Abstract
The functional association between intronic miRNAs and their host genes is still largely unknown. We found that three gene loci, which produced miR-26a and miR-26b, were embedded within introns of genes coding for the proteins of carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase (CTDSP) family, including CTDSPL, CTDSP2 and CTDSP1. We conducted serum starvation-stimulation assays in primary fibroblasts and two-thirds partial-hepatectomies in mice, which revealed that miR-26a/b and CTDSP1/2/L were expressed concomitantly during the cell cycle process. Specifically, they were increased in quiescent cells and decreased during cell proliferation. Furthermore, both miR-26 and CTDSP family members were frequently downregulated in hepatocellular carcinoma (HCC) tissues. Gain- and loss-of-function studies showed that miR-26a/b and CTDSP1/2/L synergistically decreased the phosphorylated form of pRb (ppRb), and blocked G1/S-phase progression. Further investigation disclosed that miR-26a/b directly suppressed the expression of CDK6 and cyclin E1, which resulted in reduced phosphorylation of pRb. Moreover, c-Myc, which is often upregulated in cancer cells, diminished the expression of both miR-26 and CTDSP family members, enhanced the ppRb level and promoted the G1/S-phase transition. Our findings highlight the functional association of miR-26a/b and their host genes and provide new insight into the regulatory network of the G1/S-phase transition.
INTRODUCTION
MicroRNAs (miRNAs) belong to a class of endogenously expressed, small non-coding RNAs that cause translational repression and/or mRNA destabilization by binding to the 3′-untranslated regions (3′-UTRs) of the target mRNAs (1). Approximately 40% of all miRNAs are located within intronic regions of protein-coding transcriptional units (TUs) (2). Analysis of 175 human miRNAs across 24 different human organs reveals that the expression of intronic miRNAs largely coincides with the transcription of their host TUs (3), indicating that the intronic miRNAs and their host genes may be co-regulated and are generated from a common precursor transcript.
Emerging evidence suggests that intronic miRNAs may be functionally associated with their host genes. Few reports suggest that there is an antagonizing effect of the intronic miRNA on the function of its host gene, as with miR-218 (4) and miR-10 (5,6). The secreted Slit ligands and their Robo receptors constitute a Slit–Robo signaling pathway that controls the directed migration of neurons and vascular endothelial cells during embryonic development. miR-218 is localized in the intron of the Slit gene, and similar expression patterns are observed between miR-218 and Slit in different tissues. Furthermore, miR-218 inhibits the expression of Robo1 and Robo2, which generates a negative feedback loop in response to Slit gene activation and thereby contributes to the ‘fine-tuning’ of the Slit–Robo pathway (4). Another example of miRNAs associating with their host genes are the miR-10 family members, whose gene loci are retained within the Hox clusters that encode for developmental regulators. Interestingly, Hox transcripts are targets of the miR-10 family members in several species (5,6). However, there may be a synergistic effect of miRNA with its host gene. miR-33a/b act in concert with their SREBP (sterol regulatory element-binding protein) host genes to control cholesterol homeostasis (7–10). miR-208a/b and their Myh6/7 host genes cooperate to govern myosin expression and muscle performance (11). The miR-106–25 cluster and its host gene MCM7 transforms cells synergistically (12). miR-151 and the host gene FAK work together to enhance the motility and spreading of hepatocellular carcinoma (HCC) cells (13). An extensive investigation of the functional association between intronic miRNAs and their host genes will help to disclose the elaborate regulatory network of cellular activity.
Over the course of evolution, the genomic loci of miR-26a and miR-26b have been localized to the introns of genes coding for the proteins of carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase (CTDSP) family. However, the functional association of miR-26 with CTDSP family members and the biological significance of this association remain unknown. Herein, we show that miR-26 family members are expressed concomitantly with their host genes in physiological and pathological conditions. We further disclose that miR-26a/b and their host genes cooperate to block the G1/S-phase transition by synergistically activating the pRb protein. Our findings highlight the functional association of miR-26a/b and their host genes and the potential implication of this association in physiological and pathological processes.
MATERIALS AND METHODS
Cell lines and human tissue specimens
Cancer cell lines, which were derived from the liver (MHCC-97L, HepG2 and Huh7), lung (95D), breast (MCF7) and cervix (HeLa), were maintained in Dulbecco's modified Eagle's medium (DMEM, Hyclone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS, HyClone, Thermo Fisher Scientific, Austria). Primary fibroblasts were isolated from human neonatal foreskin using an optimized enzymatic dissociation procedure (14) and grown in RPMI 1640 medium (Hyclone) supplemented with 10% FBS, penicillin and streptomycin.
Normal liver tissues were collected from patients undergoing resection of hepatic hemangiomas, and paired HCC and adjacent non-tumor liver tissues were obtained from patients undergoing HCC resection at the Cancer Center of Sun Yat-sen University. Both tumor and non-tumor tissues were histologically confirmed. No local or systemic treatments had been conducted before the operations. Informed consent was obtained from each patient, and the study was approved by the Institute Research Ethics Committee at the Cancer Center of Sun Yat-sen University.
Two-thirds partial hepatectomy in mice
Eight- to twelve-week-old male C57BL/6 mice were used. Two-thirds of the liver was surgically removed under chloral hydrate anesthesia as previously described (15). All experimental procedures involving animals were in accordance with the Guide for the Care and Use of Laboratory Animals (NIH publications Nos. 80-23, revised 1996) and were performed according to the institutional ethical guidelines for animal experiments.
RNA oligoribonucleotides and plasmids
hsa-miR-26a/b (Pre-miR miRNA Precursor Product, cat. AM17100) and NC (Pre-miR Negative Control #2, cat. AM17111) were purchased from Applied Biosystems (Foster City, CA, USA). Other RNA oligoribonucleotides (Supplementary Table S1) were obtained from Genepharma (Shanghai, P.R. China). The siRNAs targeting human CDK6 (GenBank accession no. NM_001259), CCNE1 (NM_001238), c-Myc (NM_002467), CTDSP1 (NM_021198 and NM_182642), CTDSP2 (NM_005730) and CTDSPL (NM_005808 and NM_001008392) transcripts were designated as siCDK6, siCCNE1, siMyc, siCTDSP1, siCTDSP2 and siCTDSPL, respectively. The anti-miR-26, with a sequence complementary to mature miR-26b, was a 2′-O-methyl-modified oligoribonucleotide. The anti-miR-C, which is non-homologous to any human genome sequences, was used as a negative control for anti-miR-26.
The vectors used (details in Supplementary Materials and Methods) included firefly luciferase reporter plasmids that contained a wild-type or mutant 3′-UTR segment of human CDK6 or CCNE1, and pc3-gab-CDK6, pc3-gab-CCNE1, pc3-gab-c-Myc, pc3-gab-CTDSP1, pc3-gab-CTDSP2 and pc3-gab-CTDSPL, which expressed CDK6, CCNE1, c-Myc, CTDSP1, CTDSP2 and CTDSPL, respectively.
Cell transfection
RNA oligos were transfected using Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA, USA). A final concentration of 50 nM duplex or 200 nM miRNA inhibitor was used unless indicated. RNA transfection efficiency is ∼70–80% (16), and overexpression of a miRNA mimic persists for at least 4 days (17). TurboFectTM in vitro Transfection Reagent (Fermentas, Hanover, MD, USA) was used for transfection of HepG2 with pc3-gab or pc3-gab-c-Myc. Other transfection of plasmid DNA with or without RNA oligos was performed using Lipofectamine 2000 (Invitrogen).
Cell cycle analysis
All cell cycle analyses were performed using a detergent-containing hypotonic solution (Krishan’s reagent) and fluorescence-activated cell sorting (FACS) (Gallios, Beckman Coulter, Miami, FL, USA) as previously reported (18). Nuclear debris and overlapping nuclei were gated out.
Luciferase reporter assay
MHCC-97L cells grown in a 48-well plate were co-transfected with 10 ng of firefly luciferase reporter comprising a wild-type or mutant 3′-UTR of the target gene, 2 ng of pRL-TK (Promega, Madison, WI, USA) and 10 nM of RNA oligoribonucleotides. A luciferase assay was performed as previously reported (16). The pRL-TK vector that provided the constitutive expression of Renilla luciferase was used as an internal control to correct for the differences in transfection and harvest efficiencies. Transfection was performed in duplicate and was repeated at least three times in independent experiments.
Analysis of gene expression
Semiquantitative RT–PCR and real-time quantitative RT–PCR (qPCR) were performed to evaluate the RNA level. Expression of CTDSP1/2/L was quantified by THUNDERBIRD SYBR qPCR Mix (QPS-201, TOYOBO, Japan), using β-actin and GAPDH as reference genes. Expression of miR-26a/b was quantified using the miRCURY LNA Universal RT microRNA PCR system (Exiqon, Vedbaek, Denmark) when β-actin and GAPDH were used as internal standards, or by a TaqMan MicroRNA Assay kit (Applied Biosystems, Foster City, CA, USA) when U6B was used as reference. Western blot and immunohistochemical staining were performed to detect the protein level (details in Supplementary Materials and Methods).
Bioinformatic tools and statistical analysis
The databases used for bioinformatic analysis are described under Supplementary Materials and Methods.
To search for miRNA families that reside within the introns of protein families, miRNA and protein families were first extracted from the miRBase and Ensembl Genome Browsers, respectively. The sequences of miRNA family members were mapped to hosting TUs using the Ensembl Genome Browser. We only considered those host genes whose intron or exon sequences overlapped the miRNA and were transcribed on the same strand as miRNA.
Data were expressed as the mean ± standard error of the mean (SEM) from at least three independent experiments. Unless otherwise noted, the differences between groups were analyzed using Student's t-test when only two groups were compared and using one-way ANOVA when more than two groups were compared. Correlation was explored by Spearman’s correlation coefficient. All statistical tests were two-sided. A P < 0.05 was considered statistically significant. All analyses were performed using the GraphPad Prism program (GraphPad Software Inc., San Diego, CA, USA).
RESULTS
miR-26 family members are expressed concomitantly with their host genes in physiological and pathological conditions
Since the birth and death rates of miRNA genes are rather high over the course of evolution (19), the co-localization of some miRNAs and their host genes may be a coincidence. However, if a miRNA family always resides within the gene loci of a protein family, they are more likely to be physiologically relevant. To identify miRNAs that exhibit a functional association with their host genes, we first mapped the gene loci of miRNA families to hosting TUs. Among 108 miRNA families with at least two members that have been identified in human, 13 of them were mapped to the introns of protein families (Supplementary Table S2). Except for miR-128, miR-506, miR-548 and miR-1233 families, the colocalization of the other nine miRNA families with protein-coding genes has been previously noted (3–7,9–11,20,21). Notably, miR-26, miR-103, miR-33 and miR-153 family members displayed a perfect one-to-one colocalization with the members of their host protein family (Table 1).
Table 1.
Perfect one-to-one colocalization between miRNA and protein family members
| miRNA family | miRNA family IDa | Protein family IDb | Colocalization of miRNA and protein family members |
|---|---|---|---|
| miR-26 | MIPF0000043 | ENSFM00500000269811 | hsa-miR-26a-1—CTDSPL |
| hsa-miR-26b—CTDSP1 | |||
| hsa-miR-26a-2—CTDSP2 | |||
| miR-103 | MIPF0000024 | ENSFM00260000050538 | hsa-miR-103-2—PANK2 |
| hsa-miR-103-1—PANK3 | |||
| hsa-miR-107—PANK1 | |||
| miR-33 | MIPF0000070 | ENSFM00250000001438 | hsa-miR-33a—SREBF2 |
| hsa-miR-33b—SREBF1 | |||
| miR-153 | MIPF0000050 | ENSFM00260000050522 | hsa-miR-153-1—PTPRN |
| hsa-miR-153-2—PTPRN2 |
amiRNA family ID was derived from miRBase (release 16.0).
bProtein family ID was obtained from Ensembl Genome Browser (release 60).
The miR-26 family comprises miR-26a and miR-26b; they are transcribed from three genomic loci, miR-26a-1, miR-26a-2 and miR-26b, which reside in the introns of genes coding for the CTDSPL, CTDSP2 and CTDSP1 proteins, respectively. The basic arrangement of these miRNAs in the CTDSP1/2/L genes has remained constant throughout vertebrate evolution (Supplementary Figure S1). It has been reported that CTDSPL dephosphorylates the phosphorylated form of pRb (ppRb) and thereby presumably leads to the arrest of the cell cycle at the G1/S boundary (22). Furthermore, expression of miR-26a causes increased G1-cells (23–25). More importantly, among the predicted targets of miR-26a/b, we found an enrichment of G1/S transition-related genes (3.9-fold overrepresentation, P = 0.0184; Supplementary Table S3).
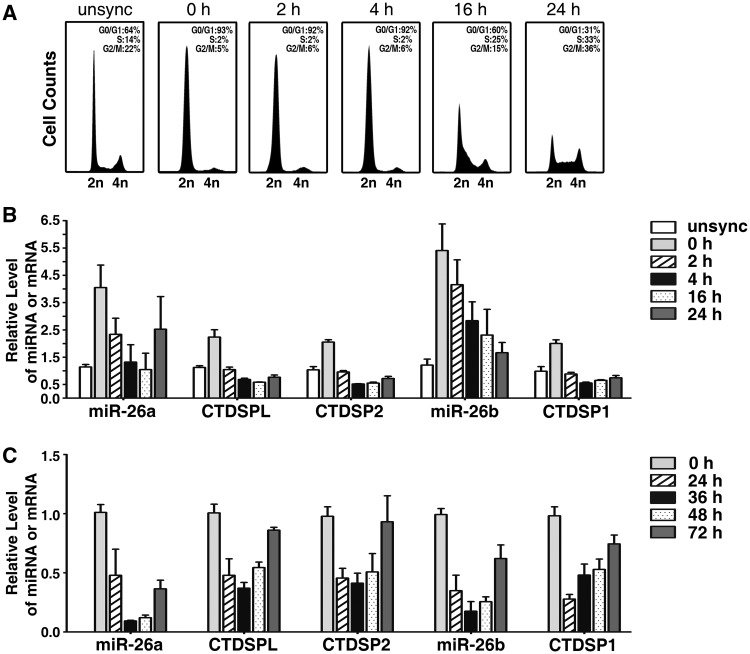
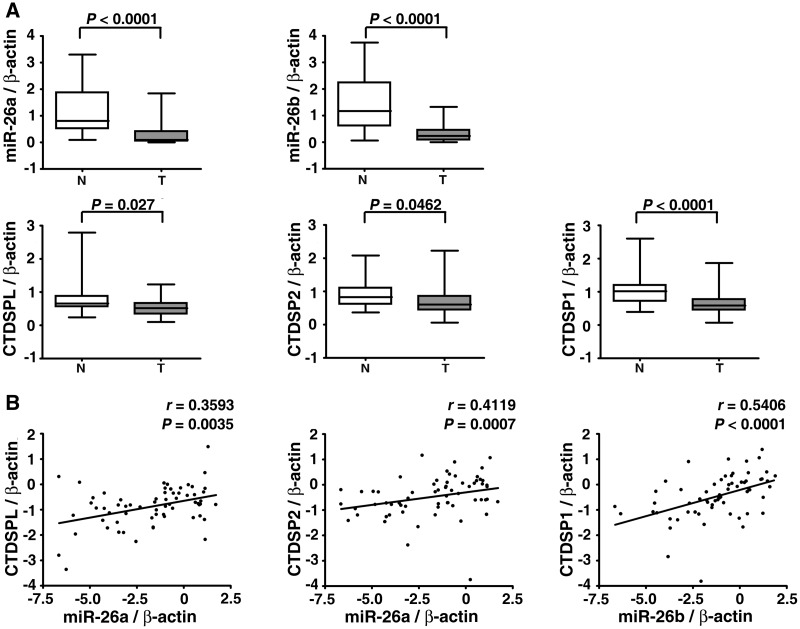
Based on the above pieces of evidence, we first examined whether miR-26a/b and CTDSP1/2/L were expressed concomitantly during the cell cycle process using a serum starvation–stimulation assay. Human primary fibroblasts were serum deprived for 48 h and stimulated to induce S-phase entry by serum re-addition. The cells were synchronized in G0-phase by serum deprivation. Within 4 h after serum addition, the cells remained in G1-phase, whereas a large proportion of cells entered S-phase at 16 h and progressed to G2/M-phase at 24 h (Figure 1A). The expression of miR-26a/b and CTDSP1/2/L increased after serum deprivation, when the cell cycle was arrested at G0-phase, and decreased quickly after serum stimulation (Figure 1B, Supplementary Figure S2A). Furthermore, miR-26a/b and CTDSP1/2/L displayed a similar trend of expression over time (Figure 1B, Supplementary Figure S2A). To confirm the findings from the in vitro study, we conducted the two-thirds partial hepatectomy in mice, a physiological liver regeneration model that is often used to study the cell cycle in vivo. It has been showed that the majority of the hepatic cells were undergoing proliferation around 36 h post-hepatectomy (15,26,27). Interestingly, we found that the expression of miR-26a/b and CTDSP1/2/L coordinately reduced at 24 h and 36 h post-hepatectomy, and began to elevate around 48 h, once cellular proliferation declined (Figure 1C, Supplementary Figure S2B). Furthermore, miR-26 and CTDSP family members were frequently down-regulated in human HCC tissues (Figure 2A, Supplementary Figure S3A), and a similar expression pattern was observed between miR-26a and miR-26b (Supplementary Figure S4) and between miR-26a/b and their respective host genes (Figure 2B, Supplementary Figure S3B).
Figure 1.
miR-26a/b are expressed concomitantly with CTDSP1/2/L under physiological conditions. (A) FACS analysis of the cell cycle distribution of human primary fibroblasts. The percentages of G0/G1-, S- and G2/M-cells are indicated in each histogram. (B) Analysis of mature miR-26a/b and CTDSP1/2/L expression in human primary fibroblasts by quantitative real-time PCR (qPCR). For (A and B), cells were serum deprived for 48 h, then stimulated to enter S-phase by serum re-addition. Unsync, exponentially growing cells without serum deprivation; 0 h denotes serum-deprived cells at the time point when serum was re-added; 2 h, 4 h, 16 h and 24 h denote cells in the indicated time points after serum re-addition. (C) Analysis of mature miR-26a/b and CTDSP1/2/L expression in mouse liver by qPCR. 0 h denotes two-thirds of the liver that was surgically removed; 24 h, 36 h, 48 h and 72 h denote livers that were obtained at the indicated time points after partial hepatectomy. The expression levels of miR-26a/b and CTDSP1/2/L were normalized to β-actin.
Figure 2.
miR-26a/b are expressed concomitantly with CTDSP1/2/L in a pathological condition. (A) Analysis of mature miR-26a/b and CTDSP1/2/L expression in 32 paired HCC and adjacent non-tumor tissues by qPCR. T, HCC tissue and N, adjacent non-cancerous tissue. The y-axis indicates the relative level of miR-26a/b and CTDSP1/2/L. The boxes represent the interquartile range (25–75th centiles). The horizontal line inside the box indicates the median. The vertical whiskers extend to the maximum and minimum values. (B) The correlation between expression levels of miR-26a/b and their respective host genes in 32 paired HCC and adjacent non-tumor tissues. The expression status of miR-26a/b and CTDSP1/2/L is shown in a log 2 scale. For (A and B), statistical analysis was performed by Wilcoxon matched-pairs signed-ranks test and Spearman’s correlation coefficient, respectively. The expression levels of miR-26a/b and CTDSP1/2/L were normalized to β-actin.
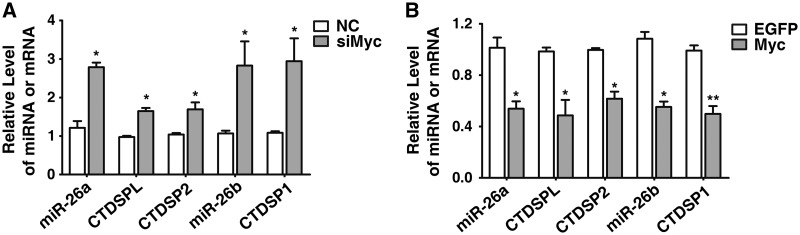
Expression of miR-26a/b can be suppressed by c-Myc in lymphoma cells (25,28). We also found that c-Myc silencing increased endogenous miR-26a/b and CTDSP1/2/L levels in HCC cells (Figure 3A), whereas overexpression of c-Myc downregulated the expression of miR-26a/b and CTDSP1/2/L (Figure 3B).
Figure 3.
The expression of miR-26a/b and CTDSP1/2/L is co-regulated by c-Myc. (A) Effect of c-Myc silencing on mature miR-26a/b and CTDSP1/2/L expression in HepG2 cells. Forty-eight hours after transfection with 20 nM indicated RNA duplex, cells were analyzed by qPCR. NC, negative control; siMyc, siRNA targets c-Myc. (B) Effect of c-Myc overexpression on mature miR-26a/b and CTDSP1/2/L levels in HepG2 cells. Cells were transfected with empty pc3-gab vector (indicated as EGFP) or pc3-gab-c-Myc (Myc) for 48 h, then subjected to qPCR. The expression levels of miR-26a/b and CTDSP1/2/L were normalized to β-actin. *P < 0.05; **P < 0.01.
These results indicate that miR-26a/b and CTDSP1/2/L may be co-regulated in both physiological and pathological processes.
miR-26a/b function synergistically with their host genes to block G1/S transition
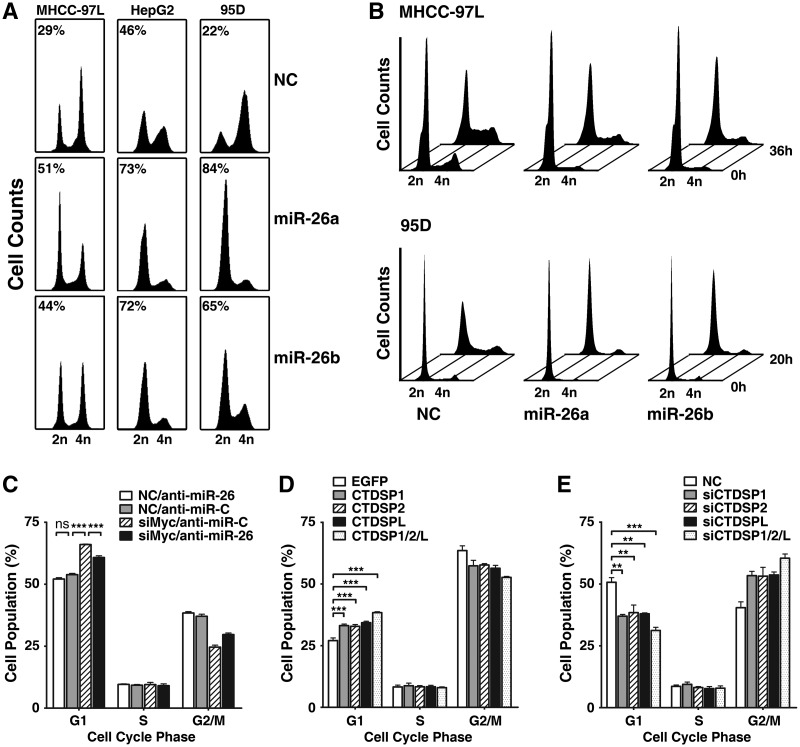
Next, we evaluated the potential roles of miR-26a/b and their host genes in the regulation of G1/S transition and determined the functional significance of their association. Overexpression of either miR-26a or miR-26b resulted in a marked accumulation of the G1-population in different cancer cell lines derived from the liver (MHCC-97L, HepG2 and Huh7), lung (95D), breast (MCF7) and cervix (HeLa) (Figure 4A, Supplementary Figure S5), indicating a ubiquitous role for miR-26a/b in cell cycle control. Serum starvation–stimulation assays further revealed that most of the miR-26a/b-transfectants remained in G1-phase (Figure 4B), while a large proportion of NC-transfected cells already entered S-phase after serum re-addition, suggesting that miR-26a/b can block G1/S transition.
Figure 4.
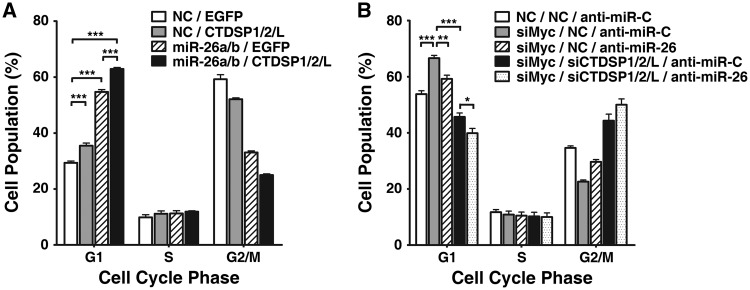
miR-26a/b and CTDSP1/2/L regulate G1/S transition. (A) miR-26a/b overexpression caused accumulation of G1-population. The percentage of the G1-population is indicated within each histogram. (B) miR-26a/b overexpression blocked S-phase entry upon serum stimulation. NC- or miR-26a/b-transfectants were serum-deprived for 48 h, followed by serum re-addition, and then were harvested at the indicated time points. The time point when serum was re-added was set as 0 h. (C) Inhibition of miR-26 promoted G1/S transition in c-Myc-knockdown cells. HepG2 cells were co-transfected with siMyc (20 nM) and anti-miR-26 (200 nM), followed by FACS analysis. The NC and anti-miR-C were used as the negative controls for siMyc and anti-miR-26, respectively. (D) Expression of CTDSP family members resulted in G1-arrest. The 95D cells were transfected with empty pc3-gab vector (EGFP), pc3-gab-CTDSP1, pc3-gab-CTDSP2, pc3-gab-CTDSPL or the mixture of CTDSP1, CTDSP2 and CTDSPL expressing vectors as indicated. (E) Knockdown of CTDSP family members promoted G1/S progression in HepG2 cells. Cells were transfected with 50 nM of siCTDSP1, siCTDSP2, siCTDSPL or a mixture of all three siRNA (16.7 nM each). In (A) and (C–E), nocodazole (40 ng/ml) was added 32 h after transfection, and the cells were cultured for an additional 16 h before harvest. ns, not significant; **P < 0.01; ***P < 0.001.
To confirm the results from the gain-of-function study, loss-of-function analysis was performed using anti-miR-26, which could knockdown the expression of miR-26a and miR-26b (Supplementary Figure S6). Compared with the control group (anti-miR-C), transfection of anti-miR-26 only caused a slight decrease in G1-cells (Figure 4C), which might be explained by the low basal level of miR-26 in these cells (Supplementary Figure S7). Thus, we evaluated the effect of miR-26 inhibition using a c-Myc-knockdown model in which c-Myc silencing caused elevated endogenous miR-26a/b (Figure 3A). As shown in Figure 4C, knockdown of c-Myc resulted in an increase in the G1-population, whereas this effect was attenuated when miR-26a/b was inhibited by anti-miR-26.
Although it has been shown that CTDSPL may arrest the cell cycle at G1-phase by dephosphorylating ppRb, the effect of the other CTDSP family members on the cell cycle is still unknown. Therefore, we examined whether all host genes of miR-26a/b play a regulatory role in G1/S transition. Expression of either of CTDSP1/2/L induced G1-arrest (Figure 4D), whereas silencing either of them promoted G1/S progression (Figure 4E). Furthermore, the effect of triple-overexpression or -knockdown of CTDSP1/2/L was more pronounced than single-expression or -knockdown of either one (Figure 4D and E).
We further determined whether a synergistic effect exists between miR-26a/b and their host genes in the context of cell cycle regulation. The G1-arrest was more evidenced when miR-26a/b and CTDSP1/2/L were co-expressed (Figure 5A). Consistently, simultaneous suppression of miR-26a/b and their host genes resulted in a more pronounced G1/S progression in siMyc-transfectants (Figure 5B).
Figure 5.
miR-26a/b function synergistically with CTDSP1/2/L to block G1/S transition. (A) Effect of miR-26a/b and CTDSP1/2/L co-expression on the cell cycle. The 95D cells were transfected with NC or miR-26a/b for 24 h, followed by transfection with the control vector (EGFP) or the mixture of the CTDSP1-, CTDSP2- and CTDSPL-expression vector. (B) Effect of miR-26a/b and CTDSP1/2/L inhibition on the cell cycle. HepG2 cells were co-transfected with 20 nM siMyc, a mixture of siCTDSP1/2/L (6.7 nM each) and 200 nM anti-miR-26. For (A and B), nocodazole was added 32 h after the last transfection, and the cells were cultured for an additional 16 h before harvesting for FACS analysis. *P < 0.05; **P < 0.01; and ***P < 0.001.
Collectively, these data implicate that miR-26a/b can function synergistically with their host genes to block G1/S transition.
miR-26a/b and their host genes block G1/S transition by synergistically activating the pRb protein
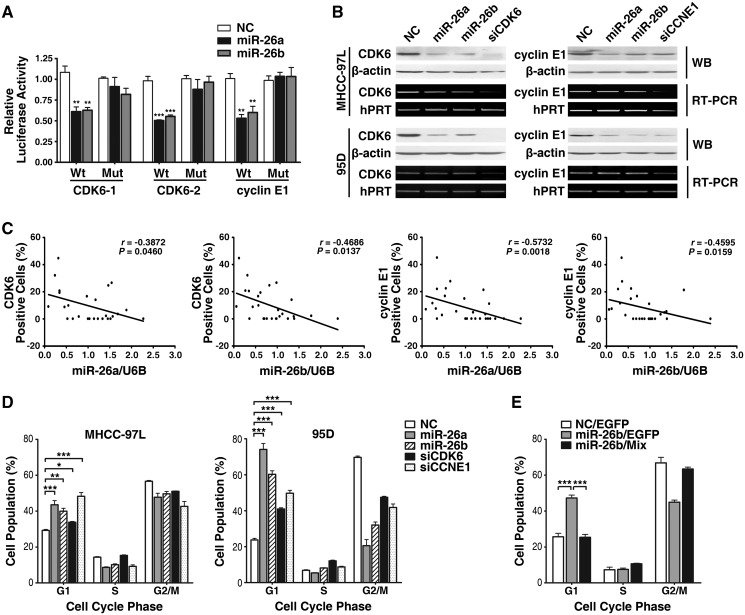
We then explored the molecular mechanisms responsible for the observed function of miR-26a/b and their host genes. The predicted targets of miR-26a/b included four G1/S transition-related genes, CDK6, cyclin E1, cyclin D2 and cyclin E2 (Supplementary Table S3), which are crucial components of the cell cycle that initiate G1/S progression. Since cyclin D2 and cyclin E2 are the known targets of miR-26 (24), we focused on CDK6 and cyclin E1. We identified one putative miR-26-binding site in the 3′-UTR of CCNE1 and two sites in the 3′-UTR of CDK6 (Supplementary Figure S8) using TargetScan. A dual-luciferase reporter system revealed that transfection of miR-26a/b significantly suppressed the firefly luciferase activity of the reporter containing the wild-type, but not the mutant binding site of the 3′-UTR (Figure 6A), indicating that miR-26 may suppress gene expression through its binding sequences at the 3′-UTR of target genes. Further investigation showed that miR-26a/b could also diminish endogenous expression of both CDK6 and cyclin E1 proteins (Figure 6B). Moreover, downregulation of miR-26a/b was statistically correlated with overexpression of CDK6 and cyclin E1 in HCC tissues (Figure 6C). These data suggest that CDK6 and cyclin E1 are direct targets of miR-26a/b.
Figure 6.
CDK6 and cyclin E1 are involved in miR-26-mediated G1-arrest. (A) miR-26a/b suppressed the activity of luciferase through its binding sequences at the 3′-UTR of target genes. MHCC-97L cells were co-transfected with miR-26a/b or NC, a firefly luciferase reporter containing wild-type or mutant 3′-UTR (indicated as Wt or Mut on the x-axis), and Renilla luciferase expressing constructs. The firefly luciferase activity of each sample was normalized to the Renilla luciferase activity. (B) miR-26a/b, siCDK6 and siCCNE1 repressed endogenous CDK6 and cyclin E1 expression. Forty-eight hours after transfection with the indicated RNA duplex (50 nM), endogenous mRNA and protein levels were assessed by RT–PCR and western blot, respectively. β-actin and hPRT were used as internal controls. (C) miR-26a/b levels were inversely correlated with CDK6/cyclin E1 expression in HCC tissues. Expression of CDK6 and cyclin E1 were analyzed by immunohistochemical staining, and the levels of mature miR-26a/b were detected by qPCR. Statistical analysis was performed using Spearman's correlation coefficient. (D) Overexpression of miR-26a/b and knockdown of either CDK6 or cyclin E1 increased the G1-population. (E) Expression of CDK6 and cyclin E1 attenuated miR-26b-induced G1-arrest. The 95D cells were first transfected with 20 nM NC (negative control) or miR-26b for 24 h, followed by transfection with the empty pc3-gab vector (indicated as EGFP) or the mixture of pc3-gab-CDK6 and pc3-gab-CCNE1 vectors (Mix), which encoded the entire coding sequence of the indicated gene, but lacked the 3′-UTR. *P < 0.05; **P < 0.01; and ***P < 0.001.
The involvement of CDK6 and cyclin E1 in miR-26-regulated G1/S transition was then addressed. Silencing of either CDK6 or cyclin E1 (Figure 6B) led to the arrest of the cell cycle in G1-phase (Figure 6D), which was the phenocopy of the result of enhanced miR-26 expression, whereas overexpression of CDK6 and cyclin E1 abrogated miR-26-induced G1-arrest (Figure 6E).
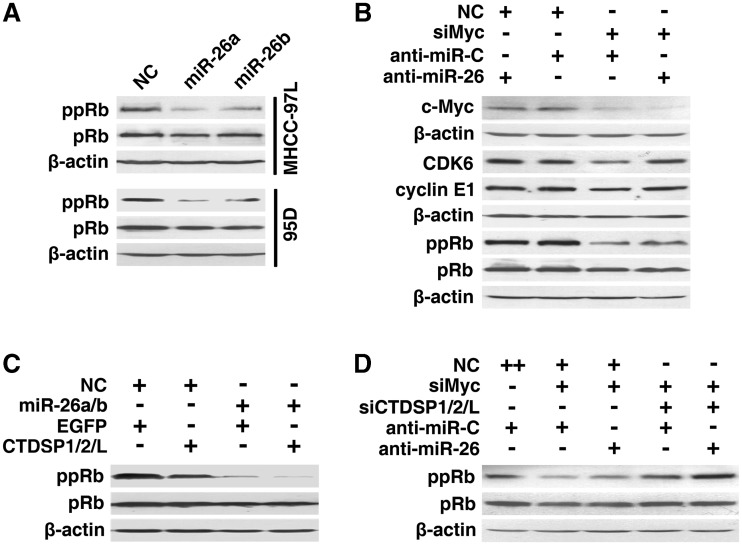
It is well known that CDK6, cyclin E1, cyclin D2 and cyclin E2 initiate the phosphorylation of pRb, which in turn, results in pRb inactivation and the release of E2F. E2F consequently transactivates the genes required for S-phase entry. Therefore, we explored whether miR-26 could attenuate these events. Introduction of miR-26 caused a prominent decrease of ppRb (Figure 7A), whereas miR-26 inhibition enhanced the expression of CDK6 and cyclin E1 and increased the level of ppRb in siMyc-transfectants (Figure 7B). These results imply that miR-26 may activate the pRb protein by suppressing CDK6 and multiple G1-phase cyclins.
Figure 7.
miR-26a/b and CTDSP1/2/L block the G1/S transition by synergistically activating the pRb protein. (A) Effect of miR-26a/b on the phosphorylation of the pRb protein. (B) Effect of anti-miR-26 on the levels of CDK6, cyclin E1 and ppRb in c-Myc-knockdown cells. HepG2 cells were co-transfected with the indicated siRNA duplex (20 nM) and miRNA inhibitor (200 nM). (C) The synergistic effect of miR-26a/b and CTDSP1/2/L on ppRb levels. The 95D cells were transfected with NC or miR-26a/b for 24 h, followed by transfection with the control vector (EGFP) or the mixture of the CTDSP1-, CTDSP2- and CTDSPL-expression vectors (CTDSP1/2/L). (D) The synergistic effect of anti-miR-26 and siCTDSP1/2/L on ppRb levels in c-Myc-knockdown cells. HepG2 cells were co-transfected with 20 nM siMyc, the mixture of siCTDSP1, siCTDSP2 and siCTDSPL (6.7 nM each) and 200 nM miRNA inhibitor. For (A–D), 48 h after the last transfection with the indicated RNA oligoribonucleotides or vectors, cells were subjected to western blot analysis. + , 20 nM of the indicated RNA duplex or 200 nM miRNA inhibitor was transfected; ++, 40 nM NC was transfected. NC and anti-miR-C were used as negative controls for RNA duplexes and anti-miR-26, respectively. pRb, total pRb protein; ppRb, Ser780-phosphorylated pRb; β-actin, internal control.
Interestingly, expression of CTDSP1/2/L significantly decreased the level of ppRb (Figure 7C, lanes 1 and 2), and a synergistic repression was observed when CTDSP1/2/L were co-expressed with miR-26a/b (Figure 7C). Consistently, knockdown of CTDSP1/2/L increased the amount of ppRb (Figure 7D, lanes 2 and 4), and the synergistic effect was also found when both miR-26a/b and their host genes were inhibited (Figure 7D).
These findings indicate that miR-26a/b and their host genes cooperate to activate pRb protein, and in turn, block the G1/S transition.
DISCUSSION
Although 40% of all miRNAs are located within intronic regions of protein-coding TUs (2), the functional association between miRNAs and their host genes remains unclear. In this study, we found that miR-26a/b were expressed concomitantly with their host genes CTDSP1/2/L in both physiological and pathological processes. Moreover, miR-26a/b and CTDSP1/2/L synergistically blocked the G1/S transition by activating the same protein, pRb.
The pRb-pathway acts as a master checkpoint in cell cycle progression. Cyclic phosphorylation and dephosphorylation of the pRb protein control the entry of cells into S-phase. Here, we found a synergistic effect of miR-26a/b and CTDSP1/2/L in decreasing the ppRb level and blocking the G1/S progression. Both miR-26a and miR-26b reduced the amount of ppRb by repressing the CDK and cyclins that mediate the phosphorylation of pRb. On the other hand, CTDSP1/2/L may directly dephosphorylate ppRb because CTDSP1/2/L possess phosphatase activity (29–31). It is intriguing to find that miR-26a/b and their host genes regulate pRb phosphorylation through different molecular mechanisms. The cooperation of miR-26 and CTDSP1/2/L in the pRb pathway may provide a ‘double guarantee system’ for the activation of the pRb protein, which guards cells from excessive proliferation. Our observations give new insight into the elaborate control of G1/S transition and suggest the existence of a single gene locus that harbors two independent, but cooperating growth-suppressive elements.
c-Myc is essential for progression from G0/G1 to S-phase (32). It has been demonstrated that c-Myc activates the positive regulator of G1/S transition, including cyclin D1, cyclin D2, cyclin E1, CDK4, CDC25A and E2F, during cell cycle progression (32). Here, we showed that c-Myc could downregulate both miR-26 and CTDSP family members, which enhanced ppRb levels and ultimately promoted the G1/S transition. Our findings disclose a novel mechanism by which c-Myc promotes cell cycle progression by suppressing one gene locus that carries both miRNA and protein-coding gene.
The serum starvation–stimulation assay in primary fibroblasts and the two-thirds partial hepatectomy in mice revealed the increased expression of miR-26a/b and CTDSP1/2/L in quiescent cells and the downregulation of both families during cell proliferation. Furthermore, the levels of miR-26a/b and CTDSP1/2/L were reduced in HCC tissues. These data suggest that miR-26a/b and CTDSP1/2/L may keep fibroblasts and hepatocytes in a non-dividing state, and their deregulation may promote malignant transformation. The underlying mechanism responsible for the decreased expression of these two families in HCC is still unknown. Notably, c-Myc activation is a common event in HCC (33). Amplification of chromosome 8q24, in which c-Myc is located, and upregulation of c-Myc and its downstream target genes are observed in viral and alcohol-related HCC (34). Studies from animal models have also revealed that c-Myc overexpression promotes HCC development (33). Thus, abnormal c-Myc activation may represent an important mechanism underlying the downregulation of miR-26a/b and CTDSP1/2/L. In addition, CTDSP1/miR-26b is located at chromosome 2q35, a region that is frequently deleted in human cancers, especially HCC (35). We were also able to computationally map CpG islands in the promoter regions (between −2000 and 600 bp) of CTDSP1/2/L and miR-26a/b. Therefore, deletion and promoter hypermethylation may also account for the reduced expression of CTDSP1/2/L and miR-26a/b in HCC.
CTDSP1, CTDSP2 and CTDSPL belong to a protein family of small C-terminal domain (CTD) phosphatases, also known as the SCP family. They can negatively regulate RNA polymerase II (RNAPII) by dephosphorylating its CTD on Ser-5 in vitro (31), and also function as transcriptional co-repressors that inhibit the transcription of neuronal gene in non-neuronal cells (36). Growing evidence also suggests that CTDSP1/2/L act as phosphatases of Smad1, Smad2/3 and snail (29,30). It has been demonstrated that CTDSPL dephosphorylates the ppRb protein and presumably regulates the cell cycle. CTDSPL is homozygously deleted in ∼15% of cervical carcinomas, and CTDSPL suppresses the ability of renal carcinoma cells to form colonies in vitro and to develop tumors in SCID mice (22). However, the effect of the other two family members, CTDSP1 and CTDSP2, on the cell cycle and tumorigenesis is still unknown. Here, we showed that CTDSPL, CTDSP1 and CTDSP2 could block G1/S progression, and all CTDSP family members were downregulated in HCC tissues, suggesting that CTDSP1/2/L dysfunction may be involved in hepatocarcinogenesis.
In summary, we disclosed the cooperative effect of miR-26a/b and their host genes CTDSP1/2/L in the cell cycle control and their potential implication in physiological and pathological processes. Our findings highlight the functional association of miR-26a/b and their host genes, provide new insight into the regulatory network of the cell cycle and open the possibility for future therapeutic intervention.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Methods, Supplementary Tables S1–S3, Supplementary Figures S1–S8.
FUNDING
This work was supported by the Ministry of Science and Technology of China (2010CB912803, 2011CB811305); National Natural Science Foundation of China (30870964, 30925036); and the Ministry of Health of China (2012ZX10002-011). Funding for open access charge: grant from the Ministry of Science and Technology of China (2010CB912803).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Qi He for the help of statistical analysis. We are grateful to the Bank of Tumor Resources and Prof. Yunfei Yuan in Department of Hepatobiliary Oncology, Cancer Center, Sun Yat-sen University for providing tissue specimens and the clinical information of patients.
REFERENCES
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 3.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Small EM, Sutherland LB, Rajagopalan KN, Wang S, Olson EN. MicroRNA-218 regulates vascular patterning by modulation of Slit-Robo signaling. Circ. Res. 2010;107:1336–1344. doi: 10.1161/CIRCRESAHA.110.227926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lund AH. miR-10 in development and cancer. Cell Death Differ. 2010;17:209–214. doi: 10.1038/cdd.2009.58. [DOI] [PubMed] [Google Scholar]
- 6.Woltering JM, Durston AJ. MiR-10 represses HoxB1a and HoxB3a in zebrafish. PLoS One. 2008;3:e1396. doi: 10.1371/journal.pone.0001396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerin I, Clerbaux LA, Haumont O, Lanthier N, Das AK, Burant CF, Leclercq IA, MacDougald OA, Bommer GT. Expression of miR-33 from an SREBP2 intron inhibits cholesterol export and fatty acid oxidation. J. Biol. Chem. 2010;285:33652–33661. doi: 10.1074/jbc.M110.152090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horie T, Ono K, Horiguchi M, Nishi H, Nakamura T, Nagao K, Kinoshita M, Kuwabara Y, Marusawa H, Iwanaga Y, et al. MicroRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 (Srebp2) regulates HDL in vivo. Proc. Natl Acad. Sci. USA. 2010;107:17321–17326. doi: 10.1073/pnas.1008499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, Naar AM. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, Moore KJ, Fernandez-Hernando C. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, Kelm RJ, Olson EN. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev. Cell. 2009;17:662–673. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poliseno L, Salmena L, Riccardi L, Fornari A, Song MS, Hobbs RM, Sportoletti P, Varmeh S, Egia A, Fedele G, et al. Identification of the miR-106b∼25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci. Signal. 2010;3:a29. doi: 10.1126/scisignal.2000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding J, Huang S, Wu S, Zhao Y, Liang L, Yan M, Ge C, Yao J, Chen T, Wan D, et al. Gain of miR-151 on chromosome 8q24.3 facilitates tumour cell migration and spreading through downregulating RhoGDIA. Nat. Cell Biol. 2010;12:390–399. doi: 10.1038/ncb2039. [DOI] [PubMed] [Google Scholar]
- 14.Veelken H, Jesuiter H, Mackensen A, Kulmburg P, Schultze J, Rosenthal F, Mertelsmann R, Lindemann A. Primary fibroblasts from human adults as target cells for ex vivo transfection and gene therapy. Hum. Gene Ther. 1994;5:1203–1210. doi: 10.1089/hum.1994.5.10-1203. [DOI] [PubMed] [Google Scholar]
- 15.Mitchell C, Willenbring H. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat. Protocols. 2008;3:1167–1170. doi: 10.1038/nprot.2008.80. [DOI] [PubMed] [Google Scholar]
- 16.Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y, Zhuang SM. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res. 2009;69:1135–1142. doi: 10.1158/0008-5472.CAN-08-2886. [DOI] [PubMed] [Google Scholar]
- 17.Xiong Y, Fang JH, Yun JP, Yang J, Zhang Y, Jia WH, Zhuang SM. Effects of microRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology. 2010;51:836–845. doi: 10.1002/hep.23380. [DOI] [PubMed] [Google Scholar]
- 18.Wang WZ, Cheng J, Luo J, Zhuang SM. Abrogation of G2/M arrest sensitizes curcumin-resistant hepatoma cells to apoptosis. FEBS Lett. 2008;582:2689–2695. doi: 10.1016/j.febslet.2008.06.048. [DOI] [PubMed] [Google Scholar]
- 19.Lu J, Shen Y, Wu Q, Kumar S, He B, Shi S, Carthew RW, Wang SM, Wu CI. The birth and death of microRNA genes in Drosophila. Nat. Genet. 2008;40:351–355. doi: 10.1038/ng.73. [DOI] [PubMed] [Google Scholar]
- 20.Wilfred BR, Wang WX, Nelson PT. Energizing miRNA research: a review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways. Mol. Genet. Metab. 2007;91:209–217. doi: 10.1016/j.ymgme.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kashuba VI, Li J, Wang F, Senchenko VN, Protopopov A, Malyukova A, Kutsenko AS, Kadyrova E, Zabarovska VI, Muravenko OV, et al. RBSP3 (HYA22) is a tumor suppressor gene implicated in major epithelial malignancies. Proc. Natl Acad. Sci. USA. 2004;101:4906–4911. doi: 10.1073/pnas.0401238101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu J, He ML, Wang L, Chen Y, Liu X, Dong Q, Chen YC, Peng Y, Yao KT, Kung HF, et al. MiR-26a inhibits cell growth and tumorigenesis of nasopharyngeal carcinoma through repression of EZH2. Cancer Res. 2011;71:225–233. doi: 10.1158/0008-5472.CAN-10-1850. [DOI] [PubMed] [Google Scholar]
- 24.Kota J, Chivukula RR, O'Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sander S, Bullinger L, Klapproth K, Fiedler K, Kestler HA, Barth TF, Moller P, Stilgenbauer S, Pollack JR, Wirth T. MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood. 2008;112:4202–4212. doi: 10.1182/blood-2008-03-147645. [DOI] [PubMed] [Google Scholar]
- 26.Taub R. Liver regeneration: from myth to mechanism. Nat. Rev. Mol. Cell Biol. 2004;5:836–847. doi: 10.1038/nrm1489. [DOI] [PubMed] [Google Scholar]
- 27.Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302:255–259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- 28.Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu Y, Evers BM, Zhou BP. Small C-terminal domain phosphatase enhances snail activity through dephosphorylation. J. Biol. Chem. 2009;284:640–648. doi: 10.1074/jbc.M806916200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sapkota G, Knockaert M, Alarcon C, Montalvo E, Brivanlou AH, Massague J. Dephosphorylation of the linker regions of Smad1 and Smad2/3 by small C-terminal domain phosphatases has distinct outcomes for bone morphogenetic protein and transforming growth factor-beta pathways. J. Biol. Chem. 2006;281:40412–40419. doi: 10.1074/jbc.M610172200. [DOI] [PubMed] [Google Scholar]
- 31.Yeo M, Lin PS, Dahmus ME, Gill GN. A novel RNA polymerase II C-terminal domain phosphatase that preferentially dephosphorylates serine 5. J. Biol. Chem. 2003;278:26078–26085. doi: 10.1074/jbc.M301791200. [DOI] [PubMed] [Google Scholar]
- 32.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat. Rev. Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 33.Shachaf CM, Kopelman AM, Arvanitis C, Karlsson A, Beer S, Mandl S, Bachmann MH, Borowsky AD, Ruebner B, Cardiff RD, et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431:1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- 34.Schlaeger C, Longerich T, Schiller C, Bewerunge P, Mehrabi A, Toedt G, Kleeff J, Ehemann V, Eils R, Lichter P, et al. Etiology-dependent molecular mechanisms in human hepatocarcinogenesis. Hepatology. 2008;47:511–520. doi: 10.1002/hep.22033. [DOI] [PubMed] [Google Scholar]
- 35.Li SP, Wang HY, Li JQ, Zhang CQ, Feng QS, Huang P, Yu XJ, Huang LX, Liang QW, Zeng YX. Genome-wide analyses on loss of heterozygosity in hepatocellular carcinoma in Southern China. J. Hepatol. 2001;34:840–849. doi: 10.1016/s0168-8278(01)00047-2. [DOI] [PubMed] [Google Scholar]
- 36.Yeo M, Lee SK, Lee B, Ruiz EC, Pfaff SL, Gill GN. Small CTD phosphatases function in silencing neuronal gene expression. Science. 2005;307:596–600. doi: 10.1126/science.1100801. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.