Abstract
Toxin-antitoxin (TA) loci encode inhibitors of translation, replication or cell wall synthesis and are common elements of prokaryotic plasmids and chromosomes. Ten TA loci of Escherichia coli K-12 encode mRNases that cumulatively contribute to persistence (multidrug tolerance) of the bacterial cells. The mechanisms underlying induction and reversion of the persistent state are not yet understood. The vapBC operon of Salmonalla enterica serovar Typhimurium LT2 encodes VapC, a tRNase that reversibly inhibits translation by site-specific cleavage of tRNAfMet. VapB is an antitoxin that interacts with and neutralizes VapC via its C-terminal tail and regulate TA operon transcription via its N-terminal DNA binding domain that recognize operators in the vapBC promoter region. We show here that transcription of the vapBC operon of S. enterica is controlled by a recently discovered regulatory theme referred to as ‘conditional cooperativity’: at low T/A ratios, the TA complex binds cooperatively to the promoter region and represses TA operon transcription whereas at high T/A ratios, the excess toxin leads to destabilization of the TA-operator complex and therefore, induction of transcription. We present evidence that an excess of VapC toxin leads to operator complex destabilization by breaking of toxin dimers.
INTRODUCTION
Prokaryotic chromosomes and plasmids encode a plethora of toxin–antitoxin (TA) loci that belong to three different types. In type I TA loci, the antitoxin is a cis-encoded antisense RNA that inhibits translation of a toxin-encoding mRNA (1,2). In type II loci, the antitoxin is a protein that combines with and neutralizes the cognate toxin (3). Finally, in type III loci, the antitoxin is an RNA that combines with and neutralizes the toxin (4). Type II TA loci are common and, based on toxin sequence similarities, have been divided into evolutionary independent gene families (5,6). Three of these families, vapBC, relBE and hicAB are common in both bacteria and archaea (5,7,8). Of these three families, vapBC loci are particularly abundant: of approximately 3825 TA loci identified in 900 prokaryotic genomes, approximately 1300 were vapBC loci (9). In some organisms, the numbers of TA loci are particularly high. Remarkably, Mycobacterium tuberculosis has at least 88 type II TA loci, 45 of which are vapBC homologues (10). The reason for the large expansions of TA loci in particular organisms is not known.
TA loci have been linked to several biological functions, such as plasmid and gene stabilization (11,12) and environmental stresses, such as amino acid starvation (13). It has also been suggested that TA loci have no biological function (14). However, recently we showed that, in the model organism Escherichia coli, TA loci were required for bacterial persistence (15). Thus, progressive deletion of type II TA loci gradually reduced the persistence level and deletion of 10 TA loci encoding RNA endonucleases reduced persistence approximately 200-fold.
Even though the different families of type II TA loci have independent evolutionary origins their regulation and genetic organization are remarkably similar. In almost all cases, a TA locus consists of two closely linked genes, one of which encodes the antitoxin and the other the toxin. Although rare exceptions exists (16,17), transcription of almost all type II TA operons is autoregulated by the TA complex that binds to one or more operators in the TA promoter region. The TA complex binds to DNA via a domains present in the antitoxin. Usually, the toxin enhances the binding of the antitoxin to the operator (3). The DNA domains belong to four different classes: Helix–Turn–Helix (HTH), Ribbon–Helix–Helix (RHH), AbrB and Phd/YefM (3). In the vapBC and relBE gene families, the antitoxins can have any of these four DNA binding motifs, indicating that domain shuffling took place during the evolution of these gene families (6).
In actively growing cells, the TA operators are occupied by cognate TA complexes and transcription of the operons is repressed (13,18–22). The antitoxins are metabolically unstable due to degradation by cellular proteases. For example, Lon degrades RelB antitoxin of E. coli (13). In growing cells, de novo synthesis of RelB replenishes the RelB pool and relBE translation stays repressed. In contrast, when translation is reduced by, e.g. amino acid starvation, the RelB level is reduced. Consequently, the TA promoter is activated (13). In turn, the increased transcription-rate of relBE leads to partial replenishment of the antitoxin pool after an initial delay (13,23). Even though the antitoxin level is restored to 50% of the initial pre-starvation level, the transcription-rate of the TA operon stays unexpectedly high during amino acid starvation (13). More in-depth analysis revealed that transcription of relBE is regulated by the RelB/RelE ratio such that when [RelB] > [RelE], two RelB2•RelE complexes bind strongly and cooperatively to two operators (relO) overlapping the relBE promoter sequences and repress transcription (23). In contrast, when [RelB] < [RelE], the excess RelE invades the RelB2•RelE•relO complex and abolishes cooperative binding, probably via the formation of a RelB2•RelE2 tetramer that does not bind cooperatively to relO. By this mechanism that we called ‘conditional cooperativity’, the RelB/RelE ratio controls the transcription-rate of relBE (23). In turn, the RelB/RelE ratio is controlled by the interplay of the relBE translation-rate and the degradation-rate of RelB that is determined by Lon. Transcription of the phd doc and ccd operons of plasmid P1 and F is also regulated by conditional cooperativity (24,36). That evolutionarily unrelated TA loci are regulated by conditional cooperativity by different molecular mechanisms indicates that this peculiar mode of transcriptional regulation is a biologically important property of TA loci.
Here we investigate if conditional cooperativity controls transcription of an enteric vapBC locus. VapC toxins from enterobacteria are tRNases that inhibit global translation by site-specific cleavage of tRNAfMet between the anticodon stem and loop (25). As with other TA loci, VapBC complexes bind to operators in the promoter regions and autoregulate transcription (26–28). Structural analysis of VapBC from Neisseria gonorrhoeae (also called FitAB) showed that an octamer of four VapB–VapC heterodimers [(VapBC)2]2 binds to operator DNA via an RHH motif in VapB (29), consistent with autoregulation of vapBC transcription by VapBC.
We show that VapB of Salmonella enterica serovar Typhimurium LT2 that contains an AbrB-like DNA-binding motif is degraded by the ATP-dependent Lon protease. When translation is inhibited, VapB decays and vapBC promoter activity increases, consistent with transcriptional autoregulation by VapB. Indeed, VapB binds to two operator sequences (vapO1 and vapO2) in the promoter region in vitro and this binding is strongly enhanced by VapC. Remarkably, an excess of VapC decreases the affinity of the VapBC complex for DNA and, consistently, increases transcription-rate in vivo. Structural modelling and mutational analysis allow us to propose a novel mechanism of conditional cooperativity in which excess VapC toxin induces vapBC transcription by breaking of VapC dimers.
MATERIALS AND METHODS
Media, antibiotics, strains and plasmids
Bacteria were grown in Luria–Bertani medium (LB) as described (30). When required, the medium was supplemented with 30 or 100 µg/ml ampicillin, 50 µg/ml chloramphenicol, 25 µg/ml kanamycin. X-gal (5-bromo-4-chloro-3-indoyl-b-d-galactoside) was added to a final concentration of 40 µg/ml. Expression of proteins from the PA1/O4/O3 or PBAD promoters was induced by 2 mM isopropylb-d-thiogalactopyranoside (IPTG) or 0.2% arabinose, respectively. Strains and plasmids are listed in Supplementary Table S1 and oligonucleotides in Supplementary Table S2, respectively.
Purification of VapB and VapC proteins
VapC and VapB were purified essentially according to (25). Strain C41 containing pKW512HB, pKW512HBL43A, pKW512HBI44A, pKW512HBY72A, pKW512HBA76S or pKW512HC was grown exponentially under aeration in LB at 37°C. At OD450 = 0.5, expression was induced by addition of 2 mM IPTG. After 3 h of growth, the culture was harvested and resuspended in ice-cold lysis buffer (50 mM NaH2PO4, 0.3 M NaCl, 10 mM imidazole, 5 mM β-mercaptoethanol pH8 supplemented with EDTA-free protease inhibitor, Roche). Cells were disrupted using a Constant Cell disruption system and lysate cleared by centrifugation at 15 000 rpm for 30 min at 4°C. The cleared lysate was then incubated with Ni-NTA agarose (Qiagen) for at least 2 h at 4°C and subsequently loaded onto a gravity column. The column was washed extensively in wash buffer (50 mM NaH2PO4, 0.3 M NaCl, 35 mM imidazole, 5 mM β-mercaptoethanol pH 8). VapC or VapB was then eluted from complex under denaturing conditions by incubating the column overnight (ON) at room-temperature in 10 column volumes of denaturing buffer (100 mM NaH2PO4, 10 mM Tris–HCl, 9.8 M Urea, pH8). Denatured protein was refolded by four-step dialysis; (i) 1 × PBS 0.1% Triton X-100 5 mM DTT, (ii) 1 × PBS 5 mM DTT, (iii) 1 × PBS 5 mM DTT and (iv) 1× PBS 20% glycerol 1 mM DTT. The authenticity and purity was verified by SDS–PAGE.
Electrophoretic mobility shift assays and DNase I footprinting
DNA fragments were constructed containing either one binding site vapO1 (using hybridized oligos VapBCbinding#small-down and VapBCbinding#small-up) or two binding sites, vapO1 and vapO2 (PCR product with oligos vapBC_EMSA_down and vapBC_EMSA_up). Prior to the hybridization and PCR reaction VapBCbinding#small-down or vapBC_EMSA_down were 5′-end-labelled with [γ-32P]ATP using T4 polynucleotide Kinase (New England Biolabs). Binding site mutations in vapO1 and vapO2 were introduced by PCR using primers PBC-10_MUT_DOWN and PBC-10_MUT_UP for site 1 and PBC-35_MUT_DOWN and PBC-35_MUT_UP for site 2. Labelled probes (0.5–2 nM) were incubated with purified proteins in binding buffer (20 mM Tris–HCl pH 7.5, 100 mM KCl, 2 mM MgCl2, 1 mM DTT, 50 µg/ml BSA and 10% glycerol) to avoid unspecific DNA binding sonicated salmon sperm DNA (ssDNA) was added to a final concentration 0.1 mg/ml. Reactions were incubated for 20 min at 37°C before DNA bound complexes were separated by native PAGE in 5% or 6% acrylamide gels with 0.5× TBE for two and one binding site probes, respectively. The separation was followed by phosphorimaging.
For DNase I footprinting, samples were incubated with 0.01 U/µl DNase I and 1 × DNase I buffer (Roche) at 37°C for 2 min, followed by addition of 100 µl of Stop buffer (2 M ammonium acetate, 20 mM EDTA, 10 mg/ml ssDNA). The resulting DNA fragments were extracted once in phenol, once in chloroform, precipitated in ethanol and separated on an 8% denaturing acrylamide gel along with a dideoxyNTP sequencing ladder. The digestion pattern was analysed by to phosphorimaging.
RESULTS
VapB autoregulates vapBC transcription and is degraded by Lon protease
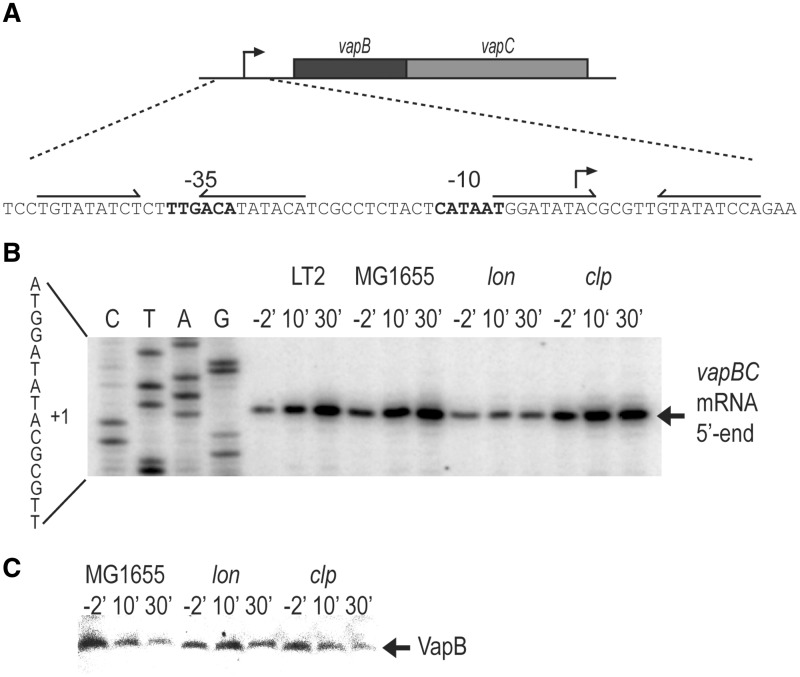
VapB of S. enterica LT2 (STM3034) contains an AbrB DNA binding domain in its N-terminus (31), raising the possibility that VapB autoregulates vapBC transcription via binding to two inverted repeats in the promoter region that we call vapO1 and vapO2 (Figure 1A). The entire operator was called vapO. Previously, we showed that amino acid starvation induces vapBC transcription (32). We examined vapBC transcription using primer extension analysis of RNA prepared from strains S. enterica LT2 (KP1001) or E. coli K-12 (MG1655, wt) carrying vapBC on a low-copy-number plasmid before and after the inhibition of translation by the addition of chloramphenicol. As seen from Figure 1B, the level of transcripts increased in both strains. Transcription also increased in strain MG1655ΔclpP but not in the isogenic Δlon strain, suggesting that Lon protease degrades VapB. To test this inference directly, we measured VapB levels. As seen from Figure 1C, VapB decayed rapidly in wt and Δclp but not in the Δlon strain. Thus, Lon degrades VapB. The increased mRNA level seen after inhibition of translation with chloramphenicol suggested that VapB autoregulates vapBC transcription, possibly via binding to vapO1 and vapO2 that overlap with the −10 and −35 promoter sequences, respectively (Figure 1A).
Figure 1.
Lon degrades VapB and is required for activation of vapBC transcription. (A) DNA sequence of the vapBC promoter region showing −10 and −35 promoter sequences and the two operators vapO1 and vapO2 as inverted repeats. (B) Primer extension analysis of the 5′-end of vapBC mRNA. Strains MG1655 (E. coli K-12, wt), KW10 (MG1655Δlon) or KW11 (MG1655ΔclpP) containing pKW71512 (pNDM71::vapBC) and KP1001 (S. enterica LT2) were grown exponentially in LB medium. At time zero, chloramphenicol (50 µg/ml) was added and cell samples were withdrawn at the time points indicated (min). Total RNA was extracted and reverse transcription was performed using primer vapB-5#PE. (C) Western blotting analysis of VapB. MG1655 KW10 or KW11 containing pKW51 (pA1/03/04::SDopt::vapB) were grown exponentially in LB medium. Ten minutes before the addition of chloramphenicol (50 µg/ml; t′ = 0), 1 mM IPTG was added to induce vapB. Samples were taken at the time points indicated and VapB detected by polyclonal antibodies directed towards VapB.
VapC enhances binding of VapB to vapO1 and vapO2
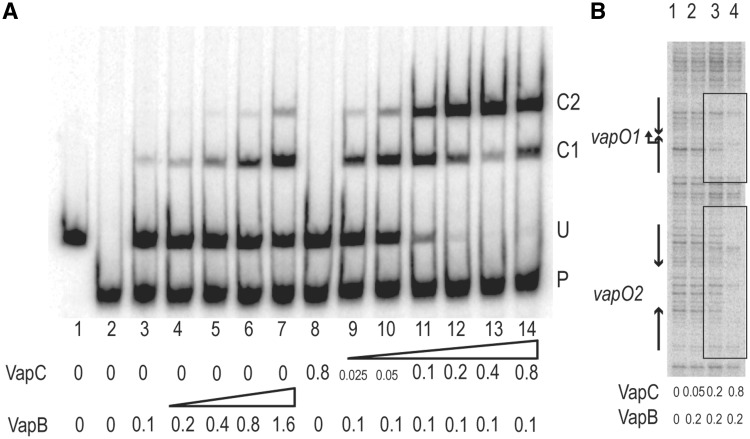
Native VapB and VapC were purified and used in gel shift analysis to probe their binding to vapO. VapB alone formed a complex with vapO-encoding DNA (C1 and C2 in Figure 2A); however only at very high concentrations more significant DNA–VapB complexes were observed (lanes 3–7). In contrast, VapB did not bind to a control DNA fragment (P in Figure 2A). Thus, VapB has low but specific affinity for vapO. In contrast, VapC alone bound neither to the vapO fragment (U) nor to the control fragment (lane 8). However, addition of VapC to a low concentration of VapB yielded an increase in complex formation that further increased dramatically with increasing concentration of VapC (lanes 9–13). Thus, VapC enhances the binding of VapB to vapO. Interestingly, at very high concentrations, VapB alone produced a complex with a mobility similar to that generated by VapBC (Figure 2A, comp. lanes 3–7 with lanes 9–14). This could be due to the presence of a small amount of VapC in the VapB preparation or because the complexes were complexes are not resolved in this experimental setup.
Figure 2.
VapC increases the affinity of VapB for vapO-encoding DNA. (A) VapB and VapBC complex binding to vapO analysed by gel shifting. Purified VapB and VapC were added to a 302-bp 32P-labelled vapO DNA probe (labelled U in the gel) and a 199-bp mock DNA fragment derived from pUC19 (labelled P). Concentrations of VapB and VapC are given below each lane (μM). C1 and C2 denote the inferred VapBC•vapO complexeses. (B) VapBC binding to a vapO-encoding DNA fragment analysed by DNase I protection. Purified VapB and VapC were added to the same DNA probe as in (A) and subsequently incubated with DNase I (lanes 1–4). The protected regions are enclosed by boxes and the positions of the transcriptional start site and the two inverted repeats are marked with arrows.
Next, we performed foot printing analysis of the complex bound to vapO. DNA was incubated with a constant concentration of VapB and increasing concentrations of VapC and digested with DNase I (Figure 2B, lanes 1–4). Two protected regions overlapping with the two inverted repeats appeared, consistent with VapBC binding to vapO1 and vapO2. This observation raised the possibility that complexes C1 and C2 observed in the gel shift analysis (Figure 2A) corresponded to one and two VapBC complexes bound to vapO, respectively. Most importantly, increasing the concentration of VapC increased VapB binding to operator DNA.
An excess of VapC releases VapBC from operator DNA
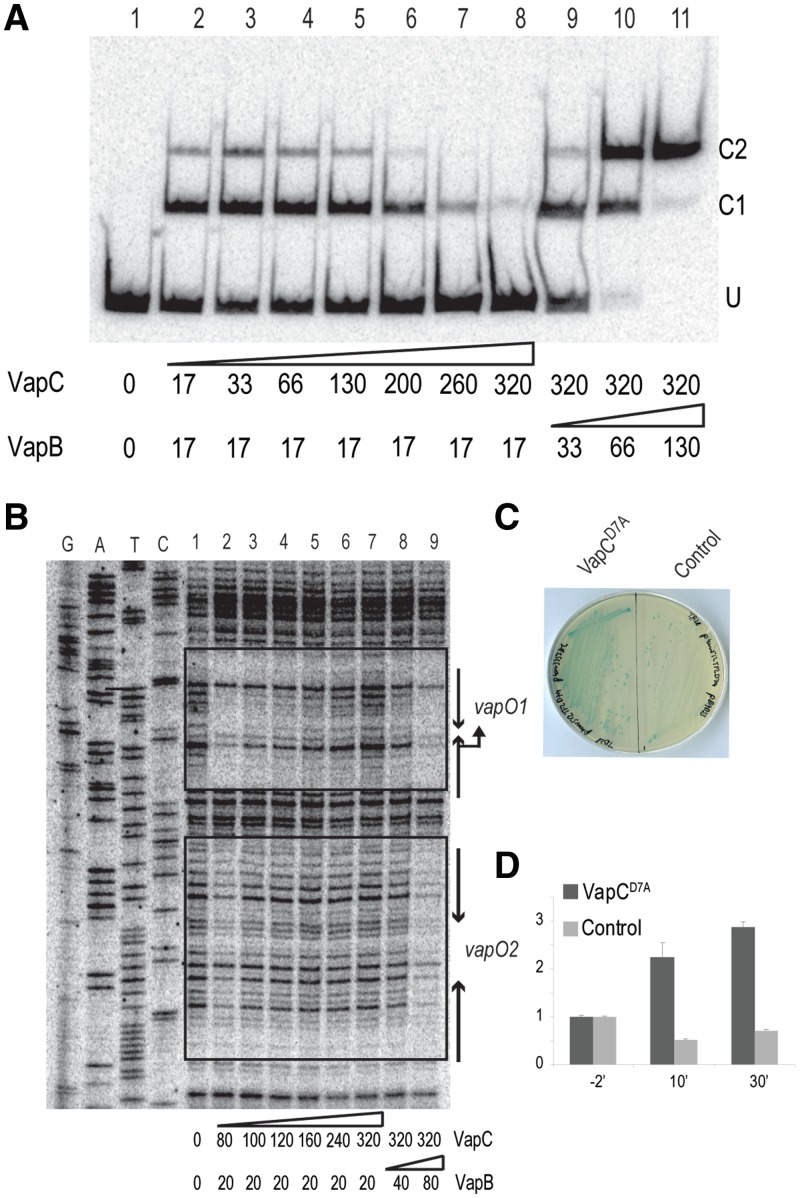
At a VapB/VapC ratio of ∼2/16 in the gel shift analysis shown in Figure 2A, the amount of C2 complex decreased modestly in intensity while that of C1 increased (lane 14), thus raising the possibility that very high concentrations of VapC destabilized the VapBCO complex. To investigate this phenomenon further, we performed gel shift and foot printing analysis with higher concentrations of VapC. As before, increasing [VapC] increased VapB specific binding to vapO (Figure 3A, lanes 2 and 3). However, increasing [VapC] further severely reduced binding (lanes 4–8) with almost no VapBC bound at a B/C ratio of 1/16 (lane 8). To investigate if the destabilization of VapBC binding to vapO was reversible, we then increased the VapB/VapC ratio by increasing [VapB] while keeping [VapC] constant (lanes 9–11). We observed a resulting increase in binding to vapO, consistent with a reversible and direct effect of VapC.
Figure 3.
The vapBC promoter is controlled by conditional cooperativity. (A) Binding of VapB and VapBC complex to a vapO-encoding DNA fragment analysed by gel shifting. Purified VapB and VapC were added to a 302-bp 32P-labelled vapO probe (lanes 1–11; numbers below the gel are in nM). Protein–DNA complexes were separated by 5% native PAGE. U denotes unbound vapO DNA and C1 and C2 VapBCO complexes. (B) DNase I protection assay of vapO. VapB and VapC were incubated with vapO DNA as in (A) and subsequently incubated with DNase I (lanes 1–9; numbers are pmol). A DNA sequencing ladder was generated using 5′-end labelled vapBC_EMSA_down primer. Inverted repeats sites 1 and 2 and promoter sequences are indicated by arrows. DNAse I protected bases are enclosed by boxes. (C) Ectopic expression of VapCD7A in vivo induces vapBC transcription. TB28 (MG1655ΔlacIZYA) pKW512TFZD7A (vapBCD7A::lacZYA) containing either pKW3353HC (pBAD::SDopt::vapCD7A) or pBAD33 were streaked to single colonies on LB plates containing X-gal and 0.2% arabinose. (D) VapCD7A induced transcription quantified by qPCR. TB28 (MG1655 ΔlacIZYA) pKW512TFZD7A (vapBCD7A::lacZYA) with pKW3353HC (pBAD::SDopt::vapCD7A-H6) or pBAD33 (empty vector plasmid) were grown exponentially in LB medium. At 0′, arabinose was added to induce transcription from the pBAD promoter. Samples were taken at time points indicated (min) and total RNA extracted. Fold-of-changes relative to house keeping gene rpsA mRNA were measured by quantitative RT-PCR.
DNase I foot printing yielded more detailed information. First, at high [VapC], vapO1 and vapO2 were both protected (Figure 3B, lane 2); however, vapO1 was clearly protected better than vapO2 and an increase of [VapC] released VapBC binding at vapO2 before vapO1 (lanes 2–7). When the VapB/VapC ratio was increased, protection was regained (lanes 8 and 9). Again, vapO1 was protected at a lower concentration of VapB (lane 8) than vapO2 (lane 9), consistent with higher affinity of the VapBC complex for vapO1.
An excess of VapC stimulates vapBC transcription
The above-described results showed that excess VapC destabilized the VapBCO complex and predicted that an excess of VapC in living cells would derepress the vapBC promoter. To test this, we fused vapBC transcriptionally to the lacZ gene. Induction in trans of vapCD7A, encoding a non-toxic VapCD7A variant, increased LacZ expression from the vapBC::lacZ transcriptional fusion (Figure 3C). Induction of vapBC by excess VapCD7A was confirmed by quantitative RT-PCR measurements (Figure 3D). After 30′ of induction of vapCD7A, the transcription rate of vapBC::lacZ had increased ∼3-fold. No increase was observed with the control plasmid. This result was consistent with the VapC-mediated destabilization of the VapBCO complex seen in vitro. Thus, vapBC operon transcription is regulated by conditional cooperativity in vivo.
One operator site is sufficient for regulation by conditional cooperativity
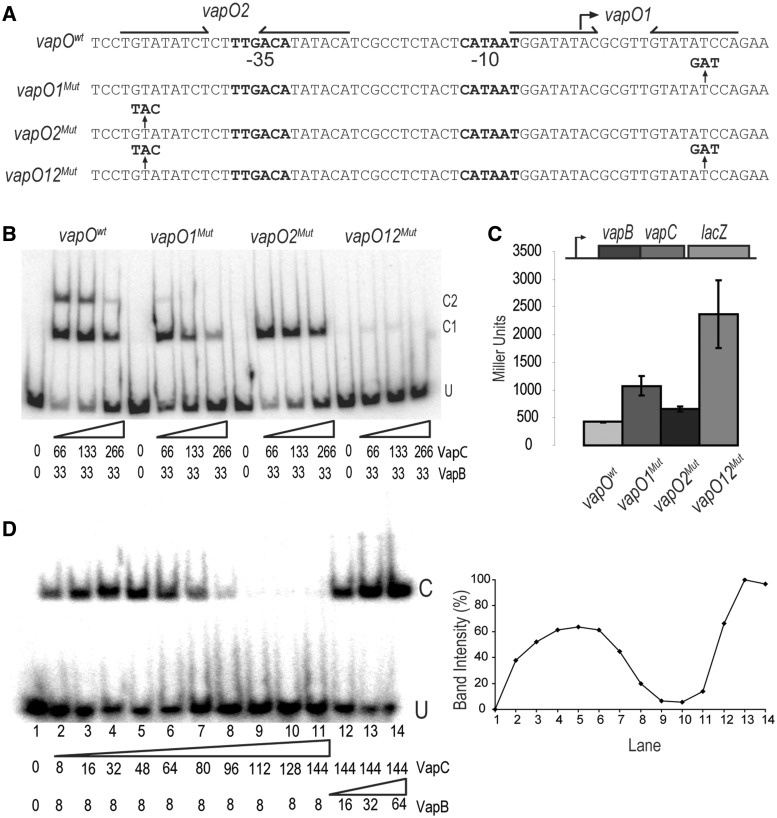
The definition of conditional cooperativity in TA locus regulation entails cooperative binding of the TA complex to the promoter when the antitoxin is in excess and release of the complex when the toxin is in excess (23,24). To investigate the function of vapO1 and vapO2 in the cooperative binding of VapBC to vapO, we introduced mutations in the half-sites of vapO1 and vapO2, respectively (Figure 4A). To avoid interference with promoter activity, the mutations were located away from the −10 and −35 promoter sequences (see below). First, we analysed vapO1Mut and vapO2Mut in gel shift assays (Figure 4B). As before, a vapOwt fragment generated two complexes (C1 and C2) that decreased in intensity with increasing concentration of VapC. The vapO1Mut and vapO2Mut fragments exhibited changed patterns: the C2 complex disappeared (vapO2Mut) or was highly reduced (vapO1Mut). This pattern supported that the C1 and C2 complexes corresponded to vapO bound with one and two VapBC complexes, respectively. Most importantly, however, complex formation was in both cases again reduced when VapC was increased. This result suggested that a single vapO operator was sufficient for transcriptional regulation by conditional cooperativity. The fact that C1 complexes formed by vapO1Mut and vapO2Mut migrate so similarly suggests that the VapBC protein complexes formed on either vapO1 or vapO2 are identical. A fragment carrying the double vapO12Mut mutation hardly bound VapBC at all, although a faint C1 complex was observed that most likely reflected incomplete disruption of VapBC bound to the vapO1Mut. This inference is also consistent with the weak C2 complex seen with the vapO1Mut fragment. Nevertheless, the vapO1 mutation decreased the amount of complex formation more than the vapO2 mutation, consistent with the observation that the VapBC complex has the highest affinity for vapO1.
Figure 4.
One vapBC operator is sufficient for regulation by conditional cooperativity. (A) DNA sequences of the vapBC promoter region showing the base substitutions in vapO1Mut, vapO2Mut and vapO12Mut. Inverted repeats are indicated by arrows and promoter sequences by −10 and −35, respectively. (B) Gel shift assay of VapBC complex binding to the DNA fragments shown in (A). VapB and VapC were incubated with DNA (numbers are in nM) . U indicates unbound DNA fragment, C1 and C2 are fragments bound by either one or two VapBC complexes, respectively. (C) vapBC promoter activity in binding site mutants. TB28 (MG1655ΔlacIZYA) containing either Pwt::vapBCD7A::lacZYA (pKW512TFZD7A), P::vapO1Mut::vapBCD7A::lacZYA (pKW512TFZD7A-1), P::vapO2Mut::vapBCD7A::lacZYA (pKW512TFZD7A-2) or P::vapO1Mut::vapO2Mut::vapBCD7A::lacZYA (pKW512TFZD7A-1-2) were grown exponentially in LB medium at 37°C. At an OD600 of approximately 0.5, samples were collected and LacZ activity measured (Miller Units). (D) Gel shift analysis as in (B) but with a promoter DNA fragment (36 bp) containing only vapO1. Numbers below the gel are protein concentrations in nM (lane 1–14). Protein–DNA complexes were separated by 6% native PAGE. U and C indicate positions of unbound and bound DNA, respectively. Insert at right: Quantification of the C band-intensities (%) in the gel-shift shown at left.
To investigate the effect of the vapO mutations (Figure 4A) on transcriptional repression in vivo, we again used transcriptional vapBC-lacZ fusions (Figure 4C). The double vapO1Mut and vapO2Mut mutant (vapO12Mut) caused a strong derepression of vapBC transcription, consistent with the lack of VapBC binding to the operator sites seen in vitro. The single vapO mutations also derepressed vapBC transcription, but to a much lesser extent. The vapO1Mut yielded stronger derepression than vapO2Mut, consistent with higher affinity of VapBC for vapO1 (Figure 4B). Importantly, these observations showed that vapO1 and vapO2 can function independently to regulate vapBC transcription.
The above-described results raised the possibility that a fragment containing one VapBC binding site would respond with conditional cooperativity at increased VapC/VapB ratios. To test this, we performed gel shift assays using a DNA fragment containing only vapO1 (Figure 4D). As before, increasing [VapC] increased VapB binding (lanes 2–5). As before, at increased VapC/VapB ratios (2/16, lane 6), binding was reduced and finally totally abolished (lanes 9–11). In these reactions, binding could also be regained by increasing the concentration of VapB (lanes 12–14). Quantification showed the dramatic changes in VapBC binding as a function of the VapC/VapB ratio (Figure 4D).
Mutations in the VapC dimer interface derepress vapBC transcription
The crystal structure of a VapBC homologous complex of N. gonorrhoea (FitAB, fast intracellular trafficking) revealed an octamer of four VapBC heterodimers [(VapBC)2]2 bound to operator DNA (Supplementary Figure S1D) (29). In this structure, VapC form bridges between two VapB dimers bound to operator half-sites. The structure readily explains why VapC mediates strong cooperative binding of VapB to operator DNA (29). More recently, the crystal structure of the VapBC complex from Shigella flexneri 2a YSH6000 virulence plasmid pMYSH6000 (VapBCS.flex) was solved, revealing a similar but more compact structure (Supplementary Figure S1C) (33). This observation implies that VapBC complexes in general form octameric complexes. For the S. enterica proteins, the theoretical molecular weight of a [(VapBC)2]2 octamer is 93.2 kD. Our estimation of the size of the VapBC complex bound to vapO1 was 92 ± 10 kD (Supplementary Figure S2A–S2D). We infer that VapBC of S. enterica also binds as an octameric [(VapBC)2]2 complex to operator DNA. A similar complex is very likely also formed at vapO2.
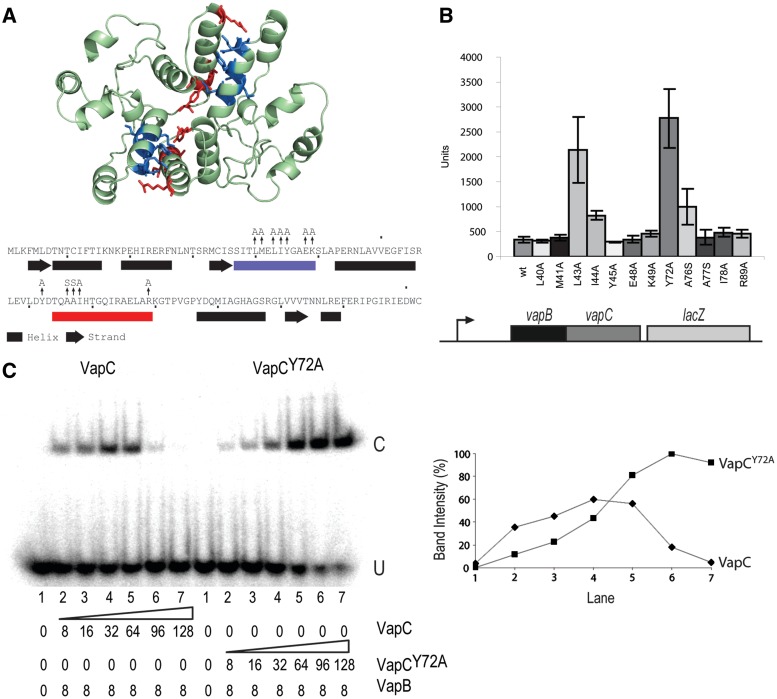
The FitAB crystal structure (Supplementary Figure S1C and S1D) predicts that VapC dimerization is essential for cooperative binding of the [(VapBC)2]2 complex to vapO. Therefore, the VapC mediated destabilization of the [(VapBC)2]2•vapO1 complex can be explained by breaking of the VapC dimer by an excess of VapC (see Discussion). We modelled the tertiary structure of VapC using Phyre (34) and aligned the predicted structure with VapCS.flex and FitB (Supplementary Figure S1B). The primary sequences of VapC and FitB exhibit low similarity (22% identical, 41% similar, Supplementary Figure S1A). However, a VapC dimer aligned well with the FitB dimer in the tetrameric [(FitAB)2]2•DNA complex and with the VapCS.flex dimer in S. flexneri [(VapBC)2]2 complex (Supplementary Figure S1C and S1D). The modelled structure of the VapC dimer is shown in Figure 5A. This model predicted two patches in VapC to be involved in dimerization. The effect of changing these residues to alanine (or to serine in one case) on vapBC transcription was measured using a vapBC::lacZ transcriptional fusion (Figure 5B). Changing four residues L43A, I44A, Y72A and A76S individually significantly increased vapBC transcription (5.2-, 1.4-, 7.0- and 1.9-fold, respectively). The activity of the vapBCY72A::lacZ fusion was similar to that of a vapBC::lacZ fusion carrying mutations in both operator sites (comp. Figures 4B and 5C), indicating that the Y72A substitution in VapC resulted in complete loss of repression by the VapBC complex. The tertiary structure alignments showed that tyrosine 72 of VapC and VapCS.flex corresponds to phenylalanine 78 in FitB, a key residue for FitB dimerization (Supplementary Figure S1B) (29). These results support that VapC dimerization is important for repression of vapBC transcription by the VapBC complex.
Figure 5.
VapC dimerization is required for conditional cooperativity. (A) Predicted tertiary structure of a VapC dimer showing positions of amino acid changes in the dimer interface. The predicted secondary structure of VapC is shown below the primary sequence. Amino acid patches involved in dimerization are shown in blue and red, and amino acid substitutions in these patches are indicated by vertical arrows. (B) LacZ activities of vapBC::lacZ transcriptional fusions carrying mutations in vapC. The genetic set-up of the transcriptional vapBC::lacZ fusion used is shown schematically below the diagram. A broken arrow pointing rightward indicates the vapBC promoter. TB28 (MG1655ΔlacZIYA) containing pKW254BC (vapBC::lacZYA) or its isogenic vapC substitution mutant derivatives (see Supplementary Table S1) were grown exponentially in LB medium at 37°C and specific LacZ activities were determined. (C) Gel shifts of vapO1 DNA with VapB and VapC (left panel) or VapCY72A (right panel). The proteins were mixed with radio labelled vapO1 DNA in given concentrations (nM) (lanes 1–7). U and C indicate positions of unbound and bond complexes, respectively. Right: Quantification of C band intensities (%) seen in (C) as a function of lane number in the gel-shift shown at left.
To challenge this inference directly, we asked if the Y72A mutation would affect VapC dimerization. As seen from Supplementary Figure S3, indeed the Y72A mutation abolished dimerization of purified VapC. Thus, we conclude that dimerization of VapC is required for repression of the vapBC promoter by the VapBC complex.
Mutations in the VapC dimer interface abolish conditional cooperativity
The VapCY72A variant that was defective in dimerization yielded a possibility to test directly, in vitro, if indeed VapC dimerization is required for efficient binding of VapBC to vapO DNA. More importantly, however, we could also test if conditional cooperativity depends on VapC dimerization. We used purified VapB, VapC and VapCY72A (Figure 5C). As expected, low concentrations of wild-type VapC increased binding of VapB to vapO1 and higher concentrations destabilized the complex. A dramatically different pattern was seen with VapCY72A. First, a much higher concentration of VapCY72A was required to yield efficient binding, consistent with the prediction that VapC dimers bridges two VapB dimers bound to operator DNA. However, strikingly, high concentrations of VapCY72A did not destabilize the VapBCY72A complex. On the contrary, increasing [VapCY72A] led to a strong increase in binding. VapC variants carrying other changes in the dimerization patches (VapCY72A, VapCL43A, VapCI44A and VapCA76S) behaved similarly although less dramatic (Supplementary Figure S4). The reduced capability of VapCL43A and VapCA76S to increase binding of VapB to vapO1 (Supplementary Figure S4) is consistent with the increased transcription rates of vapBC loci carrying the corresponding alleles (Figure 5B). These observations show that not only is dimerization of VapC crucial for cooperative binding of VapB2 to vapO but also for excess VapC to destabilize VapBCO.
DISCUSSION
We show here that vapBC of S. enterica is under complex transcriptional regulation by VapB and VapC: in the presence of an excess of VapB, VapC induced avid and cooperative binding of the VapBC complex to vapO operator DNA whereas an excess of VapC destabilized VapBCO (Figures 2–4). Band shifting with a vapO DNA fragment encoding both vapO1 and vapO2 yielded two different nucleoprotein complexes (Figure 2A) and mutations in vapO1 or vapO2 abolished VapBC binding at the mutated vapO operator but not at the other, intact operator (Figure 4B). Consistently, foot printing analysis revealed that the VapBC protected regions corresponding to vapO1 and vapO2 (Figures 2B and 3B). We conclude that the VapBC complex binds cooperatively to vapO1 and vapO2 in the presence of excess VapB.
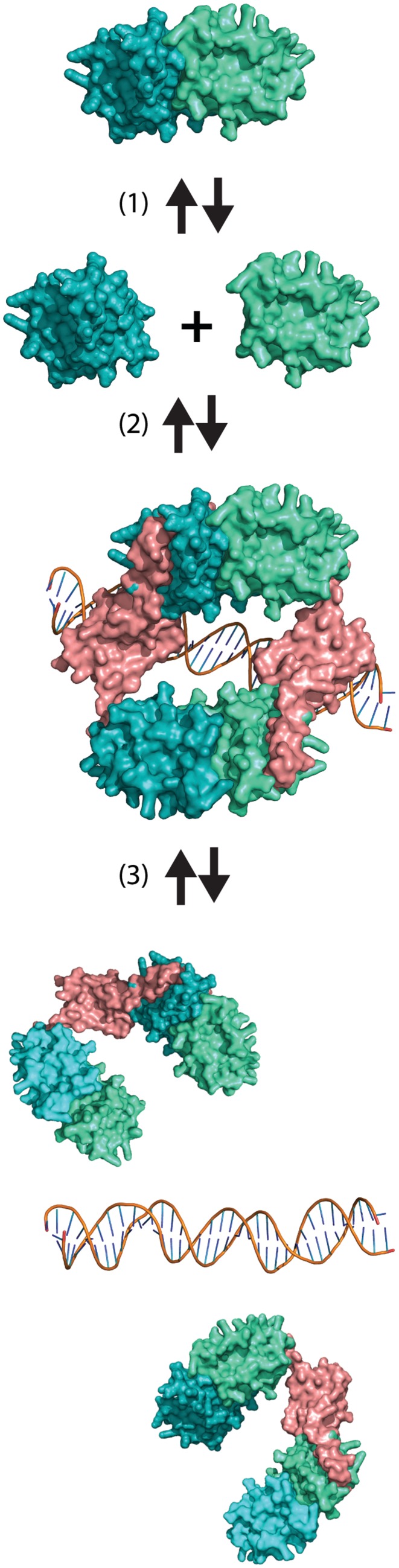
In contrast, an excess of VapC destabilized VapBC binding to both vapO1 and vapO2 (Figure 3A and B) and, consistently, overexpression of a non-toxic variant of VapC in living cells stimulated transcription (Figure 3C and D). Transcription of vapBC is thus regulated by conditional cooperativity. Remarkably, a DNA fragment carrying only one operator (vapO1) responded similarly, showing that the two operator sites function independently and that one operator site is sufficient for conditional cooperativity (Figure 4B and D). The vapO1 fragment bound a complex with a molecular weight consistent with the binding of an octamer of four VapB–VapC heterodimers [(VapBC)2]2, a stoichiometry similar to that of a VapBC (FitAB) complex from N. gonorrhoea bound to DNA (29). In this complex, VapB dimers bound to each operator half-site are bridged by VapC dimers (Supplementary Figure S1D). A similar stoichiometry has also recently been shown for the DNA-bound VapBC complex of Rickettsia felis (35). In addition, Brodersen and co-workers showed that VapBC from S. flexneri pMYSH6000 forms a complex of similar stoichiometry (Supplementary Figure S1C) (33). These scientists also showed that VapBC binds to two operator sites forming complexes of similar sizes. Figure 6 presents a model explaining how the VapB/VapC ratio controls the formation of the complex of VapBC with a single operator site by conditional cooperativity. First, the strong cooperative binding of VapBC to an operator site is explained by the two VapC dimers that bridge the two VapB dimers each recognizing the operator half-sites. Secondly, the bridging by the VapC dimers in the complex makes VapC dimerization the controlling element in the cooperative binding of the complex: with an excess of VapB, a stable [(VapBC)2]2 complex binds cooperatively to an operator site. In contrast, with an excess of VapC, VapC destabilizes VapBC bound to an operator site by invading the complex and breaking the VapC dimer.
Figure 6.
Molecular model explaining destabilization of the VapBC complex bound to a single operator site by VapC. (i) VapC dimer exist in an equilibrium with two VapC monomers. (ii) When VapC is in excess of VapB, VapC monomers ‘invade’ the VapBCO complex. (iii) Invasion abrogates cooperativity of VapBCO complex binding and decreases the affinity of VapBC for DNA. The structures shown were modelled, using the known structure of the [(FitAB)2]2 •DNA complex (29).
We challenged the model experimentally by alanine/serine scanning of the patches in VapC responsible for dimerization (Figure 5A). Four of the 12 single amino acid changes that were tested significantly reduced transcriptional repression by VapBC (Figure 5B), consistent with the model. In particular, the Y72A change in VapC completely abolished repression. The corresponding amino acid in VapC of N. gonorrhoea (P78) has previously been shown to be important for VapC dimerization (29) (Supplementary Figure S1B). We confirmed that this was also the case for VapC; VapCY72A only showed weak dimerization in vitro (Supplementary Figure S3). We conclude that VapC forms dimers, both in solution and in the VapBCO repression complex.
We then analysed the VapC variants that exhibited a reduced ability to repress transcription in vivo and in in vitro (Figure 5B and C). The changes were dramatic: The vapBCY72A operon was strongly de-repressed and, consistently, the Y72A substitution reduced VapBC complex formation with vapO1. Most importantly, however, the binding-response of VapBCY72A was non-cooperative and an excess VapCY72A did not destabilize the VapBCY72AO complex (Figure 5C). A similar abrogation of VapBCO complex destabilization was seen with the other mutants that exhibited reduced transcriptional repression in vivo (Supplementary Figure S4). Thus, VapC dimerization is key, not only to repression but also to derepression of vapBC transcription, that is, for conditional cooperativity. These results support a model in which both formation and destabilization of VapBCO is controlled by VapC dimerization. In particular, destabilization is caused by a VapC monomer switching partner by pairing with a VapC monomer within the VapBCO complex and thereby breaking the bridging dimer required for strong and cooperative binding of VapBC to vapO (Figure 6). In all cases, the amino acid changes in VapC that conferred reduced repression in vivo (Figure 5B) also abolished VapBCO destabilization in vitro (Supplementary Figure S4). These results lend further support to the model.
Conditional cooperativity has been described to control TA operon transcription in three evolutionary independent gene families, ccd of F, relBE of E. coli and phd-doc of P1 and is understood at the mechanistic level for all three families (2,3,24,36). RelB antitoxin dimers bind relO operator half-sites via their Ribbon–Ribbon–Helix motifs (37). We previously proposed a model in which RelE has two binding sites for RelB, a high-affinity and a low-affinity site and that one RelE monomer bridges two RelBs belonging to two different dimers bound to operator half-sites (23). In this model, excess RelE destabilized RelBEO by invasion of a second RelE molecule into the complex by breaking the interaction between the low-affinity-site between RelE and RelB. Thus, a high-affinity interaction replaced a low-affinity interaction and was consistent with a very high efficiency of complex destabilization—that is—RelBEO was destabilized already when the RelE/RelB ratio exceeded 1 and at a ratio of approximately 2, the entire RelBEO complex had disintegrated. In contrast, complete destabilization of VapBCO occurred at a VapC/VapB ratio >16, that is, destabilization of the VapBCO complex occurred gradually and required a much higher relative level of VapC. This considerably lower sensitivity of conditional cooperativity in vapBC regulation can be explained by the model in Figure 6: VapC is a dimer in solution that will be in equilibrium with two monomers. The monomers will also be in equilibrium with the dimers in VapBCO but competition is in this case between molecules with identical affinities, assuming that the dimer interface in a VapC dimer is the same as that of a VapC dimer in VapBCO. Therefore, in VapBCO, the VapC dimer equilibrium will be pushed towards complex destabilization and dimer breaking by high VapC concentrations. The observations made in this study support that under conditions when VapC is in excess of VapB, VapC can directly promote transcription of vapBC and thereby stimulate VapB production to help regenerate a balanced VapB/VapC ratio. Whether this mechanism is active during physiological conditions still needs to be tested.
Conditional cooperativity in the case of the phd-doc operon control is understood at a profound mechanistic level that lends support to the above-proposed model of how RelE controls relBE transcription (24). Using a direct structural approach, Remy Loris and colleagues showed that Doc toxin has both a low-affinity and high-affinity binding site for the C-terminus of the Phd antitoxin, similar to what we postulated for the RelBE interaction as described earlier. Structural analysis of Phd in the presence and absence of Doc showed that binding of Doc to Phd changed a partly disordered DNA binding domain to an ordered one and thereby increased the affinity of Phd for is operator. Due to the second, low affinity binding-site in Doc, one Doc molecule is able to bridge two Phd dimers bound to DNA and thereby confer cooperativity. Thus, Doc has two separate effects on DNA binding of Phd to its operator, both of which increase binding. At a high Doc/Phd ratio, further Doc molecules invade and destabilise the Doc–Phd–operator complex. These considerations raise the possibility that the mechanisms by which Doc and RelE mediate conditional cooperativity may be mechanistically related even though the TA loci encoding these components evolved independently. It is also clear from the results presented here, that the molecular mechanisms of conditional cooperativity in the cases of vapBC on the one hand and phd-doc/relBE on the other are entirely different.
It has now become clear that conditional cooperativity is a property common to TA loci (all TA loci investigated so far are regulated by conditional cooperativity) and it is relevant to ask how conditional cooperativity relates to the biological function of TA loci. We showed recently that TA loci that encode RNases are required for persistence of E. coli (15). We proposed a model in which Lon, in a minor fraction of the cells, degrade the antitoxins and thereby induce toxin activity, dormancy and persistence (drug tolerance). The vapBC locus has all the properties required to function in persistence: it encodes a tRNase whose activity inhibits translation reversibly (25,32) and Lon degrades VapB antitoxin (Figure 1). It is not known how bacterial cells resuscitate from the dormancy that is characteristic of the persistent state (38). Persister cells have higher TA transcription-rates than the average cell population (39,40). This suggests that persisters have increased T/A ratios and therefore raises the possibility that conditional cooperativity is operative in persister cells. It is thus possible that conditional cooperativity secures a high, on-going transcription rate of TA loci in dormant cells and thereby allow de novo synthesis of antitoxin that quench the toxins. By inference, such quenching of the toxins must be required for the persisters to escape dormancy and resuscitate. A second possibility, not mutually exclusive with first, is that conditional cooperativity functions to rapidly turn off toxin activity and TA operon transcription. The latter possibility would also imply that conditional cooperativity help reduce fortuitous events of toxin activation due to random fluctuations (noise) in the antitoxin level.
VapC activity and transcription of vapBC are both regulated by Lon because the protease degrades VapB antitoxin (Figure 1). It is not yet known if degradation of VapB by Lon is controlled by cellular signals. It is tempting to speculate that VapB degradation is actively regulated by as yet unknown factors that control Lon activity because such active regulation would enable the cell to turn TA locus activity on and off according to internal or external stimuli. We are now pursuing this question.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables S1 and S2, Supplementary Figures S1–4, Supplementary Material and Methods and Supplementary References [25,29,32,33,41–44].
FUNDING
Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ditlev Brodersen for critical reading of the manuscript and the members of the Centre for Bacterial Cell Biology for stimulating discussions.
REFERENCES
- 1.Fozo EM, Hemm MR, Storz G. Small toxic proteins and the antisense RNAs that repress them. Microbiol. Mol. Biol. Rev. 2008;72:579–589. doi: 10.1128/MMBR.00025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerdes K, Wagner EG. RNA antitoxins. Curr. Opin. Microbiol. 2007;10:117–124. doi: 10.1016/j.mib.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Gerdes K, Christensen SK, Lobner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 2005;3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 4.Fineran PC, Blower TR, Foulds IJ, Humphreys DP, Lilley KS, Salmond GP. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc. Natl Acad. Sci. USA. 2009;106:894–899. doi: 10.1073/pnas.0808832106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jørgensen MG, Pandey DP, Jaskolska M, Gerdes K. HicA of Escherichia coli defines a novel family of translation-independent mRNA interferases in bacteria and archaea. J. Bacteriol. 2009;191:1191–1199. doi: 10.1128/JB.01013-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Makarova KS, Wolf YI, Koonin EV. Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol. Direct. 2009;4:19. doi: 10.1186/1745-6150-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerdes K. Toxin-antitoxin modules may regulate synthesis of macromolecules during nutritional stress. J. Bacteriol. 2000;182:561–572. doi: 10.1128/jb.182.3.561-572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pandey DP, Gerdes K. Toxin - antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005;33:966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shao Y, Harrison EM, Bi D, Tai C, He X, Ou HY, Rajakumar K, Deng Z. TADB: a web-based resource for Type 2 toxin-antitoxin loci in bacteria and archaea. Nucleic Acids Res. 2010;39:D606–D611. doi: 10.1093/nar/gkq908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramage HR, Connolly LE, Cox JS. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet. 2009;5:e1000767. doi: 10.1371/journal.pgen.1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerdes K, Rasmussen PB, Molin S. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc. Natl Acad. Sci. USA. 1986;83:3116–3120. doi: 10.1073/pnas.83.10.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szekeres S, Dauti M, Wilde C, Mazel D, Rowe-Magnus DA. Chromosomal toxin-antitoxin loci can diminish large-scale genome reductions in the absence of selection. Mol. Microbiol. 2007;63:1588–1605. doi: 10.1111/j.1365-2958.2007.05613.x. [DOI] [PubMed] [Google Scholar]
- 13.Christensen SK, Mikkelsen M, Pedersen K, Gerdes K. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc. Natl Acad. Sci. USA. 2001;98:14328–14333. doi: 10.1073/pnas.251327898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsilibaris V, Maenhaut-Michel G, Mine N, Van ML. What is the benefit to Escherichia coli of having multiple toxin-antitoxin systems in its genome? J. Bacteriol. 2007;189:6101–6108. doi: 10.1128/JB.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maisonneuve E, Shakespeare LJ, Jorgensen MG, Gerdes K. Bacterial persistence by RNA endonucleases. Proc. Natl Acad. Sci. USA. 2011;108:13206–13211. doi: 10.1073/pnas.1100186108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Hallez R, Geeraerts D, Sterckx Y, Mine N, Loris R, Van ML. New toxins homologous to ParE belonging to three-component toxin-antitoxin systems in Escherichia coli O157:H7. Mol. Microbiol. 2010;76:719–732. doi: 10.1111/j.1365-2958.2010.07129.x. [DOI] [PubMed] [Google Scholar]
- 17.Nariya H, Inouye M. MazF, an mRNA interferase, mediates programmed cell death during multicellular Myxococcus development. Cell. 2008;132:55–66. doi: 10.1016/j.cell.2007.11.044. [DOI] [PubMed] [Google Scholar]
- 18.Christensen-Dalsgaard M, Jorgensen MG, Gerdes K. Three new RelE-homologous mRNA interferases of Escherichia coli differentially induced by environmental stresses. Mol. Microbiol. 2010;75:333–348. doi: 10.1111/j.1365-2958.2009.06969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christensen SK, Pedersen K, Hansen FG, Gerdes K. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol. Biol. 2003;332:809–819. doi: 10.1016/s0022-2836(03)00922-7. [DOI] [PubMed] [Google Scholar]
- 20.Gotfredsen M, Gerdes K. The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol. Microbiol. 1998;29:1065–1076. doi: 10.1046/j.1365-2958.1998.00993.x. [DOI] [PubMed] [Google Scholar]
- 21.Magnuson R, Lehnherr H, Mukhopadhyay G, Yarmolinsky MB. Autoregulation of the plasmid addiction operon of bacteriophage P1. J. Biol. Chem. 1996;271:18705–18710. doi: 10.1074/jbc.271.31.18705. [DOI] [PubMed] [Google Scholar]
- 22.Magnuson R, Yarmolinsky MB. Corepression of the P1 addiction operon by Phd and Doc. J. Bacteriol. 1998;180:6342–6351. doi: 10.1128/jb.180.23.6342-6351.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Overgaard M, Borch J, Jorgensen MG, Gerdes K. Messenger RNA interferase RelE controls relBE transcription by conditional cooperativity. Mol. Microbiol. 2008;69:841–857. doi: 10.1111/j.1365-2958.2008.06313.x. [DOI] [PubMed] [Google Scholar]
- 24.Garcia-Pino A, Balasubramanian S, Wyns L, Gazit E, De GH, Magnuson RD, Charlier D, van Nuland NA, Loris R. Allostery and intrinsic disorder mediate transcription regulation by conditional cooperativity. Cell. 2010;142:101–111. doi: 10.1016/j.cell.2010.05.039. [DOI] [PubMed] [Google Scholar]
- 25.Winther KS, Gerdes K. Enteric virulence associated protein VapC inhibits translation by cleavage of initiator tRNA. Proc. Natl Acad. Sci. USA. 2011;108:7403–7407. doi: 10.1073/pnas.1019587108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilbur JS, Chivers PT, Mattison K, Potter L, Brennan RG, So M. Neisseria gonorrhoeae FitA interacts with FitB to bind DNA through its ribbon-helix-helix motif. Biochemistry. 2005;44:12515–12524. doi: 10.1021/bi0511080. [DOI] [PubMed] [Google Scholar]
- 27.Bodogai M, Ferenczi S, Bashtovyy D, Miclea P, Papp P, Dusha I. The ntrPR operon of Sinorhizobium meliloti is organized and functions as a toxin-antitoxin module. Mol. Plant Microbe Interact. 2006;19:811–822. doi: 10.1094/MPMI-19-0811. [DOI] [PubMed] [Google Scholar]
- 28.Robson J, McKenzie JL, Cursons R, Cook GM, Arcus VL. The vapBC operon from Mycobacterium smegmatis is an autoregulated toxin-antitoxin module that controls growth via inhibition of translation. J. Mol. Biol. 2009;390:353–367. doi: 10.1016/j.jmb.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 29.Mattison K, Wilbur JS, So M, Brennan RG. Structure of FitAB from Neisseria gonorrhoeae bound to DNA reveals a tetramer of toxin-antitoxin heterodimers containing pin domains and ribbon-helix-helix motifs. J. Biol. Chem. 2006;281:37942–37951. doi: 10.1074/jbc.M605198200. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. New York: Laboratory Press, Cold Spring Harbor; 1989. [Google Scholar]
- 31.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winther KS, Gerdes K. Ectopic production of VapCs from Enterobacteria inhibits translation and trans-activates YoeB mRNA interferase. Mol. Microbiol. 2009;72:918–930. doi: 10.1111/j.1365-2958.2009.06694.x. [DOI] [PubMed] [Google Scholar]
- 33.Dienemann C, Boggild A, Winther KS, Gerdes K, Brodersen DE. Crystal Structure of the VapBC toxin-antitoxin complex from Shigella flexneri reveals a hetero-octameric DNA-binding assembly. J. Mol. Biol. 2011;414:713–722. doi: 10.1016/j.jmb.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 35.Mate MJ, Vincentelli R, Foos N, Raoult D, Cambillau C, Ortiz-Lombardia M. Crystal structure of the DNA-bound VapBC2 antitoxin/toxin pair from Rickettsia felis. Nucleic Acids Res. 2012;40:3245–3258. doi: 10.1093/nar/gkr1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De,Jonge N, Garcia-Pino A, Buts L, Haesaerts S, Charlier D, Zangger K, Wyns L, De GH, Loris R. Rejuvenation of CcdB-poisoned gyrase by an intrinsically disordered protein domain. Mol. Cell. 2009;35:154–163. doi: 10.1016/j.molcel.2009.05.025. [DOI] [PubMed] [Google Scholar]
- 37.Overgaard M, Borch J, Gerdes K. RelB and RelE of Escherichia coli form a tight complex that represses transcription via the ribbon-helix-helix motif in RelB. J. Mol. Biol. 2009;394:183–196. doi: 10.1016/j.jmb.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewis K. Persister cells. Annu. Rev. Microbiol. 2010;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 39.Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 2004;186:8172–8180. doi: 10.1128/JB.186.24.8172-8180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shah D, Zhang ZG, Khodursky A, Kaldalu N, Kurg K, Lewis K. Persisters: a distinct physiological state of E-coli. BMC Microbiol. 2006;6:53. doi: 10.1186/1471-2180-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller JH. Experiments in Molecular Genetics. New York: Laboratory Press, Cold Spring Harbor; 1972. [Google Scholar]
- 42.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 43.Orchard K, May GE. An EMSA-based method for determining the molecular weight of a protein–DNA complex. Nucleic Acids Res. 1993;21:3335–3336. doi: 10.1093/nar/21.14.3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.