Abstract
An ATP-dependent DNA translocase domain consisting of seven conserved motifs is a general feature of all ATP-dependent chromatin remodelers. While motifs on the ATPase domains of the yeast SWI/SNF and ISWI families of remodelers are highly conserved, the ATPase domains of these complexes appear not to be functionally interchangeable. We found one reason that may account for this is the ATPase domains interact differently with nucleosomes even though both associate with nucleosomal DNA 17–18 bp from the dyad axis. The cleft formed between the two lobes of the ISW2 ATPase domain is bound to nucleosomal DNA and Isw2 associates with the side of nucleosomal DNA away from the histone octamer. The ATPase domain of SWI/SNF binds to the same region of nucleosomal DNA, but is bound outside of the cleft region. The catalytic subunit of SWI/SNF also appears to intercalate between the DNA gyre and histone octamer. The altered interactions of SWI/SNF with DNA are specific to nucleosomes and do not occur with free DNA. These differences are likely mediated through interactions with the histone surface. The placement of SWI/SNF between the octamer and DNA could make it easier to disrupt histone–DNA interactions.
INTRODUCTION
The accessibility of DNA to different factors in the eukaryotic genome is regulated by protein complexes that can reorganize chromatin. One of two main enzymatic activities that regulate chromatin structure is ATP-dependent chromatin remodeling (1,2). ATP-dependent remodeling enzymes contain a core subunit with an ATPase or DNA translocase domain of the SF2 superfamily (3–5). The common feature of SF2 superfamily proteins is the conserved seven helicase-related sequence motifs I, Ia, II, III, IV, V and VI that are critical for nucleic acid binding and ATP hydrolysis. The ATPase domain of SNF2 family proteins possesses two RecA-like domains (6,7). The four principal families of ATP-dependent chromatin remodelers are SWI/SNF, ISWI, CHD and INO80. Distinguishing features of these remodelers are the different domains found in the catalytic subunit such as the bromo, chromo, SnAC, SANT and HSA domains. Each remodeler appears to have its distinct role and different biochemical properties.
ISWI moves and spaces nucleosomes along DNA, and is involved in the global regulation of chromatin structure (8–11). A minimal length of extranucleosomal DNA is required by ISWI to move nucleosomes and is the basis for ISWI spacing nucleosomes (12–14). SWI/SNF on the other hand does not require a minimal length of extranucleosomal DNA and disassembles rather than spaces nucleosomes in short arrays (15–17). There are two members of the SWI/SNF family (SWI/SNF and RSC) and three of the ISWI family (ISW1a, ISW1b and ISW2) in yeast. While canonical nucleosome structure is preserved after mono-nucleosomes are moved by ISW2, SWI/SNF remodeling changes the path of DNA around the nucleosome (16,18–21). It has been unclear how DNA translocation can have such different outcomes, since both ISW2 and SWI/SNF translocate on DNA at the same superhelical location (SHL) 2 in nucleosomes (22–24).
The domains and subunits unique to particular remodeling complexes are likely to facilitate and modulate their activities in crucial ways, but could there also be important differences between the helicase domains of the various remodelers? The helicase or DNA translocase domain in chromatin remodelers has several features that are not found in other members of the SNF2 family of helicases. Chromatin remodelers have an elongated region between motifs III and IV not found in other SNF2 helicases (3). The N-terminal lobe of remodelers has four additional conserved blocks E, F, A and G; and the C-terminal blocks K, D and L that are found only in chromatin remodelers (4). When comparing the helicase domains of remodelers across many different species, they were found to cluster based on their sequence similarity into groups that correlated remarkably well to the subfamilies of SWI/SNF, ISWI, INO80, SWR1, CHD1 and CHD7 remodelers (4). DNA translocase domains thus appear to be specially adapted to their particular subfamily of remodelers and these adaptations are evolutionarily conserved. The differences between subfamilies are not in the conserved helicase motifs comprising the catalytic core of the ATPase domain, but are in the linker and other parts of the DNA translocase domains.
The possibility of the DNA translocase domain helping to dictate differences in mobilizing nucleosomes has been indicated for human SWI/SNF (BRG1) and ISWI (SNF2h) (25). When the DNA translocase domain of BRG1 was replaced with that of SNF2h, the chimeric BRG1–SNF2h has remodeling activities like SNF2h either as free BRG1–SNF2h or assembled into the BRG1 containing core or complete SWI/SNF complex. The position to which BRG1–SNF2h moved nucleosomes is reminiscent of SNF2h and not BRG1, and remodeling required histone tails like SNF2h. In the reciprocal experiment with the DNA translocase domain of SNF2h replaced with that of BRG1, the SNF2h–BRG1 chimera had remodeling properties like wild-type BRG1. These differences in remodeling activities are not observed when the DNA translocase domain is swapped between SWI and SNF from different species (26,27).
We have now found that one fundamental difference between the DNA translocase domains of SWI/SNF and ISWI complexes is the manner in which they engage nucleosomal DNA at SHL2. Isw2 was found to bind to the exposed side of the DNA gyre while Snf2 intercalates between the histone octamer and DNA gyre using a DNA crosslinking approach that scans interactions in regards to whether they are facing towards or away from the histone octamer. The region of the ATPase domain of Snf2 associated with nucleosomal DNA at SHL2 is distinct from that observed for Isw2 and suggests that Snf2 wedges between DNA and histone octamer of the nucleosome. Because of the manner in which the ATPase domain is positioned between the octamer and DNA, when it starts to translocate on DNA there will be more leverage in breaking histone–DNA interactions than when the DNA translocase is bound to the outward side of nucleosomal DNA. These differences may help explain why SWI/SNF remodeling of nucleosomes is more disruptive than when remodeled by ISW2.
MATERIALS AND METHODS
SWI/SNF purification
Yeast SWI/SNF (28), ISW2, I-S-I and S-I-S complexes were purified by a one-step affinity purification using M2 agarose (anti-FLAG antibody conjugated, Sigma) as reported previously (12,29). The Snf6 subunit was tagged with a single copy of the FLAG epitope and the Snf2 subunit was tagged at the C-terminus with hemagglutinin(HA)-His6 for SWI/SNF. The construction of the I-S-I and S-I-S subunits is in the Information of Supplementary Data.
Mapping SWI/SNF interactions with DNA by Fe-BABE
Fe-EDTA was conjugated to free DNA using the sulfhydryl-specific Fe-BABE (Pierce). Fe-BABE was attached to DNA by incorporation of phosphorothioates into DNA during the PCR amplification of the desired 601 DNA sequence and then reacted with Fe-BABE (Information of Supplementary Data). After binding SWI/SNF to modified DNA, the cleavage reaction was initiated by the addition of H2O2 and ascorbate (Information of Supplementary Data). The specific location of the cleavage sites within the Snf2 polypeptide was determined by SDS–PAGE and immunoblotting with an antibody against the HA tag fused to the extreme C-terminus of Snf2. Molecular weight markers were made for this purpose by in vitro translation of different truncations of the full-length Snf2 (Supplementary Table S1 and S3).
Mapping the region of Snf2 crosslinked to nucleosomal DNA at two helical turns from the dyad axis (SHL2)
DNA probe [601 nucleosome positioning sequence (NPS)] with photoreactive nucleotides incorporated 17 and 18 bp from the dyad axis of the nucleosome was prepared as described previously (29). The 601 NPS in the probe DNA is flanked by 34 and 60 bp of extranucleosomal DNA with the two Gal4 binding sites on the 60 bp extranucleosomal DNA side. Nucleosomes were reconstituted by mixing DNA probe (4 ng/µl), salmon sperm DNA (1.5 µg/ul) and recombinant Xenopus laevis histone octamer (1 µg/µl) in 10 µl buffer containing 20 mM Tris–HCl pH 8.0, 2 M NaCl, 1 mM EDTA and 1 mM β-mercaptoethanol followed by stepwise dilution to 300 mM final salt concentration (30). SWI/SNF was recruited to nucleosomes with photoreactive nucleotides at 17 and 18 bp from the dyad axis by Gal4-VP16 as described previously and crosslinked by UV irradiation with no ATP added (29,31). The crosslinked products were analyzed on 6% SDS–PAGE and visualized by phosphorimaging. The Snf2 band was excised and electroeluted with buffer containing 50 mM NH4HCO3 and 0.1% SDS (31). The radiolabeled Snf2 was cleaved under limiting conditions at Asn-Gly (hydroxylamine), Met (cyanogen bromide) or Cys (NTCB or 2-nitro-5-thiocyanobenzoic acid) residues, resolved by SDS–PAGE and visualized by phosphorimaging to determine the location of the crosslinked site (Information of Supplementary Data). The apparent molecular weight of the labeled fragments generated by chemical cleavage were determined by comparing with Mark12® protein standard (Invitrogen) and molecular weight markers corresponding to different truncations of Snf2 (Supplementary Table S2 and S4).
Determine the helical phase of the interactions of Snf2 and Isw2 by DNA crosslinking
DNA photoaffinity probes in the 601 DNA were constructed as described previously in which a single phosphorothioate is incorporated into DNA using oligonucleotide primers extended on an immobilized DNA template (32). The oligonucleotide primer has a phosphorothioate incorporated 2–3 nt from the 5′-end and labeled with γ-32P ATP and Optikinase (USB). A different primer is used for each position and DNA crosslinking and label transfer is done as described.
RESULTS
The ATPase domains of SWI/SNF and ISW2 are not functionally interchangeable
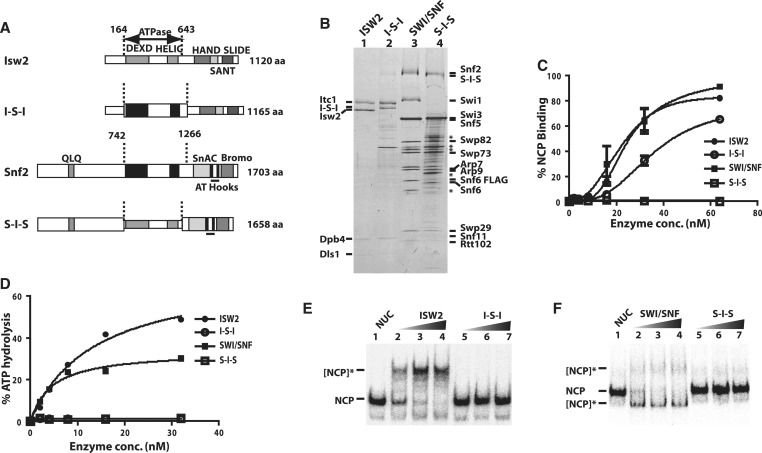
The ATPase domains of Snf2 and Isw2 were swapped such that the ATPase domain of Isw2 is in Snf2 (S-I-S) or that of Snf2 is in Isw2 (I-S-I; Figure 1A) to determine if they are functionally interchangeable. The S-I-S and I-S-I subunits were respectively assembled in vivo into the appropriate complex. I-S-I assembled with the Itc1, Dpb4 and Dls1 subunits of ISW2 (Figure 1B, lanes 1 and 2). The I-S-I complex bound to free DNA and nucleosomes with only a slightly lower affinity than ISW2 (Figure 1C and Supplementary Figure S1). Although the binding properties were not adversely affected, there was no appreciable ATPase or nucleosome movement with I-S-I as compared to ISW2 (Figures 1D and Supplementary Figure S1A–S1D). In the converse experiment with S-I-S, Swi1 was not properly assembled into SWI/SNF but all of the other SWI/SNF subunits were retained and the complex could bind to DNA but not to nucleosomes (Figure 1B, lanes 3 and 4; Figure 1C; Supplementary Figure S1). The S-I-S complex could not hydrolyze ATP or consequently mobilize nucleosomes (Figure 1D–E and S1-C). The ATPase domain of ISW2 and SWI/SNF although very similar are not interchangeable and appear to have some crucial differences that prevent them from working the same. Swapping the ATPase domains of BRG1 and SNF2h did not have as adverse an effect on ATP hydrolysis and nucleosome mobilization as it did for Snf2 and Isw2 (25). A key difference in these studies is BRG1 and SNF2h were recombinant proteins and their wild-type activities could be significantly less than the native proteins. On the other hand the lack of activity observed by swapping the ATPase domains of yeast SWI/SNF and ISW2 could be caused by unintended changes in the complexes or the ATPase domains of each could bind to nucleosomes different enough to prevent them from being interchangeable. Next, we examined the interactions of Snf2 with nucleosomes as part of the SWI/SNF complex and compared to that of Isw2 to determine if some of these distinctions could be caused by how they bind.
Figure 1.
The ATPase domains of SWI/SNF and ISW2 are not interchangeable. (A) The domain organization of Isw2 and Snf2 are shown along with the boundaries of the ATPase domains as dashed lines. The ATPase domains of Isw2 (gray) and Snf2 (black) are swapped with each other to make two hybrid catalytic subunits referred to as I-S-I and S-I-S. (B) The I-S-I and S-I-S subunits were purified along with their associated subunits and analyzed by 4–20% SDS–PAGE and Coomassie staining. The different subunits from SWI/SNF and ISW2 are indicated on either side. SWI/SNF had Snf6 tagged with two copies of FLAG epitope at its C-terminus instead of Snf2 being FLAG tagged as for the S-I-S. The asterisk indicates those protein bands which are not part of SWI/SNF, but co-purify with SWI/SNF when using M2 agarose. (C) The affinity of SWI/SNF and ISW2 for nucleosomes was compared to that of I-S-I and S-I-S by gel shift. Increasing amounts of enzyme were bound to 8 nM of end positioned 0N70 nucleosome. Error bars represent mean and standard deviation of two independent binding experiments. (D) The ATPase activity of SWI/SNF, ISW2, I-S-I and S-I-S with 0N70 nucleosomes was measured using γ-32P ATP and plotted. (E and F) The nucleosome remodeling activity of ISW2, I-S-I, SWI/SNF and S-I-S was measured by gel shift as shown with 0N70 nucleosomes (80 nM based on histone octamer). The concentrations of remodeler were 3, 10 and 15 nM. The reactions were incubated for 30 min at 30°C before stopping with competitor salmon sperm DNA and γ-S-ATP, and analyzed on 5% native polyacrylamide gel. NCP is nucleosome core particle and shows where nucleosomes migrated before remodeling. The [NCP]* indicates the position of the nucleosomes after being remodeled either by SWI/SNF or ISW2.
The catalytic subunit of SWI/SNF likely wedges between the DNA gyre and histone octamer
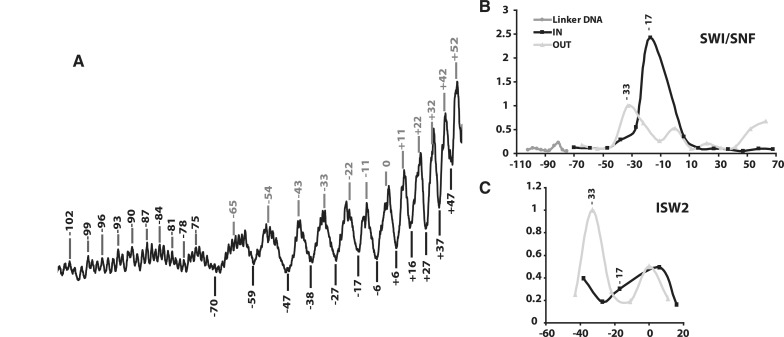
Different nucleosomes were used with SWI/SNF and ISW2 to position the ATPase domain of each over the same nucleosomal DNA region. The core nucleosome contained the 601 NPS but for ISW2 extranucleosomal DNA was at only one entry site while for SWI/SNF there was extranucleosomal DNA at both entry sites. Unique and efficient positioning of ISW2 onto nucleosomes is promoted by nucleosomes having extranucleosomal DNA at only one entry site. SWI/SNF was bound to nucleosomes in one preferred orientation through its interaction with the transcription activator Gal4-VP16 associated through one of the two extranucleosomal DNA. The position of the catalytic subunit relative to the histone octamer and DNA gyre of nucleosomes was examined by DNA crosslinking with a photoreactive group attached to the phosphate backbone of DNA (32,33). The helical periodicity of the DNA gyre inside the nucleosome has been mapped by DNA footprinting with hydroxyl radical (12,34) such that the positions are known that face in or away from the histone octamer (Figure 2A). The placement of photoreactive group on the phosphate backbone scans both the major and minor grooves of DNA due to the racemic nature of the crosslinker. A series of photoreactive nucleosomes were made in which the nucleosome surface is scanned from one entry site to the other either facing in or away from the histone octamer and in the absence of ATP (Supplementary Figure S2). The most efficient crosslinking of Snf2 to the phosphate backbone of DNA was at nucleotide −17, 17 bp from the dyad axis that would be facing in towards the histone octamer (Figure 2B and Supplementary Figure S2A). The crosslinking efficiency of Snf2 drops off dramatically half a helical turn in either direction at nucleotides −22 or −11, 22 and 11 bp from the dyad axis, and would be away from the histone octamer. The DNA crosslinking pattern of Snf2 is consistent with Snf2 binding between the DNA gyre and histone octamer and not with the DNA away from the histone octamer at SHL2. The interactions between the DNA gyre and histone octamer are primarily restricted to the SHL2 position as the efficiency of Snf2 crosslinking one helical turn from nt −17 at nt −27 is much less. It might be that SWI/SNF binding disturbs the histone–DNA interactions such that DNA positions relative to the histone octamer are altered and the rotational phasing changed. Previous DNA footprinting, however, does not show a change in rotational phasing of DNA on the nucleosome surface when SWI/SNF is bound as evident by none of the protected regions becoming more accessible after binding (29). The 601 nucleosome positioning sequence used in these experiments would also strongly disfavor a change in rotational phasing of DNA inside the nucleosome. If DNA phasing is not affected then Snf2 also associates with the outward side of nucleosomal DNA at nucleotides −33, 1.5 helical turns from nucleotides −17. Besides these two sites, there is little Snf2 interaction detected with nucleosomal DNA, except for some towards the edge of the nucleosome at base pair +52 and +65, 52 and 65 bp from the dyad axis on the other side and away from the histone octamer.
Figure 2.
Snf2 intercalates between the DNA gyre and histone octamer, and Isw2 does not. (A). The DNA footprinting pattern of the 601 nucleosome with hydroxyl radical is shown for the DNA strand in which the photoreactive group is also placed. The numbers indicate the specific site at which a photoreactive group is attached and those numbers in gray are on the exposed side of the nucleosomal DNA. (B and C) The relative efficiency of Snf2 (B) and Isw2 (C) crosslinked to DNA is plotted versus DNA location. The light gray line denotes the positions on the exposed side of the nucleosome and the black line denotes the positions on the histone octamer side of the DNA. The numbers shown on the x-axis correspond to the number of nucleotides from the dyad axis. Those positions shown as a darker gray line are in the extranucleosomal DNA region.
The interactions of Isw2 around SHL2 of the nucleosome were investigated in a similar manner, but focused on a narrower region from 43 bp upstream of the dyad axis to 16 bp downstream (−43 to +16). Isw2 does not crosslink well at nucleotides −17 and was about nine times less than that observed with Snf2 (Figure 2C). The most efficient crosslinking of Isw2 was with the part of DNA that generally faces away from the octamer at nucleotides −33 and to a lesser extent at the dyad axis. It is more evident from DNA footprinting of ISW2-nucleosome complexes than for SWI/SNF that ISW2 binding does not alter the rotational phasing of DNA on the nucleosome surface (12). ISW2 only protects 10 bp of DNA at SHL2 and ∼15 bp at SHL5–6 with no nucleosome protected regions becoming more accessible due to changes in DNA rotational phasing. DNA crosslinking indicates that Isw2 likely binds to DNA facing away from the histone octamer at SHL3 and does not intercalate between the gyre and octamer like Snf2 at SHL2. These data show there is a fundamental difference in the way which the catalytic subunit engages the nucleosome in these two remodelers. While there may be alternative interpretations it seems most likely that SWI/SNF invades the nucleosome surface between the DNA gyre and octamer, and in contrast the ATPase domain of ISW2 prefers to bind to the exposed side of nucleosomal DNA.
The C-terminal lobe between motif IVa and V of the Snf2 ATPase domain associates with nucleosomal DNA at SHL2.
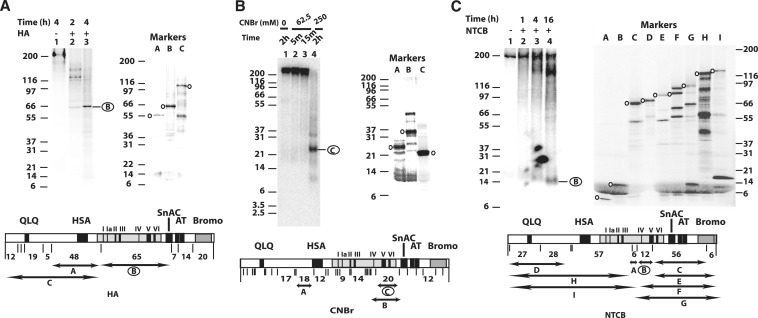
We wanted to find the region of Snf2 that is associated with nucleosomal DNA at nucleotides −17 and −18 or 17 and 18 bp from the dyad axis so that we could compare it with those of Isw2 which have been previously determined (31). In this case Snf2 was crosslinked to nucleosomal 601 DNA with the photoreactive group attached to the C-5 position of deoxythymidine in the major groove of DNA, the same as done earlier for Isw2. DNA translocation by SWI/SNF at this position or SHL2 is required for nucleosome mobilization and is a common feature of the RSC, ISW2 and NURF chromatin remodeling complexes (22–24). SWI/SNF was recruited to nucleosomes by Gal4-VP16, photo-crosslinked in the absence of ATP, and the crosslinking site determined by peptide mapping (29–31). The crosslinked Snf2 was analyzed by SDS–PAGE and subjected to sequence-specific chemical proteolysis to identify the region of Snf2 that associates with nucleosomal DNA (31). Three different cleavage conditions were used to identify the region contacting nucleosomal DNA as shown in Figure 3.
Figure 3.
The region-spanning motif IV and V of the Snf2 ATPase domain is close to nucleosomal DNA 17 and 18 bp from the dyad axis. (A) The region of Snf2 crosslinked to nucleosomal DNA was mapped by cleavage with hydroxylamine (HA), fragments separated by SDS–PAGE (4–12% Bis–Tris) and visualized by phosphorimaging. Crosslinked Snf2 was cleaved for 2 and 4 h (lanes 2 and 3). Truncated Snf2 polypeptides corresponding to proteolytic fragments that would be obtained by HA digestion were prepared by in vitro coupled transcription and translation with 35S labeled methionine (lanes A–C). The domain organization of Snf2 and location of the HA cut sites are shown with the location of the size markers shown by double-head arrows and corresponding label. (B) Crosslinked Snf2 was cleaved with CNBr, proteolytic fragments separated by SDS–PAGE (10% Bis-Tris), and visualized by phosphorimaging. The concentration of CNBr and duration of digestion are shown. Protein molecular weight markers of the CNBr proteolytic fragments were prepared and analyzed (lanes A–C) and the location of the fragments shown as in A. (C) The region of Snf2 crosslinked to DNA was determined by cleavage with NTCB and analyzed as in A. The molecular weight markers are in lanes A–I and their location shown as before. The regions shown to be crosslinked to nucleosomal DNA are circled.
After extensive digestion with hydroxylamine that cleaves at Asn-Gly, the major proteolytic fragment of the photoaffinity labeled Snf2 was ∼65 kDa (Figure 3A, lanes 1–3). Polypeptides corresponding to this and surrounding regions of Snf2 were prepared by in vitro translation with 35S methionine and used as molecular weight markers to validate the expected electrophoretic mobility of the hydroxylamine cleavage products. A 48 kDa Snf2 polypeptide synthesized by in vitro translation corresponding to amino acids 370–787 had an apparent electrophoretic mobility of 55 kDa, while the Snf2 polypeptide from amino acids 787 to 1347 (65 kDa) had an electrophoretic mobility of 68 kDa (Figure 3A, lanes A and B and Supplementary Table S2). The electrophoretic mobility of the radiolabeled band generated by extensive hydroxylamine cleavage of Snf2 indicates that the region from amino acid 787 to 1347 interacts with nucleosomal DNA 17 and 18 bp from the dyad axis and overlaps with the ATPase domain between amino acid residues 750–1250.
Peptide mapping of crosslinked Snf2 was further refined by digestion with cyanogen bromide (CNBr) which cleaves at the C-terminus of methionine. Crosslinked Snf2 cleaved with CNBr generated primarily a radiolabeled proteolytic fragment with an apparent molecular weight of ∼25 kDa (Figure 3B, lane 4). There are two regions of Snf2 that could produce such a large fragment that would be resistant to further cleavage by CNBr (Figure 3B, lanes A and C). The first region from amino acid 450 to 608 is in the N-terminal part of Snf2 and is outside the region previously observed by mapping with hydroxylamine cleavage. The second region from amino acid 1121 to 1295 is within the segment observed by hydroxylamine cleavage and is at the C-terminal end of the ATPase domain covering helicase motifs IV–VI. The in vitro translated Snf2 regions of amino acid 1121–1295 (20 kDa) had an apparent gel mobility of 21 kDa while the region spanning amino acids 450–608 (18 kDa) was 25 kDa (Figure 3B). The region of Snf2 crosslinked to nucleosomal DNA consistent with both CNBr and hydroxylamine cleavage is from amino acid 1121 to 1295 and the slower mobility of the CNBr cleavage product in comparison to the corresponding marker is likely due to the additional mass from the crosslinked DNA fragment as observed previously (35).
A third cleavage was used to further delineate the region of Snf2 that interacts with DNA at SHL2. Crosslinked Snf2 was cleaved with NTCB that cuts N-terminal to cysteine. NTCB cleavage produced a small proteolytic fragment of ∼12 kDa (Figure 3C, lane 4) corresponding to the region spanning amino acids 1058–1157. The electrophoretic mobility was confirmed using known fragments of Snf2 that were synthesized to correspond to these particular proteolytic fragments (Figure 3C, lanes A–I). Alignment of the three different cleavage patterns suggested that the region from amino acids 1121 to 1157 of Snf2 was crosslinked to DNA 17–18 bp from the dyad axis of the nucleosome (Figure 4A). This region includes part of the helicase-related motif IVa and V. Motif V of Snf2 has been shown to couple ATP hydrolysis to nucleosome remodeling (36).
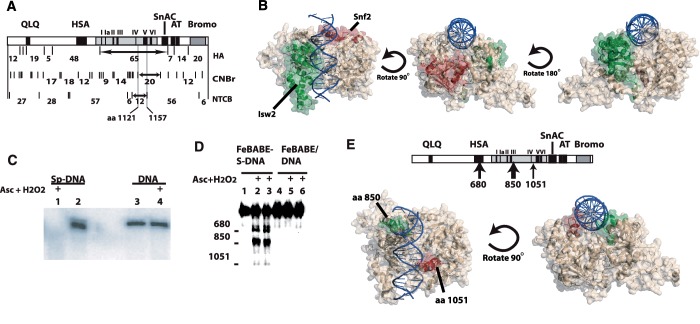
Figure 4.
Interactions of the ATPase domains of Isw2 and Snf2 with free and nucleosomal DNA. (A) A summary of the region of Snf2 crosslinked to nucleosomal DNA 17 and 18 bp from the dyad is shown for all three types of peptide mapping conditions as described in Figure 1. The final intersecting region is from residue 1121 to 1157. (B) The ATPase domains of Snf2 and Isw2 are modeled based on their sequence homology with the ATPase domain of Rad54 and the crystal structure of Sulfulobus Rad54 bound to free DNA (7,31). The region of Isw2 crosslinked to nucleosomal DNA 17 and 18 bp from the dyad axis is highlighted in green while that for Snf2 is shown in red. Three orientations of the model are shown with DNA in blue and the remainder of the ATPase domain in beige. (C) Phosphorothioate-DNA was conjugated to Fe-BABE (lanes 1 and 2) and a mock reaction was done with non-phosphorothioate containing DNA (lanes 3 and 4). Ascorbate and hydrogen peroxide was added in lanes 1 and 4. (D) SWI/SNF was bound to DNA immobilized on magnetic beads and cleavage initiated by the addition of hydrogen peroxide and ascorbate for 5 (lanes 2 and 5) and 20 s (lanes 3 and 6). Samples in lanes 1 and 4 had no hydrogen peroxide and ascorbate added. (E) The location of the cleavage sites are shown with regard to the domain organization of Snf2. The two Snf2 cleavage sites in the ATPase domain are shown in the model of the Snf2 ATPase domain similar as in (B). The green and red highlighted regions are the 20 amino acid regions in the vicinity of the two cleavage sites at amino acids 850 and 1051, respectively.
The structure of the ATPase domain of Snf2 was modeled based on its sequence homology to the ATPase domain of Rad54 and the crystal structure of Rad54 bound to DNA (Figure 4B). A similar approach had been previously used to map the region(s) of Isw2 that is associated with nucleosomal DNA at the same position as for Snf2 (31). While the region of Isw2 crosslinked to DNA corresponds to the N-terminal lobe of the ATPase domain consistent with the region of Rad54 known to bind DNA, the region of Snf2 crosslinked to nucleosomal DNA was the C-terminus of Snf2 encompassing parts of motifs IVa and V. Based on the structural model, the region of Snf2 associated with nucleosomal DNA is off to one side of the normal binding cleft formed between the two lobes of the ATPase domain and is inconsistent with the Rad54-DNA structure (Figure 4B). These data show the DNA translocase domains of SWI/SNF and ISW2 bind in different orientations relative to the nucleosomal DNA consistent with binding to either the exposed surface of nucleosomal DNA or between the gyre and octamer. These distinctions likely reflect basic differences in the mode of remodeling used by ISW2 to only mobilize nucleosomes or SWI/SNF to unravel and disassemble nucleosomes.
A different part of the ATPase domain contacts DNA along with the HSA domain when SWI/SNF binds to free DNA
Why does the ATPase domain of Snf2 bind so differently to nucleosomal DNA when compared to Isw2? It seemed unusual given the sequence homology that the ATPase domain of Snf2 would inherently bind DNA differently than Rad54 or Isw2. For this reason the interactions of SWI/SNF with free DNA was mapped by site-directed proteolysis to determine if the interactions of the ATPase domain with free DNA are the same as with nucleosomal DNA or like that seen for Rad54 and Isw2. Phosphorothioate–DNA was synthesized by PCR in which all of the deoxycytidine residues were replaced with deoxycytidine α-thiomonophosphate. Fe–EDTA was coupled to phosphorothioate using Fe-BABE, a derivative of Fe–EDTA used to conjugate to sulfhydryl groups. The successful coupling of Fe-BABE to phosphorothioate–DNA was verified by the selective degradation of modified DNA with the addition of hydrogen peroxide and ascorbate, but not of unmodified DNA (Figure 4C, compare lanes 1 and 2). DNA without phosphorothioates when modified otherwise the same as before was not able to self-cleave consistent with phosphorothioate being needed for coupling of Fe-BABE to DNA (Figure 4C, compare lanes 3 and 4 with lanes 1 and 2).
The sites in Snf2 that were cleaved by SWI/SNF association with modified DNA were mapped using Snf2 with a hemagluttin (HA) epitope tag attached to its C-terminus and immunoblotting. Snf2 was cleaved at the ATPase and HSA domains near residues 850 and 680, respectively, when SWI/SNF was bound to Fe-BABE-modified DNA in the absence of ATP (Figure 4D, lanes 2–3; Figure 4E). There was also less efficient cleavage at a second site in the ATPase domain near residue 1051. Residue 850 is in the N-terminal lobe of the ATPase domain near motif Ib and residue 1050 is located in the C-terminal lobe of the ATPase domain of Snf2 between the hinge connecting the two lobes and motif IV (Figure 4E). Cleavage of Snf2 was dependent on conjugation of Fe-BABE to DNA and was not detected when DNA without phosphorothioate was used (Figure 4D, compare lanes 2 and 3 with lanes 5 and 6). Cleavage of Snf2 was also dependent on the presence of ascorbate and hydrogen peroxide in addition to the Fe-BABE-tethered DNA (Figure 4D, lane 1). The two cleavage sites in the ATPase structure should lay along the cleft formed by the two lobes of the Snf2 ATPase domain based on our model and is consistent with the ATPase domain of Snf2 binding free DNA in the same manner as Rad54 (6). The question remains as to why SWI/SNF binding to DNA is altered when SWI/SNF binds nucleosomes? One clue comes from histone crosslinking data showing Snf2 interacting with the histone octamer around the SHL2 region (29) and points to concerted interactions between histones and Snf2 contributing to the altered binding of nucleosomal DNA.
The HSA domain is shown for the first time to interact with DNA by cleavage occurring near amino acid 680 (Figure 4E). The HSA domain of Snf2 binds the nuclear actin-related proteins (ARPs) 7 and 9, and has a role in regulating the activity of the ATPase domain (37). The HSA domain may enhance the ATPase activity of SWI/SNF and RSC by contributing to the affinity of the complex for DNA.
DISCUSSION
SWI/SNF and ISW2 have several basic catalytic activities in common such as their DNA translocase domains binding to the same location in nucleosomes and moving nucleosomes to different positions on DNA. But they also have important differences in nucleosome remodeling, such as whether they can conservatively move nucleosomes on DNA without altering the canonical structure (ISW2) or cause disruptions of histone–DNA interactions and even disassemble nucleosomes (SWI/SNF). While these differences may be orchestrated by the auxiliary subunits or domains of these complexes, we have found that even the interactions of the ATPase domains are distinct for these two complexes. Although the ATPase domains of SWI/SNF and ISW2 are highly similar at the level of their catalytic cores (motifs I–VI), they function quite differently making it seemingly impossible for one ATPase domain to replace another without serious problems. A fundamental difference is the interactions of the ATPase domain of ISW2 and Snf2 with nucleosomal DNA. The ATPase domain of ISW2 contacts the surface of nucleosomal DNA facing away from histone octamer, but SWI/SNF likely binds between nucleosomal DNA and the histone octamer. In this situation, SWI/SNF would have a strategic advantage for disrupting histone–DNA contacts with the ATPase domain positioned as a wedge that when translocating along DNA could readily break additional histone–DNA contacts.
The DNA footprint of ISW2-nucleosome shows discrete binding of ISW2 to the exposed side of one helical turn of nucleosomal DNA consistent with the ATPase domain binding to the outside surface of nucleosomal DNA (12). The interactions of SWI/SNF with nucleosomal DNA by DNA footprinting are more extensive than that indicated by DNA crosslinking. Snf2 is primarily crosslinked to ∼16 bp of nucleosomal DNA, but SWI/SNF protects a region of ∼50 bp of nucleosomal DNA. This discrepancy may be due to the limited reactivity of the aryl azide and could require a more reactive crosslinker as shown previously (29). Binding of SWI/SNF between the DNA gyre and histone octamer would require movement of the DNA and/or of the histone octamer to make space for Snf2. There is no indication as of yet that the DNA is significantly pulled off the histone octamer thereby losing its rotational phasing on the nucleosome to provide this additional flexibility, but further studies will be necessary to find how this might occur.
The DNA crosslinking experiments could be misleading if modification of the DNA altered the rotational phasing of nucleosomes. We suspect this is unlikely because the rotational phasing of nucleosomes is not readily changed, especially of strong nucleosome positioning sequences like 601. Evidence for rotational phasing likely not being altered by DNA modification comes from experiments with polyamides that bind to nucleosomal DNA. Polyamides bind to the minor groove of nucleosomes and can distort the DNA helix, but have been observed to not alter the protein–DNA interactions inside the nucleosome (38–40). While binding of polyamides to SHL6 changed the rotational phasing at SHL-6, these changes were found to be due to crystallization conditions and as observed by DNase I footprinting likely do not reflect the state in solution. The attachment of an aryl azide to phosphate is a significantly smaller and less obtrusive to nucleosomal DNA and less likely to change the rotational phasing of nucleosomes.
Another concern for the DNA crosslinking experiments is the differences in nucleosomes with one being end positioned nucleosomes and the other more central positioned nucleosomes for crosslinking to SWI/SNF and ISW2. Extranucleosomal DNA seems to influence SWI/SNF as seen by SWI/SNF moving nucleosomes to preferred sites away from nucleosome positioning sequences (41–43), but is not likely to change the way in which SWI/SNF engages nucleosomes in the absence of ATP. Unlike ISW2, SWI/SNF does not require extranucleosomal DNA in order to remodel nucleosomes and extranucleosomal DNA cannot be used to uniquely position SWI/SNF onto nucleosomes (15). The key in these experiments is that the ATPase domains of Snf2 or Isw2 are shown to engage the same region of the nucleosome under these different conditions and thus provides us the opportunity to compare their binding under structurally similar conditions although different conditions have been used to achieve this purpose.
The functional consequence of the ATPase domain being bound on the outside of the nucleosome with no apparent change in histone–DNA interactions or wedged between DNA and histone octamer could affect the way which nucleosomes are moved on DNA (Figure 5). As shown previously, after addition of ATP, SWI/SNF quickly disrupts histone–DNA contacts 54 bp from the dyad axis and they remain broken until the DNA has been moved 52 bp (24). These observations are consistent with SWI/SNF wedging between the DNA and octamer and upon translocation causing large scale disruptions of histone–DNA interactions. This same action could also cause DNA loops of significant size to form on the nucleosome surface. Extensive characterization of SWI/SNF remodeling intermediates by restriction enzyme accessibility has provided evidence for the formation of DNA loops during remodeling (19,20,44). The formation of DNA loops large enough to be detected by restriction enzyme cutting is a property reserved for SWI/SNF and is not observed for ISWI remodelers. A concern raised about the DNA looping model for SWI/SNF and RSC remodeling has been the step size of the DNA translocase being so much smaller than the loop size being created (21,45). Finding that the DNA translocase starts out wedge between the histone octamer and nucleosomal DNA accounts for how short movements on nucleosomal DNA could have longer range effects on histone–DNA interactions. A natural outcome of creating these large scale disruptions of histone–DNA interactions followed by repositioning the DNA back on to the histone octamer surface is that some differences in the path DNA are likely to occur. Distortions of this kind have been observed for SWI/SNF remodeling by high resolution mapping of specific histone–DNA contacts before and after remodeling to examine the spacing of DNA on nucleosomes (16).
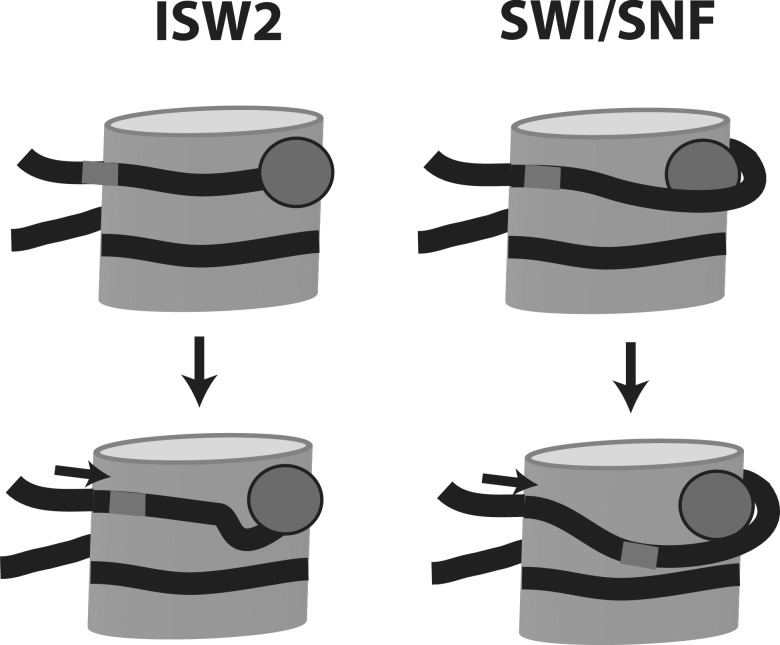
Figure 5.
Model for the two modes of SWI/SNF and ISW2 remodeling. The nucleosome is shown with DNA in black and the ATPase domains of SWI/SNF and ISW2 as a dark grey spheres. SWI/SNF remodeling with the ATPase domain intercalated between the DNA gyre and histone octamer causes a larger scale disruption of histone-DNA contact upon translocation and therefore a larger loop on the nucleosome surface than ISW2. ISW2 being bound to the outside of the DNA gyre is more prone to cause smaller distortions in the histone–DNA interface coupled with its translocation on nucleosomal DNA.
ISW2 remodels nucleosomes in a very different manner that correlates well to its ATPase domain binding to the exposed side of nucleosomal DNA. Shortly after the addition of ATP, ISW2 moves DNA short distances of only 9–11 bp on the histone octamer without ever massively or persistently disrupting histone–DNA contacts like SWI/SNF (24). The formation of DNA loops during ISW2 remodeling is also not detected by restriction enzyme cleavage. Likely due to the small and progressive changes in histone–DNA interactions that occur as part of ISW2 remodeling the pathway of DNA around the histone octamer is also well conserved after remodeling (18). Translocation of the ATPase domain on the exposed surface as expected tends to move nucleosomes in short increments without significantly disrupting histone–DNA interactions. There may be a connection with the ATPase domain being lodge between the histone octamer and DNA that causes the remodeler to be more prone to disrupting histone–DNA interactions and unraveling DNA from the histone octamer such as for SWI/SNF. When the ATPase domain is instead bound to the outside surface of nucleosomal DNA it will likely move nucleosomes in such a way as to not disturb the canonical structure such as seen for ISW2.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–4, Supplementary Figures 1 and 2, Supplementary Methods and Supplementary References [46–50].
FUNDING
Funding for open access charge: The National Institutes of Health (GM 48413 and GM 70864 to B.B.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Dr Claude Meares for providing CHA225 antibodies and Soumyadipta Kundu and Jim Persinger for their input.
REFERENCES
- 1.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 2.Gangaraju VK, Bartholomew B. Mechanisms of ATP dependent chromatin remodeling. Mutat. Res. 2007;618:3–17. doi: 10.1016/j.mrfmmm.2006.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Durr H, Flaus A, Owen-Hughes T, Hopfner KP. Snf2 family ATPases and DExx box helicases: differences and unifying concepts from high-resolution crystal structures. Nucleic Acids Res. 2006;34:4160–4167. doi: 10.1093/nar/gkl540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flaus A, Martin DM, Barton GJ, Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;34:2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richmond E, Peterson CL. Functional analysis of the DNA-stimulated ATPase domain of yeast SWI2/SNF2. Nucleic Acids Res. 1996;24:3685–3692. doi: 10.1093/nar/24.19.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Durr H, Korner C, Muller M, Hickmann V, Hopfner KP. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell. 2005;121:363–373. doi: 10.1016/j.cell.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 7.Thoma NH, Czyzewski BK, Alexeev AA, Mazin AV, Kowalczykowski SC, Pavletich NP. Structure of the SWI2/SNF2 chromatin-remodeling domain of eukaryotic Rad54. Nat. Struct. Mol. Biol. 2005;12:350–356. doi: 10.1038/nsmb919. [DOI] [PubMed] [Google Scholar]
- 8.Corona DF, Siriaco G, Armstrong JA, Snarskaya N, McClymont SA, Scott MP, Tamkun JW. ISWI regulates higher-order chromatin structure and histone H1 assembly in vivo. PLoS Biol. 2007;5:e232. doi: 10.1371/journal.pbio.0050232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vincent JA, Kwong TJ, Tsukiyama T. ATP-dependent chromatin remodeling shapes the DNA replication landscape. Nat. Struct. Mol. Biol. 2008;15:477–484. doi: 10.1038/nsmb.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fazzio TG, Tsukiyama T. Chromatin remodeling in vivo: evidence for a nucleosome sliding mechanism. Mol. Cell. 2003;12:1333–1340. doi: 10.1016/s1097-2765(03)00436-2. [DOI] [PubMed] [Google Scholar]
- 11.Sala A, Toto M, Pinello L, Gabriele A, Di Benedetto V, Ingrassia AM, Lo Bosco G, Di Gesu V, Giancarlo R, Corona DF. Genome-wide characterization of chromatin binding and nucleosome spacing activity of the nucleosome remodelling ATPase ISWI. EMBO J. 2011;30:1766–1777. doi: 10.1038/emboj.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kagalwala MN, Glaus BJ, Dang W, Zofall M, Bartholomew B. Topography of the ISW2-nucleosome complex: insights into nucleosome spacing and chromatin remodeling. EMBO J. 2004;23:2092–2104. doi: 10.1038/sj.emboj.7600220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Narlikar GJ. A proposal for kinetic proof reading by ISWI family chromatin remodeling motors. Curr. Opin. Chem. Biol. 2010;14:660–665. doi: 10.1016/j.cbpa.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prasad P, Bartholomew B. Control of nucleosome movement: to space or not to space nucleosomes? Epigenetics. 2010;5:282–286. doi: 10.4161/epi.5.4.11607. [DOI] [PubMed] [Google Scholar]
- 15.Dechassa ML, Sabri A, Pondugula S, Kassabov SR, Chatterjee N, Kladde MP, Bartholomew B. SWI/SNF has intrinsic nucleosome disassembly activity that is dependent on adjacent nucleosomes. Mol. Cell. 2010;38:590–602. doi: 10.1016/j.molcel.2010.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kassabov SR, Zhang B, Persinger J, Bartholomew B. SWI/SNF unwraps, slides, and rewraps the nucleosome. Mol. Cell. 2003;11:391–403. doi: 10.1016/s1097-2765(03)00039-x. [DOI] [PubMed] [Google Scholar]
- 17.Lorch Y, Zhang M, Kornberg RD. RSC unravels the nucleosome. Mol. Cell. 2001;7:89–95. doi: 10.1016/s1097-2765(01)00157-5. [DOI] [PubMed] [Google Scholar]
- 18.Kassabov SR, Henry NM, Zofall M, Tsukiyama T, Bartholomew B. High-resolution mapping of changes in histone-DNA contacts of nucleosomes remodeled by ISW2. Mol. Cell. Biol. 2002;22:7524–7534. doi: 10.1128/MCB.22.21.7524-7534.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Narlikar GJ, Phelan ML, Kingston RE. Generation and interconversion of multiple distinct nucleosomal states as a mechanism for catalyzing chromatin fluidity. Mol. Cell. 2001;8:1219–1230. doi: 10.1016/s1097-2765(01)00412-9. [DOI] [PubMed] [Google Scholar]
- 20.Fan HY, He X, Kingston RE, Narlikar GJ. Distinct strategies to make nucleosomal DNA accessible. Mol. Cell. 2003;11:1311–1322. doi: 10.1016/s1097-2765(03)00192-8. [DOI] [PubMed] [Google Scholar]
- 21.Cairns BR. Chromatin remodeling: insights and intrigue from single-molecule studies. Nat. Struct. Mol. Biol. 2007;14:989–996. doi: 10.1038/nsmb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling through directional DNA translocation from an internal nucleosomal site. Nat. Struct. Mol. Biol. 2005;12:747–755. doi: 10.1038/nsmb973. [DOI] [PubMed] [Google Scholar]
- 23.Schwanbeck R, Xiao H, Wu C. Spatial contacts and nucleosome step movements induced by the NURF chromatin remodeling complex. J. Biol. Chem. 2004;279:39933–39941. doi: 10.1074/jbc.M406060200. [DOI] [PubMed] [Google Scholar]
- 24.Zofall M, Persinger J, Kassabov SR, Bartholomew B. Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat. Struct. Mol. Biol. 2006;13:339–346. doi: 10.1038/nsmb1071. [DOI] [PubMed] [Google Scholar]
- 25.Fan HY, Trotter KW, Archer TK, Kingston RE. Swapping function of two chromatin remodeling complexes. Mol. Cell. 2005;17:805–815. doi: 10.1016/j.molcel.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 26.Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 27.Elfring LK, Deuring R, McCallum CM, Peterson CL, Tamkun JW. Identification and characterization of Drosophila relatives of the yeast transcriptional activator SNF2/SWI2. Mol. Cell. Biol. 1994;14:2225–2234. doi: 10.1128/mcb.14.4.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neely KE, Hassan AH, Brown CE, Howe L, Workman JL. Transcription activator interactions with multiple SWI/SNF subunits. Mol. Cell. Biol. 2002;22:1615–1625. doi: 10.1128/MCB.22.6.1615-1625.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dechassa ML, Zhang B, Horowitz-Scherer R, Persinger J, Woodcock CL, Peterson CL, Bartholomew B. Architecture of the SWI/SNF-nucleosome complex. Mol. Cell. Biol. 2008;28:6010–6021. doi: 10.1128/MCB.00693-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sengupta SM, Persinger J, Bartholomew B, Peterson CL. Use of DNA photoaffinity labeling to study nucleosome remodeling by SWI/SNF. Methods. 1999;19:434–446. doi: 10.1006/meth.1999.0880. [DOI] [PubMed] [Google Scholar]
- 31.Dang W, Bartholomew B. Domain architecture of the catalytic subunit in the ISW2-nucleosome complex. Mol. Cell. Biol. 2007;27:8306–8317. doi: 10.1128/MCB.01351-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Persinger J, Sengupta SM, Bartholomew B. Spatial organization of the core region of yeast TFIIIB-DNA complexes. Mol. Cell Biol. 1999;19:5218–5234. doi: 10.1128/mcb.19.7.5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pendergrast PS, Chen Y, Ebright YW, Ebright RH. Determination of the orientation of a DNA binding motif in a protein-DNA complex by photocrosslinking. Proc. Natl Acad. Sci. USA. 1992;89:10287–10291. doi: 10.1073/pnas.89.21.10287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gangaraju VK, Prasad P, Srour A, Kagalwala MN, Bartholomew B. Conformational changes associated with template commitment in ATP-dependent chromatin remodeling by ISW2. Mol. Cell. 2009;35:58–69. doi: 10.1016/j.molcel.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tate JJ, Persinger J, Bartholomew B. Survey of four different photoreactive moieties for DNA photoaffinity labeling of yeast RNA polymerase III transcription complexes. Nucleic Acids Res. 1998;26:1421–1426. doi: 10.1093/nar/26.6.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith CL, Peterson CL. A conserved Swi2/Snf2 ATPase motif couples ATP hydrolysis to chromatin remodeling. Mol. Cell. Biol. 2005;25:5880–5892. doi: 10.1128/MCB.25.14.5880-5892.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szerlong H, Hinata K, Viswanathan R, Erdjument-Bromage H, Tempst P, Cairns BR. The HSA domain binds nuclear actin-related proteins to regulate chromatin-remodeling ATPases. Nat. Struct. Mol. Biol. 2008;15:469–476. doi: 10.1038/nsmb.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edayathumangalam RS, Weyermann P, Dervan PB, Gottesfeld JM, Luger K. Nucleosomes in solution exist as a mixture of twist-defect states. J. Mol. Biol. 2005;345:103–114. doi: 10.1016/j.jmb.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 39.Suto RK, Edayathumangalam RS, White CL, Melander C, Gottesfeld JM, Dervan PB, Luger K. Crystal structures of nucleosome core particles in complex with minor groove DNA-binding ligands. J. Mol. Biol. 2003;326:371–380. doi: 10.1016/s0022-2836(02)01407-9. [DOI] [PubMed] [Google Scholar]
- 40.Gottesfeld JM, Melander C, Suto RK, Raviol H, Luger K, Dervan PB. Sequence-specific recognition of DNA in the nucleosome by pyrrole-imidazole polyamides. J. Mol. Biol. 2001;309:615–629. doi: 10.1006/jmbi.2001.4694. [DOI] [PubMed] [Google Scholar]
- 41.Pham CD, He X, Schnitzler GR. Divergent human remodeling complexes remove nucleosomes from strong positioning sequences. Nucleic Acids Res. 2010;38:400–413. doi: 10.1093/nar/gkp1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sims HI, Baughman CB, Schnitzler GR. Human SWI/SNF directs sequence-specific chromatin changes on promoter polynucleosomes. Nucleic Acids Res. 2008;36:6118–6131. doi: 10.1093/nar/gkn623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sims HI, Lane JM, Ulyanova NP, Schnitzler GR. Human SWI/SNF drives sequence-directed repositioning of nucleosomes on C-myc promoter DNA minicircles. Biochemistry. 2007;46:11377–11388. doi: 10.1021/bi7008823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bouazoune K, Miranda TB, Jones PA, Kingston RE. Analysis of individual remodeled nucleosomes reveals decreased histone-DNA contacts created by hSWI/SNF. Nucleic Acids Res. 2009;37:5279–5294. doi: 10.1093/nar/gkp524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sirinakis G, Clapier CR, Gao Y, Viswanathan R, Cairns BR, Zhang Y. The RSC chromatin remodelling ATPase translocates DNA with high force and small step size. EMBO J. 2011;30:2364–2372. doi: 10.1038/emboj.2011.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen HT, Hahn S. Binding of TFIIB to RNA polymerase II: mapping the binding site for the TFIIB zinc ribbon domain within the preinitiation complex. Mol. Cell. 2003;12:437–447. doi: 10.1016/s1097-2765(03)00306-x. [DOI] [PubMed] [Google Scholar]
- 47.Meares CF, Datwyler SA, Schmidt BD, Owens J, Ishihama A. Principles and methods of affinity cleavage in studying transcription. Methods Enzymol. 2003;371:82–106. doi: 10.1016/S0076-6879(03)71006-4. [DOI] [PubMed] [Google Scholar]
- 48.Schmidt BD, Meares CF. Proteolytic DNA for mapping protein-DNA interactions. Biochemistry. 2002;41:4186–4192. doi: 10.1021/bi015582r. [DOI] [PubMed] [Google Scholar]
- 49.Traviglia SL, Datwyler SA, Meares CF. Mapping protein-protein interactions with a library of tethered cutting reagents: the binding site of sigma 70 on Escherichia coli RNA polymerase. Biochemistry. 1999;38:4259–4265. doi: 10.1021/bi983016z. [DOI] [PubMed] [Google Scholar]
- 50.Traviglia SL, Datwyler SA, Yan D, Ishihama A, Meares CF. Targeted protein footprinting: where different transcription factors bind to RNA polymerase. Biochemistry. 1999;38:15774–15778. doi: 10.1021/bi9917232. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.