Abstract
The lateral transmembrane protein-protein interaction has been regarded as “undruggable” despite its importance in many biological processes. The homo-trimerization of transmembrane domain 5 (TMD-5) of latent membrane protein 1 (LMP-1) is critical for the constitutive oncogenic activation of the Epstein-Barr virus (EBV). Herein, we report a small molecule agent, NSC 259242 (compound 1), to be a TMD-5 self-association disruptor. Both the positively charged acetimidamide functional groups and the stilbene backbone of compound 1 are essential for its inhibitory activity. Furthermore, cell-based assays revealed that compound 1 inhibits full-length LMP-1 signaling in EBV infected B cells. These studies demonstrated a new strategy for identifying small molecule disruptors for investigating transmembrane protein-protein interactions.
Keywords: Epstein–Barr virus, latent membrane protein 1, transmembrane domain, protein-protein interaction, cell-based screen, small molecule inhibitor
1. Introduction
Major strides have been made in the development of small molecule inhibitors of protein-protein interactions (PPI) in the last 15 years despite the inherent challenge of targeting their expansive protein interfaces [1,2,3]. However, the development of PPI inhibitors that target transmembrane domains (TMD) of proteins has lagged behind, primarily due to the hurdle of identifying small molecules that are able to not only insert into phospholipid bilayers but also specifically recognize these highly hydrophobic TMDs [4,5,6,7].
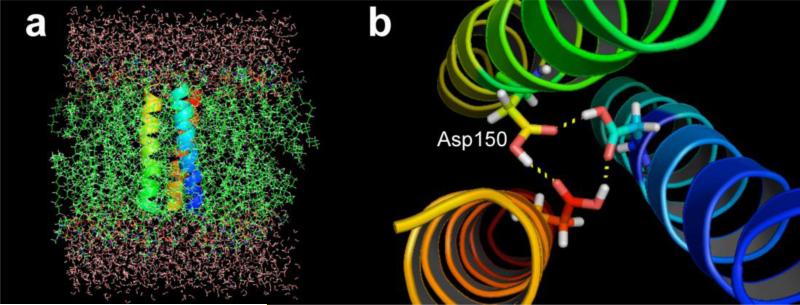
The Epstein-Barr virus (EBV) oncoprotein latent membrane protein 1 (LMP-1) is a viral integral transforming protein essential for B cell immortalization [8]. The constitutive signaling activities of LMP-1 depend on its homo-oligomerization [8]. Recently, we identified that the fifth transmembrane domain (TMD-5) of LMP-1 mediates its oligomerization [9]. A membrane-embedded aspartic acid residue (Asp 150) in TMD-5 has been shown to facilitate a hydrogen bonding network, driving LMP-1 homo-trimeric complex formation (Figure 1). Such a hydrogen-bonding network is possible because the carboxylic acid groups of the aspartic acid residues remain protonated in a phospholipid bilayer, which is consistent with previous reports that the pKa values of membrane-embedded carboxylic acid are significantly elevated [10]. Mutation of the Asp 150 residue to Ala abolishes the downstream signaling of LMP-1 (e.g. NF-κB), suggesting a novel strategy to target LMP-1 by disrupting its lateral TMD interactions. Herein, we report a small molecule inhibitor that suppresses TMD-5 oligomerization, thereby inhibiting the LMP-1-mediated oncogenic signaling.
Figure 1.
(a), Energy-optimized atomistic model [9] of homo-trimeric complex of LMP-1 TMD-5 in explicit membrane bilayers. (b), The TMD-5 association is driven by the H-bonds mediated by Asp 150.
2. Materials and Methods
2.1 High-throughput screening
ToxR7-TMD-5, ToxR7–TMD of diacylglycerol kinase (DAGK) and ToxR7-TMD of integrin αIIb plasmids were constructed as described previously [11,12,13]. Diversity Set II library (1364 compounds) was provided by the Developmental Therapeutic Program, NCI/NIH. High-throughput screening of TMD-5 disruptors was carried out as following. ToxR7-TMD-5 plasmid (200 ng) was transformed into 200 μL of FHK12 competent cells (kindly provided by D. Langosch, Technische Universität München, Germany) with heat shock at 42 °C for 90 s and incubation on ice for 2 minutes, followed by addition of 800 μL SOC media and incubation with shaking at 37 °C for 1 h. 5 μL of the transformation mixture was used to inoculate 100 μL LB + arabinose (0.0025%) and chloramphenicol (30 μg/mL) and 100 μM compound. Cultures were incubated with shaking at 37 °C for 20 h and β-galactosidase activity was measured using a Beckman Coulter DTX 880 plate reader (Beckman, Coulter, Fullerton, CA, USA) as described previously [11,12]. Briefly, 5 μL of culture was transferred to the wells of a Costar 3596 polystyrene 96-well plate (Corning, NY, USA) containing 100 μL Z buffer/chloroform (1% β-mercaptoethanol, 10% chloroform, 89% A buffer: 1 M sodium phosphate, 10 mM KCl, 1 mM MgSO4 and pH 7.0). Cells were lysed by addition of 50 μL Z buffer/ SDS (1.6% w/v sodium dodecyl sulfate (SDS) in Z buffer) and shaking at 28 °C for 10 min. 50 μL Z buffer/ONPG (0.4% w/v ONPG in Z buffer) was added and β-galactosidase activity was measured by monitoring the reaction at 405 nm for a period of 20 min at 28 °C. Miller units were calculated using the following equation: Miller units = (OD405/min)/OD600 × 1000. Compounds that decreased TMD-5 oligomerization by 50% at the concentration of 100 μM were selected as possible hits. After first round of screening, the identified possible hits were re-tested their inhibition toward TMD-5 oligomerization. 26 compounds were found as potential hits after the first and second round of screen. Next, toxicity test (measuring OD 600 nm) was further carried out to eliminate the false positive hits owing to the growth inhibition effect.
2.2 Western Blotting
The samples were first separated by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and then electroblotted to PVDF membrane. After blocking with 5% bovine serum albumin (BSA), the membranes were incubated with 0.5 μg/mL of MBP primary antibody at room temperature for 2 h. The membranes were washed 5 times in phosphate buffered saline (PBS, 137 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, 1.46 mM KH2PO4, pH 7.4) solution with 0.05% of detergent Tween 20 (PBST) for 5 min each and then incubated for 1 h at room temperature with secondary antibody-horseradish peroxidase (HRP) conjugate (50 ng/mL). After extensive washing in PBST, the protein–antibody complexes were visualized by exposure to X-ray film after reacting with Super-Signal West Pico Chemiluminescent Substrate (Pierce, Rockford, IL, USA).
2.3 Peptide synthesis and purification
Peptides were synthesized on Rink Amide MBHA resin (100-200 mesh) (Calbiochem-Novabiochem, San Diego, CA, USA) with a substitution level of 0.57 mmol/g. Peptides were synthesized using a CEM Liberty automated synthesizer (CEM, Mathews, NC, USA) with Discovery microwave module at 0.1 mmol scale. The GGPG/GPGG sequence motif was included at the N-/C-termini of the peptide to increase solubility. An additional glycine was added to the N-terminus of each peptide to act as a spacer for the addition of a fluorophore. Activation of the free amino acids was achieved using HATU (0.40 M solution in N, N-dimethylformamide). The reaction solvent was N-methyl-2-pyrrolidone (HPLC grade, Fisher, PA, USA). Side chain deprotection and simultaneous cleavage from the resin was performed using a mixture of trifluoroacetic acid (TFA)/water/1,2-ethanedithiol/triisopropylsilane (90:2.5:2.5:1,v/v) or trifluoroacetic acid/water/phenol/thioanisole/1,2-ethanedithiol/triisopropylsilane (81.5:5:5:5:2.5:1) at room temperature for two hours. The crude peptides were collected by precipitation with cold (–20 °C) diethyl ether (Sigma-Aldrich, St. Louis, MO, USA). The peptides were then purified using Agilent 1200 series semi-preparative reverse phase HPLC system (Santa Clara, CA, USA) with an Agilent Zorbax 300 SB-C8 column using a linear gradient of buffer A (10% 2-propanol, 0.1% trifluoroacetic acid in Millipore water) and buffer B (6:3:1 2-propanol/acetonitrile/water containing 0.1% trifluoroacetic acid). The identities of the purified peptides were confirmed as the monomeric species by MALDI-TOF mass spectroscopy on a Voyager DE-STR Biospectrometry Workstation (PerSeptive Biosystems, CA, USA).
Peptides were labeled with coumarin using the following method: 7-Hydroxycoumarin-3-carboxylic acid (100 mg, 0.485 mmol) and HBTU (186 mg, 0.490 mmol) were dissolved in 1:9 dimethyl sulfoxide/N,Ndimethylformamide and added to the resin. N,N'-Diisopropylethylamine (185 μL, 1.06 mmol) was added and the reaction was stirred for 2-16 h until the Kaiser test indicated that the reaction was complete. Cleavage and purifications were performed as described above.
The concentration of TMD-5 peptide (GGPG-WQLLAFFLAFFLDLILLIIALYL-GPGG) derived from LMP-1 was determined by absorbance at 280 nm using an extinction coefficient of 6990 M-1cm-1. The concentration of coumarin labeled TMD-5 peptide (coumarin -GGGPG-WQLLAFFLAFFLDLILLIIALYL-GPGG) was determined by absorbance at 400 nm using an extinction coefficient of 39300 M-1cm-1.
2.4 Chemical synthesis
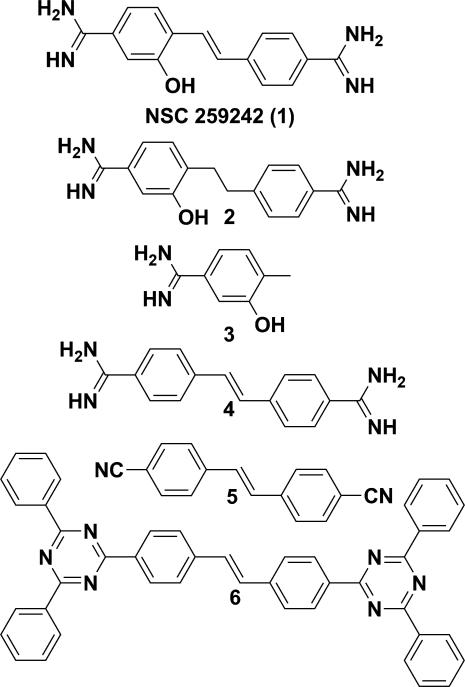
Compound 1 (Figure 2) was provided by NCI/DTP Open Chemical Repository (http://dtp.cancer.gov). Compound 6 (Figure 2) (2,2'-(1,2-ethenediyldi-4,1-phenylene)bis(4,6-diphenyl-1,3,5-triazine)) was purchased from Chembridge (Cat. No. 5255931, San Diego, CA, USA). The other compounds (Figure 2) were synthesized and characterized as shown in the Supplementary Data.
Figure 2.
Structure of NSC 259242 (compound 1) and its analogues.
2.5 Fluorescence dequenching assay
Coumarin labeled TMD-5 peptide was dissolved in trifluoroethanol (TFE) with C14 betaine (3-(N,N-dimethylmyristylammonio)propanesulfonate, Sigma-Aldrich, St. Louis, MO, USA). The organic solvent was evaporated under a stream of nitrogen to generate a thin film of peptide/detergent mixture, which was then dissolved in 50 mM HEPES (pH = 7.4) buffer. 100 μL of coumarin labeled TMD-5 peptide (100 nM) solution (50 mM HEPES, 150 μM C14 betaine, pH 7.4) with the indicated concentration of drug was pipetted into black 96-well plates in triplicate. Samples were mixed and allowed to sit at room temperature in the dark overnight to reach equilibrium and then excited at 360 nm and emission was read at 430 nm using a Beckman-Coulter DTX 880 Multimode Detector plate reader. Appropriate drug controls were subtracted from the observed coumarin fluorescence signal to eliminate the possible interference from drug fluorescence.
2.6 NMR spectroscopy on TMD-5
2.6.1 Sample preparation
Selectively 15N-Gly, Leu, Ala (15N-GLA) labeled and uniformly 13C, 15N enriched TMD-5 peptides (GGPG-WQLLAFFLAFFLDLILLIIALYL-GPGG) were synthesized and purified as described previously. Isotope labeled amino acids were purchased from Cambridge Isotope Laboratories (Andover, MA, USA) and Sigma-Aldrich (St. Lious, MO, USA). The bicelles were prepared by mixing 105 mM 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and 350 mM dihexanoylphosphadtidylcholine (DHPC) with q = 0.3 (AvantiLipids, Alabaster, AL, USA). The peptide/bicelle mixture was resuspended so that the final concentration was 0.5 mM in 10 mM phosphate buffer at pH 7.0 with 10 % (v/v) D2O. For the titration, Compound 1 was added incrementally to theTMD-5 sample to yield the molar ratio 1:0.5, 1:1, 1:2, and 1:4. The concentration of compound 1 was 100 mM in DMSO, and the final concentration of DMSO in the NMR sample was negligible - maximum 0.25 % (v/v).
2.6.2 NMR spectroscopy
For resonance assignment, 2D 1H, 15N-HSQC, 3D HNCO, HNCA, HN(CO)CA, and CBCA(CO)NH spectra [14] were collected at 37 °C on VNMRS 800 MHz spectrometer equipped with HCN z-axis gradient cold probe. For Compound 1 titrations, the amide proton and nitrogen resonances were detected using SOFAST HMQC [15] at 37 °C on Varian INOVA 500 MHz spectrometer equipped with a triple resonance z-axis gradient probe. Spectra were processed with NMRPipe [16] and analyzed with Sparky (Goddard, T. D. and Kneller, D. G., SPARKY 3, UCSF).
2.7 Bis-Tris gel electrophoresis
Electrophoresis was carried out using precast SDS–PAGE gels according to manufacture's instructions (12% NuPAGE 10-well Bis-Tris gels, Invitrogen, Grand Island, NY, USA). Briefly, after NMR titration experiment (compound 1 to TMD-5 peptide ratio 4:1), sample was incubated with equal volume of 2x Laemmli sample buffer (126 mM Tris-HCl, 20% glycerol, 4 % SDS, 0.02% bromphenol blue, pH 6.8) at 90 °C for 7 min. Electrophoresis was carried out at room temperature with NuPAGE MES SDS running buffer (Invitrogen, Grand Island, NY, USA) at 125 mV for 55 min. The resulting gel was stained using SilverXpress® Silver Staining Kit (Invitrogen, Grand Island, NY, USA).
2.8 NF-κB assay
The human B lymphoblastoid cell line 721 (Epstein–Barr virus (EBV)-positive) was kindly provided by Dr Jennifer M. Martin (University of Colorado, Boulder) and cultured in RPMI medium supplemented with 10% fetal bovine serum (FBS), penicillin (50 unit/mL) and streptomycin (50 μg/mL). NF-κB reporter cells wereconstructed by Cignal Lenti NF-κB Reporter kit (SABiosciences, MD, USA) and selected by puromycin treatment according to manufacture's instructions. The firefly luciferase gene was placed under the control the NF-κB transcriptional response element. Briefly 50 μL of B 721 cells (105 cells/mL) was added to one well of 96-well plate. 50 μL of NF-κB Reporter letivirus suspension (SABiosciences, MD, USA) was added. 8 μg/mL of SureENTRY Transduction Reagent (SABiosciences, MD, USA) was also added to increase the transduction efficiency. After further 48 h incubation, 4 μg/mL puromycin was added to select transduced cells.
B lymphoblastoid cell line 721 NF-κB reporter cells were seeded at a density of 1×104 cells/well in 96-well plates (100 μL/each well). After overnight incubation, different concentrations of compounds were added to the cells. Following 48 h of treatment, the NF-κB activity was detected by Steady-Glo Luciferase Assay System (Promega, Madison, MI, USA) according to manufacture's instructions. Briefly, 75 μL Steady-Glo Luciferase Assay reagent was added to each well and incubated at room temperature for 15 min. The luminescence was measured by Beckman Coulter DTX880 reader (Beckman Coulter, CA, USA)
2.9 Nitric oxide (NO) assay
Human B lymphoblastoid cell line 721 was seeded at a density of 1×104 cells/well in 96-well plates. After overnight incubation, indicated concentrations of compounds were added. Following an additional 48 h treatment, 20 μL of media was removed and added to flat black 96-well microfluor plates (Thermo Scientific, MA, USA). 80 μL of water was added to each well. Subsequently, 10 μL of 2, 3-diaminonaphthalene (0.05 mg/mL in 0.62 M HCl) was added to each well and incubated for 15 min. The reaction was quenched by addition of 5 μL of 3 M NaOH and the plate was read on Beckman Coulter DTX880 reader (Beckman Coulter, CA, USA) with excitation at 360 nm and emission at 430 nm.
2.10 Cell proliferation assay
Human B lymphoblastoid cell line 721 was seeded at 96 well plate with a density of 5×104 cells per mL. After overnight incubation, different concentrations of compounds were added. Following 48 h stimulation, plates were centrifuged and washed by PBS twice. 100 μL of fresh medium was added to each well along with 20 μL of Cell Proliferation Reagent WST-1 (Roche Diagnostics GmbH, Mannheim, Germany). After further incubation in a 37° C chamber for 1 h, the absorbance at 450 nm was measured on a Beckman-coulter DTX 880 microplate reader. A wavelength of 620 nm was chosen as the reference wavelength. The A450 nm-A620 nm for the control group was set as 100%.
2.11 Kinase profiling
The kinase profiling was performed by Luceome Biotechnologies, Tucson, AZ. NSC 259242 was first evaluated for false positive against split luciferase and found it did not inhibit luciferase control. NSC 259242 (50 μM) was then profiled in duplicate against the following kinases: AKT1, CAMK1, DDR2, GSK3α, MARK1, MET, PAK1, PDGFRB, PIM1, PKC-γ, PKL4, and SRC using the protocol described by Ghosh and co-workers [17]. The percent inhibition and percent activity remaining was calculated using the following equations: %inhibition= (ALUcontrol-ALUsample)/ALUcontrol × 100 and % activity remaining = 100% - % inhibitio n. Staurosporine (10 μM) was served as the positive control for kinase inhibitor.
2.12 Statistical analysis
Data are expressed as mean ± s.d. and analysis of variance was carried out using Student's t test with Origin 7.5 (OriginLab Corporation, Northampton, MA, USA), where p<0.05 was considered significant.
3. Results and Discussions
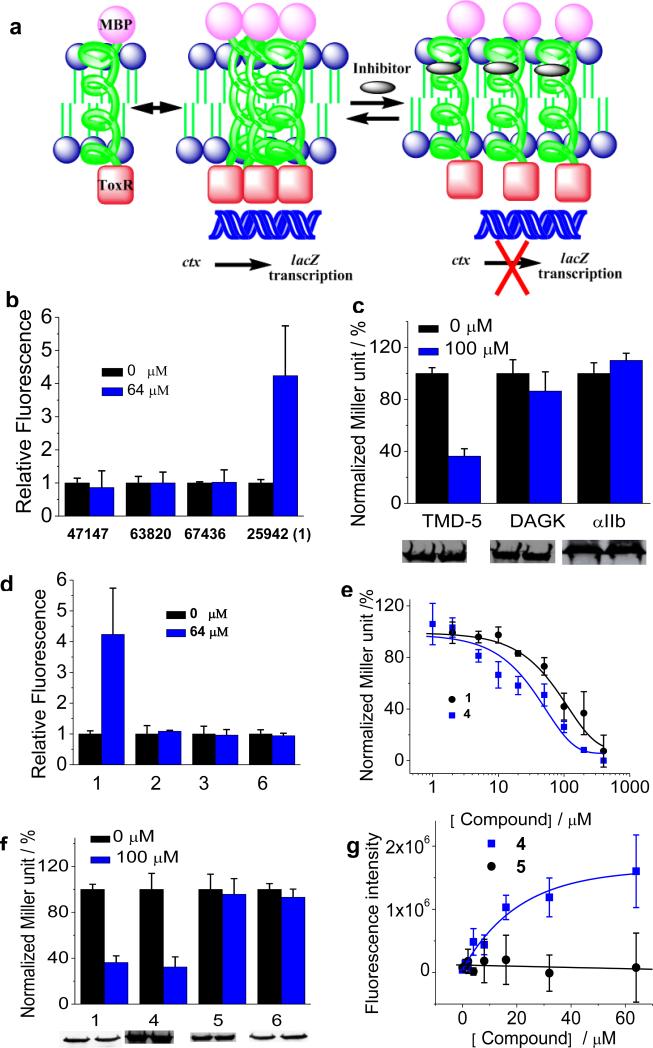
We utilized a previously established ToxR assay as the platform for screening TMD disruptors in cellular membranes (Figure 3a) [11,12,13]. This E. coli-based assay uses a chimeric construct made of the N-terminal DNA binding domain of ToxR and the monomeric anchor maltose binding protein (MBP) fused to the LMP-1 TMD-5 cytoplasmic and periplasmic termini, respectively. The chimeric plasmid was then transformed into the FHK12 E. coli competent cells. MBP localizes to the periplasm, forcing a parallel orientation of TMD-5 in the membrane. Oligomerization of TMD-5 brings the fused ToxR in proximity, promoting binding to the cholera toxin (ctx) promoter and initiation of lacZ transcription. The lacZ transcription in turn produces β-galactosidase, which is monitored by the conversion of the O-nitrophenyl ß-galactoside (ONPG) to O-nitrophyenylate by β-galactosidase. A small molecule that disrupts the TMD-5 trimerization will inhibit the LacZ activation, resulting in a decrease of the β-galactosidase production level.
Figure 3.
(a), Schematic representation of the ToxR assay used for screening LMP-1 TMD-5 disruptors. (b), Coumarin fluorescence dequeching assay. Compounds were added into 100 nM of coumarin-labeled TMD-5 solution (50 mM HEPES, 150 μM C14 betaine, pH = 7.4) and equilibrated overnight. Samples were excited at 360 nm and emission was read at 430 nm using a Beckman-Coulter DTX 880 Multimode Detector plate reader. The background correction and the fluorescence of inhibitors were subtracted from the observed coumarin fluorescence signal. The fluorescence intensity of control sample (no compound) was set as 1.0. (c), ToxR measurement of the inhibitory effect of compound 1 on the oligomerization of LMP-1 TMD-5 and TMDs of diacylglycerol kinase (DAGK) and integrin αIIb. TMD oligomerization activity in the absence of compound was normalized as 100%. Western blot showed the chimeric MBP-TMD-ToxR protein expression level and gel loading normalized by OD600 nm of cultures. (d), Fluorescence dequenching assay of compound 1 and its analogues. Experiments were carried out as described in (b). (e), Dose-dependent titration of 1 and 4 on TMD-5 oligomerization using the ToxR assay. (f), ToxR measurement of oligomerization of TMD-5 in the presence of compound 1 or its analogues. Western blot showed the chimeric MBP-TMD-5-ToxR protein expression level and gel loading normalized by OD600 nm of cultures. (g), the dose dependent fluorescence enhancement curves of coumarin-labeled TMD-5 induced by compound 4 and compound 5. Experiments were carried out as described in (b).
The NCI Diversity Set II library was screened using our ToxR reporter assay. Compounds that decreased TMD-5 oligomerization greater than 50% at 100 μM were selected as preliminary hits. After the initial screen, 26 compounds were found as potential oligomerization inhibitors. Because the stock of these compounds was in DMSO solution, the effect of DMSO on TMD-5 oligomerization was investigated. It was found that, at concentrations up to 2%, DMSO showed negligible effects (Supplementary Figure S1). Toxicity tests were also carried out to eliminate the false positives due to the compounds inhibitory effect on E. coli cell growth. Four compounds were selected after the second round of screen: NSC 259242 (compound 1, Figure 2), NSC 47147, NSC 636820 and NSC 67436 (Supplementary Figure S2).
Fluorescence dequenching assays (Supplementary Figure S3) were performed to biophysically test whether TMD-5 is the direct target of these identified small molecule inhibitors. Coumarin-labeled TMD-5 peptide (Coum-GGGPG-WQLLAFFLAFFLDLILLIIALYL-GPGG) was synthesized following standard solid phase peptide synthesis (SPPS) procedure for hydrophobic peptides. Two –GGPG- tags were appended onto either terminus of TMD-5, serving as both a flexible linker to the fluorescence dye as well as a facilitator of membrane insertion [18]. TMD-5 forms homo-trimer in the presence of 150 μM C14 betaine (critical micelle concentration=100 μM), resulting in coumarin fluorescence self-quenching [9]. Disruption of the TMD-5 oligomer dequenches the coumarin dye, leading to fluorescence enhancement. Among the four initial hits, only compound 1 was found to be able to efficiently disrupt TMD-5 homo-trimeric association, and reverse the coumarin fluorescence quenching (Supplementary Figure S4). Also importantly, in the absence of C14 betaine detergent, 1 failed to cause coumarin labeled TMD-5 fluorescence increase (Figure 3b). These results eliminate the possibility that the fluorescence increase was due to interaction between the coumarin dye and compound 1.
To demonstrate the specificity of 1, we tested its inhibitory effect on the diacylglycerol kinase (DAGK), a well studied 3-pass integral membrane protein that, like LMP-1, also forms a homo-trimer complex through TMD association [11]. As shown in Figure 3c, compound 1 did not inhibit DAGK TMD oligomerization, confirming the specificity of 1 to TMD-5. To further demonstrate the specificity of 1, we also tested the effect of 1 on the TMD of integrin αIIb, a single pass integral membrane protein [13]. No apparent oligomerization inhibition was observed (Figure 3c). The effect of compound 1 on MBP-TMD-ToxR chimeric protein expression was also investigated. Compound 1 did not affect the TMD chimeric protein expression (Figure 3c), which eliminated the possibility that the observed inhibitory effects of 1 were due to the reduction of TMD-5 chimeric protein expression.
To further understand the nature of the inhibitory effect of compound 1, we carried out a limited structure-activity relationship study. Reduction of the olefin backbone of 1 to yield compound 2 resulted in diminished TMD-5 inhibitory activity (Figure 3d). Compound 3, a fragment of 1, did not inhibit TMD-5 association either (Figure 3d). Given the fact that both compounds 2 and 3 containing a hydroxyl group along with ortho-alkyl substitution are inactive, we concluded that the meta-hydroxyl benzamidine motif alone is not sufficient to inhibit TMD-5 trimerization. Removing the hydroxyl group of compound 1 to yield compound 4 resulted in a slight increase of TMD-5 inhibitory activity (Figure 3e). Dose-dependent titrations of 1 and 4 were performed using ToxR assay. Compound 1 and 4 showed a moderate IC50 of 90.0 ± 10.5 μM and 51.9 ± 39.3 μM, respectively, to disrupt TMD-5 oligomerization in whole cells (Figure 3e). The results show the hydroxyl group of compound 1 is not important for its anti-TMD-5 activity. The cationic acetimidamide groups of 1/4 are speculated to play a critical role in its membrane insertion and disruption of the TMD-5 association. Removing the positive charged acetimidamide groups of 4 to yield compound 5 resulted in loss of TMD-5 inhibitory activity (Figure 3f and 3g). Last, we also tested commercially available compound 6, a stilbene derivative that contains the same core structure with compound 1 but with the acetimidamide substituted with 1,3,5-triazine (Figure 2). As shown in Figure 3d and 3f, compound 6 did not inhibit TMD-5 oligomerization. In conclusion, these findings imply that the acetimidamide groups and the conjugated pi system in compound 1 provide complementary electronic and conformational match with TMD-5, confirming that the substituted stilbene scaffold plays essential roles in compound 1's inhibitory activities.
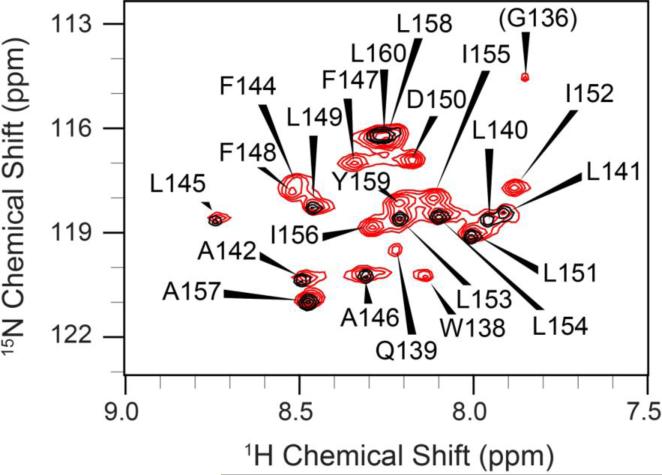
To further confirm the interaction of compound 1 with TMD-5, solution phase nuclear magnetic resonance (NMR) experiments were conducted on isotope enriched TMD-5 peptides. High-field NMR measurements were performed at 800 MHz with selectively 15N-Gly, Leu, Ala(15N-GLA) labeled and with uniformly 13C, 15N-enriched TMD-5 peptides in bicelles composed of 105 mM POPC and 350 mM DHPC (q = 0.3). Backbone resonance assignments (Supplementary Table 1) were obtained by the analysis of 2D (1H, 15N) heteronuclear single quantum coherence (HSQC) and a number of 3D spectra (Supplementary Figure S5). Furthermore, the selectively 15N-GLA-labeled sample facilitated the assignments of these residue types (Gly, Leu, Ala). The (1H, 15N)-HSQC spectrum of TMD-5 is well-dispersed (Figure 4), indicative of a well-defined protein structure in the presence of bicelles. The chemical shift indices (CSI) [19,20,21] of individual residues indicated that the TMD-5 peptide indeed adopts a native α–helical conformation (Figure 5). To confirm the binding of compound 1 to TMD-5, 2D (1H, 15N) heteronuclear multiple quantum coherence (HMQC) NMR measurements were performed at 500 MHz. Titration of 1 induced chemical shift changes at a number of sites including the deeply buried Asp 150. This is consistent with the observation that Asp 150 plays a critical role in LMP-1 homo-oligomerization [9]. Further, Bis-Tris SDS-PAGE (SDS-PAGE is widely used as a membrane mimetic gel to study TMD oligomerization [22]) of the NMR samples before and after the titration demonstrated that compound 1 indeed disrupts the TMD-5 trimer (MW. 10.2 kDa), resulting in TMD-5 monomer (3.4 kDa) formation (Figure 6). Future NMR experiments at high-field (800-900 MHz) will yield greater spectral resolution and allow site-specific identification of all residues involved in compound 1 binding.
Figure 4.
2D 1H, 15N-HSQC spectra(excluding most the Gly region) of the TMD-5 peptides solubilized in bicelles. Red and black contours correspond to the spectrum of uniformly 13C, 15N enriched TMD-5 and selectively 15N-GLA labeled TMD-5, respectively. The spectrum shows nice chemical shift dispersion with well-defined peaks and line shapes, indicative of a folded, homogenous protein tertiary structure. Residues are numbered according their position in the native LMP-1 sequence.
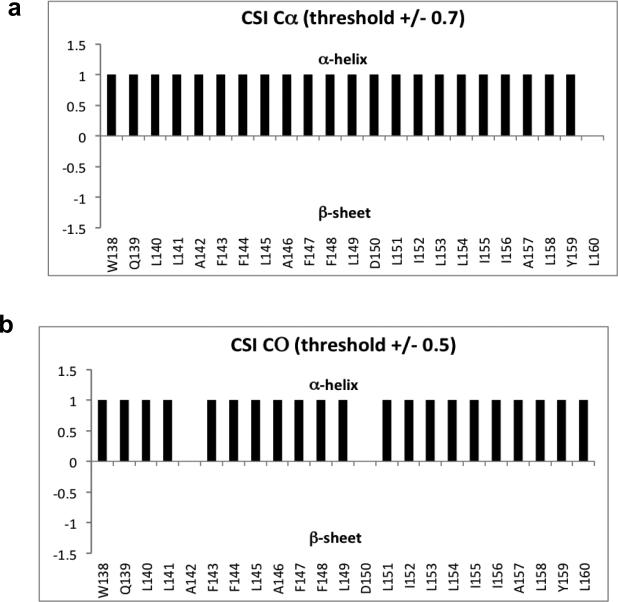
Figure 5.
The chemical shift index (CSI) values of (a) Cα and (b) CO of residues in LMP-1 TMD-5 confirmed that the TMD-5 peptide adopts a native α-helical conformation under the condition used in the NMR experiments
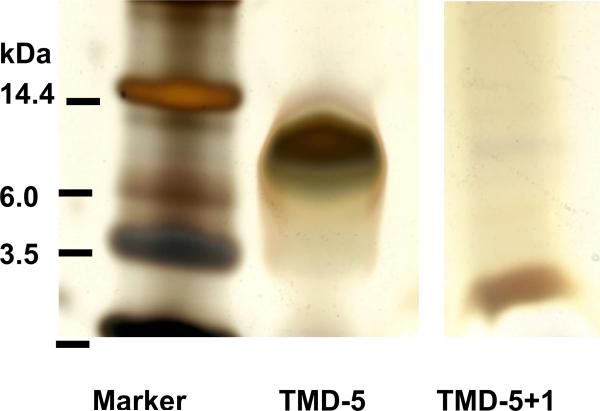
Figure 6.
12% Bis-Tris SDS-PAGE analysis of TMD-5 after NMR titration. The ratio of compound 1 to TMD-5 in TMD-5 + 1 sample shown here is 4:1. Compound 1 disrupts the TMD-5 trimer (MW 10.2 kDa ) and causes TMD-5 monomer (3.4 kDa) formation. It should be noted that the loading amount of TMD-5 in lane 2 was greater than that of lane 3.
Lastly, compound 1's ability to block full-length LMP-1 oligomerization in cells was studied by monitoring the down stream NF-κB signal transduction. LMP-1 acts as a CD40 mimic, interacts with intracellular signaling proteins TNF receptor associated factors (TRAFs) and activates the downstream effector NF-κB pathway to regulate B cell survival, proliferation, and differentiation [8,9]. The immortalized EBV positive B 721 cell line was used for investigating the effect of 1 on LMP-1 signaling. Stable NF-κB reporter cell line was constructed by transfecting B 721 cells with luciferase NF-κB reporter lentivirus and selected by antibiotic puromycin. As shown in Figure 7a, treatment of 1/4 abolished NF-κB activation in the EBV positive B 721 cell, with an IC50 of 29.6 ± 6.6 μM /19.1 ± 1.9 μM, respectively. In contrast, compound 2, 3, 5 and 6 demonstrated significantly lower inhibitory activities against NF-κB (2, IC50 = 107.9 ± 7.5 μM; 3, IC50 > 200 μM; 5, IC50 > 200; 6, IC50 = 180.4 ± 27.5 μM). It is interesting to note that 1 demonstrated elevated inhibitory potency in the EBV positive B 721 cell than in the ToxR assay in bacteria, perhaps due to a better recognition of the full-length protein in more native-like mammalian cell membranes.
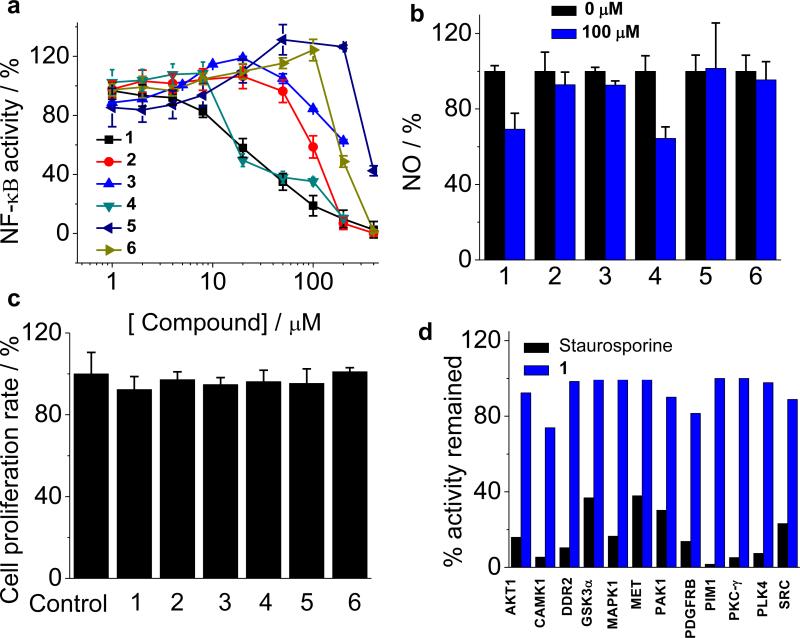
Figure 7.
(a), Dose-dependent inhibitory effects of the full-length LMP-1-mediated NF-κB activity in EBV-infected B cells by TMD-5 inhibitor 1 and its analogues. (b), Effects of compound 1 and analogues on the NF-κB downstream signaling of NO production. (c), Cell proliferation assay. No apparent growth inhibition was observed at a concentration 100 μM of compounds 1 and its analogues. (d), Kinases profile of compound 1, (50 μM) measured by KinaseSeeker™ assay. Staurosporine (10 μM) was served as the positive control for kinase inhibitor.
Nitric oxide (NO), produced by inducible NO synthase (iNOS), is a downstream signal molecule of NF-κB pathway. To further confirm the effect of compound 1 on NF-κB signaling, the production of NO was measured. Compound 1/4 (100 μM) was found to suppress NO over-production in EBV positive B 721 cells, while 2, 3, 5 and 6 show no significant inhibition activity (Figure 7b). It should be noted that none of the small molecules tested showed cytotoxicity at 100 μM (Figure 7c), which eliminated the possibility that the observed inhibition of NF-κB activity and NO production was due to non-specific cell death. Compound 1 was further examined for its specificity against TMD-5 by screening 12 representative kinases (AKT1, CAMK1, DDR2, GSK-3α, MAPK1, MET, PAK1, PDGFRB, PIM1, PKC-γ, PLK4, and SRC) [17]. Ten out of twelve kinases examined retained >95% of their activity in the presence of 50 μM compound 1 (Figure 7d). Two kinases, CAMK1 and PDGFRB, showed approximately 20% inhibition under the same condition. These results suggest that the active dose of compound 1 required to inhibit LMP-1-induced NF-κB activation did not substantially perturb the activities of these essential kinases. Taken together, these data showed that compound 1 specifically inhibits LMP-1 signaling in the EBV-infected B cells.
4. Conclusions
In summary, we have identified novel LMP-1 TMD-5 homo-trimerization disruptors. Compound 1 has been shown to disrupt the TMD-5 association. Furthermore, compound 1 inhibits LMP-1 signaling in EBV infected B cells. Even though only mild potency (~μM) was achieved, our studies provided a proof-of-concept that the lateral TMD interactions could be modulated by small molecule agents. These results might lead to a generally applicable strategy to identify PPI inhibitors of multi-pass membrane proteins by screening against isolated individual TMDs.
Supplementary Material
Highlights.
Propose a cell-based screen based strategy for identifying TMDs disruptors
Identified an LMP-1 TMD-5 self association disruptor, NSC 259242
NSC 259242 disrupts TMD-5 homotrimerization and inhibits LMP-1 signaling
Acknowledgements
We thank Prof. Arthur Pardi for the access of the 500 MHz NMR spectrometer and helpful discussion. We also thank Dr. Geoffrey Armstrong for help with NMR data collectionon the 800 MHz NMR spectrometer. We acknowledge the NCI/DTP Open Chemical Repository for providing the Diversity Set II (http://dtp.cancer.gov). We thank the National Institutes of Health (R21CA138373), Cancer League of Colorado for financial supports of this work. H.Y. is grateful for the 2009 Elion Award from the American Association of Cancer Research, a Kimmel Scholar Award from the Sidney Kimmel Foundation for Cancer Research (SKF-08-101) and a Stand Up to Cancer (SU2C) Innovative Research Award.
Abbreviations
- EBV
Epstein–Barr virus
- LMP-1
latent membrane protein 1
- PPI
protein-protein interaction
- TMD
transmembrane domain
- MBP
maltose binding protein
- ONPG
O-nitrophenyl ß-galactoside
- SPPS
standard solid phase peptide synthesis
- DAGK
diacylglycerol kinase
- BSA
bovine serum albumin
- HRP
horseradish peroxidase
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- DHPC
dihexanoylphosphadtidylcholine
- HMQC
heteronuclear multiple quantum coherence
- HSQC
heteronuclear single quantum coherence
- CSI
chemical shift index
- C14 betaine
3-(N,N-dimethylmyristylammonio)propanesulfonate
- TRAF
TNF receptor associated factor
- iNOS
inducible NO synthase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sackett DL, Sept D. Protein-protein interactions: making drug design second nature. Nat. Chem. 2009;1:596–597. doi: 10.1038/nchem.427. [DOI] [PubMed] [Google Scholar]
- 2.Arkin MR, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat. Rev. Drug Discov. 2004;3:301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 3.Wilson AJ. Inhibition of protein-protein interactions using designed molecules. Chem. Soc. Rev. 2009;38:3289–3300. doi: 10.1039/b807197g. [DOI] [PubMed] [Google Scholar]
- 4.Yin H. Exogenous agents that target transmembrane domains of proteins. Angew. Chem. Int. Ed. Engl. 2008;47:2744–2752. doi: 10.1002/anie.200704780. [DOI] [PubMed] [Google Scholar]
- 5.Zhao TX, Martinko AJ, Le VH, Zhao J, Yin H. Development of agents that modulate protein-protein interactions in membranes. Curr. Pharm. Des. 2010;16:1055–1062. doi: 10.2174/138161210790963878. [DOI] [PubMed] [Google Scholar]
- 6.Quintana FJ, Gerber D, Bloch I, Cohen IR, Shai Y. A structurally altered D,L-amino acid TCRalpha transmembrane peptide interacts with the TCRalpha and inhibits T-cell activation in vitro and in an animal model. Biochemistry. 2007;46:2317–2325. doi: 10.1021/bi061849g. [DOI] [PubMed] [Google Scholar]
- 7.Richter L, Munter LM, Ness J, Hildebrand PW, Dasari M, Unterreitmeier S, Bulic B, Beyermann M, Gust R, Reif B, Weggen S, Langosch D, Multhaup G. Amyloid beta 42 peptide (Abeta42)-lowering compounds directly bind to Abeta and interfere with amyloid precursor protein (APP) transmembrane dimerization. Proc. Natl. Acad. Sci. USA. 2010;107:14597–14602. doi: 10.1073/pnas.1003026107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat. Rev. Cancer. 2004;4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 9.Sammond DW, Joce C, Takeshita R, McQuate S, Ghosh N, Martin JM, Yin H. Transmembrane peptides used to investigate the homo-oligomeric interface and binding hot-spot of latent membrane protein 1. Biopolymers. 2011;95:772–784. doi: 10.1002/bip.21672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zscherp C, Schlesinger R, Tittor J, Oesterhelt D, Heberle J. In situ determination of transient pKa changes of internal amino acids of bacteriorhodopsin by using time-resolved attenuated total reflection Fourier-transform infrared spectroscopy. Proc. Natl. Acad. Sci. USA. 1999;96:5498–5503. doi: 10.1073/pnas.96.10.5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joce C, Wiener A, Yin H. Transmembrane domain oligomerization propensity determined by ToxR assay. J. Vis. Exp. 2011:e2721. doi: 10.3791/2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joce C, Wiener AA, Yin H. Multi-Tox: Application of the ToxR-transcriptional reporter assay to the study of multi-pass protein transmembrane domain oligomerization. Biochim. Biophys. Acta. 2011;1808:2948–2953. doi: 10.1016/j.bbamem.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin H, Slusky JS, Berger BW, Walters RS, Vilaire G, Litvinov RI, Lear JD, Caputo GA, Bennett JS, DeGrado WF. Computational design of peptides that target transmembrane helices. Science. 2007;315:1817–1822. doi: 10.1126/science.1136782. [DOI] [PubMed] [Google Scholar]
- 14.Cavanagh J, Fairbrother WJ, Palmer AG, III, Rance M, Skelton NJ. Protein NMR Spectroscopy: Principles and Practice. 2nd ed. Academic Press; San Diego, CA: 2007. [Google Scholar]
- 15.Schanda P, Kupce E, Brutscher B. SOFAST-HMQC experiments for recording two-dimensional heteronuclear correlation spectra of proteins within a few seconds. J. Biomol. NMR. 2005;33:199–211. doi: 10.1007/s10858-005-4425-x. [DOI] [PubMed] [Google Scholar]
- 16.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. Journal of Biomolecular NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 17.Jester BW, Cox KJ, Gaj A, Shomin CD, Porter JR, Ghosh I. A coiled-coil enabled split-luciferase three-hybrid system: applied toward profiling inhibitors of protein kinases. J. Am. Chem. Soc. 2010;132:11727–11735. doi: 10.1021/ja104491h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hessa T, Kim H, Bihlmaier K, Lundin C, Boekel J, Andersson H, Nilsson I, White SH, von Heijne G. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 2005;433:377–381. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- 19.Wishart DS, Sykes BD, Richards FM. Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J. Mol. Biol. 1991;222:311–333. doi: 10.1016/0022-2836(91)90214-q. [DOI] [PubMed] [Google Scholar]
- 20.Wishart DS, Sykes BD, Richards FM. The chemical shift index: a fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry. 1992;31:1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- 21.Wishart DS, Sykes BD. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J. Biomol. NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 22.Rath A, Glibowicka M, Nadeau VG, Chen G, Deber CM. Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. Proc. Natl. Acad. Sci. USA. 2009;106:1760–1765. doi: 10.1073/pnas.0813167106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.