Abstract
Most bacteria control oxidative stress through the H2O2-responsive transactivator OxyR, a member of the LTTR family (LysR Type Transcriptional Regulators), which activates the expression of defensive genes such as those encoding catalases, alkyl hydroperoxide reductases and superoxide dismutases. In the human opportunistic pathogen Pseudomonas aeruginosa, OxyR positively regulates expression of the oxidative stress response genes katA, katB, ahpB and ahpCF. To identify additional targets of OxyR in P. aeruginosa PAO1, we performed chromatin immunoprecipitation in combination with whole genome tiling array analyses (ChIP-chip). We detected 56 genes including all the previously identified defensive genes and a battery of novel direct targets of OxyR. Electrophoretic mobility shift assays (EMSAs) for selected newly identified targets indicated that ∼70% of those were bound by purified oxidized OxyR and their regulation was confirmed by quantitative real-time polymerase chain reaction. Furthermore, a thioredoxin system was identified to enzymatically reduce OxyR under oxidative stress. Functional classification analysis showed that OxyR controls a core regulon of oxidative stress defensive genes, and other genes involved in regulation of iron homeostasis (pvdS), quorum-sensing (rsaL), protein synthesis (rpsL) and oxidative phosphorylation (cyoA and snr1). Collectively, our results indicate that OxyR is involved in oxidative stress defense and regulates other aspects of cellular metabolism as well.
INTRODUCTION
One of the major challenges for living organisms is the oxidative stress derived from reactive oxygen species (ROS) produced via normal aerobic metabolism or by exposure to redox-cycling drugs, ionizing radiation or by stimulated human phagocytic cells during infections of multiple phyla. ROS in relatively low concentrations are capable of causing damage to virtually all biomolecules including DNA, RNA, lipid and protein. As a primary defense against ROS, both prokaryotes and eukaryotes have developed antioxidant defense systems to protect cells from the aforementioned lesions (1). In bacteria, antioxidant defense systems have been extensively studied in Escherichia coli (2–4). In this organism, superoxide-generating compounds such as paraquat activate SoxR-mediated transcriptional activation of SoxS, which then modulates the expression of a battery of genes including sodA (manganese superoxide dismutase), fpr (ferredoxin/flavodoxin-NADP1 reductase), zwf (glucose 6-phosphate dehydrogenase), fumC (fumarase C), nfo (endonuclease IV), acnA (aconitase A) and micF (a small regulatory RNA). In parallel, hydrogen peroxide (H2O2) activates OxyR, a LysR Type Transcriptional Regulator (LysR)-type transcriptional regulator (LTTR) via the oxidation of two conserved cysteine residues (Cys 199 and Cys 208) and the formation of an intramolecular disulfide bond (1,5). The oxidized OxyR induces the expression of a set of defensive genes including katG (hydrogen peroxidase I), ahpCF (alkyl hydroperoxidase), dps (a nonspecific DNA-binding protein), gorA (glutathione reductase), grxA (glutaredoxin I) and oxyS (a small regulatory RNA) (6,7). Oxidized E. coli OxyR possesses DNA-binding activity and recognizes a conserved motif consisting of four regularly spaced ATAG elements. Both oxidized and reduced forms of E. coli OxyR bind to the control region of the own gene, but the two forms establish different contacts with the operator. In contrast, reduced OxyR and a cysteine mutant (C199S) do not bind to other targets (8,9). Similarly to many other LTTRs, E. coli OxyR also represses its own transcription irrespective of its redox state (10). OxyR homologs have also been identified in various bacteria, including other Proteobacteria such as Neisseria gonorrhoeae, Legionella pneumophila, Xanthomonas campestris and Haemophilus influenza (www.ncbi.nlm.nih.gov). These proteins showed properties significantly different from those of E. coli OxyR with respect to the transcriptional regulation pattern, hydroperoxide-sensing behavior and conserved cysteine composition.
As a human opportunistic pathogen, Pseudomonas aeruginosa causes severe acute nosocomial infections in immunocompromised individuals or chronic lung infections in cystic fibrosis (CF) patients. Intrinsic antibiotic resistance and versatile ecological adaptability, especially as biofilms, contribute to making the eradication of this bacterium from the infection sites highly problematic (11). During both the infectious process in human disease as well as its free-living, planktonic lifestyle, endogenous and exogenous ROS are scavenged by the redundant antioxidant defense systems of P. aeruginosa. In the context of peroxide detoxification (both H2O2 and organic peroxides), most of the genes involved were found to be controlled by OxyR (12,13). Additional findings have shed light on the important roles of the regulator in the pathogenesis of P. aeruginosa (14,15) as well as in iron uptake mediated by the siderophore pyoverdine (16), and in the regulation of the phenazine compound pyocyanin production (17). In particular, extensive studies have focused on its roles in the antioxidant defense response to H2O2 by transcriptional activation of a battery of genes such as katA (catalase A) (13), katB (catalase B), ahpB (alkyl hydroperoxide reductases B) and ahpCF (alkyl hydroperoxide reductases CF) (12). However, there is a relative paucity of information concerning the association of OxyR with its target promoter regions in vivo.
Chromatin immunoprecipitation (ChIP) is a powerful technique to explore transcriptional regulatory mechanisms and to measure the association of transcription factors with their targets in vivo. It overcomes the limitations of genetics and biochemical approaches by directly determining both the position and the strength of protein–DNA interactions in vivo. When integrated with transcriptional profiling or microarrays to create the ChIP-chip technique, also known as genome-wide location analysis, this approach allows for the quantitative measurement of protein–DNA interactions on a genome-wide scale (18,19). Until recently, only a limited number of studies have taken advantage of the ChIP-chip technique in elucidating transcriptional regulatory mechanisms in prokaryotes, especially in human pathogens (20–22).
In this report, we used ChIP-chip to identify direct binding targets of OxyR on the P. aeruginosa genome and validated the results by in vitro binding assays with purified OxyR protein to selected targets. We confirm its key role in oxidative stress response by regulating mainly katA, katB, ahpB and ahpCF and report its implication in other unexpected important responses such as quorum-sensing (QS) regulation, iron homeostasis, oxidative phosphorylation and transport, in agreement with some previous phenotypic observations (14,16,17,23,24). These OxyR-mediated mounted responses to different stresses explain why P. aeruginosa is highly adaptable to environmental changes.
MATERIALS AND METHODS
Construction of his-tagged protein for ChIP analysis
Pseudomonas aeruginosa wild-type strain PAO1 was used in this study and modified as follows to construct a strain with an extrachromosomal encoded hexahistidine-tagged OxyR protein. First, a DNA fragment containing the entire coding region of the oxyR gene (PA5344) was amplified and ligated to the NdeI–HindIII site of pET24a (Novagen) to generate pET24-5344. The encoded protein contains a carboxyl-terminal His-tag sequence, KLAAALEHHHHHH, allowing Ni2+ ion affinity purification for the downstream protein analysis work, such as EMSA. Second, the entire coding sequence together with the C-terminal His-tag was amplified and ligated into the low-copy-number, broad-host-range shuttle vector, pUCP20, under the control of the lac promoter of the vector (25). Then we transformed the recombinant construct into P. aeruginosa PAO1 wild-type via electroporation. The resultant strain was designated PAO1/pUCP5344. All of the plasmids used in this study were verified by Sanger sequence analysis.
ChIP
ChIP was based on a previously described procedure (26). Briefly, bacteria were grown in Luria-Bertani (L.B.) broth until early stationary phase (OD600 = 2.0) and 1 mM H2O2 was added to treat the cultures for 10 min. Formaldehyde was then added to a final concentration of 1%. After incubation for 20 min at room temperature, glycine was added to a final concentration of 0.5 M. Cells were harvested by centrifugation, washed twice in ice-cold Tris-buffered saline (TBS, pH 7.6) and resuspended in 500 µl of lysis buffer [10 mM Tris–HCl, pH 8.0, 20% sucrose, 50 mM NaCl, 10 mM ethylenediaminetetraacetic acid (EDTA)]. Lysozyme (2.5 mg) was added and the cells were incubated for 30 min at 37°C. The DNA was sheared to an average size of 100–1000 bp by a series of pulsed sonication steps (Branson Sonicator S250 Analogue, 5 mm Standard tip, six times 60 s at 90% duty on Level 4). Insoluble material was removed by centrifugation at 13 000g for 10 min and 850 µl of the cell extract supernatant from His-tagged or non-His-tagged control bacterial strain were mixed with 750 µl of immunoprecipitation (IP) buffer [50 mM HEPES-KOH, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% (v/v) Triton X-100, 0.1% (m/v) sodium deoxycholate] and phenylmethanesulfonyl fluoride (PMSF, final concentration 1 mM) and incubated with 20 µl of protein G-Sepharose beads (Sigma) for 1 h at room temperature with gentle mixing. After removal of the Sepharose beads by gentle centrifugation, proteins were immunoprecipitated by adding 20 µl of protein G-Sepharose beads (Sigma) coupled to mouse anti-His monoclonal antibody (Novagen). The composite was incubated overnight at 4°C with gentle mixing. Samples were then washed twice with IP buffer, once with IP buffer containing 500 mM NaCl, once with washing buffer [10 mM Tris–HCl, pH 8.0, 250 mM LiCl, 1 mM EDTA, 0.5% (v/v) Nonidet-P40, 0.5% (m/v) sodium deoxycholate] and once with TE buffer (10 mM Tris–HCl, 1 mM EDTA, pH 7.5). Immunoprecipitated complexes were eluted by incubation of the beads with elution buffer [50 mM Tris–HCl, pH 7.5, 10 mM EDTA, 1% (m/v) sodium dodecyl sulfate (SDS)] at 65°C for 10 min. Forty microliters of the immunoprecipitated samples were de-cross-linked by the addition of 32 µg Pronase and incubation for 2 h at 42°C and for 6 h at 65°C. DNA was purified using polymerase chain reaction (PCR) Purification kit (Qiagen, Germany). All ChIPs were performed in duplicate.
Western blot
Fifteen microliters of cell extract was used for native SDS–polyacrylamide gel electrophoresis (PAGE). Briefly, equal amounts of sample loading buffer without the reducing agent mercaptoethanol was mixed with the cell extract from sonicated samples and incubated at 37°C for 15 min. The mixture was run on a lab-cast gel, transferred onto polyvinylidene fluoride (PVDF) membrane and analyzed by western blot using monoclonal anti-His antibody (Promega). Western blot was developed with Goat anti-mouse IgG/IgM peroxidise-linked antibody (Dianova) and Lumilight system (Roche) and photographed with the Fujifilm system (Fujifilm Image Reader 2.5).
Amplification of DNA
The precipitated DNA was amplified and used as probe in the P. aeruginosa genome tiling array. The amplification was based on previously described Affymetrix protocols (http://www.biotechniques.com/article.asp?id=112343). Briefly, DNA was incubated with dNTPs, sequenase (USB, Cleveland, USA) and random primer (GTTTCCCAGTCACGGTC(N)9) at 37°C, followed by inactivation of the sequenase at 95°C. This procedure was repeated three times. After DNA purification with PCR Purification kit (Qiagen), DNA was PCR amplified using the corresponding GTTTCCCAGTCACGGTC primer and a dNTP/dUTP mix (10 mM dATP, 10 mM dCTP, 10 mM dGTP, 8 mM dTTP and 2 mM dUTP). DNA was purified using PCR Purification kit (Qiagen).
Microarray analysis
Approximately 7.5 µg of amplified DNA from the control immunoprecipitation and the DNA-binding proteins (DBP) immunoprecipitations (IPs) were fragmented (to a size of 50–500 bp) and labeled using the GeneChip WT double-stranded DNA terminal labeling kit (Affymetrix, Santa Clara, USA) according to the instructions of the manufacturer. The biotin-labeled DNA was hybridized to an Affymetrix P. aeruginosa genome chip as described previously (27). Enrichment of hybridization signals was calculated with the Tiling Analysis Software (TAS, Affymetrix) from two independent DBP IPs compared to two independent control IPs (bandwidth parameter was set to 150 bp). For each gene (PAO1 annotation taken from www.pseudomonas.com), the promoter region was defined as the sequence from −350 to −1 bp with respect to the ATG start codon, excluding overlap to upstream coding regions. Promoter enrichment was defined as the maximum enrichment found within the promoter region.
Identification of consensus sequence for OxyR binding
Promoter regions were scanned with the web-based programs CONSENSUS and PATSER available at http://rsat.ulb.ac.be/rsat/ to detect DBP consensus sequences. Sequence logos were created with WebLogo (http://weblogo.berkeley.edu/). The detection of consensus sequences by PATSER was limited to a weight score of 6.0. The global representations of the defined consensus sequence were identified by Virtual Footprint (http://www.prodoric.de/vfp/vfp_regulon.php) for the entire genome of P. aeruginosa PAO1.
Protein expression and purification
The recombinant expression vector pET24a-5344 (for OxyR), pET24a-0849 (for TrxB2) and pET24a-5240 (for TrxA) was introduced into E. coli BL21(DE3), respectively. Transformants were grown and induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 12 h at 28°C. The harvested cells were resuspended in TGE buffer [50 mM Tris–HCl (pH 7.5), 10% glycerol, 1 mM EDTA and 10 mM imidazole] and disrupted by pulsed sonication. The clear lysate was centrifuged at 12 000g for 15 min, and the clear supernatant was loaded onto and purified with Hi-Trap FF column (GE Healthcare) integrated by AKTA™ FPLC system (GE Healthcare). The proteins were eluted using TGE buffer containing 250 mM imidazole. The purity of the His-tagged protein was verified as >95% homogeneity after 10% SDS–PAGE (Invitrogen). The purified proteins were dialyzed against 50 mM Tris–HCl (pH 7.5), 50 mM NaCl and 5% glycerol. Protein concentration was determined using a NanoDrop 1000 spectrophotometer (Thermo Scientific).
Electrophoretic mobility shift assay
EMSA experiments were performed as described previously with minor modifications (28). DNA fragments generated by PCR amplification using P. aeruginosa PAO1 genomic DNA were 5′-end labeled with [γ-32P]-ATP by T4 polynucleotide kinase. Primers used for amplification are listed in Supplementary Table S4. Separation of OxyR–DNA complexes from free DNA was performed on 6% polyacrylamide gels. All binding reactions were performed at 30°C for 20 min in OxyR binding buffer [40 mM bis–Tris borate (pH 7.5), 80 mM KCl, 10 mM MgCl2, 2 mM EDTA, 50 μg/ml BSA, 10% glycerol, 25 μg/ml sonicated herring sperm DNA]. The protein–DNA migration was carried out for 3 h at 8 V/cm in TEB buffer (89 mM Tris–HCl, 89 mM boric acid, 0.25 mM EDTA). Apparent dissociation equilibrium constants (KD) were determined by densitometry of the free DNA bands on the autoradiograph as a function of the protein concentration and are the average value of at least two independent assays.
Quantitative real-time PCR
Bacteria were harvested during the stationary phase and RNA was extracted using the RNeasy Midi Kit (Qiagen). The purity and concentration of the RNA were determined by gel electrophoresis and spectrophotometry (NanoDrop, Thermo Scientific). First-strand cDNA was reverse transcribed from 1 µg of total RNA with the First-strand cDNA Synthesis Kit (Amersham Biosciences, GE Healthcare). Quantitative real-time PCR (qRT-PCR) was performed in a Bio-Rad iCycler with Bio-Rad iQ SYBR Green Supermix. For all primer sets (Supplementary Table S4), the following cycling parameters were used: 94°C for 3 min followed by 40 cycles of 94°C for 60 s, 55°C for 45 s and 72°C for 60 s, followed by 72°C for 7 min. oprI (house-keeping gene control, outer membrane lipoprotein precursor) was used to normalize gene expression (29). Amplification products were electrophoresed on 0.8% agarose gels. For statistical analysis of relative gene expression, the 2−ΔΔCT method was used (30).
RESULTS
Genome-wide mapping of OxyR-binding regions
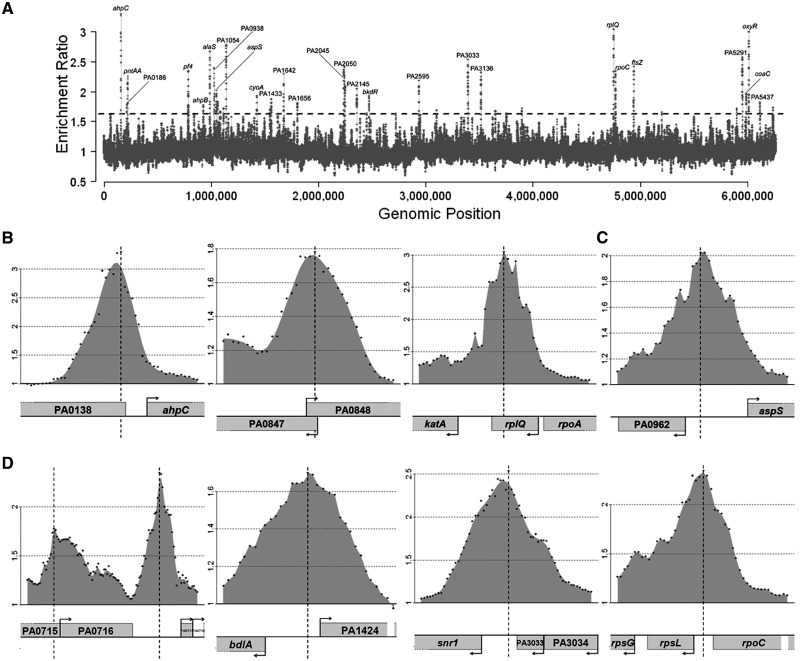
To identify all target genes of OxyR, we applied the established ChIP-chip approach (see Supplementary Figure S1 in Supplementary Data) to the H2O2-treated and in trans oxyR-transformed P. aeruginosa PAO1 bacteria. A comprehensive data set was obtained and is shown in Figure 1, Table 1 and Supplementary Table S1, respectively. As many as 122 genomic regions showed an enrichment of more than 0.5 (log2-fold) and among these regions, 56 were enriched by at least 0.7 (log2 fold) (P < 0.05). For 30 of these regions (enrichment >0.5), the binding region is located between two divergently transcribed genes, which therefore might be both controlled by OxyR. Our results show that OxyR binds mainly to intergenic regions, a trait that is a hallmark of transcriptional regulators. However, in the instances of neighboring genes with short intergenic regions, the binding site lies within the coding regions, as in the case of recG. Additionally, nine regions between convergently transcribed genes were excluded from the present analysis of the OxyR regulon in accordance with our focus on promoter regions. In total, using an enrichment threshold of 0.5 (log2 fold), 195 genes were identified; when the enrichment was set up to 0.7 or 1 (log2 fold), the number of potential affected targets was reduced to 86 and 41, respectively.
Figure 1.
Distribution of OxyR binding sites across the P. aeruginosa genome. (A) An overview of results from ChIP-chip experiments that measure the binding profiles of OxyR throughout the P. aeruginosa PAO1 chromosome during stationary growth phase in contact with 1 mM H2O2 for 10 min. Binding intensities (y-axis, log2 ratio) are plotted against their corresponding genomic locations on the chromosome (x-axis). The dashed threshold line of 1.6 (=20.7) is labeled. A complete list of all the targets can be seen from Supplementary Table S1. Some selected regions were also labeled in the graph. (B) The figure shows the known binding regions of OxyR in P. aeruginosa, PA0139 (ahpC), PA0848 (ahpB) and PA4236 (katA). (C) The figure displays OxyR binding to the previously identified potential OxyR targets in P. aeruginosa, PA0962 (dps). (D) The figure displays OxyR binding to some of the newly identified OxyR targets in P. aeruginosa, PA0717-0719 (pf4 bacteriophage genes), PA1423 (bdlA), PA3032 (snr1) and PA4268 (rpsL). Only partial results are listed here and all data can be found in Supplementary Table S1. Data shown in all panels are average values from replicates (P < 0.05). y-axis of (B), (C) and (D) shows log2 ratio number of binding intensities.
Table 1.
Whole-genome location analysis of oxidative stress sensor OxyR in Pseudomonas aeruginosa (cutoff >1.0)
| PA no.a | Genea | ChIP-chip |
Binding motif |
Gene producta | ||||
|---|---|---|---|---|---|---|---|---|
| Max. Enrich. | Distance to ATG | Genes affecteda | Conserved motifb | Distance to ATGb | PMW scoreb | |||
| PA0139 | ahpC | 1.72 | 171 | 1 | ATAGATTTAGATAAT | 101 | 7.71 | Alkyl hydroperoxide reductase subunit C |
| PA0140 | ahpF | 1.72 | 127 | 1 | Alkyl hydroperoxide reductase subunit F | |||
| PA4236 | katA | 1.61 | 260 | 2 | ATTGATCTCGCTTAT | 116 | 7.35 | Catalase |
| PA5345 | recG | 1.59 | 738 | 1 | ATP-dependent DNA helicase RecG | |||
| PA0962 | dps | 1.02 | 107 | 1 | ATAGGGAGAATCTAT | 107 | 7.66 | Probable DNA-binding stress protein |
| PA2146 | 1.04 | 287 | 1 | ATCTTTGAACCCTAT | 153 | 7.02 | Conserved hypothetical protein | |
| PA2050 | 1.27 | 202 | 2 | AAAAAGAATATTAAT | 171 | 6.26 | Probable ECF sigma-factor | |
| PA2054 | cynR | 1.07 | 13 | 1 | ATTGGTCTGGTCTAT | 15 | 7.36 | Transcriptional regulator |
| PA3135 | 1.22 | -1 | 2 | AAGTGTAGCGCCTAT | 31 | 6.09 | Probable transcriptional regulator | |
| PA0717 | 1.23 | 306 | 3 | ATAGAGCAAGACTAT | 352 | 7.59 | Hypothetical protein of bacteriophage Pf1 | |
| PA4406.1 | 1.27 | 1 | sRNA | |||||
| PA0195 | pntAA | 1.17 | 289 | 1 | ATTAAAAACATTAAT | 354 | 6.81 | Putative NAD(P) transhydrogenase. subunit alpha part 1 |
| PA0903 | alaS | 1.42 | 106 | 1 | ATAGTTATTGCCTAT | 92 | 7.77 | Alanyl-tRNA synthetase |
| PA0963 | aspS | 1.02 | 304 | 2 | ATAGGGAGAATCTAT | 319 | 7.66 | Aspartyl-tRNA synthetase |
| PA1054 | 1.48 | 271 | 6 | ATAGATTGCGATAAT | 138 | 7.45 | Probable NADH dehydrogenase | |
| PA1541 | 1.20 | 308 | 2 | ATCGATTGGCTTTTT | 237 | 6.36 | Probable drug efflux transporter | |
| PA2053 | cynT | 1.07 | 102 | 2 | ATTGGTCTGGTCTAT | 86 | 7.36 | Carbonate dehydratase |
| PA3032 | snr1 | 1.34 | 282 | 2 | ATAGACTCAGGCTAT | 201 | 7.53 | Cytochrome c Snr1 |
| PA3136 | 1.22 | 71 | 2 | AAGTGTAGCGCCTAT | 25 | 6.09 | Probable secretion protein | |
| PA4268 | rpsL | 1.23 | 76 | 1 | ATAGCTCCACTGATT | 3 | 6.17 | 30S ribosomal protein S12 |
| PA4406 | lpxC | 1.27 | 123 | 1 | ACTGTTTTCAACAAT | 30 | 6.50 | UDP-3-O-acyl-N-acetylglucosamine deacetylase |
| PA5291 | 1.36 | 103 | 1 | ATCGTGATAGCTAAT | 64 | 7.12 | Probable choline transporter | |
| PA0938 | 1.27 | 16 | 1 | ATCATCGGTAGTTAT | 96 | 6.67 | Hypothetical protein | |
| PA2044 | 1.25 | 101 | 1 | ATTGATAAAACCTAT | 95 | 7.84 | Hypothetical protein | |
| PA2145 | 1.04 | 8 | 1 | ATAGACCCATTCAAT | 38 | 7.23 | Hypothetical protein | |
| PA2595 | 1.13 | 8 | 1 | Conserved hypothetical protein | ||||
aChIP-enriched OxyR-binding regions are shown by the PA number of the associated genes; the respective gene names, operon compositions (indicated by the number of affected genes) and gene annotations are extracted from http://www.pseudomonas.com.
bConserved motifs, distance to ATG and the PMW scores were all obtained by Virtual Footprint (as described in ‘Materials and Methods’ section) for the entire genome of P. aeruginosa PAO1 using regions with cutoff >0.5 as the PMW entry; the defined motifs in Supplementary Table S1 were chosen based on those motifs with the highest PMW scores for each binding region.
As shown in Supplementary Table S1, the target genes are organized into three categories: (i) category I (seven genes) comprises the genes previously identified as being directly OxyR-regulated; (ii) category II (nine genes) corresponds to genes expected to respond to OxyR based on observations made in other bacteria; and (iii) category III contains 106 newly identified targets, which were tentatively subdivided into the six following groups: oxidative stress related, transcriptional regulators, virulence-related, non-coding RNAs, metabolism-related and genes encoding hypothetical or conserved proteins. As developed below, representative genes from each category were confirmed to be direct targets of the regulator by in vitro binding. The functional classification of these identified direct targets is shown in Supplementary Figure S2 according to the Pseudomonas Genome Database (www.pseudomonas.com) (31).
Extension of the OxyR regulon
Our ChIP-chip analyses identified an additional 115 OxyR target genes (enrichment >0.5) that were not known to belong to the OxyR regulon (Table 1 and Supplementary Table S1). Among those are several genes that are required for oxidative stress response such as dps, trxA and PA3450 (a probable antioxidant protein) and many transcriptional regulators including himA, pvdS, rsaL and mvfR (pqsR), which are involved in QS regulation and iron homeostasis in P. aeruginosa (32–34). The association of OxyR with virulence factors encoding genes including aprA, bdlA, pvdS and ppx constitutes a new insight given that some of them are additionally controlled through QS regulatory pathways (35). These findings can also provide insights into the multiple phenotypes observed for an oxyR mutant, including the overproduction of pyocyanin (17) and the reduced virulence in animal models (14).
In addition, OxyR was shown to bind to the promoter regions of genes implicated in various metabolic processes such as small molecule transport [PA0185 (putative nitrate ABC transporter), PA0186 (nitrate transport, periplasmic component), PA1541 (predicted drug efflux protein) and PA5291 (choline–betaine transporter)], protein synthesis [PA0584 (cca, tRNA nucleotidyl transferase), PA0903 (alsS, alanyl-tRNA synthetase), PA0963 (aspS, aspartyl-tRNA synthetase) and PA4268 (rpsL, ribosomal protein S12)], aerobic/anaerobic respiration PA1317 (cyoA, cytochrome o ubiquinol oxidase) and PA3032 (snr1, cytochrome c) and secretion [PA3136 (HlyD family secretion protein)]. Other OxyR targets include a large number of genes involved in energy metabolism as well as hypothetical genes. Surprisingly, as many as six non-coding RNA genes were found to be associated with OxyR under oxidative stress, implying the existence of possible indirect post-transcriptional control by OxyR.
Another interesting and unexpected finding is the binding of OxyR to the promoter region of the bacteriophage Pf1 operon (PA0717, recently designated as Pf4), which was previously found to be highly expressed in P. aeruginosa biofilms as compared to planktonic cultures (36) and was also implicated in virulence, biofilm structural integrity and phenotypic variant formation (37). The appearance of variants has been linked to the increased adaptability of the stressed bacterial population (38).
Likewise, OxyR was found to bind to the promoter region of one biofilm dispersion locus (bdlA, PA1423), which further links the biofilm formation to OxyR-mediated regulation in P. aeruginosa (39). We can postulate that, under oxidative stress conditions, OxyR will probably promote the dispersion of biofilm bacteria under stress. Evidence supporting this notion is that the oxyR mutant of P. aeruginosa demonstrated decreased swarming motility (17), as also observed in another Gram-negative pathogen Serratia marcescens (40).
Another intriguing finding is the direct binding of OxyR to the promoter region of the 30S ribosomal protein S12 (encoded by rpsL, PA4268). Protein synthesis has been shown to be inhibited in response to oxidative stress and treatment with aminoglycosides in P. aeruginosa (24,41) and to be reprogrammed in lower eukaryotes such as Saccharomyces cerevisiae (42,43). It is therefore tempting to hypothesize that OxyR-mediated inhibition of protein synthesis or ribosome reprogramming is another fundamental adaptive circuit breaker to oxidative stress.
It is also noteworthy that OxyR binds to the promoter region of a cell wall synthesis gene, which is involved in the second step of lipid A biosynthesis in P. aeruginosa (lpxC, PA4406) (44). The mechanisms implicated in this process mediated by OxyR could result in structural changes or increased outer membrane thickness to withstand the oxidative stress elicited by H2O2.
Additionally, the implication of OxyR has been shown for the first time in (i) the regulation of the extracytoplasmic (ECF) sigma factor gene, pvdS, which further connects iron homeostasis with oxidative stress as already described in other microorganisms; (ii) the regulation of cytochrome c genes (cyoA and snr1); and (iii) the regulation of small molecule transport genes (PA5291 and PA2145).
Validation of ChIP-chip results
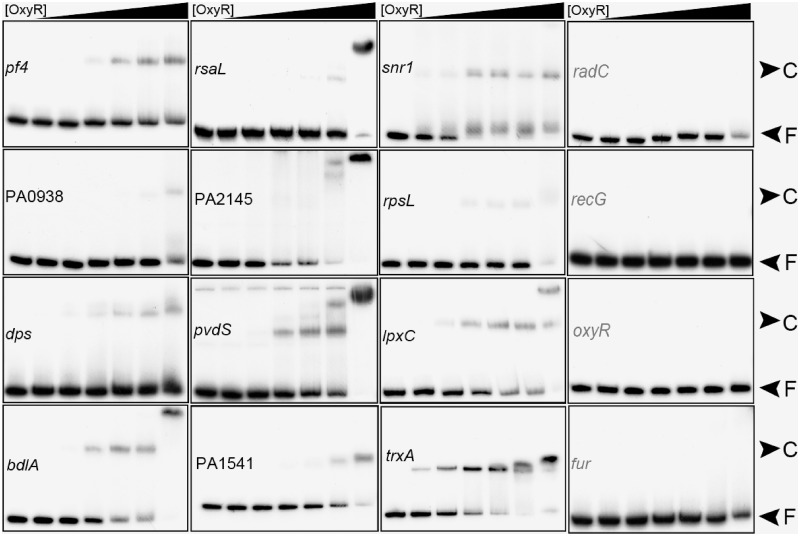
To validate the ChIP-chip data, 18 promoter regions of OxyR target genes were tested for in vitro binding of purified his6-tagged P. aeruginosa OxyR protein by EMSA. The known P. aeruginosa OxyR-regulated ahpC gene was included as a positive control (Figure 1B and Supplementary Figure S3A) and fur, which is controlled by OxyR in E. coli but not in P. aeruginosa, was used as a negative control (8,45). Out of the 18 tested potential targets, 13 showed clearly retarded migration after incubation with the purified oxidized OxyR protein, while no binding was detected to the five remaining targets (mvfR, himA, oxyR, radC and recG) (Figure 2 and Supplementary Figure S3B). The absence of binding can be due to the requirement of thus-far unappreciated factors to aid the OxyR–DNA interaction or to the difference between in vivo and in vitro conditions or to the existence of different active forms of OxyR besides the oxidized form (46). Almost all of the OxyR-bound targets harbor the conserved motif identified in this study (see subsequent section) while the non-binding targets do not contain this motif in their promoter regions (Supplementary Table S2). There are, however, exceptions such as rsaL and PA4406.1 (sRNA), which bind to OxyR but do not contain a consensus motif based on our position weight matrix (PWM) prediction, indicating a possible degenerative mechanism for OxyR binding. However, a manual search for motif-like sequence identified two possible motifs for both of them, which were not identified by Virtual Footprint analysis (Supplementary Table S2).
Figure 2.
Validation of binding of OxyR to selected target regions by EMSA. The promoter regions were chosen randomly and EMSA analyses are described in ‘Materials and Methods’ section. OxyR was purified in air conditions and used in the following concentration (from left to right) (nM): 0, 84, 168, 336, 504, 840 and 1680. Protein–DNA complex (C) and free DNA (F) are indicated. Promoter regions with positive binding results are shown in black while regions with negative results are highlighted in grey. Promoter region of fur was used as a negative control. Supplementary results using the positive control (ahpC) are shown in Supplementary Figure S3A.
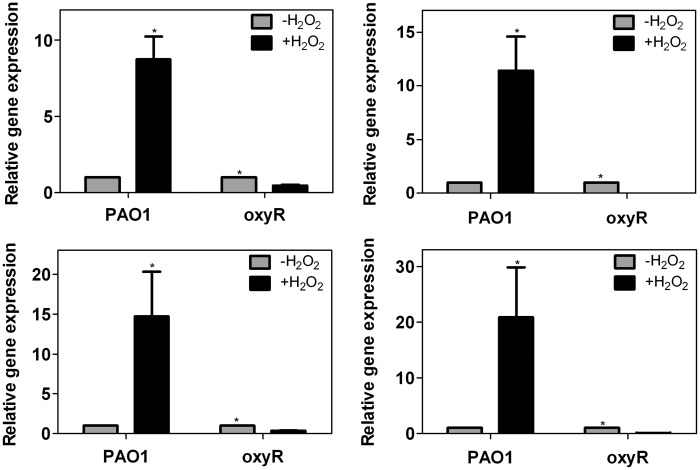
We further investigated the gene expression of OxyR-dependent targets by qRT-PCR. As shown in Figure 3, genes that for the first time were identified in this study as belonging to the OxyR regulon, such as dps, pvdS and trxB2 clearly demonstrated an OxyR-dependent activation when the cells were exposed to H2O2 (1 mM). Interestingly, disruption of oxyR abrogates the induction of those genes when exposed to oxidative stress. The positive control aphC also responded to oxidized OxyR, in agreement with previous work (12).
Figure 3.
RT-PCR validation of OxyR-dependent gene expression. (Top left) ahpC, (Top right) dps, (Bottom left) pvdS and (Bottom right) trxB2. ahpC was selected as a positive control of OxyR-dependent activation after exposure to H2O2 (23,24). All the data were obtained from at least two independent experiments with at least three replicates. Error bar indicates SD. *P < 0.05 by Student's t-test.
Features of OxyR binding
In order to correlate our results with previous findings in terms of transcriptional regulation and to identify the regulation pattern of OxyR in P. aeruginosa, we examined the data from previous whole-genome transcriptional analyses performed under oxidative stress conditions similar to our ChIP-chip analysis (23,24). All differentially regulated genes in both transcriptional analyses were compared with the output of our ChIP-chip analysis and 56 out of the identified 122 genes were found to be differentially expressed in oxidized conditions in P. aeruginosa (Supplementary Table S1). The discrepancy between our ChIP-chip and the transcriptional analyses can be explained by (i) a methodological difference between ChIP-chip and microarray analysis—the former measures the direct binding of the transcriptional activator to the DNA, while the latter reveals differences in gene expression which may be affected by secondary and tertiary effects and (ii) the presence of other transcriptional regulator(s) or co-factor(s), which cooperate with OxyR to activate or repress the target genes in a non-degenerative way and remain elusive. Interestingly, further classification of all differentially expressed genes (56 genes) according to their respective functions indicates that expression of those involved in adaptation and protection is promoted, whereas genes implicated in energy metabolism including transcription and translation are repressed during oxidative stress (Supplementary Figure S4).
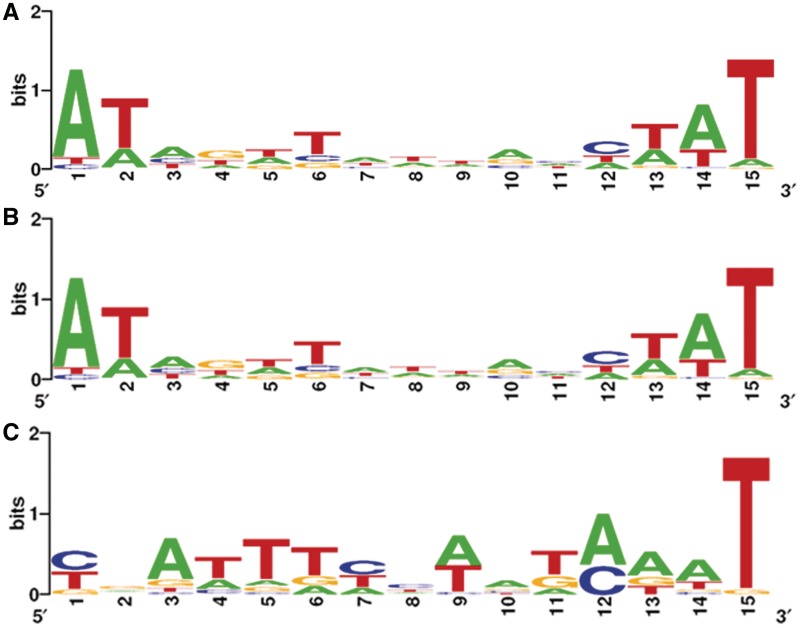
Putative DNA binding sites of OxyR in E. coli were predicted by computational analyses using the identified OxyR-binding site sequences as a model (10,11) and are shown in Supplementary Figure S5A. Furthermore, the alignment of previously identified OxyR-regulated promoters from P. aeruginasa had revealed four putative OxyR-binding tetranucleotide sequences (ATAG) spaced by heptanucleotides (12). However, a more accurate determination of the DNA-binding motif of OxyR based on a genome-wide analysis has not been performed. In particular, the identified conserved motif for OxyR displays a dyad-symmetry pattern. Therefore, we aimed at detecting the half unit of this OxyR motif in P. aeruginosa. Toward this end, three different groups of promoter regions were used. The first, second and third groups comprise promoter regions with enrichment cutoff larger than 1.0, 0.7 and 0.5, respectively. Selected promoter regions were scanned with the web-based programs CONSENSUS and PATSER and sequence logos were created with WebLogo (see ‘Materials and Methods’ section). Figure 4 displays the OxyR consensus sequence determined with selected promoters from the three groups of promoter regions. It is noteworthy that a palindrome-like sequence (ATAGATTNAATCTAT) with relatively low sequence conservation is present in all three DNA consensus sequences. In particular, the ATAG tetranucleotide typical of OxyR-binding motif in E. coli and P. aeruginosa from previous studies was confirmed by our analysis. Additionally, the T(N)11A motif typical of LTTRs was also identified (10) (Supplementary Figure S5B). It is also noteworthy that the OxyR-binding motif is extremely A+T rich.
Figure 4.
Determination of the OxyR consensus sequence. (A) The calculated OxyR consensus sequence in P. aeruginosa using ChIP-chip data with enrichment value above 1.0 (Log2 ratio). (B) The calculated OxyR consensus sequence using ChIP-chip data with enrichment value above 0.7 (Log2 ratio). (C) The calculated OxyR consensus sequence using ChIP-chip data with enrichment value above 0.5 (Log2 ratio). All logos were created using WebLogo (12).
A pattern-based search within the promoter regions was performed by Virtual Footprint (see ‘Materials and Methods’ section) for the entire genome of P. aeruginosa PAO1 with the consensus sequence shown in Figure 4A. Eighty-eight out of the ChIPchip identified 122 genes (72.13%, cutoff > 0.5, Log2 ratio) or 42 out of 57 genes (73.68%, cutoff > 0.7, Log2 ratio) were identified as having a consensus motif-like sequence in the corresponding promoter region (Table 1 and Supplementary Table S1). This strongly supports the hypothesis that OxyR binds essentially around the proposed consensus sequence to activate or repress the transcription of target genes.
To determine whether OxyR binding is conserved in bacteria other than E. coli and P. aeruginosa, we used our created PWM based on Figure 4A to search the genomes of other bacteria containing an oxyR homolog for putative OxyR-binding sites. Due to the high degeneracy of the proposed OxyR motif, most of the false positives in the original search results could not be excluded. Therefore, only those genes that are involved in oxidative stress as well as targets homologous to those identified in our study were retained. Concurrently, those targets were used to generate new species-specific motifs. The results of this search are summarized in Supplementary Table S3 and Supplementary Figure S5. It is evident that the OxyR-binding motif differs slightly in the 13 different bacteria analyzed. The strongest conservation is found for the nucleotides A1, A3, C12, A14 and T15, respectively. We conclude that these residues may play a major role in the interaction with oxidized OxyR. In addition, it is also noteworthy that genes encoding antioxidant enzymes, such as alkyl hydroperoxide reductases and/or catalases, were found to be a common target of OxyR in all examined bacterial genomes (14 hits out of 14 species analyzed). Intriguingly, cellular thiol-reducing systems including thioredoxin (Trx) and/or glutaredoxin (Grx), were also identified as a common target (11 hits out of 14 species analyzed), suggesting a general auto-reduction mechanism for OxyR.
Thioredoxin-mediated enzymatic regulation of OxyR
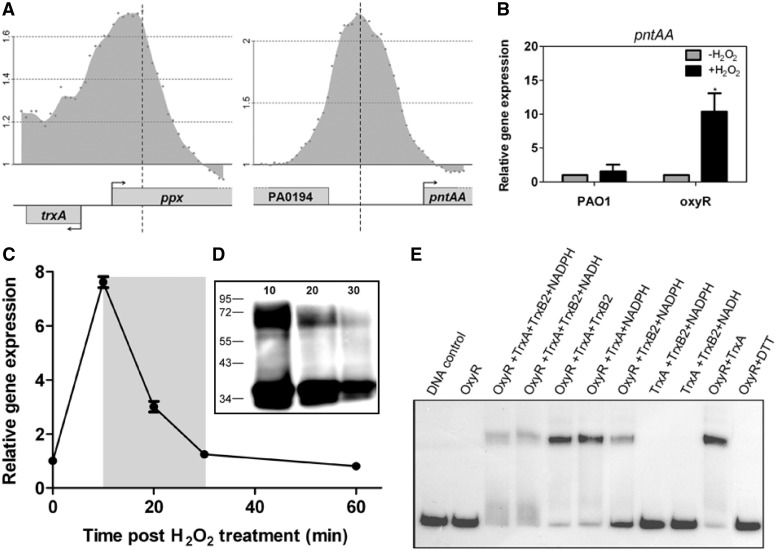
One of the most interesting findings from this study is that P. aeruginosa OxyR binds to the promoter region of a cellular disulfide reducing component, the thioredoxin (encoded by trxA, PA5240) as well as one gene involved in NAD(P)/NAD(P)H metabolism (PA0195, pntAA) (Figure 5A). This suggests a potential auto-regulatory mechanism for OxyR reduction whereby OxyR is deactivated through enzymatic disulfide bond reduction by thioredoxin (TrxA) in combination with a thioredoxin reductase at the expense of NADPH. Toward this end, we also discovered that one of the two thioredoxin reductase-encoding genes, trxB2 (PA0849), is found immediately downstream of ahpB (PA0848) that has been identified as a direct target of OxyR in P. aeruginosa (Figure 1B). Indeed, under oxidative stress conditions we could observe an OxyR-dependent activation of trxB2 (Figure 3 Top left), suggesting the auto-regulatory recruitment of thioredoxin reductase by OxyR for its subsequent reduction. In addition, we detected a negative regulation of OxyR for the expression of pntAA under oxidative stress condition (Figure 5B). The reduction of OxyR in P. aeruginosa was first observed in vivo where the H2O2-activated OxyR dimer (active and oxidized form) completely disappeared after 30 min incubation, leaving only the inactive reduced OxyR monomeric form further confirmed by the OxyR-dependent activation of ahpC after H2O2 contact (Figure 5C and D). This reaction time is consistent with that of OxyR in E. coli during reduction by glutaredoxin in the presence of glutathione, which constitutes another major thiol-reducing system in bacteria (1).
Figure 5.
TrxA/TrxB2 thioredoxin system mediates OxyR reduction. (A) The figure shows the confirmed binding of OxyR to the indicated regions involved in thioredoxin production [PA5240 (trxA)] and in NAD(P)/NAD(P)H metabolism [PA0195 (pntAA)] in P. aeruginosa. (B) OxyR-dependent repression of pntAA. The expression of pntAA in the presence of OxyR is not induced by exposure to H2O2 while the disruption of oxyR leads to a great increase in the gene expression, suggesting that OxyR works as a repressor of pntAA. All the data were obtained from at least two independent experiments with at least three replicates. Error bar indicates SD. *P < 0.05 by Student's t-test. (C) OxyR-dependent attenuation of ahpC activation after 60 min contact with H2O2. Relative gene expression of ahpC was determined as described in the ‘Materials and Methods’ section. The relative expression (y-axis) is defined as the ratio of relative expression of ahpC at time 10, 20, 30 and 60, respectively, to that of time 0 (untreated). (D) Time-dependent reduction of OxyR. Western blot analysis (see ‘Materials and Methods’ section) of cell lysates extracted from formaldehyde-treated cells at 10, 20 and 30 min after exposure to H2O2, respectively showed that 30 min after treatment with 1 mM H2O2 there is a time-dependent reduction of the number of OxyR dimers (upper band) while monomers accumulate (lower band). (E) EMSA confirmation of OxyR reduction by a thioredoxin system. Samples of purified P. aeruginosa OxyR (0.1 μM) were incubated with 10 μM thioredoxin (TrxA), 10 μM thioredoxin reductase (TrxB2) and 0.5 mM NADPH or NADH for 30 min at 30°C. A mixture missing either NADPH, TrxB2 or TrxA was used as a control to confirm the reduction effect of the TrxA/TrxB2 system. A mixture missing OxyR was used as a control to confirm the absence of binding of the thioredoxins on DNA. OxyR was also incubated with TrxA to show whether trace amount of reduced form of TrxA could reduce oxidized OxyR, which turned out to be negative, confirming the need for TrxA, TrxB2 and NAD(P)H. 0.1 M DTT was also used as a positive control to show the effect of reducing OxyR, causing an inability of the protein to bind the target DNA.NADPH and NADH were purchased from Sigma. Results shown are representative of six independent experiments.
To further test our hypothesis that a thioredoxin system could catalyze the OxyR deactivation in P. aeruginosa, we overproduced and purified the TrxA and TrxB2 proteins to electrophoretic homogeneity. When air-oxidized OxyR was incubated together with TrxA and TrxB2 in the presence of NAD(P)H for different times, we observed a rapid decrease of the binding of OxyR to the ahpC promoter region, although to a lower extent compared to the effect of DTT treatment (Figure 5E).
The mechanism of OxyR reduction in P. aeruginosa appears therefore to involve thioredoxins, whereas in E. coli both glutaredoxin and thioredoxin play an important role in enzymatic deactivation of OxyR (1). Furthermore, it was found that E. coli OxyR binds to the promoter region of one of the two thioredoxin-encoding genes, trxC (47). At this stage, we cannot exclude the participation of a glutaredoxin in the reduction of oxidized P. aeruginosa OxyR.
DISCUSSION
The ability of bacteria to alter gene expression patterns in response to environmental stresses is critical for their survival. The central transcriptional regulator of the oxidative stress response in many Gram-negative bacteria is OxyR, which, upon induction by H2O2, undergoes a conformational change allowing oxidized OxyR to acquire DNA-binding capacity and to activate the transcription of a battery of antioxidant genes including those encoding catalases, glutathione reductase and alkyl hydroperoxide reductases (2–4). The extensive studies of OxyR-mediated oxidative response in E. coli have greatly broadened our understanding of mechanisms of transcriptional regulation of OxyR and extended the OxyR regulon beyond the genes encoding antioxidants (1,6,8). In the human opportunistic pathogen P. aeruginosa, OxyR is known to activate the expression of the katA, katB, ahpB and ahpCF genes to counteract the oxidative damages caused by H2O2 (12,13). After exposure to H2O2, the expression of a large number of genes was found to be altered, including an up-regulation of protection mechanisms and a down-regulation of primary metabolism genes (23,24).
Extension and evolution of the P. aeruginosa OxyR regulon
In this study, we developed a ChIP-chip method to identify all the binding sites for OxyR in P. aeruginosa. Through this chromosome-wide DNA-binding profiling analysis, we identified for the first time the direct targets of OxyR in vivo under oxidative stress. A large number of targets (∼122) were identified and all of the previously known targets were confirmed, indicating the robustness and accuracy of this genome-wide approach. The results presented in this study revealed that under oxidative conditions, OxyR is activated to directly bind to the promoter regions of previously identified antioxidant genes (OxyR core regulon) as well as an unexpected high number of other previously unknown target genes (OxyR variable regulon), revealing other unsuspected and unidentified features of the global OxyR-mediated stress response in P. aeruginosa. Functional classification of the newly identified target genes revealed that OxyR primarily binds to the control region of genes involved in adaptation and protection, DNA recombination and repair, non-coding RNA genes, transcriptional regulators and those encoding proteins for the transport of small molecules as well as a large number of hypothetical or putative genes. This finding greatly extends the OxyR regulon to areas beyond classical antioxidant genes. Specifically, some of the OxyR targets identified in this study that have been confirmed by in vitro binding include the following functional groups: (1) antioxidant genes (katA, katB, ahpB and ahpCF); (2) biofilm formation (pf4 and bdlA); (3) QS (rsaL); (4) iron metabolism (dps and pvdS); (5) protein synthesis (rpsL); (6) aerobic/anaerobic respiration (snr1); (7) transport (PA1541); (8) cell wall synthesis (lpxC); (9) OxyR reduction (trxA, trxB2); and (10) other genes involved in regulation (sRNA, PA4406.1). Collectively, our data strongly support the notion that OxyR in P. aeruginosa possesses more regulatory functions than previously expected and behaves rather as a dramatically more versatile, global transcriptional regulator. In E. coli, it was shown that OxyR could bind to control regions of several genes including dsbG, encoding a periplasmic disulfide-bond chaperone-isomerase and fhuF, encoding a protein required for the reductive release of iron from the ferrichrome siderophore (8), strongly suggesting a versatile mode of action of OxyR also in this microorganism.
Our results further confirm and extend the composition of the OxyR core regulon of P. aeruginosa, which now includes genes coding for thiol-reducing systems in addition to the previously established antioxidant genes. Indeed, our computational analysis of OxyR-binding sites in different bacteria strongly suggests that two OxyR thiol-reducing systems exist based on either Trx or Grx. The variable regulon, also known as the species-specific regulon or accessory regulon, constitutes a variable set of genes that define the plasticity of the transcription factor binding sites. As a human pathogen, P. aeruginosa will ensure its efficient invasion and infection of the host and will otherwise constantly encounter oxidative stress and likely damage generated from phagocytic cells. In its defensive role, OxyR performs its detoxification function as well as other processes that may facilitate the proliferation and invasion. This study also generates an evolutionary perspective of gene regulation mediated by OxyR in different bacteria, indicating that pathogenic species invest energy not only in the regulation of detoxification but also in virulence. We also demonstrate an important overlap between oxidative stress response, QS, biofilm formation and iron metabolism (45,48–50).
OxyR involvement in iron homeostasis, QS and other processes
Of major interest is the undeniable link between oxidative stress and iron homeostasis since our study for the first time demonstrates that OxyR activates the expression of the pvdS gene which encodes an ECF sigma factor that is required for the expression of the pyoverdine siderophore biosynthesis genes (34). Interestingly, pvdS is also under the control of the Fur regulator, which means that its expression is repressed by Fur-Fe2+ and activated by OxyR (34,51). In addition, in E. coli OxyR is known to activate the transcription of the fur gene encoding the ferric uptake regulator, firstly establishing a clear link with the control of iron homeostasis (7,52). However, our study suggests that in P. aeruginosa OxyR does not directly control the expression of fur.
Very recently, we found that in both an oxyR mutant and an OxyR-overexpression strain, the production of the redox antibiotic pyocyanin increased even without the addition of H2O2, indicating the protective role of this phenazine compound in the oxidative response (17). Interestingly, we also found that OxyR could bind to the promoter region of mvfR in vivo under oxidative stress condition, although we did not confirm the direct binding of OxyR to this region in an in vitro binding assay (even though the in silico approach predicted an OxyR binding motif). MvfR together with its two cognate ligands, HHQ (4-hydroxy-2-heptyquinoline) and PQS (3,4-dihydroxy-2-heptyquinoline), forms a QS regulatory circuit that controls the production of PQS, pyocyanin, elastase and is required for full virulence of P. aeruginosa (53). Additionally, a LasR-based genome-wide location analysis also identified mvfR as a direct target of LasR (22), suggesting the dual control of mvfR by OxyR and LasR. Possibly, the binding of LasR to the promoter region of mvfR will facilitate the OxyR binding in an unknown way. In bacteria, including P. aeruginosa, it has been shown that the QS system(s) and oxidative stress response are linked (49,54).
Another intriguing finding is the binding of OxyR to the control region of non-coding RNA genes in P. aeruginosa, raising the possibility of post-transcriptional regulation involving small regulatory RNAs (sRNA). In E. coli, oxyS, a small and abundant RNA that is induced by oxidative stress and controlled by OxyR, regulates numerous genes including those encoding the transcriptional regulators FhlA and RpoS (55). Although no homolog of this sRNA has been found in P. aeruginosa, its genome contains more than 29 non-coding sRNAs (31). Interestingly, one possible counterpart found in a recent study is PhrS sRNA, which has recently been demonstrated to be involved in post-transcriptional regulation of MvfR and thus potentiate the production of pyocyanin (56).
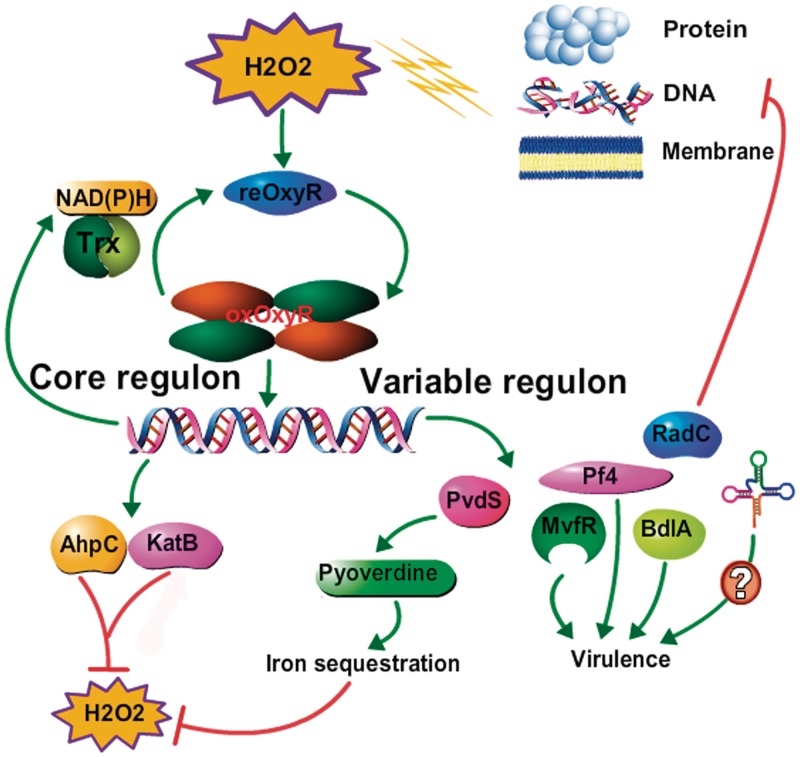
The multiple OxyR-dependent cellular processes suggest a multilayered defense against oxidative stress whereby the oxidized OxyR regulator binds to the promoter regions of the respective genes to (i) activate the expression of antioxidants to detoxify ROS (mainly H2O2); (ii) inhibit the primary metabolism including protein synthesis, and oxidative respiration to lower the production of ROS; (iii) decrease the expression of iron-uptake related proteins to minimize the occurrence of the Fe/Cu-dependent Fenton reaction; (iv) enhance the production of sulfur compounds to repair the damaged Fe-S cluster containing proteins including SoxR; (v) switch to a less motile lifestyle (biofilm) or microaerobiosis; and (vi) unknown ways involving the regulation of non-coding RNA genes (six were identified in this study). The combination of these data led us to formulate a hypothetical model, which is summarized in Figure 6.
Figure 6.
Proposed model for OxyR-mediated oxidative stress responses in P. aeruginosa. In contact with H2O2, bacterial cells respond rapidly to the stress and alter the gene expression patterns to prevent the damages caused by H2O2. OxyR plays central roles in these processes by activating both core and variable regulons to detoxify and eliminate the toxic effect of H2O2. The activity of OxyR itself is controlled through reduction of the newly identified thioredoxin system TrxA/TrxB2. Green arrow means an activation effect; red arrow means an inhibition effect.
Regulation of OxyR activity by thioredoxin
Arguably, the most intriguing findings of this study is the discovery of a novel thioredoxin system in P. aeruginosa (TrxA/TrxB2) that can reduce oxidized OxyR in the presence of NAD(P)H and, the activation of this thioredoxin system by oxidized OxyR, indicative of a clear negative feedback and auto-regulatory loop of OxyR. When we searched various genomes for OxyR targets based on our PWM, we identified two types of cellular thiol-reducing systems either involving thioredoxin (Trx) and/or glutaredoxin (Grx) among the common OxyR targets (11 hits out of 14 species). The choice of the OxyR-reduction system is probably dependent on functional redundancy of each system in diverse bacteria. For example, in P. aeruginosa, at least four thioredoxins are found while only one glutaredoxin has been identified so far (31). In contrast, three glutaredoxins and only two thioredoxins were identified in E. coli, which uses glutaredoxin to reduce oxidized OxyR (57). Furthermore, it is worth noting that the oxidative stress response in P. aeruginosa is also coupled with the physiological status of pathways that generate NAD(P)H in an OxyR-dependent manner. In addition, the OxyR-mediated thioredoxin regulation could also contribute to the reduction of other oxidized proteins in addition to OxyR itself under oxidative stress, suggesting another level of protection featured by OxyR.
An evident conclusion from our data is that in P. aeruginosa the oxidative stress response directed by OxyR is not simply a process dedicated to detoxification performed by members of the core OxyR regulon, but also implicates a far more complex cellular response than previously envisioned. This complex stress response orchestrated by P. aeruginosa OxyR and its orthologs might be a common theme in other bacteria.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–4, Supplementary Figures 1–5.
FUNDING
Fonds voor Wetenschappelijk Onderzoek (FWO-Vlaanderen) (G.0119.10N to P.C. and D.C.); Research Council of the Vrije Universiteit Brussel; Q.W. is a CSC-Vrije Universiteit Brussel scholarship receiver and supported by a FEMS research fellowship to perform ChIP-chip experiments in the laboratory of Dr Susanne Haüssler (HZI, Germany); Veteran's Administration Research Program (to D.J.H., partial); FWO-Vlaanderen (post-doctoral fellow, P.N.L.M.). Funding for open access charge: Fonds voor Wetenschappelijk Onderzoek (FWO-Vlaanderen).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all the members of our group and of Dr Susanne Haüssler's team for the generous assistance and helpful discussion. We are very grateful to Prof. Luis E. N. Quadri for sharing the microarray results.
REFERENCES
- 1.Zheng M, Aslund F, Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279:1718–1721. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]
- 2.Pomposiello PJ, Demple B. Redox-operated genetic switches: the SoxR and OxyR transcription factors. Trends Biotechnol. 2001;19:109–114. doi: 10.1016/s0167-7799(00)01542-0. [DOI] [PubMed] [Google Scholar]
- 3.Storz G, Imlay JA. Oxidative stress. Curr. Opin. Microbiol. 1999;2:188–194. doi: 10.1016/s1369-5274(99)80033-2. [DOI] [PubMed] [Google Scholar]
- 4.Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev. Biochem. 2008;77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee C, Lee SM, Mukhopadhyay P, Kim SJ, Lee SC, Ahn WS, Yu MH, Storz G, Ryu SE. Redox regulation of OxyR requires specific disulfide bond formation involving a rapid kinetic reaction path. Nat. Struct. Mol. Biol. 2004;11:1179–1185. doi: 10.1038/nsmb856. [DOI] [PubMed] [Google Scholar]
- 6.Zheng M, Wang X, Templeton LJ, Smulski DR, LaRossa RA, Storz G. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 2001;183:4562–4570. doi: 10.1128/JB.183.15.4562-4570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng M, Doan B, Schneider TD, Storz G. OxyR and SoxRS regulation of fur. J. Bacteriol. 1999;181:4639–4643. doi: 10.1128/jb.181.15.4639-4643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng M, Wang X, Doan B, Lewis KA, Schneider TD, Storz G. Computation-directed identification of OxyR DNA binding sites in Escherichia coli. J. Bacteriol. 2001;183:4571–4579. doi: 10.1128/JB.183.15.4571-4579.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toledano MB, Kullik I, Trinh F, Baird PT, Schneider TD, Storz G. Redox-dependent shift of OxyR–DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell. 1994;78:897–909. doi: 10.1016/s0092-8674(94)90702-1. [DOI] [PubMed] [Google Scholar]
- 10.Schell MA. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 11.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 12.Ochsner UA, Vasil ML, Alsabbagh E, Parvatiyar K, Hassett DJ. Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: OxyR-dependent regulation of katB-ankB, ahpB, and ahpC-ahpF. J. Bacteriol. 2000;182:4533–4544. doi: 10.1128/jb.182.16.4533-4544.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heo YJ, Chung IY, Cho WJ, Lee BY, Kim JH, Choi KH, Lee JW, Hassett DJ, Cho YH. The major catalase gene (katA) of Pseudomonas aeruginosa PA14 is under both positive and negative control of the global transactivator OxyR in response to hydrogen peroxide. J. Bacteriol. 2010;192:381–390. doi: 10.1128/JB.00980-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lau GW, Britigan BE, Hassett DJ. Pseudomonas aeruginosa OxyR is required for full virulence in rodent and insect models of infection and for resistance to human neutrophils. Infect. Immun. 2005;73:2550–2553. doi: 10.1128/IAI.73.4.2550-2553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Melstrom KA, Jr, Kozlowski R, Hassett DJ, Suzuki H, Bates DM, Gamelli RL, Shankar R. Cytotoxicity of Pseudomonas secreted exotoxins requires OxyR expression. J. Surg. Res. 2007;143:50–57. doi: 10.1016/j.jss.2007.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vinckx T, Matthijs S, Cornelis P. Loss of the oxidative stress regulator OxyR in Pseudomonas aeruginosa PAO1 impairs growth under iron-limited conditions. FEMS Microbiol. Lett. 2008;288:258–265. doi: 10.1111/j.1574-6968.2008.01360.x. [DOI] [PubMed] [Google Scholar]
- 17.Vinckx T, Wei Q, Matthijs S, Cornelis P. The Pseudomonas aeruginosa oxidative stress regulator OxyR influences production of pyocyanin and rhamnolipids: protective role of pyocyanin. Microbiology. 2010;156:678–686. doi: 10.1099/mic.0.031971-0. [DOI] [PubMed] [Google Scholar]
- 18.Kuo MH, Allis CD. In vivo cross-linking and immunoprecipitation for studying dynamic protein:DNA associations in a chromatin environment. Methods. 1999;19:425–433. doi: 10.1006/meth.1999.0879. [DOI] [PubMed] [Google Scholar]
- 19.Kurdistani SK, Grunstein M. In vivo protein–protein and protein–DNA crosslinking for genomewide binding microarray. Methods. 2003;31:90–95. doi: 10.1016/s1046-2023(03)00092-6. [DOI] [PubMed] [Google Scholar]
- 20.Wade JT, Struhl K, Busby SJ, Grainger DC. Genomic analysis of protein–DNA interactions in bacteria: insights into transcription and chromosome organization. Mol. Microbiol. 2007;65:21–26. doi: 10.1111/j.1365-2958.2007.05781.x. [DOI] [PubMed] [Google Scholar]
- 21.Sala C, Grainger DC, Cole ST. Dissecting regulatory networks in host–pathogen interaction using ChIP-on-chip technology. Cell Host Microbe. 2009;5:430–437. doi: 10.1016/j.chom.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 22.Gilbert KB, Kim TH, Gupta R, Greenberg EP, Schuster M. Global position analysis of the Pseudomonas aeruginosa quorum-sensing transcription factor LasR. Mol. Microbiol. 2009;73:1072–1085. doi: 10.1111/j.1365-2958.2009.06832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang W, Small DA, Toghrol F, Bentley WE. Microarray analysis of Pseudomonas aeruginosa reveals induction of pyocin genes in response to hydrogen peroxide. BMC Genomics. 2005;6:115. doi: 10.1186/1471-2164-6-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palma M, DeLuca D, Worgall S, Quadri LE. Transcriptome analysis of the response of Pseudomonas aeruginosa to hydrogen peroxide. J. Bacteriol. 2004;186:248–252. doi: 10.1128/JB.186.1.248-252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.West SE, Schweizer HP, Dall C, Sample AK, Runyen-Janecky LJ. Construction of improved Escherichia–Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene. 1994;148:81–86. doi: 10.1016/0378-1119(94)90237-2. [DOI] [PubMed] [Google Scholar]
- 26.Wade JT, Reppas NB, Church GM, Struhl K. Genomic analysis of LexA binding reveals the permissive nature of the Escherichia coli genome and identifies unconventional target sites. Genes Development. 2005;19:2619–2630. doi: 10.1101/gad.1355605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dotsch A, Pommerenke C, Bredenbruch F, Geffers R, Haussler S. Evaluation of a microarray-hybridization based method applicable for discovery of single nucleotide polymorphisms (SNPs) in the Pseudomonas aeruginosa genome. BMC Genomics. 2009;10:29. doi: 10.1186/1471-2164-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caldara M, Minh PN, Bostoen S, Massant J, Charlier D. ArgR-dependent repression of arginine and histidine transport genes in Escherichia coli K-12. J. Mol. Biol. 2007;373:251–267. doi: 10.1016/j.jmb.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 29.Cornelis P, Bouia A, Belarbi A, Guyonvarch A, Kammerer B, Hannaert V, Hubert JC. Cloning and analysis of the gene for the major outer membrane lipoprotein from Pseudomonas aeruginosa. Mol. Microbiol. 1989;3:421–428. doi: 10.1111/j.1365-2958.1989.tb00187.x. [DOI] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Winsor GL, Van Rossum T, Lo R, Khaira B, Whiteside MD, Hancock RE, Brinkman FS. Pseudomonas Genome Database: facilitating user-friendly, comprehensive comparisons of microbial genomes. Nucleic Acids Res. 2009;37:D483–D488. doi: 10.1093/nar/gkn861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao H, Krishnan G, Goumnerov B, Tsongalis J, Tompkins R, Rahme LG. A quorum sensing-associated virulence gene of Pseudomonas aeruginosa encodes a LysR-like transcription regulator with a unique self-regulatory mechanism. Proc. Natl Acad. Sci. USA. 2001;98:14613–14618. doi: 10.1073/pnas.251465298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cornelis P. The ‘core’ and ‘accessory’ regulons of Pseudomonas-specific extracytoplasmic sigma factors. Mol. Microbiol. 2008;68:810–812. doi: 10.1111/j.1365-2958.2008.06223.x. [DOI] [PubMed] [Google Scholar]
- 34.Ravel J, Cornelis P. Genomics of pyoverdine-mediated iron uptake in pseudomonads. Trends Microbiol. 2003;11:195–200. doi: 10.1016/s0966-842x(03)00076-3. [DOI] [PubMed] [Google Scholar]
- 35.Schuster M, Lostroh CP, Ogi T, Greenberg EP. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 2003;185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whiteley M, Bangera MG, Bumgarner RE, Parsek MR, Teitzel GM, Lory S, Greenberg EP. Gene expression in Pseudomonas aeruginosa biofilms. Nature. 2001;413:860–864. doi: 10.1038/35101627. [DOI] [PubMed] [Google Scholar]
- 37.Rice SA, Tan CH, Mikkelsen PJ, Kung V, Woo J, Tay M, Hauser A, McDougald D, Webb JS, Kjelleberg S. The biofilm life cycle and virulence of Pseudomonas aeruginosa are dependent on a filamentous prophage. ISME J. 2009;3:271–282. doi: 10.1038/ismej.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boles BR, Thoendel M, Singh PK. Self-generated diversity produces “insurance effects” in biofilm communities. Proc. Natl Acad. Sci. USA. 2004;101:16630–16635. doi: 10.1073/pnas.0407460101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morgan R, Kohn S, Hwang SH, Hassett DJ, Sauer K. BdlA, a chemotaxis regulator essential for biofilm dispersion in Pseudomonas aeruginosa. J. Bacteriol. 2006;188:7335–7343. doi: 10.1128/JB.00599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shanks RM, Stella NA, Kalivoda EJ, Doe MR, O'Dee DM, Lathrop KL, Guo FL, Nau GJ. A Serratia marcescens OxyR homolog mediates surface attachment and biofilm formation. J. Bacteriol. 2007;189:7262–7272. doi: 10.1128/JB.00859-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hassett DJ, Alsabbagh E, Parvatiyar K, Howell ML, Wilmott RW, Ochsner UA. A protease-resistant catalase, KatA, released upon cell lysis during stationary phase is essential for aerobic survival of a Pseudomonas aeruginosa oxyR mutant at low cell densities. J. Bacteriol. 2000;182:4557–4563. doi: 10.1128/jb.182.16.4557-4563.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swaminathan S, Masek T, Molin C, Pospisek M, Sunnerhagen P. Rck2 is required for reprogramming of ribosomes during oxidative stress. Mol. Biol. Cell. 2006;17:1472–1482. doi: 10.1091/mbc.E05-07-0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shenton D, Smirnova JB, Selley JN, Carroll K, Hubbard SJ, Pavitt GD, Ashe MP, Grant CM. Global translational responses to oxidative stress impact upon multiple levels of protein synthesis. J. Biol. Chem. 2006;281:29011–29021. doi: 10.1074/jbc.M601545200. [DOI] [PubMed] [Google Scholar]
- 44.Hyland SA, Eveland SS, Anderson MS. Cloning, expression, and purification of UDP-3-O-acyl-GlcNAc deacetylase from Pseudomonas aeruginosa: a metalloamidase of the lipid A biosynthesis pathway. J. Bacteriol. 1997;179:2029–2037. doi: 10.1128/jb.179.6.2029-2037.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cornelis P, Wei Q, Andrews SC, Vinckx T. Iron homeostasis and management of oxidative stress response in bacteria. Metallomics. 2011;3:540–549. doi: 10.1039/c1mt00022e. [DOI] [PubMed] [Google Scholar]
- 46.Hausladen A, Privalle CT, Keng T, DeAngelo J, Stamler JS. Nitrosative stress: activation of the transcription factor OxyR. Cell. 1996;86:719–729. doi: 10.1016/s0092-8674(00)80147-6. [DOI] [PubMed] [Google Scholar]
- 47.Ritz D, Patel H, Doan B, Zheng M, Aslund F, Storz G, Beckwith J. Thioredoxin 2 is involved in the oxidative stress response in Escherichia coli. J. Biol. Chem. 2000;275:2505–2512. doi: 10.1074/jbc.275.4.2505. [DOI] [PubMed] [Google Scholar]
- 48.Diggle SP, Matthijs S, Wright VJ, Fletcher MP, Chhabra SR, Lamont IL, Kong X, Hider RC, Cornelis P, Camara M, et al. The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chem. Biol. 2007;14:87–96. doi: 10.1016/j.chembiol.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 49.Hassett DJ, Ma JF, Elkins JG, McDermott TR, Ochsner UA, West SE, Huang CT, Fredericks J, Burnett S, Stewart PS, et al. Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Mol. Microbiol. 1999;34:1082–1093. doi: 10.1046/j.1365-2958.1999.01672.x. [DOI] [PubMed] [Google Scholar]
- 50.Boles BR, Singh PK. Endogenous oxidative stress produces diversity and adaptability in biofilm communities. Proc. Natl Acad. Sci. USA. 2008;105:12503–12508. doi: 10.1073/pnas.0801499105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cornelis P, Matthijs S, Van Oeffelen L. Iron uptake regulation in Pseudomonas aeruginosa. Biometals. 2009;22:15–22. doi: 10.1007/s10534-008-9193-0. [DOI] [PubMed] [Google Scholar]
- 52.Cornelis P, Wei Q, Andrews SC, Vinckx T. Iron homeostasis and management of oxidative stress response in bacteria. Metallomics. 2011;3:540–549. doi: 10.1039/c1mt00022e. [DOI] [PubMed] [Google Scholar]
- 53.Xiao G, Deziel E, He J, Lepine F, Lesic B, Castonguay MH, Milot S, Tampakaki AP, Stachel SE, Rahme LG. MvfR, a key Pseudomonas aeruginosa pathogenicity LTTR-class regulatory protein, has dual ligands. Mol. Microbiol. 2006;62:1689–1699. doi: 10.1111/j.1365-2958.2006.05462.x. [DOI] [PubMed] [Google Scholar]
- 54.Lumjiaktase P, Diggle SP, Loprasert S, Tungpradabkul S, Daykin M, Camara M, Williams P, Kunakorn M. Quorum sensing regulates dpsA and the oxidative stress response in Burkholderia pseudomallei. Microbiology. 2006;152:3651–3659. doi: 10.1099/mic.0.29226-0. [DOI] [PubMed] [Google Scholar]
- 55.Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G. A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell. 1997;90:43–53. doi: 10.1016/s0092-8674(00)80312-8. [DOI] [PubMed] [Google Scholar]
- 56.Sonnleitner E, Gonzalez N, Sorger-Domenigg T, Heeb S, Richter AS, Backofen R, Williams P, Huttenhofer A, Haas D, Blasi U. The small RNA PhrS stimulates synthesis of the Pseudomonas aeruginosa quinolone signal. Mol. Microbiol. 2011;80:868–885. doi: 10.1111/j.1365-2958.2011.07620.x. [DOI] [PubMed] [Google Scholar]
- 57.Aslund F, Zheng M, Beckwith J, Storz G. Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thiol-disulfide status. Proc. Natl Acad. Sci. USA. 1999;96:6161–6165. doi: 10.1073/pnas.96.11.6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.