Abstract
During the last two decades, the importance of human genome copy number variation (CNV) in disease has become widely recognized. However, much is not understood about underlying mechanisms. We show how, although model organism research guides molecular understanding, important insights are gained from study of the wealth of information available in the clinic. We describe progress in explaining nonallelic homologous recombination (NAHR), a major cause of copy number change occurring when control of allelic recombination fails, highlight the growing importance of replicative mechanisms to explain complex events, and describe progress in understanding extreme chromosome reorganization (chromothripsis). Both non-homologous end-joining and aberrant replication have significant roles in chromothripsis. As we study CNV, the processes underlying human genome evolution are revealed.
Keywords: NAHR, FoSTeS, MMBIR, ectopic synapsis, PRDM9, triplication, chromothripsis
Introduction
Genome instability contributes tremendously to mutational processes during human genome evolution [1], in association with human diseases [2] and manifesting as polymorphic variation in populations [3–5]. Recent knowledge gained through studies of genomic disorders [2,6] and the development and clinical implementation of genome-wide assays for copy number variation (CNV) detection [7,8] enabled large-scale fine mapping and nucleotide level ascertainment of rearrangement breakpoint junctions in human populations, providing an invaluable resource with which to study rearrangement mechanisms. Several major mechanisms have been proposed for human genome rearrangements and these include nonallelic homologous recombination (NAHR), nonhomologous end-joining (NHEJ), replicative mechanisms, and long interspersed element (LINE) - mediated retrotransposition or mobile element insertions (MEIs). Of these, NAHR and replicative mechanisms have figured prominently in explaining a wide variety of germline and somatic rearrangement events.
NAHR results in recurrent rearrangements, i.e. rearrangements that include the same genomic interval occurring in unrelated individuals. Such recurrence is mediated by a common genomic structure or architecture in which the rearranged interval is flanked by paralogous repeat sequences or low-copy repeats (LCRs, also known as segmental duplications [9]). Since the NAHR model was proposed ten years ago [10], dozens of NAHR-mediated genomic disorders have been documented. Replication mechanisms, on the other hand, are a major contributor to nonrecurrent genomic rearrangements wherein the rearrangement size, genomic extent, and breakpoint position at a genetic locus can differ amongst unrelated subjects. Although replication mechanisms can explain both simple (usually a single deletion, duplication, inversion or translocation) and complex (a combination of more than one simple event) nonrecurrent rearrangements, perhaps their major contribution has been to provide a parsimonious explanation for complex human genomic rearrangements that cannot be readily explained by alternative mechanisms such as NHEJ, because of their characteristics.
Here we provide an overview of insights accumulated and knowledge gained from recent studies of both recurrent and complex rearrangements, summarize known recurrent genomic disorders, and highlight lessons learnt regarding NAHR and replication mechanisms.
NAHR mediates recurrent CNVs and chromosomal rearrangements
Nonallelic homologous recombination, or ectopic recombination, was one of the earliest mechanisms identified to be responsible for genomic disorders [2,6,10]. Recombination between paralogous LCRs in direct orientation can result in deletions and duplications, often occurring as de novo mutations and associated with recurrent sporadic genomic disorders, whereas inverted repeats can mediate inversions (Figure 1A). The NAHR mechanism favors deletions over duplications, because deletions can result from crossovers both in cis and in trans, whereas duplications can only result from crossovers in trans (Figure 1B). Turner et al. measured germline rates of de novo deletions and duplications directly at three autosomal and one Y chromosome loci and showed that, at least in male meiosis, deletions are observed to occur approximately twice as frequently as duplications on autosomes [11].
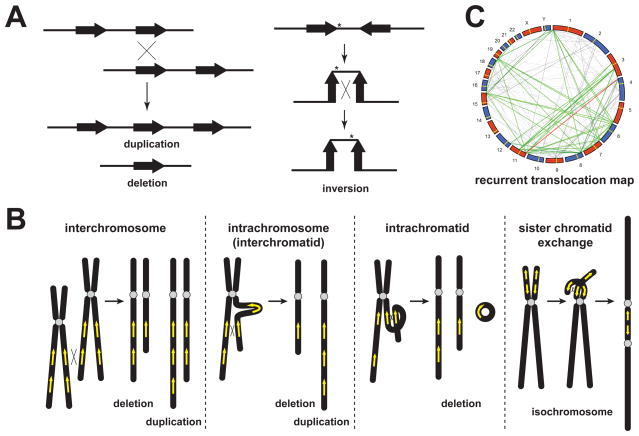
Figure 1.
NAHR as the mechanism for recurrent genomic rearrangements. (A) Ectopic crossing-over between directly oriented repeats in trans can lead to deletion and reciprocal duplication; whereas ectopic crossing-over between inversely oriented repeats in cis can result in an inversion. (B) NAHR can produce deletion or duplication in three ways, interchromosomal crossover, intrachromosomal (or interchromatidal) crossover, and intrachromatidal crossover. Note that intrachromatidal recombination can only produce deletion, not duplication. NAHR between inverted LCRs on sister chromatids can also result in isochromosome formation. (C) This panel is adapted from Ou et al [12] showing a genome-wide recurrent translocation map mediated by NAHR. In this circularized genome-wide view, LCRs fulfilling the criteria for recurrent translocations are connected by lines. The red line denotes recurrent translocations that are observed and experimentally verified, whereas the grey and green lines (olfactory receptor factor gene families) denote predicted recurrent translocations that could be mediated by paralogous LCRs.
If crossing-over occurs between LCRs in nonhomologous chromosomes, recurrent translocation may be produced (Figure 1C). Ou et al. demonstrated experimentally that NAHR mediates constitutional recurrent translocations t(4;11) and t(8;12) [12]. They also predicted computationally that 1143 LCR pairs fulfill empirically derived criteria (LCR pairs of > 5 kb in length, > 94% identity, proper substrate orientation dependent on centromere position) to mediate recurrent translocations [12].
NAHR can also act on paralogous LCR substrates located on sister chromatids leading to isochromosome formation (Figure 1B), i.e. a structurally abnormal chromosome with one arm partially deleted and the other arm duplicated. Such NAHR-generated isochromosomes are isodicentric (idic) and are observed as somatic events (usually in tumors) first described for i(17q) [13] or as constitutional events including i(Xq) [14] and idic(Y) [15].
With knowledge of human genome-wide LCR architecture, one can potentially predict regions prone to NAHR. Sharp et al. analyzed the NCBI34/hg16 build of the human reference genome and identified 130 regions of hypothesized genomic instability (flanked by LCR pairs of > 10 kb in length, > 95% identity, with intervening sequence between 50 kb and 10 Mb), spanning 274 Mb of non-redundant sequence that could predispose to interstitial deletion/duplication [16]. Since then, many of the predicted regions have been reported to be associated with genomic disorders. We now construct an updated map of genome-wide NAHR-prone regions using the aforementioned parameters plus one additional requirement that the predicted region should not span centromeres. LCR data were downloaded from the Segmental_Dup track in the UCSC genome browser (CRCh37/hg19). A total of 89 merged, non-redundant regions were identified, spanning ~189 Mb (Figure 2). Until now, 37 genomic regions have been reported to be associated with genomic disorders caused by NAHR (Figure 2). The majority of these regions (32/37) are predicted by this computational NAHR map. Amongst the most recent, is a chromosome 12 locus that deletes DPY19L2 and causes male infertility due to globozoospermia [17].
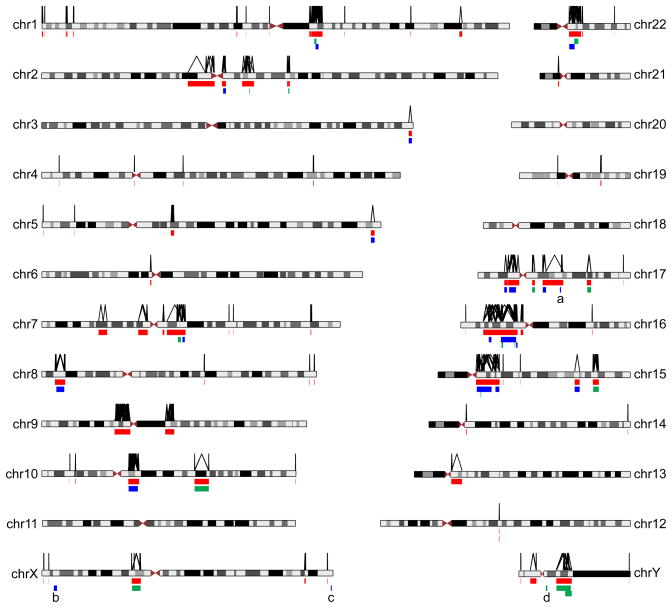
Figure 2.
Genome-wide map of computationally predicted NAHR-prone regions and empirically verified NAHR-associated disease regions. LCR pairs fulfilling chosen criteria (> 10 kb in length, > 95% in identity, directly oriented, with intervening sequence between 50 kb and 10 Mb, not spanning the centromere) are considered as potential substrates for NAHR. They are linked by an inverted V-shaped line as illustrated above the chromosome ideograms. Genomic regions flanked by such lines are merged into non-redundant sites, and illustrated as the 89 red bars below the ideograms. Regions of known genomic disorders are shown as green (only deletion associated with disease) or blue (both deletion and duplication associated with disease) bars. a, the 17q21.31 rearrangement occurs on an alternative haplotype [73]. b, the Xp22.31 rearrangement was not predicted by the NAHR map because the flanking LCR substrate is ~ 9 kb in length (< 10 kb) [26,74]. c, the Xq28 rearrangement was not predicted by the NAHR map because the flanking LCR is ~ 9.5 kb in length (< 10 kb) [75]. d, the rearrangement involving AZFa [76] was not predicted by the NAHR map because the deletion is mediated by a pair of HERV repetitive elements.
Ectopic synapsis – a prerequisite for NAHR?
Despite the fact that the phenomenon of NAHR in association with genomic disorders has been defined for twenty years [18,19], limited efforts have focused on understanding why crossing-over can occur at ectopic positions. Studies exploring the frequencies of different types of recurrent de novo deletions and duplications at 17p11.2 revealed that NAHR frequency is highly correlated with length of flanking repeats and influenced by the distance between repeats [20]. The length of flanking LCR (hundreds of kilobases) is much greater than the length of homology required to implement a homologous recombination event (hundreds of bp); i.e. the so-called Minimum Efficient Processing Segment or MEPS [21,22]. Thus, such an observed length dependence correlation with crossover frequency suggests that pairing of the flanking repeats may be a novel rate-limiting step prior to ectopic crossing-over. If NAHR predominantly occurs in meiosis, this pairing may be achieved by a faulty synapsis between nonallelic sequences, or ectopic synapsis [20]. One prediction of the ectopic synapsis model is that mutations in proteins required for synapsis might affect the rates of ectopic crossing-over or NAHR. Interestingly, recent research from yeast showed that deletion of a key component of the synaptonemal complex almost completely abolishes ectopic crossing-over (M Shinohara et al., personal communication), reinforcing the idea that ectopic crossing-over depends on synapsis. If ectopic synapsis is also required for NAHR events at genomic loci other than 17p11.2, investigations to pinpoint specific regions prone to abnormal synapsis, or specific factors involved in this process may further improve prediction of NAHR hotspots.
PRDM9 as a global regulator contributing to NAHR hotspot specification
Evidence suggests that specific nucleotide sequence level features may have a role in stimulating homologous recombination, i.e. recombination hotspots may be facilitated by DNA sequence motifs in cis. When examining historical recombination sites in HapMap samples, Myers et al. identified a degenerate 13-mer motif (5′-CCNCCNTNNCCNC-3′) that is crucial for recruiting crossovers in 40% of all human hotspots [23]. This cis-acting sequence motif is potentially analogous to an E. coli chi sequence [24]. This human HR hotspot motif, although identified from allelic recombination data, is also observed in proximity to crossover regions in NAHR products [25,26]; it has been well established that, as might have been anticipated [27], NAHR and allelic homologous recombination (AHR) hotspots coincide [28–30]. Following the discovery of this motif, three groups, using evidence from human and mouse experiments, independently reported that the motif is likely the binding site of a zinc-finger protein, H3K4 trimethylase, PR domain-containing protein 9; PRDM9 [31–33]. PRDM9 may be a major trans determinant of meiotic recombination hotspots. Berg et al. reported that variation in the zinc-finger repeat array in PRDM9 can strongly affect meiotic recombination hotspot activity, minisatellite instability and genomic instability resulting from NAHR [34,35]. These findings suggest that variation in PRDM9 may be one of the major regulators of certain human pathological genomic rearrangements. Intriguingly, the DNA sequence motif that encodes the hotspot binding element of PRDM9 is itself a minisatellite sequence; such DNA sequences are highly variable, a property that led to DNA fingerprinting and a human personal identification revolution [36].
Recurrent and complex triplications in the human genome
Improved CNV detection methods and lower cost, higher resolution arrays in clinical screening assays are unveiling an increasing number of triplications associated with disease phenotypes. Like duplications, triplications can be recurrent or nonrecurrent and may fall into two general structural categories (Figure 3A). The first category, which we designate type I, is represented by a recurrent structure of three copies of genomic segments in tandem, each with a head to tail orientation, separated and flanked by LCRs (three copies of the identical genomic interval that is triplicated and four copies of the flanking LCR on the rearranged chromosome in total). The general features of the type I triplication structure, tandem triplication due to unequal crossing-over, were outlined and proposed 75 years ago to explain the ‘double Bar’-phenotype studied by Calvin Bridges [37], decades before DNA was elucidated. The molecular details of this structure were reported recently in subjects with Xp22.31 triplications, potentially formed by a two-step NAHR event as evidenced by the double crossover mapped at breakpoints [26]. The second category (type II) consists of a triplicated segment inserted in an inverted orientation between two copies of the duplicated segments: i.e. DUP-TRP/INV-DUP. This DUP-TRP/INV-DUP configuration is found to contribute to most of the triplications observed at the MECP2 and PLP1 loci reported thus far [38]. By inverting a segment of the genome, breakpoints can be brought within spatial proximity and a remarkably complex structure is generated with only two breakpoint junctions. Furthermore, novel gene functions can be created by breakpoint junctions due to the fusions generated and the involvement of the reverse complement DNA strand in the rearrangement. A common genomic architecture, wherein a pair of inverted LCRs flanks one of the genomic intervals subsequently duplicated, is proposed to underlie formation of the complex type II triplications by both a homology-driven step (between inverted repeat substrates) and a second nonhomologous or microhomologous step [38]. This simple configuration of inverted repeats, frequently observed throughout the human genome, is proposed to predispose other genomic regions to formation of the complex type II triplication structure.
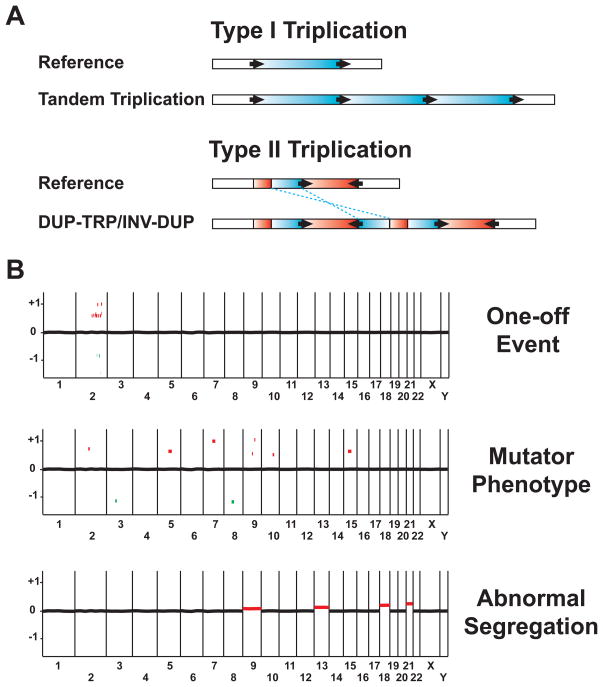
Figure 3.
Triplications and genome-scale complex rearrangements. (A) Two types of structures of triplications; the reference structure is illustrated above the novel structure formed upon triplication below. Red or blue horizontal bars represent regions that are duplicated or triplicated in the novel structure, respectively. Black arrows denote LCRs involved in this process with the orientation indicated by the arrowhead. Note that in type II triplication, the triplicated segment in the middle is inserted in an inverted orientation with respect to the flanking regions. The inversion is indicated by blue dashed lines. (B) Three distinct types of genome-level complex rearrangements. Shown in the figure are three hypothetical array CGH genome-view results representing three types of complex rearrangements. The labels on the X-axis denote chromosome numbers. The numbers on the Y-axis denote the log2 fluorescence intensity ratio of the hypothetical aCGH results. Red dots represent de novo copy number gains; green dots represent de novo copy number losses. Black dots at zero on the X-axis represent hybridizing oligonucleotide signals with no copy number change.
Replication mechanisms and complex rearrangements
Complex genomic rearrangements (CGR) are those that consist of more than one simple rearrangement, and have two or more breakpoint junctions. The phenomenon was initially reported when complex patterns of duplications (showing discontinuities, mixed with deletions, triplications or inversions, etc.) were identified from studying nonrecurrent duplications at the PLP1 locus [39]. Features including multiple copy number changes, evidence for long-distance template switching, insertion of short sequences at breakpoints apparently ‘templated’ from nearby genomic intervals, and microhomology at the breakpoint junctions all were consistent with such rearrangements being generated by a DNA replication mechanism. The mechanism describing this phenomenon was termed Fork Stalling and Template Switching or FoSTeS [39,40]. Based on experimental observations from bacteria, yeast and human studies, microhomology-mediated break-induced replication, or MMBIR, was proposed to explain the molecular details that contribute to the origin of human copy number variation [40,41]. This model expanded on the lessons learnt from break-induced replication or BIR [42–45] and even earlier work first elucidated in the bacteriophage T4 system [46]. Other similar replication models, such as microhomology-mediated replication-dependent recombination (MMRDR), have also been proposed [47].
Microhomology is a characteristic trace left at the breakpoint by the proposed replicative repair mechanisms. Breakpoint sequencing of both locus-specific rearrangements and genome-wide benign or pathological CNV revealed that a substantial proportion of CNV events showed microhomology (Table 1). Other experimental evidence to support replicative mechanisms comes from studies on the effect of replication stress and replication timing on de novo structural variation formation. Arlt et al. showed that treatment of human cells with aphidicolin or hydroxyurea, two chemicals that inhibit DNA replication through distinct mechanisms, resulted in increased CNV formation. Such ‘induced-CNVs’ have breakpoint and other features that mimic those observed in association with polymorphic and pathogenic CNV formation [48,49]. It has recently been shown that end points of somatic CNVs tend to be in three-dimensional spatial proximity, and they tend to replicate simultaneously [50,51]. All of these findings indicate that replicative mechanisms contribute to human genomic rearrangements far more than anticipated. Recent work suggests that such ‘replicative repair’ might be ‘mutagenic’ beyond CNV mutagenesis as it can be associated with a 1000 fold increase of point mutations [52]. Such an increase in point mutations stems from a low fidelity replication during BIR in comparison to a ‘normal’ DNA replication.
Table 1.
Microhomology at sequenced breakpoint junctions in the human genome.
| Number of rearrangements with breakpoints sequenced | Number of rearrangements with breakpoint microhomology | Microhomology length range* | Reference | ||
|---|---|---|---|---|---|
| Locus specific studies | PLP1 (Xq22.2) | 19 | 15 (79%) | 2–18 | [39,68,69] |
| LIS1 (17p13.3) | 6 | 6 (100%) | 2–27 | [54] | |
| RAI1, PMP22 (17p11.2p12) | 36 | 26 (72%) | 2–33 | [20,25,55,56] | |
| STS (Xp22.31) | 13 | 10 (77%) | 2–4 | [26] | |
| Genome- wide studies | Vissers et al. | 38 | 29 (76%) | 2–30 | [70] |
| Conrad et al.# | 324 | 168 (52%) | 2–30 | [71] | |
| Kidd et al.# | 973 | 289 (30%) | 2–20 | [72] | |
| Mills et al.# | 10871 | 7166 (66%) | 2–50 | [3] |
, Microhomology is arbitrarily defined as a length of perfect matching sequence of 2 – 50 bp. Kidd et al. used a range of 2 – 20 bp as the length of microhomology.
, Different approaches and methods, such as next generation sequencing filters and matching to a consensus haploid human genome reference, might introduce bias to the interpretation of some studies.
The locus specific studies and the Vissers et al. study analyzed nonrecurrent rearrangements only, whereas the remaining genome-wide studies did not specifically focus on nonrecurrent rearrangements, and also these studies appear to be biased towards deletion events.
Although replication mechanisms have been used to explain a wide range of complex rearrangements, especially copy number gains, on the X chromosome [26,53] and autosomes [54–56], nonhomologous mechanisms, such as multiple NHEJ, may still account for some portion of complex rearrangements [57].
Chromosome catastrophe: chromosome shattering or replication collapse?
How can the complexity of rearrangements increase in both range and scale, to a chromosome-wide level? Such highly complex genomic rearrangements were revealed in cancer samples by next-generation sequencing techniques [58,59]. However, the fact that the observed complexity in cancers apparently consists of a mixture of progressively altering rearrangements hindered further characterization of rearrangement mechanisms.
Whole genome sequencing of hundreds of cancer genomes has unexpectedly revealed that massive cancer-associated genomic rearrangements can occur in a very short time interval as a ‘one-off’ event. This novel phenomenon, termed chromothripsis, was reported to occur in 2–3% of the cancers studied [60]. Strikingly, multiple cancer genes can be disrupted by a single complex genomic rearrangement in chromothripsis, thus representing multigenic events. Although it was neither clear what causes such chromosome catastrophes nor what repair mechanisms might be operative, Stephens et al. proposed that perhaps chromosome shattering and re-ligation by NHEJ were involved [60]. Recent evidence has suggested potential involvement of TP53 in generating such complexities [61].
In parallel with the elucidation of the phenomenon of chromothripsis in association with many cancers, a similar chromosome catastrophe phenomenon was reported to occur as a constitutional event [62]. Using high-density oligonucleotide array comparative genomic hybridization, chromosome analysis, and breakpoint-sequencing methods, Liu et al. identified region-focused, multiple copy number change, highly complex rearrangement events occurring de novo in several patients with developmental disabilities [63]. Several characteristic features of the rearrangements indicate that the observed constitutional chromosome catastrophe was likely generated by a replication mechanism involving multiple template switches such as FoSTeS or MMBIR. First, rearranged genomic regions are localized to a single chromosome or confined to a chromosome arm, rather than scattered throughout the genome, suggesting that the affected region may be the site where a replication fork collapse occurred. Second, multiple copy number and structural changes are observed, including deletion, duplication, triplication, inversion and insertion, which can be explained by multiple strand switches during MMBIR [41]. In particular, the presence of a triplication amongst the complexity can be readily explained by MMBIR, not by NHEJ as generation of new DNA material is involved. Third, junction clustering is observed, i.e. chromosomal segments bounded by widely scattered breakpoints are joined together in a single complex arrangement, leaving their original loci untouched [63]. The interrelatedness of multiple breakpoints suggests a simple underlying mechanism MMBIR, instead of NHEJ occurring multiple times. Fourth, small templated insertions and microhomologies are frequently found at breakpoint junctions. These distinctive features or patterns suggest that the increases in copy number (duplication and triplication) observed in the subjects may be generated by the MMBIR repair.
In contrast, Kloosterman et al. using next-generation sequencing techniques identified a single case with a germline chromosome catastrophe showing extensive translocations and inversions [62]. A key feature from this complex rearrangement is that breakpoints found in different junctions can be paired with each other to reconstitute sites where a double-strand break is proposed to have occurred. The same phenomenon has been described in highly complex events in prostate cancer: multiple endpoints relate together as though they arose from two-ended double-strand breaks [64], in contrast to the one-ended double-strand breaks that result from replication fork collapse and are repaired by BIR or MMBIR. Such two-ended double-strand breaks would necessarily be repaired by NHEJ or by homologous recombination. Thus, there appear to be at least two distinct mechanisms underlying chromothripsis-like complex events. A possible explanation for the extreme rearrangement of a few chromosomes while others remain unaltered is provided by observations on events occurring in micronuclei [65]. Micronuclei are formed when some chromosomes lag at anaphase and are excluded from the nucleus. Chromosomes are very unstable in micronuclei. The altered chromosomes can later be reincorporated into the nucleus which has not experienced instability [65].
Genome-level complex rearrangements
Investigations into constitutional complex rearrangements stimulate ideas regarding potential mechanisms beyond locus specific events and allow for genome - scale complexity. In addition to the region-focused one-off event illustrated by chromosome catastrophes discussed in the previous section, two other hypothetical types of genome-level complex rearrangements may exist (Figure 3B). In a potential ‘mutator phenotype’ model, perhaps analogous to the microsatellite instability that accompanies mutation in mismatch repair genes, multiple rearrangements might occur scattered throughout the genome. In contrast to the one-off event, these rearrangements might arise independently from each other via individual events. Such a rearrangement phenotype may be caused by a stochastic environmental stimulus or a genetic impairment in key processes related to CNV formation. Similar phenomena have been observed in genetic syndromes, including Bloom syndrome, Fanconi anemia and ataxia telangiectasis, which are caused by mutations in DNA damage and repair pathways. However, the multiple de novo CNV model may reflect mutations in proteins involved in novel replication pathways related to CNV formation, such as the human ortholog of yeast Pol32 [66]. Exploring the primary genetic lesion/mutation in this type of rearrangements may allow identification of genes important for DNA repair, recombination, or replication. In an ‘abnormal segregation’ model (Figure 3B), genomic defects are represented by multiple mosaic aneuploidies. The cause of such an outcome could be mutations in genes important for chromosome segregation.
Conclusion
In summary, recent experimental findings from studies of disease associated recurrent and complex rearrangements reveal further insights into NAHR and DNA replication mechanisms for generating DNA rearrangements. NAHR is a well-established mechanism that explains and predicts a growing number of recurrent genomic disorders and even selected recurrent chromosomal rearrangements in association with both genomic disorders and cancers. Recent research unveils factors required for facilitating NAHR, pointing to a possibility of improved prediction of NAHR hotspots in the human genome, or even in personal genomes. Whilst a role for the genomic architecture of direct repeats is now firmly established in the NAHR mechanism, the importance of inverted repeats in susceptibility to human genomic rearrangements other than inversions [67] is only beginning to be unveiled [38]. Replication mechanisms are demonstrated to provide a parsimonious explanation for a wide range of rearrangement types, particularly complex genomic rearrangements. However, it is becoming clear that multiple mechanisms are operating. Understanding these mechanisms may provide insights into not only human genome evolution, genomic disorders, and cancers, but potentially various other biological processes.
Acknowledgments
We thank Dr. Pawel Stankiewicz and Claudia Gonzaga-Jauregui for help in making the figures, Dr. Akira Shinohara for sharing his unpublished data regarding ectopic crossing-over, and Dr. Neil Hunter for helpful discussions. This work was supported in part by the National Institute of Neurological Disorders and Stroke (National Institutes of Health) grant R01NS058529 to J.R.L., Texas Children’s Hospital General Clinical Research Center grant M01RR00188, and Intellectual and Developmental Disabilities Research Centers grant P30HD024064. J.R.L. is a consultant for Athena Diagnostics, holds stock ownership of 23andMe and Ion Torrent Systems, and is a coinventor on multiple United States and European patents for DNA diagnostics. The Baylor College of Medicine and Department of Molecular and Human Genetics derive revenue from molecular genetics testing clinical services provided by the Medical Genetics Laboratories; MGL, https://www.bcm.edu/geneticlabs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
*Papers of special interest
**Papers of outstanding interest
- 1.Carvalho CM, Zhang F, Lupski JR. Evolution in health and medicine Sackler colloquium: Genomic disorders: a window into human gene and genome evolution. Proc Natl Acad Sci U S A. 2010;107 (Suppl 1):1765–1771. doi: 10.1073/pnas.0906222107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lupski JR. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–422. doi: 10.1016/s0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- *3.Mills RE, Walter K, Stewart C, Handsaker RE, Chen K, Alkan C, Abyzov A, Yoon SC, Ye K, Cheetham RK, et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. This study summarized structural variation findings in 185 individuals from the 1000 Genome Project pilot study. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *4.Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. This study provided a comprehensive map of human genome CNV. They used high resolution array CGH to discover CNV, genotyped 4978 CNVs in different populations and assessed their functional impacts. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *5.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. Constructed a first-generation human genome CNV map from 270 HapMap individuals using SNP genotyping array and clone-based CGH. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *6.Lupski JR. Genomic disorders ten years on. Genome Med. 2009;1:42. doi: 10.1186/gm42. Provided an historical perspective and reviewed lessons learned from studying genomic disorders since the term was defined ten years earlier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung SW, Shaw CA, Yu W, Li J, Ou Z, Patel A, Yatsenko SA, Cooper ML, Furman P, Stankiewicz P, et al. Development and validation of a CGH microarray for clinical cytogenetic diagnosis. Genet Med. 2005;7:422–432. doi: 10.1097/01.gim.0000170992.63691.32. [DOI] [PubMed] [Google Scholar]
- 8.Shaffer LG, Kashork CD, Saleki R, Rorem E, Sundin K, Ballif BC, Bejjani BA. Targeted genomic microarray analysis for identification of chromosome abnormalities in 1500 consecutive clinical cases. J Pediatr. 2006;149:98–102. doi: 10.1016/j.jpeds.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, Adams MD, Myers EW, Li PW, Eichler EE. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 10.Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- **11.Turner DJ, Miretti M, Rajan D, Fiegler H, Carter NP, Blayney ML, Beck S, Hurles ME. Germline rates of de novo meiotic deletions and duplications causing several genomic disorders. Nat Genet. 2008;40:90–95. doi: 10.1038/ng.2007.40. Sperm-PCR assays were used to measure rates of de novo NAHR-mediated rearrangements directly at four genomic disorder loci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *12.Ou Z, Stankiewicz P, Xia Z, Breman AM, Dawson B, Wiszniewska J, Szafranski P, Cooper ML, Rao M, Shao L, et al. Observation and prediction of recurrent human translocations mediated by NAHR between nonhomologous chromosomes. Genome Res. 2011;21:33–46. doi: 10.1101/gr.111609.110. The authors reported molecular details of two NAHR driven recurrent translocations and constructed a computational genome-wide recurrent translocation map. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barbouti A, Stankiewicz P, Nusbaum C, Cuomo C, Cook A, Hoglund M, Johansson B, Hagemeijer A, Park SS, Mitelman F, et al. The breakpoint region of the most common isochromosome, i(17q), in human neoplasia is characterized by a complex genomic architecture with large, palindromic, low-copy repeats. Am J Hum Genet. 2004;74:1–10. doi: 10.1086/380648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koumbaris G, Hatzisevastou-Loukidou H, Alexandrou A, Ioannides M, Christodoulou C, Fitzgerald T, Rajan D, Clayton S, Kitsiou-Tzeli S, Vermeesch JR, et al. FoSTeS, MMBIR and NAHR at the human proximal Xp region and the mechanisms of human Xq isochromosome formation. Hum Mol Genet. 2011;20:1925–1936. doi: 10.1093/hmg/ddr074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lange J, Skaletsky H, van Daalen SK, Embry SL, Korver CM, Brown LG, Oates RD, Silber S, Repping S, Page DC. Isodicentric Y chromosomes and sex disorders as byproducts of homologous recombination that maintains palindromes. Cell. 2009;138:855–869. doi: 10.1016/j.cell.2009.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **16.Sharp AJ, Hansen S, Selzer RR, Cheng Z, Regan R, Hurst JA, Stewart H, Price SM, Blair E, Hennekam RC, et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat Genet. 2006;38:1038–1042. doi: 10.1038/ng1862. Using a microarray design based on computational analysis of segmental duplication contents in the human genome, regions of genomic instability and pathogenic CNV were predicted to discover new genomic disorders. [DOI] [PubMed] [Google Scholar]
- 17.Koscinski I, Elinati E, Fossard C, Redin C, Muller J, Velez de la Calle J, Schmitt F, Ben Khelifa M, Ray PF, Kilani Z, et al. DPY19L2 deletion as a major cause of globozoospermia. Am J Hum Genet. 2011;88:344–350. doi: 10.1016/j.ajhg.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pentao L, Wise CA, Chinault AC, Patel PI, Lupski JR. Charcot-Marie-Tooth type 1A duplication appears to arise from recombination at repeat sequences flanking the 1. 5 Mb monomer unit. Nat Genet. 1992;2:292–300. doi: 10.1038/ng1292-292. [DOI] [PubMed] [Google Scholar]
- 19.Chen KS, Manian P, Koeuth T, Potocki L, Zhao Q, Chinault AC, Lee CC, Lupski JR. Homologous recombination of a flanking repeat gene cluster is a mechanism for a common contiguous gene deletion syndrome. Nat Genet. 1997;17:154–163. doi: 10.1038/ng1097-154. [DOI] [PubMed] [Google Scholar]
- **20.Liu P, Lacaria M, Zhang F, Withers M, Hastings PJ, Lupski JR. Frequency of nonallelic homologous recombination is correlated with length of homology: evidence that ectopic synapsis precedes ectopic crossing-over. Am J Hum Genet. 2011;89:580–588. doi: 10.1016/j.ajhg.2011.09.009. The authors reported a correlation between NAHR frequency and flanking DNA repeat length. They proposed that ectopic synapsis precedes ectopic crossing-over to explain such observed correlation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jinks-Robertson S, Michelitch M, Ramcharan S. Substrate length requirements for efficient mitotic recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:3937–3950. doi: 10.1128/mcb.13.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liskay RM, Letsou A, Stachelek JL. Homology requirement for efficient gene conversion between duplicated chromosomal sequences in mammalian cells. Genetics. 1987;115:161–167. doi: 10.1093/genetics/115.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **23.Myers S, Freeman C, Auton A, Donnelly P, McVean G. A common sequence motif associated with recombination hot spots and genome instability in humans. Nat Genet. 2008;40:1124–1129. doi: 10.1038/ng.213. A 13-mer DNA sequence motif was identified to be associated with allelic recombination hotspots in HapMap individuals. [DOI] [PubMed] [Google Scholar]
- 24.Spies M, Bianco PR, Dillingham MS, Handa N, Baskin RJ, Kowalczykowski SC. A molecular throttle: the recombination hotspot chi controls DNA translocation by the RecBCD helicase. Cell. 2003;114:647–654. doi: 10.1016/s0092-8674(03)00681-0. [DOI] [PubMed] [Google Scholar]
- 25.Zhang F, Potocki L, Sampson JB, Liu P, Sanchez-Valle A, Robbins-Furman P, Navarro AD, Wheeler PG, Spence JE, Brasington CK, et al. Identification of uncommon recurrent Potocki-Lupski syndrome-associated duplications and the distribution of rearrangement types and mechanisms in PTLS. Am J Hum Genet. 2010;86:462–470. doi: 10.1016/j.ajhg.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu P, Erez A, Nagamani SC, Bi W, Carvalho CM, Simmons AD, Wiszniewska J, Fang P, Eng PA, Cooper ML, et al. Copy number gain at Xp22. 31 includes complex duplication rearrangements and recurrent triplications. Hum Mol Genet. 2011;20:1975–1988. doi: 10.1093/hmg/ddr078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lupski JR. Hotspots of homologous recombination in the human genome: not all homologous sequences are equal. Genome Biol. 2004;5:242. doi: 10.1186/gb-2004-5-10-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindsay SJ, Khajavi M, Lupski JR, Hurles ME. A chromosomal rearrangement hotspot can be identified from population genetic variation and is coincident with a hotspot for allelic recombination. Am J Hum Genet. 2006;79:890–902. doi: 10.1086/508709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Myers SR, McCarroll SA. New insights into the biological basis of genomic disorders. Nat Genet. 2006;38:1363–1364. doi: 10.1038/ng1206-1363. [DOI] [PubMed] [Google Scholar]
- 30.Raedt TD, Stephens M, Heyns I, Brems H, Thijs D, Messiaen L, Stephens K, Lazaro C, Wimmer K, Kehrer-Sawatzki H, et al. Conservation of hotspots for recombination in low-copy repeats associated with the NF1 microdeletion. Nat Genet. 2006;38:1419–1423. doi: 10.1038/ng1920. [DOI] [PubMed] [Google Scholar]
- 31.Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, de Massy B. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Myers S, Bowden R, Tumian A, Bontrop RE, Freeman C, MacFie TS, McVean G, Donnelly P. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 2010;327:876–879. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parvanov ED, Petkov PM, Paigen K. Prdm9 controls activation of mammalian recombination hotspots. Science. 2010;327:835. doi: 10.1126/science.1181495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **34.Berg IL, Neumann R, Lam KW, Sarbajna S, Odenthal-Hesse L, May CA, Jeffreys AJ. PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat Genet. 2010;42:859–863. doi: 10.1038/ng.658. The authors demonstrated that variation in the minisatellite sequence within PRDM9 affects individual’s recombination hotspot activity and specification. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berg IL, Neumann R, Sarbajna S, Odenthal-Hesse L, Butler NJ, Jeffreys AJ. Variants of the protein PRDM9 differentially regulate a set of human meiotic recombination hotspots highly active in African populations. Proc Natl Acad Sci U S A. 2011;108:12378–12383. doi: 10.1073/pnas.1109531108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jeffreys AJ, Wilson V, Thein SL. Hypervariable ‘minisatellite’ regions in human DNA. Nature. 1985;314:67–73. doi: 10.1038/314067a0. [DOI] [PubMed] [Google Scholar]
- 37.Bridges CB. The Bar “Gene” a Duplication. Science. 1936;83:210–211. doi: 10.1126/science.83.2148.210. [DOI] [PubMed] [Google Scholar]
- **38.Carvalho CM, Ramocki MB, Pehlivan D, Franco LM, Gonzaga-Jauregui C, Fang P, McCall A, Pivnick EK, Hines-Dowell S, Seaver LH, et al. Inverted genomic segments and complex triplication rearrangements are mediated by inverted repeats in the human genome. Nat Genet. 2011;43:1074–1081. doi: 10.1038/ng.944. A common genomic structure of DUP-TRP/INV-DUP was observed in complex triplications involving MECP2 and PLP1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *39.Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. Non-recurrent genomic disorder associated rearrangements were shown to be more complex than anticipated. A DNA replication mechanism, Fork Stallng and Template Switching or FoSTeS, was proposed to explain human complex rearrangements. [DOI] [PubMed] [Google Scholar]
- 40.Slack A, Thornton PC, Magner DB, Rosenberg SM, Hastings PJ. On the mechanism of gene amplification induced under stress in Escherichia coli. PLoS Genet. 2006;2:e48. doi: 10.1371/journal.pgen.0020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *41.Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. The MMBIR model was proposed, as a detailed molecular model expanding on the phenomenology of FoSTeS and explaining a diverse range of human copy number variations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith CE, Llorente B, Symington LS. Template switching during break-induced replication. Nature. 2007;447:102–105. doi: 10.1038/nature05723. [DOI] [PubMed] [Google Scholar]
- 43.Lydeard JR, Jain S, Yamaguchi M, Haber JE. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature. 2007;448:820–823. doi: 10.1038/nature06047. [DOI] [PubMed] [Google Scholar]
- 44.Morrow DM, Connelly C, Hieter P. “Break copy” duplication: a model for chromosome fragment formation in Saccharomyces cerevisiae. Genetics. 1997;147:371–382. doi: 10.1093/genetics/147.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malkova A, Ivanov EL, Haber JE. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc Natl Acad Sci U S A. 1996;93:7131–7136. doi: 10.1073/pnas.93.14.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mosig G. Recombination and recombination-dependent DNA replication in bacteriophage T4. Annu Rev Genet. 1998;32:379–413. doi: 10.1146/annurev.genet.32.1.379. [DOI] [PubMed] [Google Scholar]
- 47.Chen JM, Cooper DN, Ferec C, Kehrer-Sawatzki H, Patrinos GP. Genomic rearrangements in inherited disease and cancer. Semin Cancer Biol. 2011;20:222–233. doi: 10.1016/j.semcancer.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 48.Arlt MF, Ozdemir AC, Birkeland SR, Wilson TE, Glover TW. Hydroxyurea induces de novo copy number variants in human cells. Proc Natl Acad Sci U S A. 2011;108:17360–17365. doi: 10.1073/pnas.1109272108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arlt MF, Mulle JG, Schaibley VM, Ragland RL, Durkin SG, Warren ST, Glover TW. Replication stress induces genome-wide copy number changes in human cells that resemble polymorphic and pathogenic variants. Am J Hum Genet. 2009;84:339–350. doi: 10.1016/j.ajhg.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.De S, Michor F. Dna replication timing and long-range Dna interactions predict mutational landscapes of cancer genomes. Nat Biotech. 2011;29:1103–1108. doi: 10.1038/nbt.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fudenberg G, Getz G, Meyerson M, Mirny L. High order chromatin architecture shapes the landscape of chromosomal alterations in cancer. Nat Biotech. 2011;29:1109–1113. doi: 10.1038/nbt.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Deem A, Keszthelyi A, Blackgrove T, Vayl A, Coffey B, Mathur R, Chabes A, Malkova A. Break-induced replication is highly inaccurate. PLoS Biol. 2011;9:e1000594. doi: 10.1371/journal.pbio.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carvalho CM, Zhang F, Liu P, Patel A, Sahoo T, Bacino CA, Shaw C, Peacock S, Pursley A, Tavyev YJ, et al. Complex rearrangements in patients with duplications of MECP2 can occur by fork stalling and template switching. Hum Mol Genet. 2009;18:2188–2203. doi: 10.1093/hmg/ddp151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bi W, Sapir T, Shchelochkov OA, Zhang F, Withers MA, Hunter JV, Levy T, Shinder V, Peiffer DA, Gunderson KL, et al. Increased LIS1 expression affects human and mouse brain development. Nat Genet. 2009;41:168–177. doi: 10.1038/ng.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang F, Khajavi M, Connolly AM, Towne CF, Batish SD, Lupski JR. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat Genet. 2009;41:849–853. doi: 10.1038/ng.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang F, Seeman P, Liu P, Weterman MA, Gonzaga-Jauregui C, Towne CF, Batish SD, De Vriendt E, De Jonghe P, Rautenstrauss B, et al. Mechanisms for nonrecurrent genomic rearrangements associated with CMT1A or HNPP. rare CNVs as a cause for missing heritability. Am J Hum Genet. 2010;86:892–903. doi: 10.1016/j.ajhg.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oh SD, Lao JP, Hwang PY, Taylor AF, Smith GR, Hunter N. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell. 2007;130:259–272. doi: 10.1016/j.cell.2007.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Campbell PJ, Stephens PJ, Pleasance ED, O’Meara S, Li H, Santarius T, Stebbings LA, Leroy C, Edkins S, Hardy C, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, Stebbings LA, Leroy C, Edkins S, Mudie LJ, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **60.Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. Chromothripsis, a ‘one-off’ event causing massive genomic rearrangements in cancers, was documented. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rausch T, Jones DT, Zapatka M, Stutz AM, Zichner T, Weischenfeldt J, Jager N, Remke M, Shih D, Northcott PA, et al. Genome Sequencing of Pediatric Medulloblastoma Links Catastrophic DNA Rearrangements with TP53 Mutations. Cell. 2012;148:59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *62.Kloosterman WP, Guryev V, van Roosmalen M, Duran KJ, de Bruijn E, Bakker SC, Letteboer T, van Nesselrooij B, Hochstenbach R, Poot M, et al. Chromothripsis as a mechanism driving complex de novo structural rearrangements in the germline. Hum Mol Genet. 2011;20:1916–1924. doi: 10.1093/hmg/ddr073. A subject with multiple chromosomal translocations and inversions was reported and chromothripsis was proposed as the underlying mechanism. [DOI] [PubMed] [Google Scholar]
- **63.Liu P, Erez A, Nagamani SC, Dhar SU, Kolodziejska KE, Dharmadhikari AV, Cooper ML, Wiszniewska J, Zhang F, Withers MA, et al. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell. 2011;146:889–903. doi: 10.1016/j.cell.2011.07.042. Highly complex human genomic rearrangements were reported to occur in human germ cells and during early development. A DNA replication mechanism was proposed to explain the observed chromosome catastrophe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, Pellman D. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Payen C, Koszul R, Dujon B, Fischer G. Segmental duplications arise from Pol32-dependent repair of broken forks through two alternative replication-based mechanisms. PLoS Genet. 2008;4:e1000175. doi: 10.1371/journal.pgen.1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lakich D, Kazazian HH, Jr, Antonarakis SE, Gitschier J. Inversions disrupting the factor VIII gene are a common cause of severe haemophilia A. Nat Genet. 1993;5:236–241. doi: 10.1038/ng1193-236. [DOI] [PubMed] [Google Scholar]
- 68.Inoue K, Osaka H, Thurston VC, Clarke JT, Yoneyama A, Rosenbarker L, Bird TD, Hodes ME, Shaffer LG, Lupski JR. Genomic rearrangements resulting in PLP1 deletion occur by nonhomologous end joining and cause different dysmyelinating phenotypes in males and females. Am J Hum Genet. 2002;71:838–853. doi: 10.1086/342728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Woodward KJ, Cundall M, Sperle K, Sistermans EA, Ross M, Howell G, Gribble SM, Burford DC, Carter NP, Hobson DL, et al. Heterogeneous duplications in patients with Pelizaeus-Merzbacher disease suggest a mechanism of coupled homologous and nonhomologous recombination. Am J Hum Genet. 2005;77:966–987. doi: 10.1086/498048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *70.Vissers LE, Bhatt SS, Janssen IM, Xia Z, Lalani SR, Pfundt R, Derwinska K, de Vries BB, Gilissen C, Hoischen A, et al. Rare pathogenic microdeletions and tandem duplications are microhomology-mediated and stimulated by local genomic architecture. Hum Mol Genet. 2009;18:3579–3593. doi: 10.1093/hmg/ddp306. The authors characterized breakpoint features of pathogenic deletions and duplications from throughout the human genome. [DOI] [PubMed] [Google Scholar]
- *71.Conrad DF, Bird C, Blackburne B, Lindsay S, Mamanova L, Lee C, Turner DJ, Hurles ME. Mutation spectrum revealed by breakpoint sequencing of human germline CNVs. Nat Genet. 2010;42:385–391. doi: 10.1038/ng.564. The authors used targeted hybridization-based DNA capture and next generation sequencing to characterize breakpoint features of human germline CNVs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *72.Kidd JM, Graves T, Newman TL, Fulton R, Hayden HS, Malig M, Kallicki J, Kaul R, Wilson RK, Eichler EE. A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell. 2010;143:837–847. doi: 10.1016/j.cell.2010.10.027. The authors studied mechanisms for germline CNVs using sequencing data obtained from fosmid clone libraries. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shaw-Smith C, Pittman AM, Willatt L, Martin H, Rickman L, Gribble S, Curley R, Cumming S, Dunn C, Kalaitzopoulos D, et al. Microdeletion encompassing MAPT at chromosome 17q21. 3 is associated with developmental delay and learning disability. Nat Genet. 2006;38:1032–1037. doi: 10.1038/ng1858. [DOI] [PubMed] [Google Scholar]
- 74.Van Esch H, Hollanders K, Badisco L, Melotte C, Van Hummelen P, Vermeesch JR, Devriendt K, Fryns JP, Marynen P, Froyen G. Deletion of VCX-A due to NAHR plays a major role in the occurrence of mental retardation in patients with X-linked ichthyosis. Hum Mol Genet. 2005;14:1795–1803. doi: 10.1093/hmg/ddi186. [DOI] [PubMed] [Google Scholar]
- 75.El-Hattab AW, Fang P, Jin W, Hughes JR, Gibson JB, Patel GS, Grange DK, Manwaring LP, Patel A, Stankiewicz P, et al. Int22h-1/int22h-2-mediated Xq28 rearrangements: intellectual disability associated with duplications and in utero male lethality with deletions. J Med Genet. 2011;48:840–850. doi: 10.1136/jmedgenet-2011-100125. [DOI] [PubMed] [Google Scholar]
- 76.Kamp C, Hirschmann P, Voss H, Huellen K, Vogt PH. Two long homologous retroviral sequence blocks in proximal Yq11 cause AZFa microdeletions as a result of intrachromosomal recombination events. Hum Mol Genet. 2000;9:2563–2572. doi: 10.1093/hmg/9.17.2563. [DOI] [PubMed] [Google Scholar]