Madison, WI).
Figure 1.
Construction of bicistronic Neil3 expression plasmids based on (a) pET30a and (b) pETDuet1. In both constructs, T7/lac, ORF6, His, and T7 term denote T7/lac promoter, ORF6 leading sequence, hexa-histidine tag, and T7 terminator, respectively. The Neil3 gene represents the MmuNeil3, MmuNeil3Δ324 or NEIL3Δ324 gene. The last nucleotide in the stop codon of the ORF6 leader sequence overlaps with the initiation codon of the Neil3 gene, thus the translation of the Neil3 gene is coupled to the ORF6 leader sequence and no fusion protein will be produced. The restriction sites are indicated by the lines and the EcoRV site in the parentheses was used to clone the MmuNeil3 gene into the pET30a-ORF6 plasmid, yet it no longer exists after ligation. The antibiotic resistance markers are shown in the parentheses at the end of each construct.
Cloning of the MmuNeil3 and NEIL3 genes into the expression vectors
The full-length MmuNeil3 gene was amplified from the cDNA clone by PCR using two primers: Neil3Fwd-5(5′ggttgaaggtccaggctgtactctg3′) and Neil3Rev (5′ccgctcgaggca accaggaacaatttccataccaggtcc3′). The PCR product was digested with the restriction enzyme XhoI and ligated into EcoRV-XhoI-cut pET30a-ORF6 resulting in the pET30a-ORF6-MmuNeil3 plasmid. To construct the pETDuet2-T7EcoMap-ORF6- MmuNeil3 plasmid, the NdeI-XhoI-cut fragment from pET30a-ORF6-MmuNeil3 plasmid, which contains the ORF6 leader sequence and full-length MmuNeil3 gene, was ligated into NdeI-XhoI-cut pETDuet2-T7EcoMap-ORF6 plasmid. We then used the pET30a-ORF6-MmuNeil3 plasmid as a template to amplify MmuNeil3Δ324 using two primers: ORF6Fwd (5′ggaattccatatgaaaatcgaagcaggtaaactgg3′) and N3C324Rev (5′ccgctcgagtttctgacagtgaggacag3′). The PCR product was digested with NdeI and XhoI and ligated into NdeI-XhoI-cut linear pET30a-ORF6 and pETDuet2-T7EcoMap-ORF6 plasmids, resulting in pET30a-ORF6-MmuNeil3Δ324 and pETDuet2-T7EcoMap -ORF6-MmuNeil3Δ324 expression plasmids, respectively. The MmuNeil3Δ259 and MmuNeil3Δ210 expression plasmids were constructed in a similar way using the primer sets: ORF6Fwd with N3C259Rev (5′ccgctcgagcagaggccttgtggtcaag3′), and ORF6Fwd with N3C210Rev (5′ ccgctcgagtaaatcagtcaaaacaagggtcg3′), respectively. NEIL3Δ324 was amplified from the cDNA clone using Neil3Fwd-5 and N3C324Rev primers and then cloned into the EcoRV-XhoI-cut pET30a-ORF6 vector to generate the pET30a-ORF6-NEIL3Δ324 plasmid. To construct the pETDuet2-T7EcoMap-ORF6- NEIL3Δ324 plasmid, the NdeI-XhoI fragment from the pET30a-ORF6-NEIL3Δ324 plasmid was amplified by PCR using the primers ORF6Fwd and N3C324Rev. PCR product was digested with NdeI and XhoI and ligated into NdeI-XhoI-cut linear pETDuet2-T7EcoMap-ORF6 plasmid, resulting in the pETDuet2-T7EcoMap-ORF6-NEIL3Δ324 expression plasmid. The sequences of all the expression plasmids constructed above were verified by DNA sequencing (Vermont Cancer Center DNA core facility, Burlington, VT) and confirmed using Sequencher software (Gene Codes Corporation, Ann Arbor, MI).
Expression and purification of full-length MmuNeil3
The two MmuNeil3 expression plasmids described above were individually co-transformed with a pRARE2 plasmid (Novagen) into competent BL21-Star (DE3) cells (Stratagene). The expression of MmuNeil3 was induced with 0.2 mM isopropyl β-D-thiogalactoside (IPTG) in Luria Broth (LB) medium supplemented with antibiotics (34 μg/ml chloramphenicol with either 30 μg/ml kanamycin or 50 μg/ml ampicillin) and 100 μM ZnSO4 at an OD600 of 0.5. The cultures were allowed to grow at 16°C for 20 hours before harvesting. To purify MmuNeil3, cell pellets were re-suspended in chelating column Buffer A (20 mM Tris-Cl pH 8.0, 300 mM NaCl, 10% glycerol, 5 mM β-mercaptoethanol, 0.01% NP-40 and 20 mM imidazole) supplemented with 1 mM phenylmethanesulfonylfluoride and 10 mM benzamidine. The clear lysates were harvested after sonication and centrifugation then loaded onto a 5 ml HiTrap chelating HP column (GE Healthcare, Piscataway, NJ) pre-charged with 100 mM ZnSO4, using an ÄKTA FPLC system (GE Healthcare). Proteins were then eluted with a linear gradient of 0–100% chelating column Buffer B (20 mM Tris-Cl pH 8.0, 300 mM NaCl, 10% glycerol, 5 mM β-mercaptoethanol, and 500 mM imidazole) in 20 column volumes. Fractions containing MmuNeil3 were identified by SDS-PAGE and by glycosylase/lyase activity assays. The protein was concentrated using an Amicon Ultra-4 centrifugal filter unit with an Ultracel-10 membrane (Millipore, Billerica, MA), and eluted on a gel filtration column (Superdex 200 10/30; GE Healthcare) in the buffer (40 mM Hepes-NaOH pH 7.0, 300 mM NaCl, 10% glycerol, 5 mM β-mercaptoethanol). Purified MmuNeil3 protein was dialyzed and stored in MmuNeil3 storage buffer (40 mM Hepes-NaOH pH 7.0, 300 mM NaCl, 50% glycerol, 10 mM tris(2-carboxyethyl)phosphine pH 7.0) at −20°C. Protein concentration was determined using the Bradford protein assay (Bio-Rad).
Expression and purification of MmuNeil3Δ324 and NEIL3Δ324
MmuNeil3Δ324 was expressed by auto-induction using the Studier method [36]. Briefly, the cultures were allowed to auto-induce in 2 L Tartoff-Hobbs Broth (Terrific Broth, Fisher Scientific, Pittsburgh, PA) supplemented with 5052 solution (0.5% glycerol, 0.05% glucose, 0.2% α-lactose), 100 mM ZnSO4, and antibiotics (34 μg/ml chloramphenicol with either 30 μg/ml kanamycin or 50 μg/ml ampicillin) at 20°C for about 72 hours. NEIL3Δ324 was expressed using the MmuNeil3 expression method described above. Both proteins were first purified with a 5 ml HiTrap chelating HP column pre-charged with 100 mM NiSO4 in chelating column buffer A and B described above. After dialyzing into buffer (40 mM Hepes-NaOH pH 7.0, 300 mM NaCl, 10% glycerol, 5 mM β-mercaptoethanol) and diluting 2-fold with buffer (40 mM Hepes-NaOH pH 7.0, 10% glycerol, 5 mM β-mercaptoethanol), both proteins were further purified by a 5 ml HiTrap Heparin column (Buffer A: 40 mM Hepes-NaOH pH 7.0, 150 mM NaCl, 10% glycerol, 5 mM β-mercaptoethanol; Buffer B: 40 mM Hepes-NaOH pH 7.0, 1 M NaCl, 10% glycerol, 5 mM β-mercaptoethanol). For enzyme activity assays, the purified proteins were stored in MmuNeil3Δ324 storage buffer (40 mM Hepes-NaOH pH 7.0, 150 mM NaCl, 50% glycerol, 1 mM dithiothreitol) at −20°C. For crystallization purposes, MmuNeil3Δ324 was further purified by size exclusion chromatography, concentrated and stored at −80°C as small aliquots in crystallization buffer (20 mM Hepes-NaOH pH 7.0, 100 mM NaCl, 10% glycerol, 10 mM tris(2-carboxyethyl)phosphine pH 7.0).
Protein N-terminal sequencing
Protein N-terminal sequencing using the Edman degradation method was performed by the Biomolecular Resource Facility, The University of Texas Medical Branch, Galveston, TX. Samples were prepared by resolving the purified MmuNeil3 and MmuNeil3 Δ324 proteins on a 12% SDS-PAGE gel and then transferring onto a PVDF membrane.
DNA substrates
The 35mer oligodeoxynucleotides 5′ TGTCAATAGCAAGXGGAGAAGTCAATCGT GAGTCT 3′, where X = thymine glycol (Tg), 5,6-dihydrothymine (DHT) or uracil (U), and their complementary strands were chemically synthesized by Midland Certified Reagent Company, Inc. (Midland, TX). A 30-mer oligodeoxynucleotide 5′ TGTTCATCATGCGTCSpTCGGTATATCCCAT 3′ carrying spiroiminodihydantoin (Sp) was kindly provided by Dr. Cynthia Burrows, University of Utah, and was synthesized as previously described [37]. 1 pmol of each damage-containing strand was 5′ end labeled with [γ-32P] ATP by T4 polynucleotide kinase (New England Biolabs) at 37°C for 15 min. The reaction was terminated by adding 25 mM EDTA followed by heat inactivation. The labeled oligodeoxynucleotide was then ethanol precipitated and diluted with 9 pmol of the unlabeled lesion-containing oligodeoxynucleotide in the substrate buffer (20 mM Tris-Cl pH 8.0, 50 mM NaCl and 1 mM EDTA) and used as single-stranded substrate. To create the duplex substrate, the lesion-containing strand was annealed to its complementary strand in a 1:1.2 ratio by heating to 95°C then slowly cooled down to room temperature. To create the substrate with an apurinic or apyrimidinic (AP) site, duplex or single-stranded oligodeoxynucleotides containing U were treated with E. coli uracil DNA glycosylase (New England Biolabs) at room temperature for 15 min.
Glycosylase/lyase activity assays
For the buffer optimization process, we first screened the glycosylase activity of both MmuNeil3 and MmuNeil3Δ324 on a single-stranded DHT substrate using the following buffer and pH conditions: 20 mM Hepes-NaOH pH 7.0 or 7.5, or 20 mM Tris-Cl pH 8.0 in addition to 100 mM NaCl, 10 μg/ml BSA, 1 mM dithiothreitol and 5% glycerol. To screen for salt concentration, 50 mM, 150 mM, and 300 mM NaCl were used in addition to buffer containing 20 mM Hepes-NaOH pH 7.0, 10 μg/ml BSA, 1 mM dithiothreitol and 5% glycerol. The optimum buffer condition, 20 mM Hepes-NaOH pH 7.0, 50 mM NaCl, 10 μg/ml BSA, 1 mM dithiothreitol and 5% glycerol, was used to compare the glycosylase/lyase activity of MmuNeil3Δ324 to Neil3Δ324. Reaction buffers for control enzymes, EcoNth and EcoNei, were described previously [11]. In each reaction, 25 nM active enzyme was incubated with 25 nM substrate at 37°C for 30 min. To measure glycosylase activity, reactions were terminated by adding 1 μl of 1 N NaOH, heated at 94°C for 2 min, followed by adding 10 μl formamide stop buffer (98% formamide, 10 mM EDTA, 0.1% bromophenol blue and 0.1% xylene cyanol). Reactions with substrates containing an AP site were terminated by adding 10 μl formamide stop buffer. The reaction products were separated on a 12% (w/v) polyacrylamide sequencing gel and quantitated with an isotope imaging system (Molecular Imaging System, Bio-Rad).
Determining the fraction of active enzyme in the preparations
The Schiff base assay was used to determine the fraction of active enzyme in each preparation as previously described [38]. Briefly, an increasing amount of each enzyme preparation was incubated with 10 μM of γ-32P labeled single-stranded substrate containing an AP site in the presence of 50 mM NaCNBH3. The reaction buffer contained 20 mM Hepes-NaOH pH 7.0, 50 mM NaCl, 10 μg/ml BSA, 1 mM dithiothreitol and 5% glycerol. Reactions were incubated at 37°C for 30 min, stopped by adding 10 μl SDS loading buffer (62.5 mM Tris-HCl pH 6.8, 2% sodium dodecyl sulfate, 10% glycerol, 0.01% bromophenol blue and 50 mM β-mercaptoethanol), heated at 95°C for 10 min and then resolved on a 12% SDS-PAGE gel. The gel bands were detected by using a phosphoimage screen (Bio-Rad, Hercules, CA) and quantified with Quantity-one software (Bio-Rad). We plotted the quantity of trapped substrate formed in the Schiff base assay versus the total enzyme concentration used, and the slope of the linear region before the plateau is taken as the fraction of active enzyme in the preparation.
Dynamic light scattering (DLS)
DLS experiments were performed on a DynaPro MS-X instrument (Protein Solutions. Inc.) using 0.4 mg/ml MmuNeil3Δ324 in the glycosylase/lyase reaction buffers screened for buffer optimization as described above. The scan was performed at 5°C, 10°C, 15°C, 20°C and 25°C with three repeats at each temperature.
Results
Expression and purification of full-length MmuNeil3
It has been shown that the secondary structure of mRNA surrounding the translation initiation region can inhibit efficient translation by reducing the accessibility of the ribosome binding site and/or the translation initiation codon [16]. In order to overcome translational inhibition of mRNAs with stable secondary structures, a bicistronic expression system developed in our lab [34] was used to achieve high-level expression of the mouse and human Neil3 genes in E. coli. First, an expression vector based on the pET30a plasmid in which a leader sequence, ORF6, preceded the cloned full-length MmuNeil3 gene was constructed (Fig. 1a). The ORF6 leader sequence prevents stable secondary structure formation, and it contains a ribosome binding site to facilitate efficient translation initiation of the downstream MmuNeil3 gene. Next, a C-terminal hexa-histidine tag was fused to MmuNeil3 to allow protein purification by affinity chromatography. Codon usage analysis of the MmuNeil3 gene had identified a significant number of rare codons for the arginine (AGG and AGA), proline (CCC), leucine (CUA) and isoleucine (AUA) codons. To avoid the potential inhibitory effect of rare codons during translation, a pRARE2 plasmid carrying seven rare-codon tRNA genes was co-transformed with the MmuNeil3 expression plasmid. In addition, because MmuNeil3 protein contains four zinc finger motifs, the culture medium was supplemented with 100 μM ZnSO4 to allow adequate amounts of Zn ion to be incorporated into the expressed MmuNeil3 protein for proper folding. Using these strategies, a moderate level of MmuNeil3 protein expression was achieved using E. coli BL21-Star (DE3) cells (compare lanes 1 and 2 of Fig. 2a). The induction conditions were optimized, and the highest level of soluble MmuNeil3 protein expression was obtained with 0.2 mM IPTG induction at 16°C for 20 hours (Fig. 2a, lane 2). Soluble MmuNeil3 protein was not obtained either when over-expressed during induction with a higher concentration of IPTG or by using an auto-induction method (data not shown).
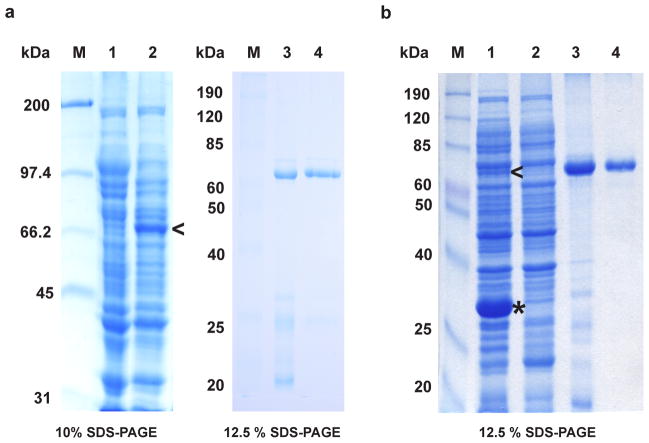
Figure 2.
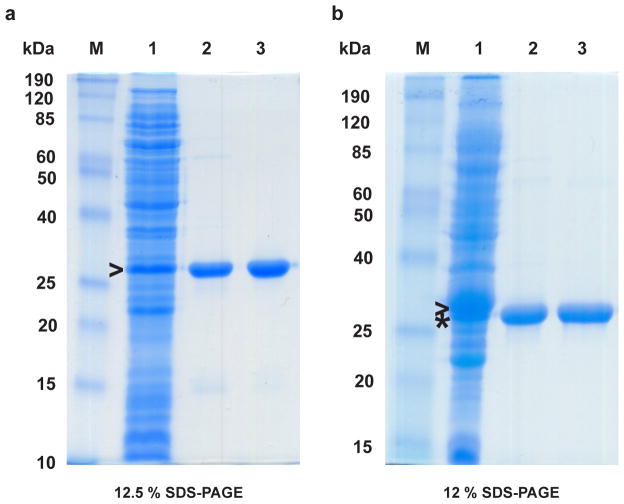
Expression and purification of full-length MmuNeil3. (a) MmuNeil3 expressed and purified using the pET30a-ORF6-MmuNeil3 plasmid. Lane 1, crude cell extract before IPTG induction. Lane 2, soluble lysate induced with IPTG. Lane 3, elute from the HiTrap Chelating column charged with ZnSO4. Lane 4, elute from the gel filtration column. (b) MmuNeil3 expressed and purified using the pETDuet2 -T7EcoMap-ORF6-MmuNeil3 plasmid. Lane 1, soluble lysate induced with IPTG. Lane 2, crude cell extract before IPTG induction. Lane 3, elute from HiTrap Chelating column charged with ZnSO4. Lane 4, elute from gel filtration column. M, molecular-weight standards. The expected MW of MmuNeil3 and EcoMapY168A are 68 kDa, and 30 kDa, respectively. Arrow head indicates the band of MmuNeil3 expressed, and the asterisk indicates the band of EcoMapY168A expressed.
To facilitate N-terminal methionine processing, a second bicistronic vector was designed to co-express EcoMapY168A with MmuNeil3. The resulting plasmid, pETDuet2- T7EcoMap-ORF6-MmuNeil3, is similar to the pET30a-ORF6-MmuNeil3 vector described above in that the MmuNeil3 gene is preceded by an ORF6 leader sequence and fused with a C-terminal his-tag (Fig. 1b). However, the new plasmid contains an additional EcoMapY168A expression unit driven by a separate T7 promoter/lac operator. Thus, the expression of both the EcoMapY168A and MmuNeil3 genes was achieved using a single expression plasmid under the same culturing and induction conditions described above (Fig. 2b, lane 1).
MmuNeil3 protein was then purified using both pET30a-ORF6-MmuNeil3 and pETDuet2-T7EcoMap-ORF6-MmuNeil3 expression plasmids. His-tagged MmuNeil3 was readily purified by affinity chromatography using a HiTrap Chelating column charged with ZnSO4. Results show that this first step eliminates most of the E. coli proteins (Fig. 2a, lane 3 and Fig. 2b, lane 3). However, further purification of MmuNeil3 using a HiTrap Heparin column, a HiTrap SP column or a HiTrap Phenyl column did not significantly reduce containments (data not shown). Protein precipitates were found before and after these columns, indicating that MmuNeil3 is unstable in the buffers used. Moreover, no MmuNeil3 glycosylase or lyase activity was detected when the elutes from these columns were assayed using single stranded substrates containing Tg or an AP site. Based on Western blot analysis of the MmuNeil3 fractions eluted from the chelating column, degradation products were detected by reaction with antibody specific for MmuNeil3 (data not shown). These degradation products were not separated from MmuNeil3 using the heparin column, the SP column or the phenyl column. Therefore, size exclusion chromatography was used to purify MmuNeil3 after the chelating column. MmuNeil3 eluted at the expected mass position (about 68 kDa). After the gel filtration step, most of the contaminants or degradation products were removed, and MmuNeil3 protein was purified to near homogeneity (Fig. 2a, lane 4 and Fig. 2b, lane 4). The identity of the purified protein was further confirmed by 12 cycles of N-terminal protein sequencing. Typically, 2–3 mg of MmuNeil3 was purified from a liter of induced bacterial culture using pET30a-ORF6-MmuNeil3 expression plasmid, and 1–2 mg of MmuNeil3 per liter of culture was obtained using pETDuet2-T7EcoMap-ORF6-MmuNeil3 plasmid.
Expression and purification of the glycosylase domain of MmuNeil3
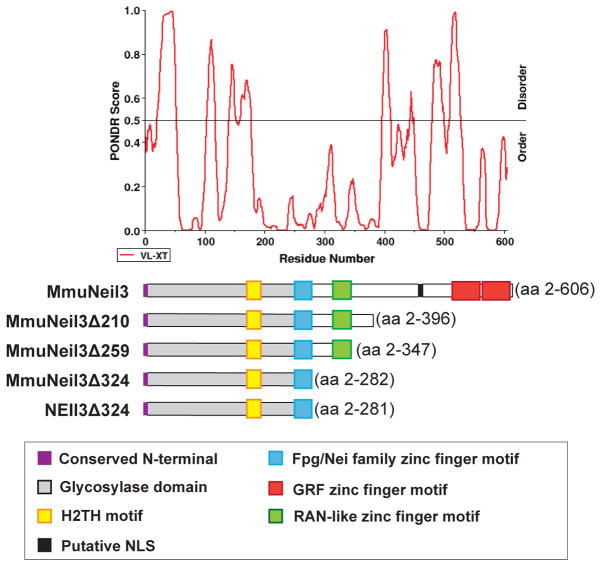
Neil3 is the largest DNA glycosylase among the Fpg/Nei family of DNA glycosylases. A sequence alignment of MmuNeil3 with other Fpg/Nei family members reveals that the N-terminal 282 amino acids of MmuNeil3 possess the complete Fpg/Nei-like core protein, including the signature helix-2-turns-helix motif and the zinc finger DNA-binding motif (Fig. 3). In addition to the glycosylase domain, Neil3 proteins also have a RAN binding protein (RANbp)-like zinc finger motif and a duplicated GRF-zinc finger motif at their extended C-terminus [6–8, 10, 11, 39]. All mammalian DNA glycosylases, including Neil3, contain extra domains or motifs in addition to their catalytic glycosylase domain, which mediate protein-protein interactions, allow post-translational modifications and localize the glycosylases to the nucleus or mitochondria in the cell [40–45]. Strong evidence shows that removing those domains or motifs gives rise to more stable glycosylases with the same or even improved activities [46, 47]. Therefore, several truncation variants of MmuNeil3 were made based on sequence alignment and Predictor of Naturally Disordered Regions (PONDR) [48] results (Fig. 3). The first truncation variant, MmuNeil3Δ324, spans the entire glycosylase domain and lacks 324 amino acid residues at the C-terminus of MmuNeil3. The second variant, MmuNeil3Δ259, contains the RANbp-like zinc finger motif in addition to the glycosylase domain. The third variant, MmuNeil3Δ210, lacks the C-terminal disordered region of MmuNeil3 predicted by PONDR. Expression and purification trials were performed to produce all three variants. However, only MmuNeil3Δ324 was able to be purified in large quantities as soluble protein. Most of the expressed MmuNeil3Δ210 protein was highly insoluble, and adding detergents and high concentrations of salt to the lysis buffer failed to solubilize the aggregated protein. On the other hand, a small amount of MmuNeil3Δ259 could be purified to homogeneity, but no glycosylase or lyase activity was detected (data not shown).
Figure 3.
PONDR analysis of MmuNeil3 aligned with full-length MmuNeil3 and the truncation variants of MmuNeil3 and NEIL3. Domains and motifs in the scheme are described in the box below the Figure.
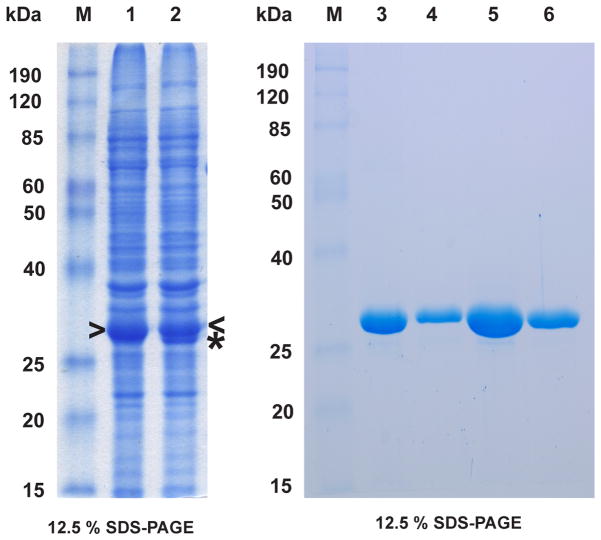
We then optimized the expression and purification conditions of MmuNeil3Δ324 transformed with both the pET30a-ORF6-MmuNeil3Δ324 and pETDuet2-T7EcoMap- ORF6-MmuNeil3Δ324 plasmids. High levels of MmuNeil3Δ324 expression were achieved by 0.5 mM IPTG induction at 16°C for 18 hours (Fig. 4, lanes 1 and 2). In contrast to full-length MmuNeil3, which aggregates when over-expressed, MmuNeil3Δ324 was soluble when over-expressed. The expression level of MmuNeil3Δ324 was further enhanced by using an auto-induction method, which allows the cells to grow to high density at which point they are automatically induced by the lactose present in the medium [36]. Typically, from 1 L of induced culture transformed with either expression plasmid, about 30–50 mg MmuNeil3Δ324 were obtained after a two-step purification using a Ni column (Fig. 4, lanes 3 and 5) followed by a heparin column (Fig. 4, lanes 4 and 6).
Figure 4.
Expression and purification of MmuNeil3Δ324. Lanes 1, 3 and 4 show expression and purification of MmuNeil3Δ324 using the pET30a-ORF6-MmuNeil3Δ324 plasmid. Lanes 2, 5 and 6 show expression and purification of MmuNeil3Δ324 using the pETDuet2-T7EcoMap-ORF6-MmuNeil3Δ324 plasmid. Lanes 1 and 2, soluble lysate induced with IPTG. Lanes 3 and 5, elute from Ni column. Lanes 4 and 6, elute from heparin column. Arrow head indicates the band of MmuNeil3Δ324 expressed, and the asterisk indicates the band of EcoMapY168A expressed. The expected MW of MmuNeil3Δ324 is 32 kDa.
Buffer conditions for optimal activity and stability of MmuNeil3 proteins
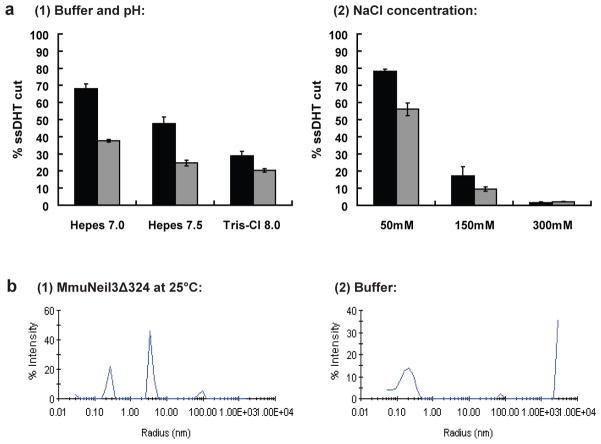
Buffer conditions are likely to be critical for the activity and stability of DNA glycosylases [35]. To optimize the buffer conditions, DNA glycosylase/lyase activity of both MmuNeil3 and MmuNeil3Δ324 was measured at varying pH and salt concentrations. MmuNeil3 proteins showed optimal glycosylase/lyase activity on a ssDHT substrate at pH 7.0 in the presence of 50 mM salt (Fig. 5a). The results of the DLS experiments also confirmed the stability of MmuNeil3Δ324 under the optimal buffer conditions (Fig. 5b). However, the full-length MmuNeil3 proved to be unstable under the low salt conditions because protein precipitates were found 24 hours after storing MmuNeil3 in buffer containing 100 mM NaCl at 4°C. To help stabilize MmuNeil3 in solution, a higher concentration of salt (300 mM) was used in the storage buffer. In addition, to prevent oxidation of the MmuNeil3 proteins, a reducing agent (1–2 mM dithiothreitol or 10 mM tris(2-carboxyethyl)phosphine) was added to storage and reaction buffers.
Figure 5.
Optimization of buffer conditions for the glycosylase/lyase reaction and stability of the MmuNeil3 proteins. (a) Optimization of buffer conditions: 25 nM active MmuNeil3Δ324 (black) and MmuNeil3 (grey) were incubated in 10 μl reactions with 25 nM 32P-labeled single-stranded substrate containing DHT at 37°C for 30 min. Reactions were stopped by formamide stop buffer to measure glycosylase plus lyase activities. Data are expressed as the mean of three independent measurements. Uncertainties are standard deviations. (1) Reaction buffers containing 20 mM Hepes-NaOH pH 7.0 or 7.5, or 20 mM Tris-Cl pH 8.0 in addition to 100 mM NaCl, 10 μg/ml BSA, 1 mM DTT and 5% glycerol. (2) Reaction buffers containing 50 mM, 150 mM, and 300 mM NaCl in addition to 20 mM Hepes-NaOH pH 7.0, 10 μg/ml BSA, 1 mM DTT and 5% glycerol. (b) DLS scanning: (1) MmuNeil3Δ324 at 25°C in buffer: 20 mM Hepes-NaOH pH 7.0, 50 mM NaCl, 1 mM DTT and 10% glycerol. (2) DLS scanning of buffer at 25°C.
NEIL3Δ324 expressed using the bicistronic vector exhibits similar activity to MmuNeil3Δ324
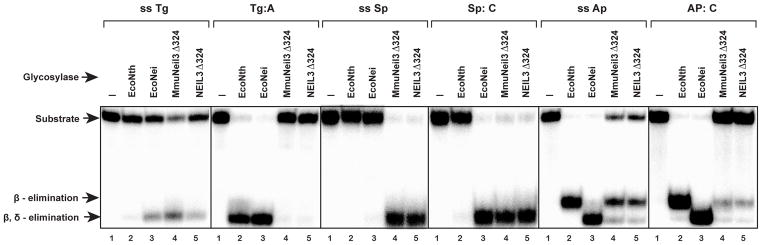
Because the BLAST search results showed that NEIL3 shares substantial sequence identity (74%) with MmuNeil3, NEIL3 was predicted to exhibit similar glycosylase/lyase activity as MmuNeil3. The glycosylase domain of NEIL3 (NEIL3Δ324) was therefore cloned, expressed and purified, and its glycosylase/lyase activity was tested using both single-stranded and double-stranded substrates containing Tg, Sp or an AP site. Fig. 6a and b shows the results of the expression (lane 1) and purification (lane 2, using a Ni column; lane 3, followed by a Heparin column) of NEIL3Δ324. About 10 mg of NEIL3Δ324 protein were obtained from 1 liter of induced culture using both pET30a-ORF6-NEIL3Δ324 (Fig. 6a) and pETDuet2-T7EcoMap-ORF6-NEIL3Δ324 (Fig. 6b) expression plasmids. The glycosylase/lyase activity assay confirmed that NEIL3Δ324 and MmuNeil3Δ324 exhibit similar activity on the substrates tested (Fig. 7).
Figure 6.
Expression and purification of NEIL3Δ324 using (a) pET30a-ORF6- NEIL3Δ324 plasmid and (b) pETDuet2-T7EcoMap-ORF6-NEIL3Δ324 plasmid. Lane 1, soluble lysate induced with IPTG. Lane 2, elute from Ni column. Lane 3, elute from heparin column. Arrow head indicates the band of NEIL3Δ324 expressed, and the asterisk indicates the band of EcoMapY168A expressed. The expected MW of NEIL3Δ324 is 32 kDa.
Figure 7.
DNA glycosylase/lyase activity of MmuNeil3Δ324 and NEIL3Δ324. Single- and double-stranded substrates containing Tg, Sp, or an AP site (25 nM) were incubated with 25 nM active MmuNeil3Δ324 (lane 4) and NEIL3Δ324 (lane 5), and control enzymes EcoNei (lane 3) and EcoNth (lane 2) at 37°C for 30 min. Lane 1, control with no enzyme.
Co-expression of EcoMapY168A improves N-terminal methionine processing and increases the percentage of active Neil3 proteins in the preparation
The ability to co-express EcoMapY168A with the full-length MmuNeil3, MmuNeil3Δ324 and NEIL3Δ324 proteins in order to improve N-terminal methionine processing was assessed first. The extent of N-terminal methionine excision was estimated by protein N-terminal sequencing using the Edman degradation method. The results showed that two sequences are present in the preparation of MmuNeil3 expressed using the pET30a-ORF6-MmuNeil3 plasmid: MVEGP and VEGPG. The relative amounts of methionine and valine in the first sequencing cycle were determined to be 2.95 pmol and 1.16 pmol, respectively, by the height of the peaks shown in the chromatograph. Thus about 28.2% of the N-terminal methionine was processed in this preparation (Table 1). However, with the expression of exogenous EcoMapY168A protein, the extent of N-terminal methionine processing increased to 54.2%. Similarly, when overexpressing EcoMapY168A, the N-terminal methionine processing of MmuNeil3Δ324 in the preparation increased from 14.6% to 57.9% (Table 1), and the N-terminal methionine processing of NEIL3Δ324 increased from 34.9% to 79.6% (Table 1). Although the extent of N-terminal methionine excision varied with different preparations, co-expression of EcoMapY168A with Neil3 proteins significantly enhanced the percentage of N-terminal methionine processing (p< 0.05, Table 1).
Table 1.
N-terminal methionine processing and activity of mouse and human Neil3 proteins.
| Protein | Expression plasmid | EcoMap | % Met processed | % active protein |
|---|---|---|---|---|
| MmuNeil3 | pET30a-ORF6-MmuNeil3 | − | 28.2% | 5% |
| MmuNeil3 | pETDuet2-T7EcoMap-ORF6-MmuNeil3 | + | 54.2% | 15% |
| MmuNeil3Δ324 | pET30a-ORF6-MmuNeil3Δ324 | − | 14.6% | 15% |
| MmuNeil3Δ324 | pETDuet2-T7EcoMap-ORF6-MmuNeil3Δ324 | + | 57.9% | 66% |
| NEIL3Δ324 | pET30a-ORF6-NEIL3 Δ324 | − | 34.9% | 17% |
| NEIL3Δ324 | pETDuet2-T7EcoMap-ORF6-NEIL3 Δ324 | + | 79.6% | 44% |
A paired student’s t-test was conducted to compare both the % Met processed and the % active protein of purified Neil3 proteins with and without co-expression of EcoMapY168A. There was a significant increase (p<0.05) in the % Met processed with EcoMapY168A co-expression (M=63.9%, SD=13.7%) compared to expression of the protein alone (M=25.9%, SD=10.3%; t(3)=6.32, p=0.012). Using the same methodology, there was a less significant increase (0.05<p<0.1) in the % active protein with EcoMapY168A co-expression (M=41.7%, SD=25.6%) compared to the expression of the protein alone (M=12.3%, SD=6.4%; t(3)=2.47, p=0.066).
A Schiff base assay was then used to determine the percentage of active Neil3 proteins in the preparations. Results showed that increasing the N-terminal methionine processing resulted in more active MmuNeil3, MmuNeil3Δ324 and NEIL3Δ324 protein products. When overexpressing EcoMapY168A, the percentage of active MmuNeil3 increased from 5% to 15% (Table 1), the percentage of active MmuNeil3Δ324 increased from 15% to 66% (Table 1), and the percentage of active NEIL3Δ324 increased from 17% to 44% (Table 1). On average, co-expression of EcoMapY168A yielded a more than three-fold increase in the percentage of active Neil3 proteins, although a paired student’s t-test showed the results to be less than significant (p=0.066, Table 1). Thus, co-expression of EcoMapY168A improves N-terminal methionine processing and enhances the percentage of active Neil3 proteins in the preparation.
Discussion
Expression and purification of both mouse and human orthologs of Neil3 have been reported previously. However, those trials failed to produce purified Neil3 proteins with both glycosylase and lyase activity [6–10]. In this study, we describe a method for expressing and purifying active mouse and human Neil3 proteins with high purity and in large quantities. We also show that N-terminal methionine processing is critical for producing active Neil3 proteins, which can be facilitated by co-expression of exogenous EcoMapY168A with the Neil3 proteins. In our experiments, co-expression of EcoMapY168A with the full-length MmuNeil3 enhances the percentage of N-terminal methionine processing about two-fold, resulting in about a three-fold increase in the active fraction of MmuNeil3 (Table 1). Similarly, co-expression of EcoMapY168A with MmuNeil3Δ324 and NEIL3Δ324 enhances the percentage of N-terminal methionine processing about four-fold and two-fold, respectively, with a greater than four-fold and two-fold increase in the fraction of active MmuNeil3Δ324 and NEIL3Δ324 proteins (Table 1). This is consistent with the previous observation by Takao and co-workers that truncation of two amino acid residues (V2 and E3) at the N-terminus of a NEIL3 variant (amino acids 1–290) completely abolished its lyase activity [6], and is in keeping with the fact that the important catalytic residues of the Fpg/Nei family members are the N-terminal proline, or valine in the case of Neil3, and the adjacent glutamic acid. Taken together our findings suggest that the lack of N-terminal methionine processing could explain why in vitro transcription/translation failed to produce both full-length MmuNeil3 and NEIL3 proteins in their active form [7, 9].
It is worth noting that even with the expression of the EcoMapY168A protein and with more than 50% of the N-terminal methionine being processed, the majority of the full-length MmuNeil3 proteins remain inactive (85% inactive, Table 1). This could result if some misfolded MmuNeil3 protein co-purified with the active MmuNeil3 protein. The long C-terminal tail of the full-length Neil3 is predicted to be highly disordered and would have difficulty folding correctly in E. coli. Moreover, Neil3 proteins are rich in cysteine residues (about 5% in both NEIL3 and MmuNeil3) which could further compromise correct folding. It is also possible that because the full-length MmuNeil3 protein is unstable, some of the MmuNeil3 proteins lose activity under the conditions used for the activity assay
The truncation variants of NEIL3, NEIL3GD (residues 1- 290) constructed by Takao and coworkers [6] and core-NEIL3 (residues 1–301) constructed by Krokeide and coworkers [7], showed substantially reduced activity compared to NEIL3Δ324 (residues 2- 281) constructed in this report. This could be due to the differences in N-terminal methionine processing, the substrates (for example, different lesions, different lengths and different sequence contexts), and the buffer conditions used. We also observed that the truncation variants of MmuNeil3 containing the glycosylase domain as well as some part of the C-terminal domain (MmuNeil3Δ259 and MmuNeil3Δ210) show reduced stability and lose glycosylase/lyase activity compared to the variant containing only the glycosylase domain (MmuNeil3Δ324), suggesting that the C-terminal extension may affect the activity of these MmuNeil3 truncation variants.
Co-expression of EcoMapY168A with MmuNeil3 under separate T7/lac promoters yielded substantially higher levels of EcoMapY168A than MmuNeil3 (Fig. 2b, lane 1). In contrast, higher levels of MmuNeil3Δ324 (Fig. 4, lane 2) and NEIL3Δ324 (Fig. 6b, lane 1) compared to EcoMapY168A were observed when EcoMapY168A was co-expressed with these proteins. It has previously been shown that E. coli cells can preferentially express one protein over the other when two proteins are co-expressed under two identical promoters [15, 49, 50]. For example, co-expression of human hemoglobin with an EcoMap protein under separate tac promoters yielded higher levels of EcoMap than the hemoglobin subunits [15, 49]. Similarly, predominant expression of EcoMap protein was observed when the EcoMap protein was co-expressed with a GSTM1 gene under the control of separate phoA promoters [50]. The reason for the preference is unknown, although it seems to depend on the solubility and stability of the proteins been expressed.
Conclusions
Co-expression of the EcoMapY168A protein with the full-length MmuNeil3, MmuNeil3Δ324 and NEIL3Δ324 proteins yielded large quantities of active MmuNeil3, MmuNeil3Δ324 and NEIL3Δ324 proteins enriched in the N-terminal methionine processed form. The purified Neil3 proteins are all active, and NEIL3Δ324 exhibits similar glycosylase/lyase activity as MmuNeil3Δ324 on both single-stranded and double-stranded substrates containing thymine glycol (Tg), spiroiminodihydantoin (Sp) or an abasic site (AP). Our method for expressing and purifying the MmuNeil3 proteins has led to the successful characterization of the mouse Neil3 protein [11], and the purified Neil3 proteins are now available for further biochemical, structural and functional studies.
Highlights.
We expressed and purified active mouse and human Neil3 proteins with high purity.
Co-expression of EcoMapY168A improves N-terminal methionine processing.
Co-expression of EcoMapY168A increases the percentage of active Neil3 proteins.
NEIL3Δ324 exhibits similar glycosylase/lyase activity to MmuNeil3Δ324.
This protocol is valuable for future structural and functional studies of Neil3.
Acknowledgments
We would like to thank Dr. Cynthia J. Burrows for providing oligonucleotides containing spiroiminodihydantoin, Dr. Ming F. Tam for providing the pBS-Map(Y168A) -cGSTM1 plasmid, Dr. Stéphanie Duclos for help with the protein purification, and Dr. Scott Kathe and Heather Galick for helpful discussions. In addition, we thank the DNA Analysis Facility at the Vermont Cancer Center for performing DNA sequencing, and the Biomolecular Resource Facility at the University of Texas Medical Branch for performing protein N-terminal sequencing. This research was supported by NIH grant PO1 CA098993 (to SSW) awarded by the National Cancer Institute.
Abbreviations used
- BER
base excision repair
- EcoMap
Escherichia coli methionine aminopeptidase
- Neil3
endonuclease VIII-Like 3
- Nei
endonuclease VIII
- Nth
endonuclease III
- Sp
spiroiminodihydantoin
- Tg
thymine glycol
- DHT
5,6-dihydrothymine
- U
uracil
- AP
apurinic or apyrimidinic site
- ORF
open reading frame
- EDTA
ethylenediaminetetraacetic acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wallace SS. Biological consequences of free radical-damaged DNA bases. Free Radic Biol Med. 2002;33:1–14. doi: 10.1016/s0891-5849(02)00827-4. [DOI] [PubMed] [Google Scholar]
- 2.Fromme JC, Verdine GL. Base excision repair. Adv Protein Chem. 2004;69:1–41. doi: 10.1016/S0065-3233(04)69001-2. [DOI] [PubMed] [Google Scholar]
- 3.Grin IR, Zharkov DO. Eukaryotic endonuclease VIII-like proteins: new components of the base excision DNA repair system. Biochemistry (Mosc) 2011;76:80–93. doi: 10.1134/s000629791101010x. [DOI] [PubMed] [Google Scholar]
- 4.Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008;18:27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.David SS, O’Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takao M, Oohata Y, Kitadokoro K, Kobayashi K, Iwai S, Yasui A, Yonei S, Zhang QM. Human Nei-like protein NEIL3 has AP lyase activity specific for single-stranded DNA and confers oxidative stress resistance in Escherichia coli mutant. Genes Cells. 2009;14:261–270. doi: 10.1111/j.1365-2443.2008.01271.x. [DOI] [PubMed] [Google Scholar]
- 7.Krokeide SZ, Bolstad N, Laerdahl JK, Bjoras M, Luna L. Expression and purification of NEIL3, a human DNA glycosylase homolog. Protein Expr Purif. 2009;65:160–164. doi: 10.1016/j.pep.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 8.Torisu K, Tsuchimoto D, Ohnishi Y, Nakabeppu Y. Hematopoietic tissue-specific expression of mouse Neil3 for endonuclease VIII-like protein. J Biochem. 2005;138:763–772. doi: 10.1093/jb/mvi168. [DOI] [PubMed] [Google Scholar]
- 9.Takao M, Kanno S, Shiromoto T, Hasegawa R, Ide H, Ikeda S, Sarker AH, Seki S, Xing JZ, Le XC, Weinfeld M, Kobayashi K, Miyazaki J, Muijtjens M, Hoeijmakers JH, van der Horst G, Yasui A. Novel nuclear and mitochondrial glycosylases revealed by disruption of the mouse Nth1 gene encoding an endonuclease III homolog for repair of thymine glycols. EMBO J. 2002;21:3486–3493. doi: 10.1093/emboj/cdf350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morland I, Rolseth V, Luna L, Rognes T, Bjoras M, Seeberg E. Human DNA glycosylases of the bacterial Fpg/MutM superfamily: an alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA. Nucleic Acids Res. 2002;30:4926–4936. doi: 10.1093/nar/gkf618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu M, Bandaru V, Bond JP, Jaruga P, Zhao X, Christov PP, Burrows CJ, Rizzo CJ, Dizdaroglu M, Wallace SS. The mouse ortholog of NEIL3 is a functional DNA glycosylase in vitro and in vivo. Proc Natl Acad Sci U S A. 2010;107:4925–4930. doi: 10.1073/pnas.0908307107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffman SJ, Looker DL, Roehrich JM, Cozart PE, Durfee SL, Tedesco JL, Stetler GL. Expression of fully functional tetrameric human hemoglobin in Escherichia coli. Proc Natl Acad Sci U S A. 1990;87:8521–8525. doi: 10.1073/pnas.87.21.8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newton DL, Boque L, Wlodawer A, Huang CY, Rybak SM. Single amino acid substitutions at the N-terminus of a recombinant cytotoxic ribonuclease markedly influence biochemical and biological properties. Biochemistry. 1998;37:5173–5183. doi: 10.1021/bi972147h. [DOI] [PubMed] [Google Scholar]
- 14.Proudfoot AE, Power CA, Hoogewerf AJ, Montjovent MO, Borlat F, Offord RE, Wells TN. Extension of recombinant human RANTES by the retention of the initiating methionine produces a potent antagonist. J Biol Chem. 1996;271:2599–2603. doi: 10.1074/jbc.271.5.2599. [DOI] [PubMed] [Google Scholar]
- 15.Shen TJ, Ho NT, Simplaceanu V, Zou M, Green BN, Tam MF, Ho C. Production of unmodified human adult hemoglobin in Escherichia coli. Proc Natl Acad Sci U S A. 1993;90:8108–8112. doi: 10.1073/pnas.90.17.8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makrides SC. Strategies for achieving high-level expression of genes in Escherichia coli. Microbiol Rev. 1996;60:512–538. doi: 10.1128/mr.60.3.512-538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirel PH, Schmitter MJ, Dessen P, Fayat G, Blanquet S. Extent of N-terminal methionine excision from Escherichia coli proteins is governed by the side-chain length of the penultimate amino acid. Proc Natl Acad Sci U S A. 1989;86:8247–8251. doi: 10.1073/pnas.86.21.8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frottin F, Martinez A, Peynot P, Mitra S, Holz RC, Giglione C, Meinnel T. The proteomics of N-terminal methionine cleavage. Mol Cell Proteomics. 2006;5:2336–2349. doi: 10.1074/mcp.M600225-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Dalboge H, Bayne S, Pedersen J. In vivo processing of N-terminal methionine in E. coli. FEBS Lett. 1990;266:1–3. doi: 10.1016/0014-5793(90)90001-b. [DOI] [PubMed] [Google Scholar]
- 20.Wallace SS, Bandaru V, Kathe SD, Bond JP. The enigma of endonuclease VIII. DNA Repair (Amst) 2003;2:441–453. doi: 10.1016/s1568-7864(02)00182-9. [DOI] [PubMed] [Google Scholar]
- 21.Zharkov DO, Shoham G, Grollman AP. Structural characterization of the Fpg family of DNA glycosylases. DNA Repair (Amst) 2003;2:839–862. doi: 10.1016/s1568-7864(03)00084-3. [DOI] [PubMed] [Google Scholar]
- 22.Chetsanga CJ, Lindahl T. Release of 7-methylguanine residues whose imidazole rings have been opened from damaged DNA by a DNA glycosylase from Escherichia coli. Nucleic Acids Res. 1979;6:3673–3684. doi: 10.1093/nar/6.11.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boiteux S, O’Connor TR, Laval J. Formamidopyrimidine-DNA glycosylase of Escherichia coli: cloning and sequencing of the fpg structural gene and overproduction of the protein. EMBO J. 1987;6:3177–3183. doi: 10.1002/j.1460-2075.1987.tb02629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cabrera M, Nghiem Y, Miller JH. mutM, a second mutator locus in Escherichia coli that generates G.C----T.A transversions. J Bacteriol. 1988;170:5405–5407. doi: 10.1128/jb.170.11.5405-5407.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michaels ML, Pham L, Cruz C, Miller JH. MutM, a protein that prevents G.C----T.A transversions, is formamidopyrimidine-DNA glycosylase. Nucleic Acids Res. 1991;19:3629–3632. doi: 10.1093/nar/19.13.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melamede RJ, Hatahet Z, Kow YW, Ide H, Wallace SS. Isolation and characterization of endonuclease VIII from Escherichia coli. Biochemistry. 1994;33:1255–1264. doi: 10.1021/bi00171a028. [DOI] [PubMed] [Google Scholar]
- 27.Jiang D, Hatahet Z, Melamede RJ, Kow YW, Wallace SS. Characterization of Escherichia coli endonuclease VIII. J Biol Chem. 1997;272:32230–32239. doi: 10.1074/jbc.272.51.32230. [DOI] [PubMed] [Google Scholar]
- 28.Jiang D, Hatahet Z, Blaisdell JO, Melamede RJ, Wallace SS. Escherichia coli endonuclease VIII: cloning, sequencing, and overexpression of the nei structural gene and characterization of nei and nei nth mutants. J Bacteriol. 1997;179:3773–3782. doi: 10.1128/jb.179.11.3773-3782.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berti PJ, McCann JA. Toward a detailed understanding of base excision repair enzymes: transition state and mechanistic analyses of N-glycoside hydrolysis and N-glycoside transfer. Chem Rev. 2006;106:506–555. doi: 10.1021/cr040461t. [DOI] [PubMed] [Google Scholar]
- 30.Sandman K, Grayling RA, Reeve JN. Improved N-terminal processing of recombinant proteins synthesized in Escherichia coli. Biotechnology (N Y) 1995;13:504–506. doi: 10.1038/nbt0595-504. [DOI] [PubMed] [Google Scholar]
- 31.Van Valkenburgh HA, Kahn RA. Coexpression of proteins with methionine aminopeptidase and/or N-myristoyltransferase in Escherichia coli to increase acylation and homogeneity of protein preparations. Methods in enzymology. 2002;344:186–193. doi: 10.1016/s0076-6879(02)44715-5. [DOI] [PubMed] [Google Scholar]
- 32.Liu LF, Hsieh CH, Liu PF, Tsai SP, Tam MF. Escherichia coli methionine aminopeptidase with Tyr168 to alanine substitution can improve the N-terminal processing of recombinant proteins with valine at the penultimate position. Anal Biochem. 2004;329:345–347. doi: 10.1016/j.ab.2004.03.031. [DOI] [PubMed] [Google Scholar]
- 33.Liao YD, Jeng JC, Wang CF, Wang SC, Chang ST. Removal of N-terminal methionine from recombinant proteins by engineered E. coli methionine aminopeptidase. Protein Sci. 2004;13:1802–1810. doi: 10.1110/ps.04679104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo Y, Wallace SS, Bandaru V. A novel bicistronic vector for overexpressing Mycobacterium tuberculosis proteins in Escherichia coli. Protein Expr Purif. 2009;65:230–237. doi: 10.1016/j.pep.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bandaru V, Blaisdell JO, Wallace SS. Oxidative DNA glycosylases: recipes from cloning to characterization. Methods Enzymol. 2006;408:15–33. doi: 10.1016/S0076-6879(06)08002-5. [DOI] [PubMed] [Google Scholar]
- 36.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 37.Kornyushyna O, Berges AM, Muller JG, Burrows CJ. In vitro nucleotide misinsertion opposite the oxidized guanosine lesions spiroiminodihydantoin and guanidinohydantoin and DNA synthesis past the lesions using Escherichia coli DNA polymerase I (Klenow Fragment) Biochemistry. 2002;41:15304–15314. doi: 10.1021/bi0264925. [DOI] [PubMed] [Google Scholar]
- 38.Blaisdell JO, Wallace SS. Rapid determination of the active fraction of DNA repair glycosylases: a novel fluorescence assay for trapped intermediates. Nucleic Acids Res. 2007;35:1601–1611. doi: 10.1093/nar/gkm021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bandaru V, Sunkara S, Wallace SS, Bond JP. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair (Amst) 2002;1:517–529. doi: 10.1016/s1568-7864(02)00036-8. [DOI] [PubMed] [Google Scholar]
- 40.Dou H, Theriot CA, Das A, Hegde ML, Matsumoto Y, Boldogh I, Hazra TK, Bhakat KK, Mitra S. Interaction of the human DNA glycosylase NEIL1 with proliferating cell nuclear antigen. The potential for replication-associated repair of oxidized bases in mammalian genomes. J Biol Chem. 2008;283:3130–3140. doi: 10.1074/jbc.M709186200. [DOI] [PubMed] [Google Scholar]
- 41.Marenstein DR, Ocampo MT, Chan MK, Altamirano A, Basu AK, Boorstein RJ, Cunningham RP, Teebor GW. Stimulation of human endonuclease III by Y box-binding protein 1 (DNA-binding protein B). Interaction between a base excision repair enzyme and a transcription factor. J Biol Chem. 2001;276:21242–21249. doi: 10.1074/jbc.M101594200. [DOI] [PubMed] [Google Scholar]
- 42.Luna L, Bjoras M, Hoff E, Rognes T, Seeberg E. Cell-cycle regulation, intracellular sorting and induced overexpression of the human NTH1 DNA glycosylase involved in removal of formamidopyrimidine residues from DNA. Mutat Res. 2000;460:95–104. doi: 10.1016/s0921-8777(00)00015-x. [DOI] [PubMed] [Google Scholar]
- 43.Otterlei M, Haug T, Nagelhus TA, Slupphaug G, Lindmo T, Krokan HE. Nuclear and mitochondrial splice forms of human uracil-DNA glycosylase contain a complex nuclear localisation signal and a strong classical mitochondrial localisation signal, respectively. Nucleic Acids Res. 1998;26:4611–4617. doi: 10.1093/nar/26.20.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagelhus TA, Haug T, Singh KK, Keshav KF, Skorpen F, Otterlei M, Bharati S, Lindmo T, Benichou S, Benarous R, Krokan HE. A sequence in the N-terminal region of human uracil-DNA glycosylase with homology to XPA interacts with the C-terminal part of the 34-kDa subunit of replication protein A. J Biol Chem. 1997;272:6561–6566. doi: 10.1074/jbc.272.10.6561. [DOI] [PubMed] [Google Scholar]
- 45.Hardeland U, Steinacher R, Jiricny J, Schar P. Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J. 2002;21:1456–1464. doi: 10.1093/emboj/21.6.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bandaru V, Cooper W, Wallace SS, Doublie S. Overproduction, crystallization and preliminary crystallographic analysis of a novel human DNA-repair enzyme that recognizes oxidative DNA damage. Acta crystallographica. 2004;60:1142–1144. doi: 10.1107/S0907444904007929. [DOI] [PubMed] [Google Scholar]
- 47.Liu X, Roy R. Truncation of amino-terminal tail stimulates activity of human endonuclease III (hNTH1) J Mol Biol. 2002;321:265–276. doi: 10.1016/s0022-2836(02)00623-x. [DOI] [PubMed] [Google Scholar]
- 48.Li X, Romero P, Rani M, Dunker AK, Obradovic Z. Predicting Protein Disorder for N-, C-, and Internal Regions. Genome Inform Ser Workshop Genome Inform. 1999;10:30–40. [PubMed] [Google Scholar]
- 49.Shen TJ, Ho NT, Zou M, Sun DP, Cottam PF, Simplaceanu V, Tam MF, Bell DA, Jr, Ho C. Production of human normal adult and fetal hemoglobins in Escherichia coli. Protein Eng. 1997;10:1085–1097. doi: 10.1093/protein/10.9.1085. [DOI] [PubMed] [Google Scholar]
- 50.Hwang DD, Liu LF, Kuan IC, Lin LY, Tam TC, Tam MF. Co-expression of glutathione S-transferase with methionine aminopeptidase: a system of producing enriched N-terminal processed proteins in Escherichia coli. Biochem J. 1999;338(Pt 2):335–342. [PMC free article] [PubMed] [Google Scholar]