Abstract
Background and rationale
Ubiquitin-conjugating enzyme 9 (Ubc9) is required for sumoylation and overexpressed in several malignancies but its expression in hepatocellular carcinoma (HCC) is unknown. Hepatic S-adenosylmethionine (SAMe) level falls in methionine adenosyltransferase 1A (Mat1a) knockout (KO) mice, which develop HCC, and in ethanol fed mice. Here we examined regulation of Ubc9 by SAMe in murine liver and human HCC, breast and colon carcinoma cell lines and specimens.
Methods
Real-time PCR and Western blotting measured gene and protein expression, respectively. Immunoprecipitation followed by Western blotting examined protein-protein interactions.
Results
Ubc9 expression is increased in HCC and when hepatic SAMe level falls. SAMe treatment in Mat1a KO mice reduced Ubc9 protein but not mRNA level and lowered sumoylation. Similarly, treatment of liver cancer cell lines HepG2 and Huh-7, colon cancer cell line RKO and breast cancer cell line MCF-7 with SAMe or its metabolite methylthioadenosine (MTA) reduced only Ubc9 protein level. Ubc9 post-translational regulation is unknown. Ubc9 sequence predicts a possible phosphorylation site by cell division cycle 2 (Cdc2), which directly phosphorylated recombinant Ubc9. Mat1a KO mice have higher phospho-Ubc9 level, which normalized after SAMe treatment. SAMe and MTA treatment lowered Cdc2 mRNA and protein levels, phospho-Ubc9 and protein sumoylation in liver, colon and breast cancer cells. Serine 71 of Ubc9 is required for phosphorylation, interaction with Cdc2 and protein stability. Cdc2, Ubc9 and phospho-Ubc9 levels are increased in human liver, breast and colon cancers.
Conclusions
Cdc2 expression is increased and Ubc9 is hyperphosphorylated in several cancers and this represents a novel mechanism to maintain high Ubc9 protein expression that can be inhibited by SAMe and MTA.
Keywords: cell cycle division 2, methylthioadenosine, hepatocellular carcinoma, methionine adenosyltransferase 1A knockout, ethanol fed mice
INTRODUCTION
Posttranslational modification regulates the activity, localization, intracellular levels, and interaction of proteins with other molecules (1). Sumoylation is small ubiquitin (Ub)-related modifier (SUMO) conjugation that is similar to ubiquitination in the conjugation process and attachment to target proteins. However, unlike ubiquitination that normally targets proteins for degradation through proteasome pathways, sumoylation regulates protein stability, protein-protein interactions, transcriptional activity and trafficking (2).
Sumoylation is a multi-step process catalyzed by E1, E2 and E3 enzymes that include activating SUMO, conjugation ending with the formation of an isopeptide bond between the C-terminal Gly residue of SUMO and the ε-amino group of a Lys residue in the acceptor protein (2). Ubiquitin-conjugating enzyme 9 (Ubc9) is the only well characterized E2 enzyme in the sumoylation cycle that transfers the activated SUMO to the target protein (3).
Ubc9 and SUMO are highly expressed in human premalignant conditions in response to low-grade, long-term oxidative stress, suggesting that up-regulation of sumoylation may be an adaptive response to oxidative stress (4, 5). Ubc9 may be fundamental for tumorigenesis and tumor progression by minimizing the acute cellular stress response associated with the accumulating DNA damage of tumor progression (5). Consistently, Ubc9 is overexpressed in several malignancies, such as lung adenocarcinoma, colorectal cancer, breast adenocarcinoma, and melanoma (6–8). Ubc9 expression in hepatocellular carcinoma (HCC) has not been reported.
Chronic hepatic S-adenosylmethionine (SAMe) deficiency generates oxidative stress (9, 10) and predisposes to injury and malignant transformation (11). SAMe, generated in the first reaction of methionine metabolism catalyzed by methionine adenosyltransferase (MAT), is the principal biological methyl donor and a precursor for glutathione and polyamines (11). In the biosynthesis of polyamines, 5′-methylthioadenosine (MTA) is generated as an end product (12). SAMe is also degraded spontaneously to MTA intra- and extracellularly (13). The catalytic subunit of MAT is the product of two different genes: MAT1A, which is expressed mainly in the adult liver, and MAT2A, which is widely distributed, expressed in the fetal liver, and replaces MAT1A in HCC (11). Consequences of hepatic SAMe deficiency as illustrated by the Mat1a knockout (KO) and intragastric ethanol infusion mouse models include increased susceptibility to oxidative liver injury and HCC (9, 10, 14).
Here, we report that Ubc9 expression and hence sumoylation are increased when hepatic SAMe level fall. Our findings reveal for the first time that Ubc9 stability is regulated by SAMe and MTA, through a change in Ubc9 phosphorylation. In addition, we identified Ubc9 as a substrate of cell division cycle 2 (Cdc2), which is overexpressed in many cancers and when SAMe level fall. These changes occur in many types of cancers, including HCC, colon and breast. Our findings have important implications in the regulation of Ubc9 and sumoylation.
MATERIALS AND METHODS
Materials
SAMe, in the stable form of disulfate p-toluene sulfonate dried powder was generously provided by Gnosis SRL (Cairate, Italy), MTA was from Sigma-Aldrich (St. Louis, MO) and roscovitine was from Calbiochem (San Diego, CA) and phosphatase inhibitor cocktail was from Thermo Scientific (Rockford, IL). [γ-32P]ATP (3,000 Ci/mmol) was from Perkin Elmer (Walltham, MA). Recombinant Cdc2/Cyclin B complex was from New England Biolabs (Ipswish, MA), while Ubc9 recombinant protein was from Prospec (Rehovot, Israel).
Cell culture and treatments
HepG2, Huh-7 and RKO cells were from the Cell Culture Core of the USC Research Center for Liver Diseases. MCF-7 cells were purchased from American Type Cell Collection (ATCC, Manassas, VA). All cell lines were grown according to instructions provided by ATCC. Prior to treatment with SAMe or MTA, medium was changed to 1% fetal bovine serum overnight. Cells were then treated with SAMe (2mmol/L) or MTA (1mmol/L) for up to 24 hours. Because of the instability of SAMe, medium was changed every 6 hours for all conditions. In other experiments, RKO cells were treated with phosphatase inhibitors cocktail (1:10,000 – 104mM AEBSF-[4-(2-Aminoethyl)benzenesulfonyl fluoride hydrochloride], 80μM aprotinin, 4mM bestatin hydrochloride, 1.4mM E-64-[N-(trans-Epoxysuccinyl)-L-leucine 4-guanidinobutylamide], 2mM leupeptin hemisulfate salt, 1.5mM pepstatin A) for 24 hours.
Mat1a KO mice and treatment
Male Mat1a KO and wild-type (WT) littermates were previously described (14). Mice were administered daily by oral gavage 100 mg/kg SAMe dissolved in water or equal volume of water and were sacrificed after seven days. Liver tissues were removed, snap frozen in liquid nitrogen, and stored at −80°C until analysis. Animals were treated humanely and all procedures were in compliance with our institutions guidelines for the use of laboratory animals.
Intragastric ethanol infusion
The intragastric ethanol infusion model was described previously (15). Male C57/B6 mice received high-fat diet (35% calories as corn oil) along with an increasing dose of ethanol (22.7–35g/Kg per day for mice) or isocaloric dextrose solutions for four weeks. Iron dextran (25 or 75mg/Kg) was injected subcutaneously two weeks before four-weeks alcohol feeding. The animals were treated in accordance with our institutions guidelines for the use of laboratory animals.
Tissue specimens
Twelve each of colon, HCC, and breast adenocarcinoma and corresponding surrounding non-tumorous tissues were used. Colon tissues were obtained by Dr. P Giordano (Whipps Cross University Hospital, London, UK). Liver and breast tissues were obtained by Dr. RM Pascale (University of Sassari, Italy). These tissues were immediately frozen in liquid nitrogen for subsequent RNA and protein extraction.
Written informed consent was obtained from each patient. The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki as reflected in a prior approval by Keck School of Medicine University of Southern California’s human research review committee.
RNA extraction and real-time polymerase chain reaction (PCR) analysis
Total RNA isolated from cells and livers as described (16) was subjected to reverse transcription (RT) by using M-MLV Reverse Transcriptase (Invitrogen, Carlsbad, CA). One μl of RT product was subjected to quantitative real-time PCR analysis. The primers and TaqMan probes for Ubc9, Cdc2, PP1 and Universal PCR Master Mix were purchased from ABI (Foster City, CA). 18SrRNA was used as housekeeping gene as described (17). The delta Ct (ΔCt) obtained was used to find the relative expression of genes according to the formula: relative expression=2−ΔΔCt, where ΔΔCt= ΔCt of respective genes in experimental groups – ΔCt of the same genes in control group.
RNA interference
The predesigned small interfering RNA (siRNA) targeting human Cdc2 (sense sequence: CCUAGUACUGCAAUUCGGGAAAUUU) (Invitrogen, Carlsbad, CA), PP1 (sense sequence: CCGCAATTCCGCCAAAGCCAA) (Qiagen, Valencia, CA), and negative control siRNA (Ambion, Austin, TX) were purchased. HepG2, RKO and MCF-7 cells were cultured in 100mm2-dish (1× 106 cells/dish) and transfected using RNAiMax (18μl/dish) from Invitrogen (Carlsbad, CA) with Cdc2 siRNA (30nM) or negative control siRNA for 48 hours for mRNA or protein expression, following the manufacturer’s manual.
Overexpression of WT and mutant Ubc9
Ubc9 overexpression vector (Ubc9-pCMV4-HA) and negative control empty vector (pCMV4-HA) were purchased from Addgene (Cambridge, MA). Mutation of Ser70 or Ser71 of Ubc9 to Phenylalanine was performed by Quick Change II Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) with pCMV4 as the template. RKO cells were cultured in 100mm2-dish (1× 106 cells/dish), transfected using 60μl of Superfect from Qiagen (Valencia, CA) and 5μg of target plasmid per dish. After 4 hours, the transfection medium was changed to normal medium. Protein expression and interaction were examined 48 hours later.
Western blots and co-immunoprecipitation (co-IP)
Protein extracts from cells and tissue samples were prepared as described (18) and immunoprecipitated by specific Ubc9 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) and processes as reported (19). Immunoprecipitated proteins were subjected to Western blotting following standard protocols (Amersham BioSciences, Piscataway, NJ), and the membrane were probed with the anti-PP1, anti-Cdc2 (Santa Cruz Biotechnology, Santa Cruz, CA), anti-SUMO-1 that detects recombinant SUMO-1 and endogenous levels of sumoylated proteins (e.g. SUMO-1 RanGAP) (Cell Signaling, Danvers, MA), and anti-phosphorylated proteins (Pan)[pS/pT/pY] (Invitrogen, Carlsbad, CA) antibodies. Blots were developed using enhanced chemoluminescence.
In vitro assay of kinase activity
Recombinant Ubc9 (2μg) was phosphorylated by the purified recombinant Cdc2/Cyclin B complex (10ng) in a final volume of 30μl containing 50mM Tris-HCl (pH7.5), 10mM MgCl2, 1mM EGTA, 1mM dithiothreitol, 100μM ATP, and 10μCi of [γ-32P]ATP. The reaction mixture was incubated for the indicated times at 30°C, after which proteins were separated by SDS-PAGE on a 12% gel. The radioactivity associated with protein bands was quantified by Quantity One (Biorad, Hercules, CA).
Protein stability assay and half-life determination
Cycloheximide (5μg/ml) was added to RKO or Huh-7 cells 48 hours after transfection with 2μg of wild type Ubc9-pCMV4 or Ubc9-F71 mutant plasmids per well. Protein levels were determined at indicated time points by Western blotting as described above using the anti-HA antibody. The relative amount of Ubc9-HA protein was evaluated by densitometry and normalized to actin. Protein half-life was determined for each experiment using linear regression analysis.
Statistical analysis
Data are expressed as mean±SEM. Statistical analysis was performed using ANOVA and Fisher’s test. For mRNA and protein levels, ratios of genes and proteins to respective housekeeping densitometric values were compared. Significance was defined by p<0.05.
RESULTS
Ubc9 is upregulated in human HCC
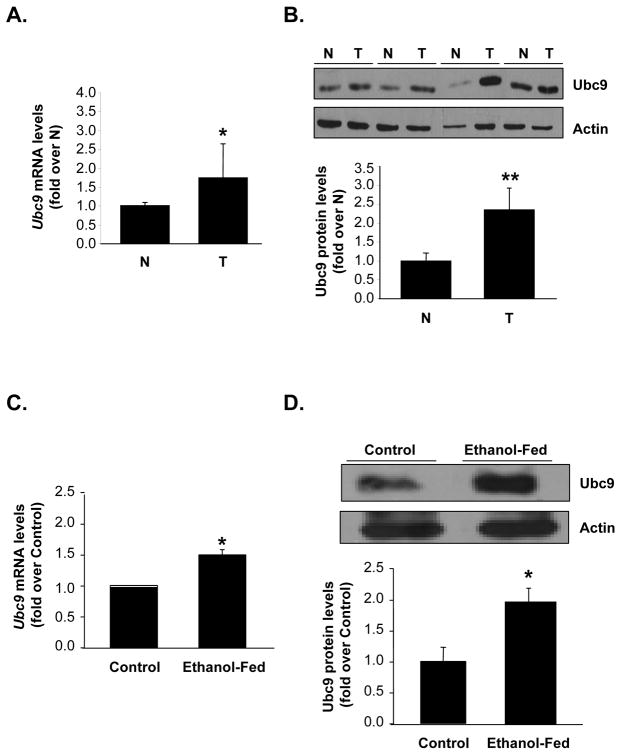
Ubc9 is overexpressed in several cancers (6–8, 20) but its expression in HCC is unknown. We found Ubc9 mRNA levels increased by 60% while Ubc9 protein levels increased by 140% in HCC as compared to surrounding non-tumorous tissue (Figure 1A&B).
Fig. 1. Ubc9 expression is up-regulated in HCC and ethanol fed mouse livers.
Expression was measured using quantitative real-time PCR and Western Blotting analysis. (A, B) Results from 18 pairs of matched non-tumorous (N) and HCC (T) tissues are expressed as fold of non-tumorous tissue (mean ± SEM). *P<0.05 and **p<0.002 vs.. non-tumorous tissues. (C, D) Ethanol fed mice were treated with ethanol or isocaloric dextrose as described in Methods. Results are expressed as fold of control (mean ±SEM) from n=4 for each group. *P<0.02 vs. control mice.
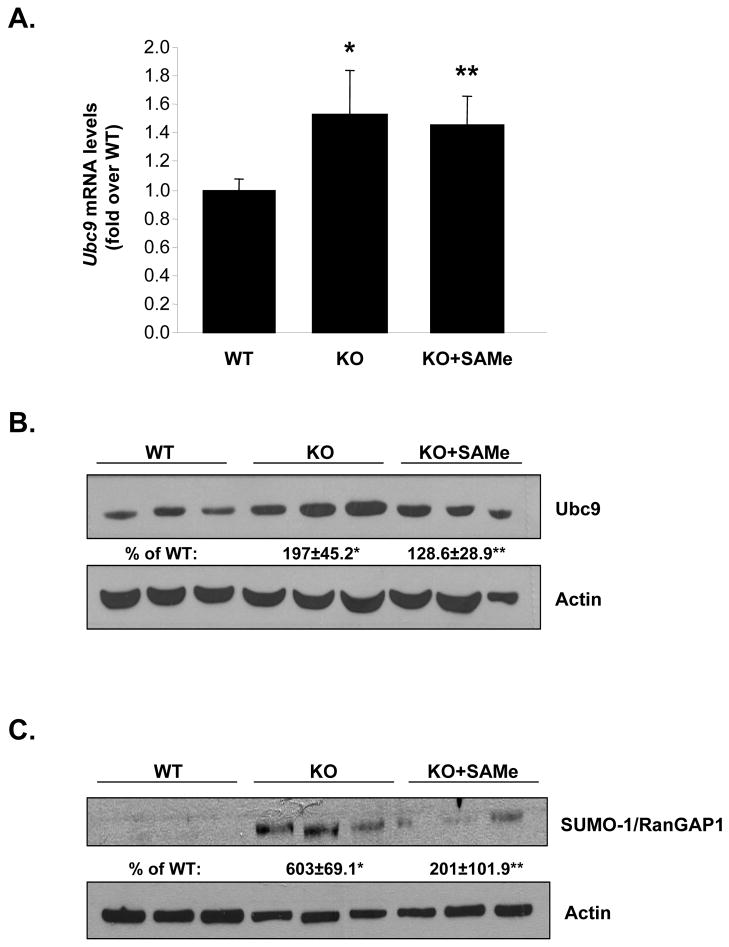
Ubc9 expression is up-regulated in Mat1a KO and ethanol fed
Mat1a KO and ethanol fed mice exhibit increased oxidative stress, which increases Ubc9 expression and sumoylation (5, 9, 21). Figure 1C and 1D show that mRNA and protein levels of Ubc9 increased by 50% and 100%, respectively, in livers of ethanol fed as compared to control mice. Similarly, in Mat1a KO mice livers Ubc9 mRNA and protein levels increased by 44% and 97%, respectively (Figure 2A and 2B). Protein sumoylation increased by 500% in the KO mice livers (Figure 2C). Since hepatic SAMe level is reduced in both Mat1a KO and ethanol fed mice, we examined the effect of SAMe treatment in Mat1a KO mice. SAMe treatment for one week, which normalized hepatic SAMe level in Mat1a KO mice (22), had no effect on Ubc9 mRNA (Figure 2A), but nearly normalized Ubc9 protein levels (Figure 2B) and protein sumoylation (Figure 2C).
Fig. 2. Effects of SAMe treatment on Ubc9 expression and sumoylation in Mat1a KO mice.
(A) Real-time PCR analysis measured Ubc9 mRNA levels. Results are expressed as fold of WT mice (mean ± SEM) from four mice per group. *P<0.03 vs. WT; **p<0.04 vs. WT. (B) Western Blotting analysis measured Ubc9 protein levels. Results are expressed as % of WT mice (mean ± SEM) from four mice per group. *P<0.01 vs. WT; **p<0.04 vs. KO. (C) SAMe treatment normalized the increase in sumoylated RanGAP1 levels in Mat1a KO mice livers. Results are expressed as % of WT mice (mean ± SEM) from four each of WT, untreated KO, and SAMe-treated KO mice. *P<0.001 vs. WT; **p<0.001 vs. KO.
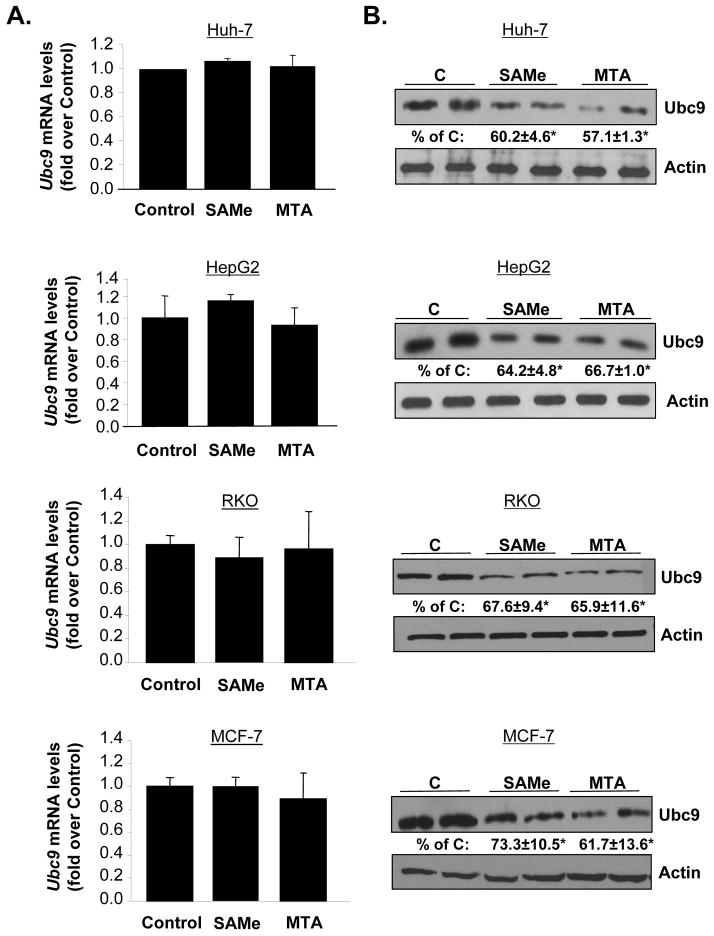
Effects of SAMe and MTA on expression of Ubc9 in HepG2, Huh-7, RKO and MCF-7 cells
We next examined effects of SAMe and its metabolite MTA on Ubc9 expression in liver, colon and breast cancer cell lines as Ubc9 is overexpressed in colon and breast cancers (8, 20). We included MTA because MTA recapitulates many of the effects of SAMe (13, 17, 23). Consistent with the effect of SAMe in Mat1a KO livers, SAMe and MTA treatment for 24 hours had no effect on Ubc9 mRNA levels in HepG2, Huh-7, RKO and MCF-7 cells (Figure 3A), but lowered Ubc9 protein levels by about 30 to 40% in all of these cells (Figure 3B). Treatment with SAMe or MTA for 12 hours had no effect on Ubc9 mRNA levels and the reduction in Ubc9 protein levels was much less (data not shown).
Fig. 3. SAMe and MTA treatment lowered Ubc9 protein levels in HepG2, Huh-7, RKO and MCF-7 cells.
Cells were treated with SAMe (2mM) or MTA (1mM) for 24 hours. (A) Quantitative real-time PCR measured mRNA levels of Ubc9. Results are expressed as fold of control (mean ± SEM) from 3 independent experiments. (B) Western blot analysis using whole cell lysates (10μg) measured Ubc9 protein levels. The values are expressed as % of control (mean ± SEM) from 3 independent experiments. *P<0.001 vs. control of HepG2, *p<0.001 vs. control of Huh-7, *p<0.01 vs. control of RKO and *p< 0.01 vs. control of MCF-7 cells.
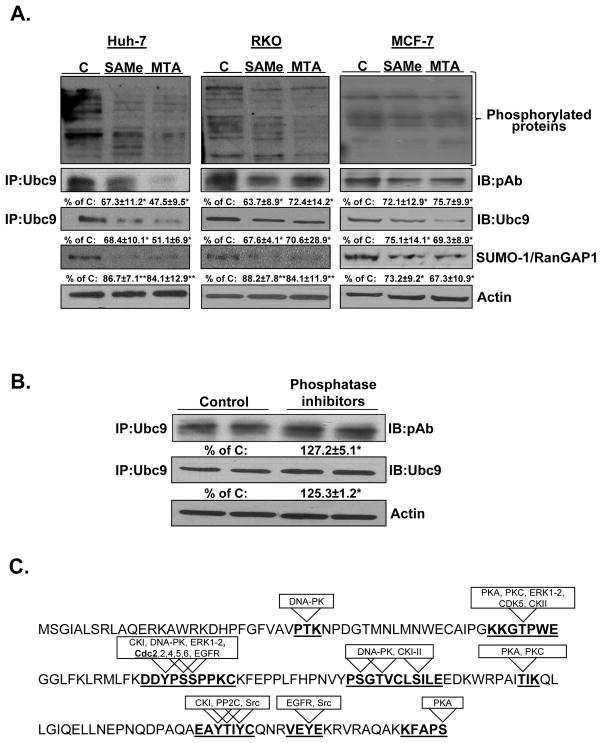
SAMe and MTA treatment alter Ubc9’s state of phosphorylation and sumoylation
Phosphorylation can alter protein stability (24). SAMe and MTA inhibited protein phosphorylation in HepG2 cells (23), which prompted us to examine whether they affect Ubc9 phosphorylation. Total protein phosphorylation was examined by Western blotting using anti-phospho antibody, while phospho-Ubc9 was determined by first immunoprecipitating total Ubc9, followed by Western blotting with anti-phospho antibody. SAMe and MTA treatment resulted in decreased levels of total phosphoproteins and phospho-Ubc9 in all cell lines (Figure 4A). Total Ubc9 levels decreased in parallel to phospho-Ubc9 levels. In addition, SAMe and MTA suppressed the level of sumoylated RanGAP1 (Figure 4A), a major SUMO substrate (25). To confirm that the phosphorylation of Ubc9 influences the protein stability, we examined the effect of phosphatase inhibitors on Ubc9 protein expression in RKO cells. Figure 4B shows that total Ubc9 and phospho-Ubc9 levels increased after treatment with phosphatase inhibitors, which is consistent with the notion that Ubc9 phosphorylation enhances its stability.
Fig. 4. SAMe and MTA treatment reduced Ubc9’s state of phosphorylation and sumoylation.
(A) Huh-7, RKO and MCF-7 cells were treated with SAMe (2mM) or MTA (1mM) for 24 hours. Total protein phosphorylation was examined by Western blotting using anti-phospho antibody, while phospho-Ubc9 was determined by first immunoprecipitating total Ubc9, followed by Western blotting with anti-phospho antibody. Sumoylation was measured using anti-SUMO-1 antibody in Western blotting. The values are expressed as % of control cells (mean ± SEM) from 3 independent experiments. *P<0.02, **p<0.05 vs. control cells. (B) RKO cells were cultured and treated with phosphatase inhibitor cocktail (1:10000) for 24 hours and processed for immunoprecipitation using anti-Ubc9 antibody followed by Western blotting with anti-phospho antibody or anti-Ubc9 antibody. Results are expressed as % of control (mean ± SEM) from 3 independent experiments. *P<0.02 vs. control cells. (C) Prediction of potential kinases that can phosphorylate Ubc9 protein sequence using the KinasePhos, PostMod, MotifScan, Phosphonet analysis tools.
Phosphorylation of Ubc9 by Cdc2/CyclinB complex in vitro assay
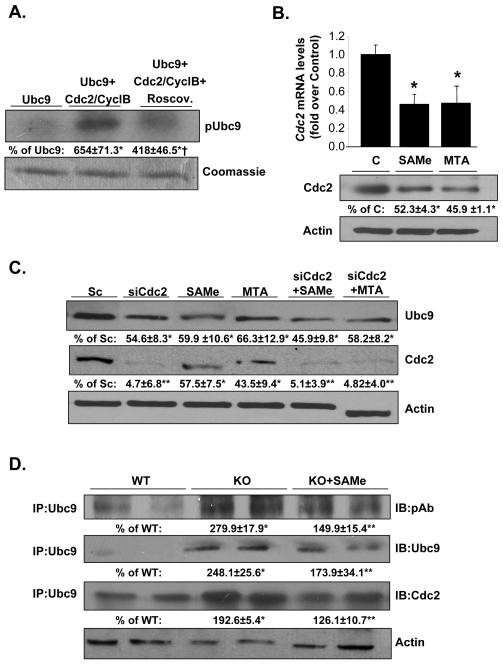
Using DISPHOS 1.3 program (Disorder-Enhanced Phosphorylation Sites Predictor), we identified two potential phosphorylated sites at Serine 70 and Serine 71. We searched for potential protein kinases that may target these sites using the KinasePhos, PostMod, MotifScan, Phosphonet analysis tools. These four prediction programs all identified several potential protein kinases (Figure 4C) such as Cdc2 and CDK5. Cdc2 shows a good match to the matrix with a similarity score of 0.8. Matrix scores lower than 0.8 were rejected. To test the possibility that Cdc2 can phosphorylate Ubc9, we carried out in vitro kinase assays using highly purified Ubc9 recombinant protein and Cdc2-Cyclin B complex in combination with roscovitine, a potent inhibitor of kinase activity. Figure 5A shows that Cdc2 is able to phosphorylate Ubc9 in vitro and this can be inhibited by roscovitine.
Fig. 5. Cdc2 phosphorylates Ubc9 in vitro, interacts with Ubc9 in vivo and its expression is inhibited by SAMe and MTA.
(A) Cdc2/Cyclin B phosphorylates recombinant Ubc9 in vitro. Results are expressed as % control (mean ± SEM) from 3 independent experiments. *P<0.001 vs. Ubc9, †p<0.05 vs. Ubc9+Cdc2/CyclB. (B) RKO cells were treated with SAMe and MTA for 24 hours. Ubc9 expression was examined by real-time PCR and Western blotting. Results are expressed as fold or % control (mean ± SEM) from 3 independent experiments. *P<0.001 vs. control cells. (C) Effect of Cdc2 RNAi, SAMe and MTA treatments on Ubc9 and Cdc2 protein levels. RKO cells were treated with scrambled or Cdc2 RNAi or with 2mM SAMe or 1mM MTA as described in Methods. Results are expressed as % of scrambled control (mean ± SEM) from 3 independent experiments. *P<0.01, **p<0.001 vs. scrambled control. (D) Ubc9 interacts with Cdc2 in vivo. Immunoprecipitation with anti-Ubc9 antibody followed by Western blotting for phospho-Ubc9, total Ubc9, and Cdc2 in Mat1a KO livers was done as described in Methods. Results are expressed as % of WT mice (mean ± SEM) from 4 mice per group. *P < 0.001 vs. WT; **P < 0.04 vs. KO.
Effect of SAMe and MTA on Cdc2 expression in vitro and in vivo
Cdc2 overexpression occurs at high frequency in several types of cancer (26). Since the above finding suggests Cdc2 may be involved in the phosphorylation of Ubc9, we next examine the effect of SAMe and MTA on Cdc2 expression. Both SAMe and MTA treatment resulted in a decrease of Cdc2 mRNA and protein levels after 24 hours in RKO cells by 50% (Figure 5B). To determine whether SAMe and MTA lower Ubc9 protein level by inhibiting Cdc2 expression, we used Cdc2 siRNA in combination with SAMe (2mM) and MTA (1mM) treatment in RKO cells. Knockdown of Cdc2 resulted in a 55% reduction in Ubc9 protein level (Figure 5C). SAMe and MTA have similar effects on Ubc9 protein level as Cdc2 knockdown, but combining SAMe or MTA with Cdc2 knockdown did not further reduce Ubc9 protein level (Figure 5C). To verify the relevance of Cdc2 on Ubc9 phosphorylation under in vivo conditions, we examined Mat1a KO livers. Figure 5D shows increased protein levels of total Ubc9, phospho-Ubc9 and Cdc2 that is associated with Ubc9 in Mat1a KO mice livers and these were nearly corrected by SAMe treatment. These results also demonstrate interaction of Cdc2 with Ubc9 in vivo that can be modulated by SAMe treatment.
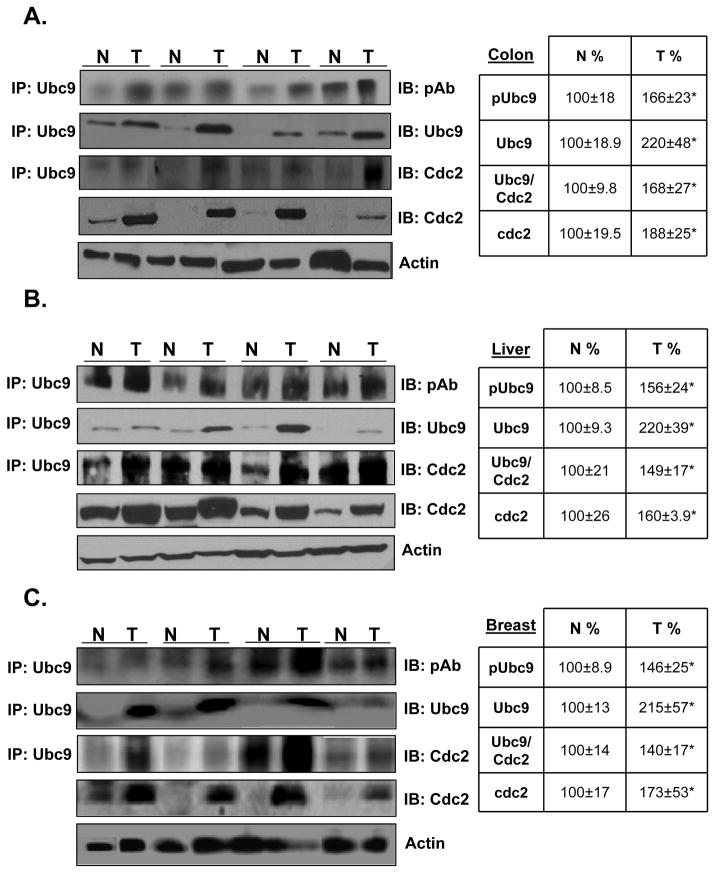
Phosphorylation of Ubc9 in human cancer specimens
Phosphorylation status of Ubc9 and interaction of Ubc9 with Cdc2 in human cancer specimens have not been reported; we next investigated this in 18 HCC, 12 colorectal cancer and 12 breast cancer and corresponding surrounding non-tumorous tissues by co-IP analysis. Figure 6A, 6B and 6C show that the levels of total Ubc9, phospho-Ubc9, Cdc2 and Cdc2 that is associated with Ubc9 are increased in all of these cancers as compared to the non-transformed tissues. These results suggest that Cdc2-mediated phosphorylation of Ubc9 may contribute to higher Ubc9 protein level in these cancers.
Fig. 6. Levels of phospho-Ubc9, total Ubc9, Cdc2 and Cdc2 that interacts with Ubc9 are increased in human colon, liver and breast cancers.
Representative Western Blots of colorectal (A), HCC (B), breast (C) cancer specimens (T) and corresponding non–tumorous (N) tissues are shown. The immunoprecipitation was done as described in Methods for phosphorylated, total Ubc9 and Cdc2. Results represent mean ± SEM from 12 colorectal, 12 HCC, and 12 breast cancers expressed as % of corresponding non-tumorous (N) tissues. *P<0.01 vs. non-tumorous tissue.
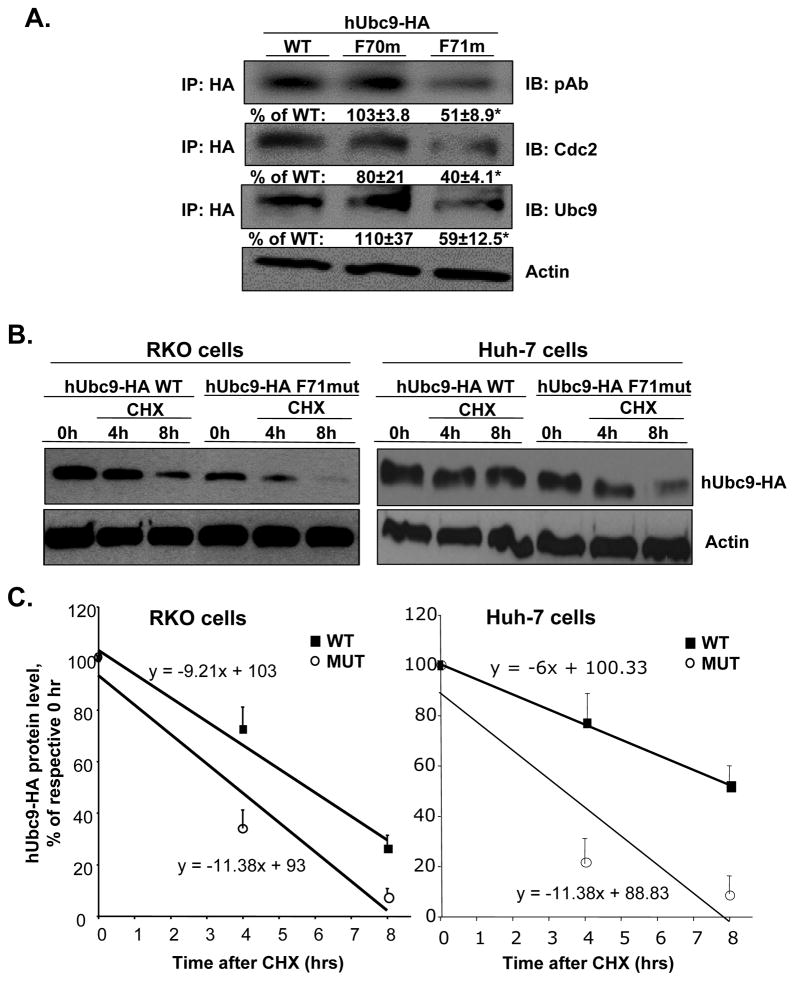
Serine 71 of Ubc9 is critical for interaction with Cdc2, phosphorylation of Ubc9 and Ubc9 protein stability in RKO cells
Finally, to examine whether serine 70 and serine 71 are critical sites of phosphorylation by Cdc2 in native Ubc9 protein and protein stability, we examined whether mutation of S70 and S71 (to F70 and F71) affected phosphorylation of Ubc9 in RKO cells. For this purpose, we created mutations in the overexpression vector hUbc9 HA-tagged at S70 and S71 and examined total Ubc9 protein and phospho-Ubc9 levels and Cdc2 interaction using co-IP analysis. Figure 7A shows that mutation at S71 (but not S70) reduced significantly interaction with Cdc2, phospho-Ubc9, and total Ubc9 levels. Mutation at S71 resulted in a less stable Ubc9 as demonstrated by measurement of protein half-life, which decreased from 5.8±0.5 hours in WT to 3.8±0.2 in mutant in RKO cells. Similarly, mutation at S71 also lowered Ubc9 half-life from 8.7±1.8 hours to 3.5±0.5 hours in Huh-7 cells (results are mean±SEM from three independent experiments for each cell type, p<0.05) (Fig. 7B and C).
Fig. 7. Mutation at Ubc9 S71 reduced interaction of Cdc2 with Ubc9 and Ubc9 protein stability.
(A) RKO cells were transfected with the vector, hUbc9-WT, hUbc9-F70mut, and hUbc9-F71mut for 48 hours. Co-IP analysis was done as described in Methods for phosphorylated, total Ubc9 and Cdc2. Results are expressed as % of WT (mean ± SEM) from 4 independent experiments. *P<0.02 vs. WT. (B) Ubc9 protein stability was determined in RKO and Huh-7 cells by Western blot analyses of HA tag following cycloheximide (CHX) treatment as described in Methods. Representative blots are shown. (C) Protein stability was determined using linear regression and half-life calculated using equation indicated. Results represent mean±SEM from 3 independent experiments expressed as % of respective 0 hr level for both cell types (p<0.05 between WT and MUT).
DISCUSSION
Ubc9 is overexpressed in several malignancies (8, 20). Suppression of Ubc9 function inhibits tumor growth in a breast cancer animal model (20), suggesting that Ubc9 plays a role in tumorigenesis. This is likely due to the fact that Ubc9 is an essential enzyme for sumoylation, and deregulation of Ubc9 could lead to alterations of sumoylation pathways, ultimately impacting cell growth and cancer development.
Ubc9 expression and sumoylation of a broad range of proteins is induced by oxidative stress (4). Oxidative stress is a key component in the induction of HCC (27). However, Ubc9 expression in HCC has not been reported. We found that Ubc9 expression is also increased in HCC (Fig. 1A), but interestingly the protein level is elevated to a higher extent than the mRNA level, suggesting Ubc9 is post-translationally regulated. Despite the importance of Ubc9 in sumoylation and tumorigenesis, factors that control its regulation remain largely unknown and in particular, post-translational regulation of Ubc9 has not been reported to our knowledge.
Two other models of increased oxidative stress and predisposition to malignant degeneration are the Mat1a KO mouse model and the ethanol infusion (EI) model of alcoholic liver disease (ALD). The EI model of ALD has been utilized to study various aspects to ALD pathogenesis (28). However, the effect of EI on Ubc9 expression and hence sumoylation are unknown. We found that hepatic Ubc9 expression is higher at both the mRNA and protein levels in EI mice, and the protein level is increased to a higher degree than the mRNA level (Fig. 1B).
Similar to HCC and ethanol fed mice, Ubc9 expression is increased in Mat1a KO livers, with the protein level increased to a higher degree than the mRNA level. We previously demonstrated that exogenous SAMe treatment normalized hepatic SAMe level in Mat1a KO mice (22), which prompted us to examine the effect of SAMe treatment in Mat1a KO mice on Ubc9 expression.
Interestingly, SAMe treatment normalized the Ubc9 protein level but had no influence on its mRNA level, which suggests SAMe de-stabilized Ubc9 protein. This is an intriguing finding because we had previously shown that SAMe can stabilize dual-specificity of MAPK phosphatase protein by inhibiting proteasomal activity (22). Thus, the effect of SAMe on Ubc9 seems to contradict this previous finding. This led us to explore mechanisms that might regulate Ubc9 protein stability besides proteasomal activity. Consistent with reduced Ubc9 expression, SAMe treatment in Mat1a KO mice reduced protein sumoylation in the livers (Fig. 2).
We next used cancer cell lines to further investigate the molecular mechanism of SAMe’s effect on Ubc9 protein stability. We chose two liver (HepG2 and Huh7), one colon (RKO) and one breast (MCF-7) cancer cell lines because Ubc9 is overexpressed in breast and colon cancers (8, 20). We included MTA in our studies because SAMe is converted to MTA spontaneously at a significant rate (13) and many of SAMe’s actions may actually be due to MTA (17, 23, 29). SAMe and MTA down-regulated Ubc9 protein levels, without any change in mRNA levels, and protein sumoylation in all cell lines (Fig. 3 and Fig. 4A).
We previously demonstrated that SAMe and MTA treatment in HepG2 cells led to increased the expression of protein phosphatase 1 (PP1) (23). Protein phosphorylation can either enhance or inhibit protein stability (24, 30). Treatment of Huh-7, RKO and MCF-7 cells with SAMe or MTA lowered overall protein phosphorylation, consistent with our previous results (Fig. 4A). We examined whether the effect of SAMe and MTA is via enhancing PP1 expression by knocking down PP1 prior to SAMe or MTA treatment. However, this had no effect on SAMe and MTA-mediated down-regulation of Ubc9 protein level (data not shown) so that the mechanism is not via PP1. Ubc9 contains multiple potential phosphorylation sites but it has not been reported to be phosphorylated. Interestingly, we found Ubc9 is phosphorylated at baseline and SAMe and MTA treatment lowered Ubc9 phosphorylation in parallel to its total protein level. The finding that inhibiting phosphatases raised total Ubc9 and phospho-Ubc9 levels further supports the notion that Ubc9 phosphorylation stabilizes Ubc9 protein.
Protein phosphorylation predominantly occurs in regions of intrinsic disorder (31). Accordingly, we identified two potential phosphorylated sites at Serine 70 and Serine 71 in Ubc9 protein sequence. This region contains the -P-X3-P-X2-P-P- motif that is conserved among all members of the Ubc protein family. Mutations occurring in this motif interfered with the in vivo activity of Ubc9 at high temperature and caused a reduction in the abundance of the mutated proteins in yeast (32). In this region we identified Cdc2 as a potential kinase to phosphorylate the S70-S71 motif. Interaction of Ubc9 with Cdc2 agrees with the matrix similarity score of 0.8, which suggests a high probability that Ubc9 is a Cdc2 substrate. Our in vitro kinase findings verified this and showed that Cdc2 phosphorylated Ubc9 recombinant protein and roscovitine, an inhibitor of Cdc2, was able to significantly reduce Cdc2-mediated phosphorylation (Fig. 5A). Cdc2 is implicated in the genesis and/or progression of breast cancers (26) and overexpressed in the majority of diffuse large B-cell lymphoma specimens (33). To investigate whether SAMe and MTA’s effect on Ubc9 protein may involve Cdc2, we examined the effect of SAMe and MTA on Cdc2 expression. Surprisingly, we found that Cdc2 expression decreased at both mRNA and protein levels after SAMe and MTA treatment. Furthermore, silencing Cdc2 reduced Ubc9 protein levels by a similar magnitude as SAMe and MTA treatments. Combining Cdc2 knockdown with either SAMe or MTA did not reduce Ubc9 protein level or phospho-Ubc9 further, suggesting that the inhibitory effect of SAMe and MTA on Ubc9 protein level is mediated largely via down-regulation of Cdc2 expression (Fig. 5C). Physical interaction of Ubc9 and Cdc2 was demonstrated in Mat1a KO livers and this was reduced by SAMe treatment (Fig. 5D). Taken together, our results support the notion that Cdc2 can interact with Ubc9 to phosphorylate it and increase its protein stability.
Since Ubc9 phosphorylation and interaction with Cdc2 in human cancers have not been reported, we next examined this in HCC, colorectal and breast cancer specimens. We found that the levels of total Ubc9, phospho-Ubc9, Cdc2 and Cdc2 that is associated with Ubc9 are all increased in these cancers. The degree of increase in total Ubc9 exceeds that of phospho-Ubc9 in cancer specimens, suggesting that other factors also contribute to a higher total Ubc9 protein expression. Consistent with this, Ubc9 has been shown to be a target of miR-30e, which is down-regulated in several tumors (6). Taken together, it is likely that higher level of Cdc2 that is associated with Ubc9 resulted in higher phospho-Ubc9 in these cancer specimens and this along with other contributing mechanism(s) resulted in higher Ubc9 expression.
To see if Cdc2 interaction and phosphorylation of Ubc9 occurs at S70–71 motif, we mutated these amino acids. Mutating S71 led to a reduction of Cdc2 binding to Ubc9, Ubc9 phosphorylation and total Ubc9 protein level. Furthermore, mutating S71 resulted in a less stable Ubc9 as demonstrated by protein stability assay. These results directly demonstrate that Cdc2 plays an important role as a positive regulator of Ubc9 expression in human carcinogenesis. They also demonstrate that post-translational regulation of Ubc9 by Cdc2 contributes to the higher Ubc9 protein than mRNA levels in HCC.
There are multiple SUMO proteins (SUMO-1, 2, 3 and 4) and since Ubc9 (SUMO E2) is required for sumoylation, we focused on Ubc9. This work is the first to demonstrate that sumoylation is increased when cellular SAMe level falls and decreased by exogenous treatment with SAMe or MTA. Interestingly, SUMO E1 enzyme is also elevated in HCC and correlates with lower survival rate (34). Whether SAMe and MTA also regulate SUMO proteins and SUMO E1 and E3 enzymes remain to be examined.
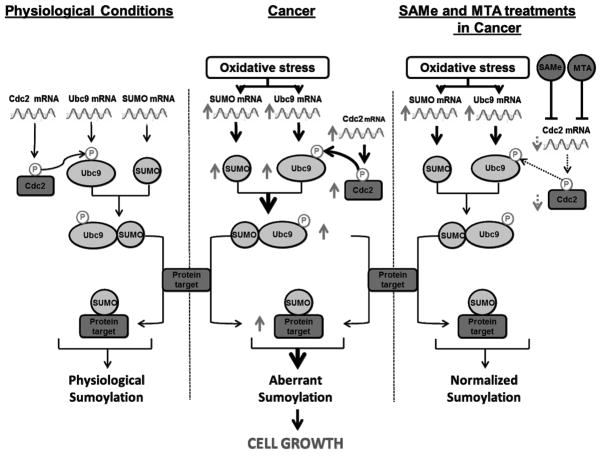
In summary, we have revealed a novel mechanism of Ubc9 post-translational regulation by Cdc2. We have also uncovered a novel mechanism of SAMe’s action. Exogenous treatment of liver, colon and breast cancer cells with SAMe and its metabolite MTA lowered Cdc2 expression, Ubc9 phosphorylation and total Ubc9 protein level. Figure 8 summarizes key findings from this work. SAMe has been shown to be chemopreventive in several animal models of HCC (35, 36). This may represent a new mechanism of that action. Given the importance of Ubc9 in sumoylation, our findings have important implications in biology and pathology. They also support SAMe and MTA as chemopreventive agents.
Fig. 8. Simplified schematic of key findings from this work.
Under physiological conditions, Cdc2 phosphorylates Ubc9 at a basal level. In cancer and conditions of increased oxidative stress (which occurs when hepatic SAMe level is reduced), expression of Ubc9 and Cdc2 is increased leading to higher phospho-Ubc9 level and increased Ubc9 stability, resulting in increased protein sumoylation. This pathway can be disrupted by SAMe or MTA treatment, which can inhibit Cdc2 at the mRNA level to lower Cdc2-mediated Ubc9 phosphorylation, resulting in reduced Ubc9 protein level and protein sumoylation.
Acknowledgments
Financial support:
This work was supported by NIH grants R01DK51719 (SC Lu), R01AT1576 (SC Lu and JM Mato), R01AT004896 (SC Lu and JM Mato), F32AA020150 (ML Tomasi), and Plan Nacional of I+D SAF 2011-29851 (JM Mato). HepG2, Huh-7, and RKO cells were provided by the Cell Culture Core of the USC Research Center for Liver Diseases (P30DK48522). Liver specimens from ethanol fed mice were provided by the Southern California Research Center for Alcoholic and Pancreatic Diseases & Cirrhosis (P50AA11999).
Abbreviations used (alphabetical order)
- ALD
alcoholic liver disease
- ATCC
American Type Cell Collection
- Cdc2
cell division cycle 2
- Co-IP
co-immunoprecipitation
- EI
ethanol infusion
- ERK
extracellular signal-regulated kinase
- GAPDH
glyceraldehydes 3-phosphate dehydrogenase
- HCC
hepatocellular carcinoma
- JNK
c-Jun-N-terminal kinase
- KO
knockout
- MAPK
mitogen activated protein kinase
- MAT
methionine adenosyltransferase
- MTA
methylthioadenosine
- NF-κB
nuclear factor kappa B
- PCR
polymerase chain reaction
- PI3K
phosphoinositide 3-kinase
- PP1
protein phosphatase 1
- SAMe
S-adenosylmethionine
- siRNA
small interfering RNA
- STAT3
signal transducer and activator of transcription 3
- SUMO
ubiquitin-related modifier
- Ub
ubiquitin
- Ubc9
ubiquitin-conjugating enzyme 9
- WT
wild type
References
- 1.Johnson ES. Protein modification by SUMO. Ann Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 2.Muller S, Hoege C, Pyrowalakis G, Jentsch S. SUMO, ubiquitin’s mysterious cousin. Nat Rev Mol Cell Biol. 2001;2:202–210. doi: 10.1038/35056591. [DOI] [PubMed] [Google Scholar]
- 3.Johnson ES, Blogel G. Ubc9 is the conjugating enzyme for the ubiquitin-like protein Smt3p. J Biol Chem. 1997;272:26799–26702. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- 4.Manza LL, Codreanu SG, Stamer SL, Smith DL, Wells KS, Roberts RL, et al. Global shifts in protein sumoylation in response to electrophile and oxidative stress. Chem Res Toxicol. 2004;17:1706–1715. doi: 10.1021/tx049767l. [DOI] [PubMed] [Google Scholar]
- 5.Romanenko AM, Kinoshita A, Wanibuchi H, Wei M, Zaparin WK, Vinnichenko WI, et al. Involvement of ubiquitination and sumoylation in bladder lesions induced by persistent long-term low dose ionizing radiation in humans. J Urol. 2006;175:739–743. doi: 10.1016/S0022-5347(05)00172-2. [DOI] [PubMed] [Google Scholar]
- 6.Wu F, Zhu S, Ding Y, Beck WT, Mo YY. MicroRNA-mediated regulation of Ubc9 expression in cancer cells. Clin Cancer Res. 2009;15:1550–1557. doi: 10.1158/1078-0432.CCR-08-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moschos SJ, Smith AP, Mandic M, Athanassiou C, Watson-Hurst K, Jukic DM, et al. SAGE and antibody array analysis of melanoma-infiltrated lymph nodes: identification of Ubc9 as an important molecule in advanced-stage melanoma. Oncogene. 2007;26:4216–4225. doi: 10.1038/sj.onc.1210216. [DOI] [PubMed] [Google Scholar]
- 8.Moschos SJ, Jukic DM, Athanassiou C, Bhargava R, Dacic S, Wang X, et al. Expression analysis of Ubc9, the single small ubiquitin-like modifier (SUMO) E2 conjugating enzyme, in normal and malignant tissues. Hum Pathol. 2010;41:1286–1298. doi: 10.1016/j.humpath.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 9.Tomasi ML, Iglesias–Ara A, Yang A, Ramani K, Feo F, Pascale MR, et al. S-adenosylmethionine regulates apurinic/apyrimidinic endonuclease 1 stability: implication in hepatocarcinogenesis. Gastroenterology. 2009;136:1025–1036. doi: 10.1053/j.gastro.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martínez-Chantar ML, Corrales FJ, Martínez-Cruz LA, Garcia-Trevijano ER, Huang ZZ, Chen L, et al. Spontaneous oxidative stress and liver tumors in mice lacking methionine adenosyltransferase 1A. FASEB J. 2002;16:1292–1294. doi: 10.1096/fj.02-0078fje. [DOI] [PubMed] [Google Scholar]
- 11.Mato JM, Lu SC. Role of S-adenosyl-L-methionine in liver health and injury. Hepatology. 2007;45:1306–1312. doi: 10.1002/hep.21650. [DOI] [PubMed] [Google Scholar]
- 12.Pegg AE. Polyamine metabolism and its importance in neoplastic growth and a target for chemotherapy. Cancer Res. 1988;15:759–774. [PubMed] [Google Scholar]
- 13.Chen H, Xia M, Lin M, Yang H, Kuhlenkamp J, Li T, et al. Role of methionine adenosyltransferase 2A and S-adenosylmethionine in mitogen-induced growth of human colon cancer cells. Gastroenterology. 2007;133:207–218. doi: 10.1053/j.gastro.2007.03.114. [DOI] [PubMed] [Google Scholar]
- 14.Lu SC, Alvarez L, Huang ZZ, Chen L, An W, Corrales FJ, et al. Methionine adenosyltransferase 1A knockout mice are predisposed to liver injury and exhibit increased expression of genes involved in proliferation. Proc Natl Acad Sci USA. 2001;98:5560–5565. doi: 10.1073/pnas.091016398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ji C, Kaplowitz N. Betaine decreases hyperhomocysteinemia, endoplasmic reticulum stress, and liver injury in alcohol-fed mice. Gastroenterology. 2003;124:1488–1499. doi: 10.1016/s0016-5085(03)00276-2. [DOI] [PubMed] [Google Scholar]
- 16.Yang H, Ara AI, Magilnick N, Xia M, Ramani K, Chen H, et al. Expression pattern, regulation, and functions of methionine adenosyltransferase 2beta splicing variants in hepatoma cells. Gastroenterology. 2008;134:281–291. doi: 10.1053/j.gastro.2007.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Veal N, Hsieh CL, Xiong S, Mato JM, Lu SC, Tsukamoto H. Inhibition of lipopolysaccharide-stimulated TNF-alpha promoter activity by S-adenosylmethionine and 5′-methylthioadenosine. Am J Physiol Gastrointest Liver Physiol. 2004;287:G352–G362. doi: 10.1152/ajpgi.00316.2003. [DOI] [PubMed] [Google Scholar]
- 18.Ramani K, Yang H, Xia M, Iglesias-Ara A, Mato JM, Lu SC. Leptin’s mitogenic effect in human liver cancer cells requires induction of both methionine adenosyltransferase 2A and 2beta. Hepatology. 2008;4:521–531. doi: 10.1002/hep.22064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pascale RM, Simile MM, De Miglio MR, Muroni MR, Calvasi DF, Asara G, et al. Cell Cycle deregulation in liver lesions of rats with and without genetic predisposition to hepatocarcinogenesis. Hepatology. 2002;35:1341–1350. doi: 10.1053/jhep.2002.33682. [DOI] [PubMed] [Google Scholar]
- 20.Mo YY, Yu Y, Theodosiou E, Ee PL, Beck WT. A role for Ubc9 in tumorigenesis. Oncogene. 2005;24:2677–2683. doi: 10.1038/sj.onc.1208210. [DOI] [PubMed] [Google Scholar]
- 21.Lu Y, Wu D, Wang X, Ward SC, Cedarbaum AI. Chronic alcohol-induced liver injury and oxidant stress are decreased in cytochrome P4502E1 knockout mice and restored in humanized cytochrome P4502E1 knock-in mice. Free Radic Biol Med. 2010;15:1406–1416. doi: 10.1016/j.freeradbiomed.2010.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomasi ML, Ramani K, Lopitz-Otsoa F, Rodriguez MS, Li TW, Ko K, et al. S-adenosylmethionine regulates dual-specificity mitogen-activated protein kinase phosphatase expression in mouse and human hepatocytes. Hepatology. 2010;51:2152–2161. doi: 10.1002/hep.23530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang H, Sadda MR, Li M, Zeng Y, Chen L, Bae W, et al. S-adenosylmethionine and its metabolite induce apoptosis in HepG2 cells: role of protein phosphatase 1 and Bcl-xs. Hepatology. 2004;40:221–231. doi: 10.1002/hep.20274. [DOI] [PubMed] [Google Scholar]
- 24.Wang SC, Nakajima Y, Yu YL, Chen CT, Yang CC, McIntush EW, et al. Tyrosine phosphorylation controls PCNA function through protein stability. Nature Cell Biology. 2006;8:1359–1368. doi: 10.1038/ncb1501. [DOI] [PubMed] [Google Scholar]
- 25.Mahajan R, Gerace L, Melchior F. Molecular characterization of the SUMO-1 modification of RanGAP1 and its role in nuclear envelope association. J Cell Biol. 1998;140:259–270. doi: 10.1083/jcb.140.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chae SW, Sohn JH, Kim DH, Choi YJ, Park YL, Kim K, et al. Overexpressions of Cyclin B1, cdc2, p16 and p53 in human breast cancer: the clinicopathologic correlations and prognostic implications. Yonsei Med J. 2011;52:445–453. doi: 10.3349/ymj.2011.52.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caraglia M, Giuberti G, Marra M, Addeo R, Montella L, Murolo M, et al. Oxidative stress and ERK1/2 phosphorylation as predictors of outcome in hepatocellular carcinoma patients treated with sorafenib plus octreotide LAR. Cell Death and Disease. 2011;2(4):e150. doi: 10.1038/cddis.2011.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.French SW. Intragastric ethanol infusion model for cellular and molecular studies of alcoholic liver disease. J Biomed Sci. 2001;8:20–27. doi: 10.1007/BF02255967. [DOI] [PubMed] [Google Scholar]
- 29.Ara AI, Xia M, Ramani K, Mato JM, Lu SC. S-adenosylmethionine inhibits lipopolysaccharide-induced gene expression via modulation of histone methylation. Hepatology. 2008;47:1655–1666. doi: 10.1002/hep.22231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Westermarck J. Regulation of transcription factor function by targeted protein degradation: an overview focusing on p53, c-Myc, and c-Jun. Methods Mol Biol. 2010;64:31–36. doi: 10.1007/978-1-60761-738-9_2. [DOI] [PubMed] [Google Scholar]
- 31.Iakoucheva LM, Radivojac P, Brown CJ, O’Conner TR, Sikes JG, Obradovic Z, et al. The importance of intrinsic disorder for protein phosphorylation. Nucl Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Betting J, Seufert W. A yeast Ubc9 mutant protein with temperature-sensitive in vivo function is subject to conditional proteolysis by a ubiquitin-and proteasome-dependent pathway. J Biol Chem. 1996;271:25790–25796. doi: 10.1074/jbc.271.42.25790. [DOI] [PubMed] [Google Scholar]
- 33.Zhao XF, Gartenhaus RB. Phospho-p70S6K and cdc2/cdk1 as therapeutic targets for diffuse large B-cell lymphoma. Expert Opin Ther Targets. 2009;13:1085–1093. doi: 10.1517/14728220903103833. [DOI] [PubMed] [Google Scholar]
- 34.Sarge KD, Park-Sarge OK. Sumoylation and human disease pathogenesis. Trends Biochem Sci. 2009;34:200–205. doi: 10.1016/j.tibs.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu SC, Ramani K, Ou X, Lin M, Yu V, Ko K, et al. S-adenosylmethionine in the chemoprevention and treatment of hepatocellular carcinoma in a rat model. Hepatology. 2009;50:462–471. doi: 10.1002/hep.22990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pascale RM, Simile MM, De Miglio MR, Nufris A, Daino L, Seddaiu MA, et al. Chemoprevention by S-adenosyl-L-methionine of rat liver carcinogenesis initiated by 1,2-dimethylhydrazine and promoted by orotic acid. Carcinogenesis. 1995;16:427–430. doi: 10.1093/carcin/16.2.427. [DOI] [PubMed] [Google Scholar]