Abstract
This study was conducted to determine whether the ratio of estrogen-DNA adducts to their respective metabolites and conjugates in serum differed between women with early-onset breast cancer and those with average or high risk of developing breast cancer.
Serum samples from women at average risk (n = 63) or high risk (n = 80) for breast cancer (using Gail model) and women newly diagnosed with early breast cancer (n = 79) were analyzed using UPLC-MS/MS. Adduct ratios were statistically compared among the three groups, and the Area Under the Receiver Operating Characteristic Curve (AUC) was used to identify a diagnostic cut-off point.
The median adduct ratio in the average-risk group was significantly lower than that of both the high-risk group and the breast cancer group (p values <0.0001), and provided good discrimination between those at average versus high risk of breast cancer (AUC = 0.84, 95% CI 0.77–0.90). Sensitivity and specificity were maximized at an adduct ratio of 77. For women in the same age and BMI group, the odds of being at high risk for breast cancer was 8.03 (95% CI 3.46–18.7) times higher for those with a ratio of at least 77 compared to those with a ratio less than 77.
The likelihood of being at high risk for breast cancer was significantly increased for those with a high adduct ratio relative to those with a low adduct ratio. These findings suggest that estrogen-DNA adducts deserve further study as potential biomarkers for risk of developing breast cancer.
Keywords: breast cancer, depurinating estrogen-DNA adducts, biomarker, breast cancer prevention
1. Introduction
In the United States, breast cancer is the most common cancer in women, with an estimated 230,480 cases and 39,520 deaths in 2011 [1]. It has been postulated that endogenous and exogenous estrogens are cancer-causing agents. The ability to identify specific biomarkers associated with breast carcinogenesis would be of great value in the clinical setting, permitting improved breast cancer risk assessment and selection of patients for intervention. The ultimate goal is to use biomarker analysis to develop individualized risk reduction/prevention strategies that will prevent the development of tumors.
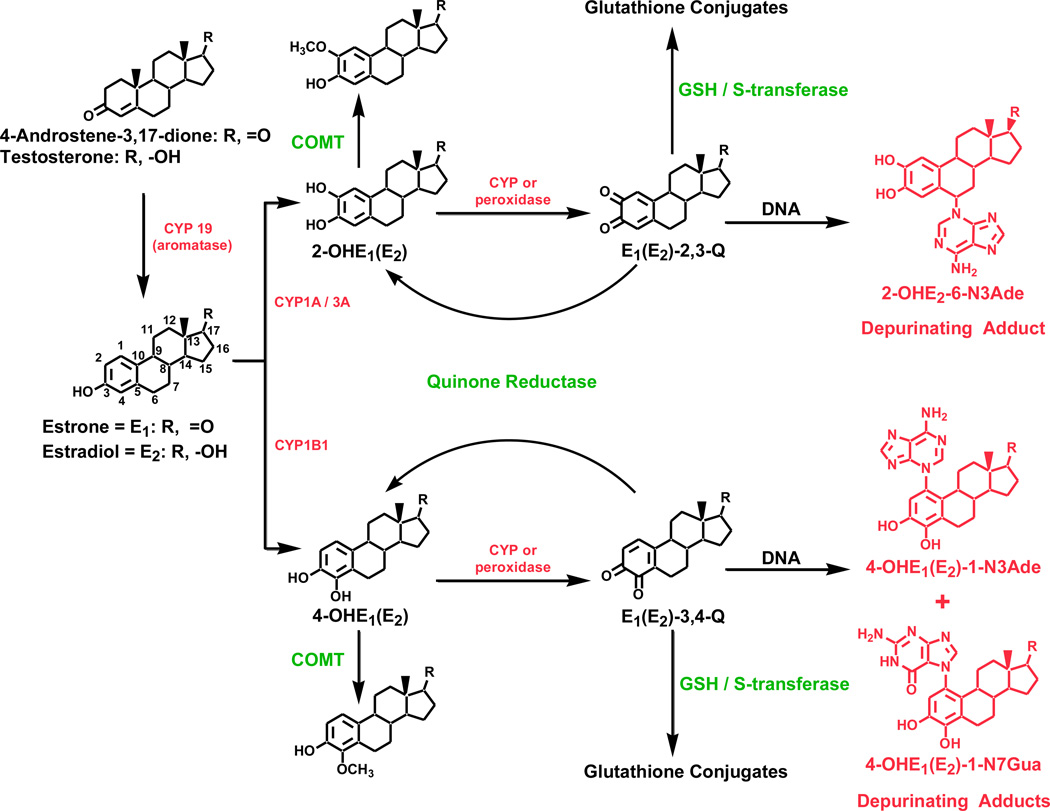
Compelling evidence supports the hypothesis that specific estrogen metabolites, predominantly catechol estrogen-3,4-quinones, react with DNA to form depurinating estrogen-DNA adducts. Estrogens are oxidized to the catechol estrogens (CE), 2-hydroxyestrone (estradiol) [2-OHE1(E2)] and 4-OHE1(E2), which are further oxidized to CE quinones (Fig. 1). This is the activating pathway of estrogen metabolism. When E1(E2)-3,4-quinones react with DNA, the depurinating estrogen-DNA adducts 4-OHE1(E2)-1-N3Ade and 4-OHE1(E2)-1-N7Gua are formed. Formation of depurinating estrogen-DNA adducts can initiate the cancer process through mutations in critical genes leading to abnormal cell proliferation and cell transformation [2,3]. These depurinating DNA adducts are rapidly lost from DNA by cleavage of the glycosyl bond leaving apurinic sites that can lead to DNA mutations by error-prone DNA repair [4–6]. These mutations can lead to the initiation of cancer.
Fig. 1. Estrogen Biosysnthesis and Metabolism.
Biosynthesis of estrogens, their metabolism via the catechol estrogen pathway, and formation of their depurinating DNA adducts.
Several deactivating pathways protect against the activating pathway of estrogen metabolism. The CEs can be deactivated by O-methylation catalyzed by catechol-O-methyltransferase (COMT) [7,8], and CE quinones can be deactivated by reduction with the quinone reductases NQO1 and NQO2 [9,10] and/or conjugation with glutathione (Fig. 1) [11,12]. Estrogen metabolism is normally balanced, and formation of estrogen-DNA adducts is relatively low. It is hypothesized that when the balance between the activating and deactivating pathways is disrupted and the activating pathway prevails, excess DNA damage results, increasing the risk of breast cancer [2,3].
Expression of cytochrome P450 (CYP) 19 and CYP1B1, two key enzymes in the activating pathway of estrogen metabolism, is higher in breast tissue from women with breast cancer than in breast tissue from healthy women. Furthermore, expression of COMT and NQO1, two key enzymes in the deactivating (protective) pathway of estrogen metabolism, is lower in women with breast cancer than in healthy women [13]. These expression profiles were shown to be associated with increased breast cancer risk in a large case-control study [14].
Depurinating estrogen-DNA adducts are shed into blood and excreted in urine after being released from DNA. They and the corresponding estrogen metabolites and conjugates are measurable in serum (this article) and urine [15,16], providing easily accessible sources from which these compounds can be isolated, identified, and measured. In two independent studies, the ratio of estrogen-DNA adducts to estrogen metabolites and conjugates was significantly higher in urine from women at high risk for breast cancer or diagnosed with the disease than in urine from women at average risk for breast cancer [15,16].
Based on the above evidence, we hypothesized that the profiles of estrogen metabolites, conjugates, and depurinating estrogen-DNA adducts in serum differ between women with breast cancer and women at various levels of risk for breast cancer. The goal of this study was to investigate the imbalance of estrogen metabolism in serum expressed as the estrogen-DNA adduct ratio and to examine its potential as a biomarker for increased breast cancer risk.
2. Methods
2.1. Study design and participants
The study participants were recruited from women aged 18 to 80 years who presented at the Breast Diagnostic Clinic or the Breast Cancer Clinic at Mayo Clinic in Rochester, MN, for new breast symptoms, routine breast health care, counseling regarding a family history of breast cancer or elevated risk for developing breast cancer or a new diagnosis of noninvasive or invasive breast cancer. The participants were asked to provide access to their medical record to verify eligibility, to determine the Gail model score [17,18], and to provide 20 mL of blood. The study protocol was approved by the Institutional Review Boards at Mayo Clinic and the University of Nebraska Medical Center (UNMC). Written informed consent was obtained from all study subjects.
To be eligible for participation, a woman had to fulfill the criteria of one of the three following groups: newly diagnosed breast cancer, high risk of breast cancer, or average risk of breast cancer. Exclusion criteria included a history of chemotherapeutic agents for breast or other cancers prior to study enrollment; current advanced breast cancer (stage III or IV); prior endocrine therapy in the prevention, neo-adjuvant, or adjuvant setting; exogenous hormone use (estrogen-containing hormone replacement therapy or hormonal contraception) within the 3 months prior to enrollment; current pregnancy; and current lactation.
The breast cancer (BC) group consisted of women with histopathologically and/or cytologically confirmed invasive breast cancer (stage I or II, TMN Staging System by the American Joint Committee on Cancer) or ductal carcinoma in-situ (stage 0) diagnosed within 30 calendar days prior to study enrollment who may have undergone definitive breast surgery. The high-risk (HR) group consisted of women with one of the following traits: (1) a Gail model score estimated 5-year risk of >1.66% or lifetime risk of >20%, (2) history of lobular carcinoma in-situ (LCIS), (3) history of atypical hyperplasia (ductal or lobular), or (4) known deleterious BRCA-1 or -2 gene mutation. The average-risk (AR) group consisted of women with a Gail model score estimated 5-year risk of ≤1.66% and lifetime risk of ≤20% with no significant risk factor for breast cancer. In addition to a personal history of breast biopsies, the Gail model score includes current age, age at menarche, age at first birth, history of first degree relatives with breast cancer, and race/ethnicity to calculate the 5-year and lifetime risk of breast cancer.
2.2. Determination of estrogen-DNA adduct ratios
Serum samples were drawn, de-identified, aliquoted, frozen, and transported on dry ice to the Eppley Institute, UNMC. The estrogen metabolites, conjugates, and depurinating DNA adducts in the serum samples were analyzed by ultraperformance liquid chromatography/tandem mass spectrometry (UPLC/MS-MS) blinded to patient characteristics. The analytical results were subsequently paired with the subject IDs for statistical analysis.
Upon arrival, the aliquots were stored at −80°C until analyzed. Samples were thawed only once prior to analysis. One-milliliter aliquots of serum underwent partial purification by solid-phase extraction with a phenyl cartridge, similar to the previously described method for urine samples [15]. Before being loaded onto cartridges preconditioned with methanol and distilled water, the serum samples were diluted with an equal volume of 0.01 M ammonium acetate buffer, pH 7, and adjusted to pH 7. After equilibrating the cartridges with loading buffer (0.01 M ammonium acetate, pH 7), the samples were loaded and the target compounds were eluted using elution buffer (methanol/0.01 M ammonium acetate, pH 7 [90:10] with 2% acetic acid). The collected eluates were concentrated, reconstituted, and analyzed by UPLC/MS-MS [15]. Analytes were identified by their retention time and fragmentation pattern. All of the analyses were carried out without glucuronidase/sulfatase treatment because treatment of serum with glucuronidase/sulfatase led to significant decreases in the levels of estrogen metabolites, conjugates, and estrogen-DNA adducts after the incubation overnight at 37°C. UPLC/MS-MS analyses were carried out using an Acquity UPLC system (Waters; Milford, MA) connected with a high-performance Quattro Micro triple quadrupole mass spectrometer (Waters; Milford, MA) [15]. The resulting data were processed by using QuanLynx software (Waters; Milford, MA) to quantify estrogen-DNA adducts, estrogen metabolites, and conjugates. The adduct ratio was defined as:
In this equation, the adducts of 4-OHE1(E2) play the predominant role (~98%), while the adducts of 2-OHE1(E2) are minimal, less than 2% [15]. The estrogen metabolites include 2-OHE1(E2) and 4-OHE1(E2). The conjugates include 2-OCH3E1(E2), 4-OCH3E1(E2), 2-OHE1(E2)-1-SG, 2-OHE1(E2)-4-SG, 2-OHE1(E2)-1-Cys, 2-OHE1(E2)-4-Cys, 2-OHE1(E2)-1-NAcCys, 2-OHE1(E2)-4-NAcCys, 4-OHE1(E2)-2-SG, 4-OHE1(E2)-2-Cys, and 4-OHE1(E2)-2-NAcCys. The adducts include 4-OHE1(E2)-1-N3Ade, 4-OHE1(E2)-1-N7Gua, and 2-OHE1(E2)-6-N3Ade.
The concentration of each of the 38 compounds and the ratio of depurinating N3Ade and N7Gua adducts to the sum of their respective estrogen metabolites and conjugates in each serum sample provides a ratio which reflects the degree of imbalance in estrogen metabolism that can lead to cancer initiation (Fig. 1).
2.3. Statistical design and analysis plan
This study was designed to assess whether the mean DNA adduct ratio differed with respect to the three patient groups. A sample size of 100 patients per study group was chosen so that for each of the 3 pairwise comparisons to be performed, a two-sided alpha = 0.01 two sample t-test would have at least an 80% chance of detecting a difference of 0.5 standard deviations or more.
2.4. Assessment of covariates
Covariates assessed included age (dummy coded as <50, 50–59, 60–69, 70+); age at menarche (continuous); age at first parity (continuous); body mass index (BMI: normal, <25; overweight, ≥25 and <30; obese, ≥30); menopausal status (pre-, peri-, postmenopausal); first degree relative with breast cancer; prior hysterectomy; prior oophorectomy; nulliparous; and history of breast disease (0 = no, 1 = yes for all five variables). Age was normally distributed so the Pearson correlation coefficient was used to assess correlation between the adduct ratio and age. The Spearman correlation coefficient was used to assess correlation between the adduct ratio and the continuous variable for BMI. Group differences in covariates were assessed using the chi-square test for categorical variables, analysis of variance for continuous, normally-distributed variables, and the Kruskal-Wallis test for differences in medians of non-normally distributed continuous variables (BMI, age at menarche, adduct ratio). The Wilcoxon rank sum test was used in a pairwise manner to assess whether the adduct ratios differed among the three patient groups.
2.5. Sensitivity and specificity analysis
The Receiver Operating Characteristic (ROC) curve was used to evaluate the potential utility of the adduct ratio as a biomarker and to find an “optimal” cut-off point for discriminating between women at average risk and women at high risk of developing breast cancer. The sensitivity and specificity associated with every observed value of the DNA adduct ratio were calculated and the results visualized by plotting the two curves on the same graph. The optimal cut-off point was chosen to be the DNA adduct ratio value at which the sensitivity-adduct ratio curve and the specificity-adduct ratio curve cross. A point and interval estimate of the true sensitivity and specificity at the optimal cut-off point was constructed using the properties of the binomial distribution.
2.6. Validation of adduct ratio cut-off point
Multivariate logistic regression modeling was used to assess whether the likelihood of a woman being at high risk for developing breast cancer relative to a woman being at average risk for developing breast cancer differs with respect to either the DNA adduct ratio (<77 vs. ≥77; see Results section) or BMI in age-adjusted models. Risk factors that were used to compute the Gail model score were not included in the analysis of the HR group. Additionally, in order to validate the cut-off value of 77, a multivariate model comparing the BC group to the AR group was used to assess whether the cut-off value of 77 showed similar odds ratios (OR) for the HR and BC groups. The BC versus AR groups were adjusted for having a first degree relative with breast cancer and a history of breast disease, important determinants of the Gail model score in the HR group.
All analyses were done in SAS v. 9.2 (SAS Institute; Cary, NC), and results were considered significant at p = 0.05; regression results are presented as OR with 95% confidence intervals (CI).
3. Results
3.1. Patient characteristics
Of 319 subjects enrolled between May 2005 and December 2007, 97 failed to meet the eligibility criteria due to age, stage of breast cancer, refusal to provide a blood sample, loss of specimen during shipping, insufficient sample volume for analysis, or exogenous hormone use within 3 months of study enrollment. Thus, the study cohort consisted of 222 women (BC: n = 79; HR: n = 80; AR: n = 63). An example of the data obtained from an individual HR subject is presented in Table 1.
Table 1.
Concentration of the 38 compounds from an individual HR subject
| No. | Compound | pmole/ml mean, n = 3 |
Total pmole/ml |
|---|---|---|---|
| 1 | Androstenedione | 1.47 ± 0.63 | 1.47 |
| 2 | Testosterone | 0.29 ± 0.19 | 0.29 |
| 3 | E2 | 1.40 ± 0.97 | 10.69 |
| 4 | E1 | 9.29 ± 6.55 | |
| 5 | E1-Sulfate | 7.30 ± 6.97 | 7.30 |
| 6 | 2-OHE2 | 0.27 ± 0.34 | 0.96 |
| 7 | 2-OHE1 | 0.69 ± 0.71 | |
| 8 | 4-OHE2 | 0.09 ± 0.06 | 2.70 |
| 9 | 4-OHE1 | 2.60 ± 2.71 | |
| 10 | 16α-E2 | 0.60 ± 0.45 | 6.22 |
| 11 | 16α-E1 | 5.62 ± 3.74 | |
| 12 | 2-OCH3E2 | 0.42 ± 0.10 | 2.76 |
| 13 | 2-OCH3E1 | 2.34 ± 2.53 | |
| 14 | 4-OCH3E2 | 0.15 ± 0.12 | 1.02 |
| 15 | 4-OCH3E1 | 0.87 ± 0.56 | |
| 16 | 2-OHE2-1-SG | 0.00 ± 0.00 | 0.32 |
| 17 | 2-OCH2-4-SG | 0.00 ± 0.00 | |
| 18 | 2-OHE1-1-SG | 0.00 ± 0.00 | |
| 19 | 2-OHE1-4-SG | 0.00 ± 0.00 | |
| 20 | 2-OHE2-1+4-Cys | 0.13 ± 0.11 | |
| 21 | 2-OHE1-1-Cys | 0.08 ± 0.07 | |
| 22 | 2-OHE1-4-Cys | 0.08 ± 0.07 | |
| 23 | 2-OHE2-1-NAcCys | 0.00 ± 0.00 | |
| 24 | 2-OHE2-4-NAcCys | 0.00 ± 0.00 | |
| 25 | 2-OHE1-1-NAcCys | 0.01 ± 0.01 | |
| 26 | 2-OHE1-4-NAcCys | 0.01 ± 0.01 | |
| 27 | 4-OHE2-2-SG | 0.20 ± 0.08 | 0.40 |
| 28 | 4-OHE1-2-SG | 0.05 ± 0.04 | |
| 29 | 4-OHE2-2-Cys | 0.12 ± 0.06 | |
| 30 | 4-OHE1-2-Cys | 0.01 ± 0.01 | |
| 31 | 4-OHE2-2-NAcCys | 0.03 ± 0.02 | |
| 32 | 4-OHE1-2-NAcCys | 0.00 ± 0.00 | |
| 33 | 4-OHE2-1-N7Gua | 0.08 ± 0.04 | 0.16 |
| 34 | 4-OHE1-1-N7Gua | 0.08 ± 0.07 | |
| 35 | 4-OHE2-1-N3Ade | 1.63 ± 2.20 | 1.65 |
| 36 | 4-OHE1-1-N3Ade | 0.02 ± 0.02 | |
| 37 | 2-OHE2-6-N3Ade | 0.08 ± 0.08 | 0.08 |
| 38 | 2-OHE1-6-N3Ade | 0.00 ± 0.00 | |
| Ratio | 461 | ||
3.2. Assessment of risk factors and covariates
The HR and BC groups were significantly older than the AR group (p < 0.0001), and thus the groups differed by menopausal status (p = 0.04) and were more likely to report a history of hysterectomy (p = 0.04). Age at menarche was lower in the HR group compared to the AR and BC groups (p = 0.02). Having a first degree relative with breast cancer (p value < 0.0001) and/or a history of breast disease (p value = 0.001) was significantly more likely in the HR group compared to the AR and BC groups. These three risk factors are used to compute the Gail model score and would be expected to be more frequent in the HR group. The three groups did not differ by BMI, prior oophorectomy, or being nulliparous in univariate analyses. Patient characteristics are shown in Table 2.
Table 2.
Individual characteristics of average risk, high risk and women diagnosed with early breast cancer
| Characteristic | Average risk (n = 63) |
High risk (n = 80) |
Breast cancer (n = 79) |
|---|---|---|---|
| Continuous variables | |||
| Mean age (SD)a | 50 (9) | 56 (9) | 57 (10) |
| Median age at menarche (min-max)c | 13 (9–17) | 12 (9–17) | 13 (10–17) |
| Median BMI (min-max) | 26 (19–46) | 28 (18–52) | 28 (18–44) |
| Median age of 1st parity (min-max) | 24 (16–40) | 26 (16–39) | 24 (13–38) |
| Median DNA adduct ratio (min-max)a | 26 (3–414) | 125 (10–919) | 107 (3–1126) |
| Categorical variables n (%) | |||
| Age (years)b | n = 63 | n = 80 | n = 79 |
| <50 | 35 (55.6) | 20 (25.0) | 19 (24.0) |
| 50–59 | 18 (28.6) | 28 (35.0) | 30 (38.0) |
| 60–69 | 9 (14.3) | 24 (30.0) | 18 (22.8) |
| 70+ | 1 (1.6) | 8 (10.0) | 12 (15.2) |
| BMI | n = 63 | n = 78 | n = 78 |
| Normal (<25 kg/m2) | 27 (42.8) | 23 (29.5) | 23 (29.5) |
| Overweight (≥25 and <30 kg/m2) | 12 (19.1) | 25 (32.0) | 23 (29.5) |
| Obese (≥30 kg/m2) | 24 (38.1) | 30 (38.5) | 32 (41.0) |
| Menopausal statusb | n = 61 | n = 80 | n = 79 |
| Premenopausal | 27 (44.3) | 24 (30.0) | 20 (25.3) |
| Perimenopausal | 8 (13.1) | 5 (6.3) | 6 (7.6) |
| Postmenopausal | 26 (42.6) | 51 (63.7) | 53 (67.1) |
| Family history of breast cancera | n = 62 | n = 79 | n = 77 |
| No | 57 (91.9) | 23 (29.1) | 55 (71.4) |
| Yes | 5 (8.1) | 56 (70.9) | 22 (28.6) |
| History of benign breast diseaseb | n = 62 | n = 79 | n = 77 |
| No | 47 (75.8) | 36 (45.6) | 43 (55.8) |
| Yes | 15 (24.2) | 43 (54.4) | 34 (44.2) |
| Prior hysterectomyb | n = 63 | n = 80 | n = 79 |
| No | 54 (85.7) | 54 (67.5) | 60 (76.0) |
| Yes | 9 (14.3) | 26 (32.5) | 19 (24.0) |
| Prior oophorectomy | n = 63 | n = 80 | n = 79 |
| No | 57 (90.5) | 62 (77.5) | 64 (81.0) |
| Yes | 6 (9.5) | 18 (22.5) | 15 (19.0) |
| Nulliparous | n = 63 | n = 79 | n = 79 |
| No | 53 (84.1) | 63 (79.8) | 72 (91.1) |
| Yes | 10 (15.9) | 16 (20.2) | 7 (8.9) |
p < 0.0001
p < 0.001
p < 0.05
The DNA adduct ratio was significantly correlated with age (r = 0.23, p = 0.0007), but after categorizing into four age categories, the association between the ratio and age was greatly reduced (p = 0.01). Menopausal status (p = 0.002), history of breast disease (p = 0.003), family history of breast cancer (p = 0.001), and history of hysterectomy (p = 0.01) were significantly associated with the adduct ratio. History of oophorectomy, being nulliparous, and the categorical BMI variable were not.
3.3. DNA adduct ratio by group
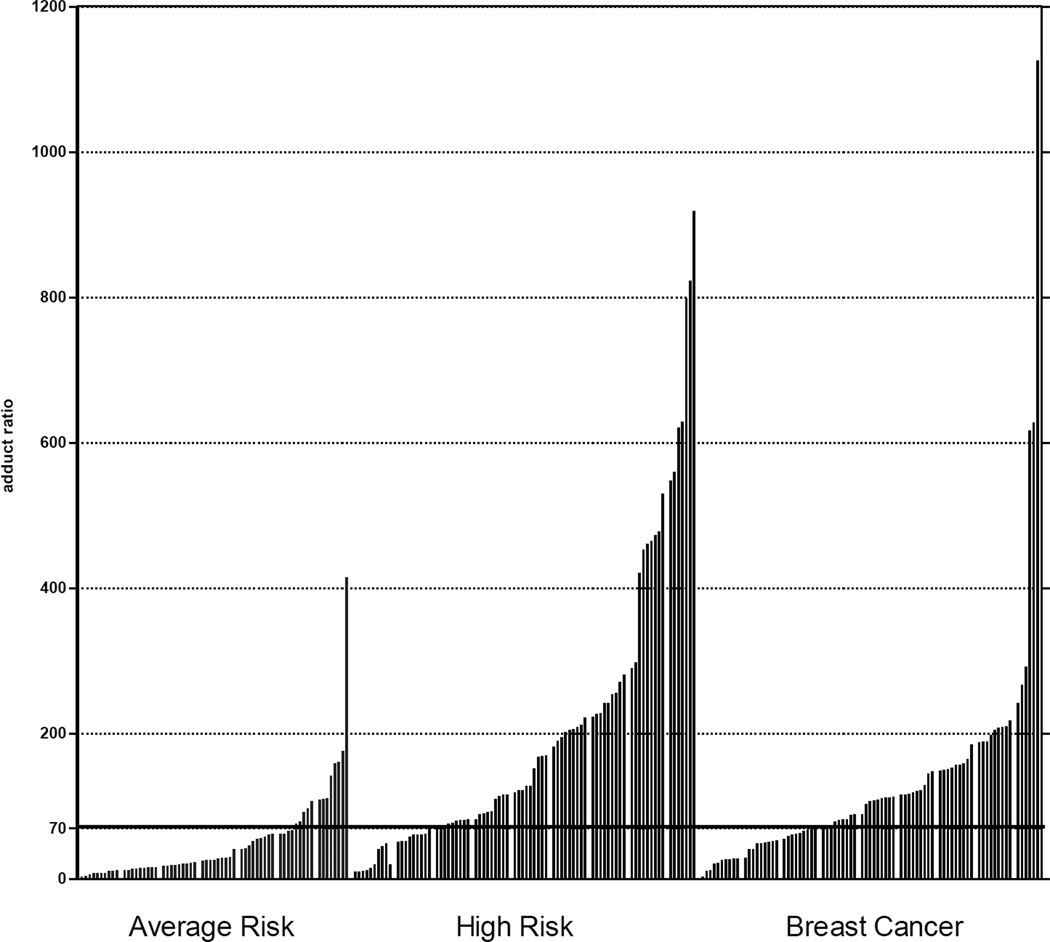
The ratios in the AR group were generally low (Fig. 2), but high ratios of adducts to metabolites and conjugates were observed in the serum of the HR and BC groups. High ratios generally came from high levels of adducts and low levels of metabolites and conjugates. In some cases a high ratio came from moderate adduct levels, combined with very low levels of metabolites and conjugates. In contrast, low ratios generally came from low levels of adducts and higher levels of metabolites and conjugates but were also associated with moderate levels of adducts with very high levels of metabolites and conjugates. In addition, the 2-OHE1(E2)-6-N3Ade adduct played an insignificant role in the total ratio, whereas the 4-OHE1(E2)-1-N3Ade and 4-OHE1(E2)-1-N7Gua adducts played the predominant role.
Fig. 2. Estrogen DNA Adduct Ratios.
Ratios of estrogen-DNA adducts to their respective estrogen metabolites and conjugates in the AR, HR, and BC groups.
The median DNA adduct ratio (interquartile range) was 25.9 (15.1–62.1) in the AR group; 124.6 (71.5–248.1) in the HR group; and 107.0 (54.8–156.5) in the BC group (p < 0.0001). The median adduct ratio of the HR group was found to be significantly higher than that of the AR group (p < 0.0001), as well as that of the BC group (p = 0.009). After excluding those with a family history of breast cancer, the median adduct ratio in the 24 women in the HR group did not differ significantly from the 57 breast cancer cases (115 vs. 108 respectively, p = 0.09). In addition, the median adduct ratio of the BC group was found to be significantly higher than that of the AR group (p < 0.0001).
3.4. Identification of an “optimal” cut-off point
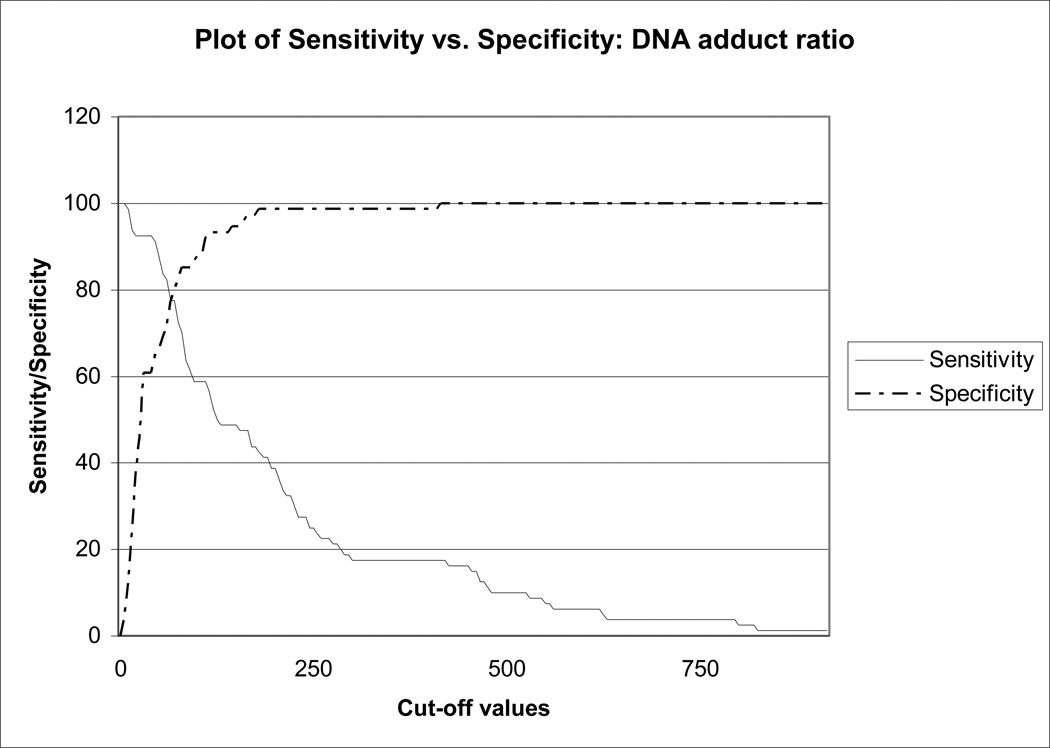
The area under the curve (AUC), a measure of discrimination between women with low risk of breast cancer and women with high risk of breast cancer, fell into the range considered good for a biomarker of risk (AUC = 0.84; 95% CI 0.77–0.90). Figure 3 provides an overlay of the plots of sensitivity-DNA adduct values and specificity-DNA adduct values. The point at which these curves intersect is the point where the sensitivity and specificity pair are at their maximum and is associated with the DNA adduct ratio value of 77, where sensitivity = 70.0%, and specificity = 81.0%.
Fig. 3. Plot of Sensitivity versus Specificity: DNA Adduct Ratio.
Sensitivity and specificity plotted against cut-off values between women with low risk of breast cancer (Gail model score ≤1.66%) and women with high risk of breast cancer (Gail model score ≥1.67%). Note: The two curves cross at a cut-off value of 77.
The results of multivariate logistic regression modeling indicated that the odds of being at high risk of developing breast cancer differed significantly with respect to age and the adduct ratio (Table 3). For example, with women in the same age and BMI group, the odds of being at high risk of breast cancer was eightfold higher for women with adduct ratios at or above 77 relative to women with adduct ratios below 77 (OR 8.03; 95% CI 3.46–18.7). Age was the only covariate that remained significant in multivariable models; however, BMI showed an elevated OR and was retained in the final model. Since having a first degree relative with breast cancer and having a history of breast disease are components of the Gail model score, they could not be included in the adjusted models of the AR and HR groups but could be included in models comparing the BC group to the AR group. In logistic regression models adjusted for age, being overweight or obese, having a family history of breast cancer and history of breast disease, and having an adduct ratio at or above 77, the effect size of the adduct ratio on breast cancer was similar to that for the HR group (OR 7.66; 95% CI 3.26–18.0).
Table 3.
Factors associated with the likelihood of being at high risk of developing breast cancer from logistic regression models.
| Characteristic | Univariate analyses OR (95% CI) |
Multivariable analysesa OR (95% CI) |
|---|---|---|
| Age (years) | ||
| <50 | 1.00 | 1.00 |
| 50–59 | 2.72 (1.21–6.11) | 2.43 (0.96–6.19) |
| 60–69 | 4.67 (1.82–12.0) | 3.85 (1.31–11.3) |
| 70+ | 14.0 (1.63–120) | 6.93 (0.71–67.5) |
| BMI | ||
| Normal (<25 kg/m2) | 1.00 | 1.00 |
| Overweight (≥25 and <30 kg/m2) | 2.45 (1.01–5.93) | 2.51 (0.86–7.29) |
| Obese (≥30 kg/m2) | 1.47 (0.68–3.18) | 1.88 (0.73–4.85) |
| DNA adduct ratio | ||
| <77 | 1.00 | 1.00 |
| ≥77 | 9.92 (4.50–21.8) | 8.03 (3.46–18.7) |
The model included age, body mass index, and DNA adduct ratio
4. Discussion
Prolonged exposure to endogenous estrogens, E2 and E1, has been linked to increased risk of breast cancer in women. The goal of this study was to determine the imbalance in estrogen metabolism as measured by the ratio of estrogen-DNA adducts to their respective metabolites and conjugates in human serum and to examine its potential as a biomarker for breast cancer risk identification and stratification. In this study we showed that the adduct ratio is significantly associated with the commonly accepted risk factors for breast cancer including family history of breast cancer (a surrogate for genetic factors), history of benign breast disease, and age. Additionally, the adduct ratio was strongly associated with being in the high-risk category for breast cancer and for having a recent breast cancer diagnosis.
After adjusting for age and BMI, the likelihood of being at high risk for breast cancer was significantly increased for those with a high adduct ratio relative to those with a low adduct ratio. This supports our hypothesis that the formation of estrogen-DNA adducts may be a potential causative factor in the etiology of breast cancer. It is well accepted that stable DNA mutations accumulate with increasing age. The DNA adduct ratio was also significantly associated with age. If mutations develop in critical genes involved in cell cycle control and tumor suppression through DNA adduct formation and subsequent error-prone DNA repair, dysregulated cellular transformation and proliferation result, increasing the risk of malignancy.
Our study revealed significantly higher levels of depurinating estrogen-DNA adducts in the women at high risk for breast cancer relative to both women at average risk for breast cancer and women with breast cancer; however, this is likely due to the HR group having more first degree relatives with breast cancer. This result illustrates the importance of family history beyond the BRCA-1 and -2 gene mutations and has implications for gene mutations in the protective, deactivating enzymes COMT and NQO1. The level of adducts was also significantly higher in women with breast cancer than in average-risk women (p < 0.0001). Of interest, it was noted that there were some patients in the HR group who had ratios higher than the BC group. These patients represent a group that deserves further study.
Today, breast cancer risk assessment is limited to evaluation of a woman’s reproductive and menstrual history (indicators of the duration of endogenous estrogen exposure) and family history. Most commonly, this information is used to calculate an estimated 5-year and lifetime risk of developing invasive breast cancer using the Gail model [17,18]. In recent years, it has become evident that the Gail model risk estimation tool in its current format has significant limitations. These limitations include the potential to underestimate risk for women with (1) multiple affected relatives, (2) affected non-first degree relatives, and (3) women with a strong family history of early-onset breast cancer (age <50 years), and possible overestimation of risk for women with both atypical hyperplasia and a first degree family history of breast cancer [19]. The Gail model also does not incorporate other known significant risk factors for breast cancer such as LCIS and high mammographic breast density.
It is becoming increasingly challenging for clinicians to accurately estimate breast cancer risk. There is a need in clinical practice for readily available tools and biomarkers to provide more definitive, individualized risk estimates that can assist in decision making regarding chemoprevention and other breast cancer risk-reduction options. It is evident that there is significant need for an objective tool by which to assess response to targeted interventions intended to reduce breast cancer risk.
One possible approach to chemoprevention of breast cancer derives from the hypothesis underpinning this study; i.e., specific estrogen metabolites react with DNA to form depurinating estrogen-DNA adducts. The resulting apurinic sites in the DNA can generate critical mutations to initiate the series of events leading to breast cancer [2,3]. Following this hypothesis, formation of higher levels of estrogen-DNA adducts would increase the risk of developing breast cancer. Chemoprevention that inhibits formation of these estrogen-DNA adducts would presumably reduce the risk of breast cancer developing [3]. If estrogen-DNA adduct ratios decline during chemopreventive therapy, this would provide an extremely valuable clinical tool for individualized care and an indicator of breast cancer risk modification. Additional investigation is warranted to evaluate this possibility.
Limitations of the current study include small sample size and lack of diversity in the study population. Also, the impact of the unbalanced estrogen metabolism on the clinical outcome of women at high risk for breast cancer or with early breast cancer could not be addressed with this study cohort.
In conclusion, significantly higher levels of depurinating estrogen-DNA adducts were observed in women at high risk for breast cancer, compared to women at average risk. The results of our study suggest that serum estrogen-DNA adducts are a potential biomarker for determining a woman’s risk for developing breast cancer, and potentially, for monitoring the effects of therapy These findings are highly novel but need further study and validation.
Highlights.
The identification of biomarkers associated with breast cancer risk is needed
Estrogen metabolites react with DNA to form depurinating estrogen DNA adducts
Depurinating estrogen-DNA adducts can initiate the cancer process
Validation of potential serum biomarkers aid cancer risk assessment
Acknowledgments
Support for this research was provided at the Eppley Institute and by the Avon Progress for Patients Awards Program grant P30 CA36727 from the National Cancer Institute at the National Institutes of Health.
Abbreviations
- CE
catechol estrogens
- COMT
catechol-O-methyltransferase
- CYP
cytochrome P450
- UPLC/MS-MS
ultraperformance liquid chromatography/tandem mass spectrometry
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors declare they have no conflict of interest.
References
- 1.American Cancer Society, Cancer Facts & Figures 2011. [Accessed November 15, 2011]; http://www.cancer.org/Research/CancerFactsFigures/CancerFactsFigures/cancer-facts-figures-2011.
- 2.Cavalieri E, Chakravarti D, Guttenplan J, Hart E, Ingle J, Jankowiak R, Muti P, Rogan E, Russo J, Santen R, Sutter T. Catechol estrogen quinones as initiators of breast and other human cancers: implications for biomarkers of susceptibility and cancer prevention. Biochim. Biophys. Acta. 2006;1766:63–78. doi: 10.1016/j.bbcan.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Cavalieri EL, Rogan EG. Depurinating estrogen-DNA adducts in the etiology and prevention of breast and other human cancers. Future Oncol. 2010;6:75–91. doi: 10.2217/fon.09.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakravarti D, Mailander PC, Li KM, Higginbotham S, Zhang HL, Gross ML, Meza JL, Cavalieri EL, Rogan EG. Evidence that a burst of DNA depurination in SENCAR mouse skin induces error-prone repair and forms mutations in the H-ras gene. Oncogene. 2001;20:7945–7953. doi: 10.1038/sj.onc.1204969. [DOI] [PubMed] [Google Scholar]
- 5.Mailander PC, Meza JL, Higginbotham S, Chakravarti D. Induction of A.T to G.C mutations by erroneous repair of depurinated DNA following estrogen treatment of the mammary gland of ACI rats. J. Steroid Biochem. Mol. Biol. 2006;101:204–215. doi: 10.1016/j.jsbmb.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 6.Zhao Z, Kosinska W, Khmelnitsky M, Cavalieri EL, Rogan EG, Chakravarti D, Sacks PG, Guttenplan JB. Mutagenic activity of 4-hydroxyestradiol, but not 2-hydroxyestradiol, in BB rat2 embryonic cells, and the mutational spectrum of 4-hydroxyestradiol. Chem. Res. Toxicol. 2006;19:475–479. doi: 10.1021/tx0502645. [DOI] [PubMed] [Google Scholar]
- 7.Lu F, Zahid M, Saeed M, Cavalieri EL, Rogan EG. Estrogen metabolism and formation of estrogen-DNA adducts in estradiol-treated MCF-10F cells. The effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin induction and catechol-O-methyltransferase inhibition. J. Steroid Biochem. Mol. Biol. 2007;105:150–158. doi: 10.1016/j.jsbmb.2006.12.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zahid M, Saeed M, Lu F, Gaikwad N, Rogan E, Cavalieri E. Inhibition of catechol-O-methyltransferase increases estrogen-DNA adduct formation. Free Radic. Biol. Med. 2007;43:1534–1540. doi: 10.1016/j.freeradbiomed.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaikwad NW, Rogan EG, Cavalieri EL. Evidence from ESI-MS for NQO1-catalyzed reduction of estrogen ortho-quinones. Free Radic. Biol. Med. 2007;43:1289–1298. doi: 10.1016/j.freeradbiomed.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaikwad NW, Yang L, Rogan EG, Cavalieri EL. Evidence for NQO2-mediated reduction of the carcinogenic estrogen ortho-quinones. Free Radic. Biol. Med. 2009;46:253–262. doi: 10.1016/j.freeradbiomed.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cavalieri EL, Kumar S, Todorovic R, Higginbotham S, Badawi AF, Rogan EG. Imbalance of estrogen homeostasis in kidney and liver of hamsters treated with estradiol: implications for estrogen-induced initiation of renal tumors. Chem. Res. Toxicol. 2001;14:1041–1050. doi: 10.1021/tx010042g. [DOI] [PubMed] [Google Scholar]
- 12.Rogan EG, Badawi AF, Devanesan PD, Meza JL, Edney JA, West WW, Higginbotham SM, Cavalieri EL. Relative imbalances in estrogen metabolism and conjugation in breast tissue of women with carcinoma: potential biomarkers of susceptibility to cancer. Carcinogenesis. 2003;24:697–702. doi: 10.1093/carcin/bgg004. [DOI] [PubMed] [Google Scholar]
- 13.Singh S, Chakravarti D, Edney JA, Hollins RR, Johnson PJ, West WW, Higginbotham SM, Cavalieri EL, Rogan EG. Relative imbalances in the expression of estrogen-metabolizing enzymes in the breast tissue of women with breast carcinoma. Oncol. Rep. 2005;14:1091–1096. [PubMed] [Google Scholar]
- 14.Wen W, Ren Z, Shu XO, Cai Q, Ye C, Gao YT, Zheng W. Expression of cytochrome P450 1B1 and catechol-O-methyltransferase in breast tissue and their associations with breast cancer risk. Cancer Epidemiol. Biomarkers Prev. 2007;16:917–920. doi: 10.1158/1055-9965.EPI-06-1032. [DOI] [PubMed] [Google Scholar]
- 15.Gaikwad NW, Yang L, Muti P, Meza JL, Pruthi S, Ingle JN, Rogan EG, Cavalieri EL. The molecular etiology of breast cancer: evidence from biomarkers of risk. Int. J. Cancer. 2008;122:1949–1957. doi: 10.1002/ijc.23329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaikwad N, Yang L, Pruthi S, Ingle J, Sandhu N, Rogan E, Cavalieri E. Urine biomarkers of risk in the molecular etiology of breast cancer. Breast Cancer: Basic and Clin. Res. 2009;3:1–8. doi: 10.4137/bcbcr.s2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gail MH, Brinton LA, Byar DP, Corle DK, Green SB, Schairer C, Mulvihill JJ. Projecting individualized probabilities of developing breast cancer for white females who are being examined annually. J. Natl. Cancer Inst. 1989;81:1879–1886. doi: 10.1093/jnci/81.24.1879. [DOI] [PubMed] [Google Scholar]
- 18.Euhus DM, Leitch AM, Huth JF, Peters GN. Limitations of the Gail model in the specialized breast cancer risk assessment clinic. Breast J. 2002;8:23–27. doi: 10.1046/j.1524-4741.2002.08005.x. [DOI] [PubMed] [Google Scholar]
- 19.Pankratz VS, Hartmann LC, Degnim AC, Vierkant RA, Ghosh K, Vachon CM, Frost MH, Maloney SD, Reynolds C, Boughey JC. Assessment of the accuracy of the Gail model in women with atypical hyperplasia. J. Clin. Oncol. 2008;26:5374–5379. doi: 10.1200/JCO.2007.14.8833. [DOI] [PMC free article] [PubMed] [Google Scholar]