Abstract
Compared to transcriptional activation, other mechanisms of gene regulation have not been widely exploited for the control of transgenes. One barrier to the general use and application of alternative splicing is that splicing-regulated transgenes have not been shown to be reliably and simply designed. Here, we demonstrate that a cassette bearing a suicide exon can be inserted into a variety of open reading frames (ORFs), generating transgenes whose expression is activated by exon skipping in response to a specific protein inducer. The surprisingly minimal sequence requirements for the maintenance of splicing fidelity and regulation indicate that this splicing cassette can be used to regulate any ORF containing one of the amino acids Glu, Gln or Lys. Furthermore, a single copy of the splicing cassette was optimized by rational design to confer robust gene activation with no background expression in plants. Thus, conditional splicing has the potential to be generally useful for transgene regulation.
INTRODUCTION
Alternative splicing of precursor mRNAs (pre-mRNAs) is an important and widely conserved mechanism for increasing protein diversity and for gene regulation. In plants and other eukaryotes, tissue-specific and development-specific expression of genes is regulated by alternative splicing (1–3). Changes in pre-mRNA splicing of plant transcripts have been observed in response to stress conditions, including growth in cold temperature or under drought conditions (4). Alternative splicing also has been shown to contribute to diverse physiological processes in plants, including regulation of circadian rhythm and the defense response to pathogens (5,6). Altogether, it has been estimated that between 20% and 30% of expressed genes are alternatively spliced in Arabidopsis thaliana and Oryza sativa (rice) (7,8).
While alternative splicing appears to be extensively employed in natural regulatory systems, it has not been generally applied to the conditional expression of transgenes. Currently, transgene regulation is based almost exclusively on transcriptional activation. This is due, in no small part, to the ease of placing a promoter sequence upstream of any gene of interest, which requires little to no characterization of promoter elements and no alteration of the coding sequence. However, many conditional promoters suffer from issues such as leaky basal expression, pleiotropic effects and species specificity (9). Promoters are also difficult to combine serially in order to generate complex regulatory patterns, as cross-talk between different promoter elements often leads to unpredictable effects on gene expression (10). Furthermore, use of multiple copies of identical promoters to coordinate regulation of several genes can trigger gene silencing (11). Thus, one motivation for developing a method for transgene regulation based on alternative splicing is that problems with existing conditional promoters may be ameliorated by combining DNA- and RNA-level regulation. Another advantage of a conditional splicing system is that the gene can be regulated and still remain under the control of its endogenous promoter. There may be additional benefits to cotranscriptional mechanisms of regulation which have yet to be explored because a general conditional splicing system has not been developed.
An important advantage to the use of conditional promoters is that it does not change the sequence of the translated open reading frame (ORF). However, alternative splicing of a cassette harboring a suicide exon can also be considered to operate in a traceless manner (Figure 1A). Exon skipping would generate a productively translated spliced product (SP-I) with the suicide exon cleanly excised from the sequence of the ORF. Alternatively, exon inclusion would introduce a premature termination codon that targets the spliced product (SP-II) for nonsense-mediated decay (NMD) instead of undergoing translation (12). Thus, the presence of the suicide exon effectively eliminates gene expression, and its conditional splicing regulates expression of the encoded ORF. The coupling of alternative splicing to mRNA quality control pathways is conserved as a regulatory mechanism in diverse eukaryotic organisms, including plants, fungi and metazoans (13), so this method for transgene regulation could have broad applicability.
Figure 1.
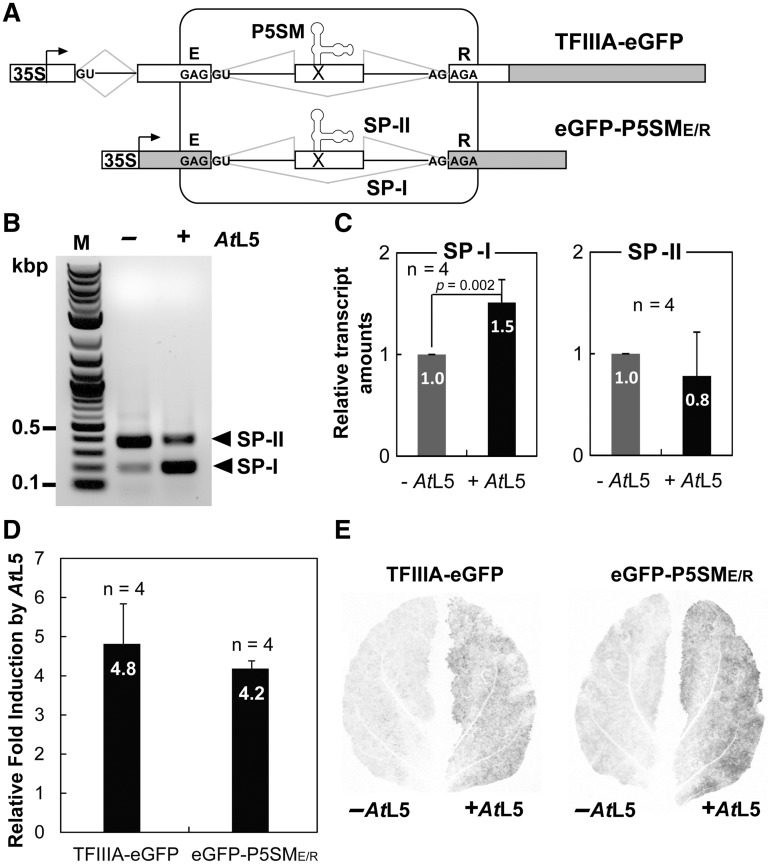
The P5SM splicing cassette functions in the eGFP coding sequence. (A) Schematic representation of the P5SM-splicing cassette (boxed) within gene constructs TFIIIA-eGFP (native context, top) and eGFP-P5SME/R (foreign context, bottom). The full sequence maps for these constructs are shown in Supplementary Figure S6. The eGFP coding sequence is shaded. The cassette exon contains the P5SM RNA element and an in-frame premature termination codon (X). The splicing reactions that generate spliced products SP-I and SP-II are shown. (B) eGFP-P5SME/R spliced products detected by RT–PCR upon induction with AtL5 (+) or LUC as a control (−). (C) Relative amounts of the spliced products of eGFP-P5SME/R determined by RT–qPCR. Data are averaged with number of biological replicates (n) and standard deviations shown. The p-value was determined using the paired t-test. (D) Fold induction of protein expression quantified by eGFP fluorescence for each construct. Induction with AtL5 was measured in comparison to induction with LUC as a control. (E) Representative whole leaf scan of eGFP fluorescence 3 days post-infiltration for each construct. The right leaf halves coexpressed AtL5 and the left halves coexpressed LUC (control).
So far, a few conditional splicing systems have been constructed that regulate gene expression in budding yeast (14) and mammalian cells (15,16). These studies were performed either on single reporter constructs or in the context of gene fusions which introduced extraneous sequences to the N-terminus of the ORF, similar to minigene reporters used in splicing assays (17). In addition, a natural riboswitch has been found that regulates gene expression through alternative splicing in response to thiamine pyrophosphate (TPP) in plants and filamentous fungi (18–21). The untranslated regions (UTRs) containing the TPP riboswitch have been appended to reporter constructs. However, the level of gene activation was modest even in thiamine-deficient plant lines, as levels of the spliced product which gives higher gene expression was increased ∼7-fold upon thiamine depletion (20). Thus, it has remained unclear whether conditionally spliced transgenes can be reliably designed for robust gene activation, such that this method is generalizable to any gene of interest.
Pre-mRNA splicing reactions can be sensitive to sequence context, as even single nucleotide polymorphisms have been shown to cause aberrant splicing in some genes (22). Thus, maintaining the fidelity and regulation of alternative splicing within diverse coding sequences could be quite challenging. In this study, we have found that a natural splicing cassette with a suicide exon could be inserted into a variety of coding sequences to regulate gene expression in plants. Further analysis has revealed that cassette function apparently requires conservation of only two upstream nucleotides in the coding sequence, making this conditional splicing system quite general. Finally, we demonstrate that strong gene activation (∼97-fold) can be accomplished by alternative splicing using an optimized version of the suicide exon.
MATERIALS AND METHODS
Oligonucleotides and DNA constructs
Sequences of all synthetic primers used for making and mutagenizing DNA constructs, performing overlap extension PCR, RT–PCR and RT–qPCR are described in Supplementary Table S1. All constructs confirmed by sequencing are shown in Supplementary Data. TFIIIA-eGFP (formerly called Pre-EGFP), LUC (used as a control except when assaying the fLUC reporter), AtL5 and DsRed2 constructs were described previously (23). The coding sequence of OsL5 (Os01g0896800) also was previously isolated and cloned into the pBinAR vector (23). The fLUC coding sequence was derived from the pGL3 vector (Promega). A. thaliana coding sequences were amplified by PCR from cDNA (see RNA isolation methods) and cloned using the TOPO TA Cloning kit (Invitrogen). Mutations to TFIIIA-eGFP were made using the QuikChange II Site-Directed Mutagenesis (Stratagene) or QuikChange Lightning Site-Directed Mutagenesis kits (Stratagene) according to the manufacturer's instructions.
The P5SM splicing cassette sequence was inserted into constructs by overlap extension PCR (24). Briefly, the coding sequence to be modified was amplified as two DNA fragments using PCR primers that introduce a region that overlaps with the sequence of the P5SM-splicing cassette. For the front fragment, the overlapping region is at the 3′-end, and for the back fragment, the overlapping region is at the 5′-end. The P5SM-splicing cassette was amplified by PCR using TFIIIA-eGFP as the DNA template, generating a third DNA fragment. The DNA fragments were purified by agarose gel electrophoresis and then mixed in a 1:1:1 ratio as partial templates for overlap extension PCR, which uses forward and reverse primers that anneal to the 5′- and 3′-ends of the desired full-length construct. The PCR product containing the P5SM cassette was purified by agarose gel electrophoresis before being cloned into the TOPO vector for sequence confirmation. To generate constructs for use in plant infiltration experiments, they were re-amplified to introduce restriction sites at the ends (Supplementary Table S1), digested with the appropriate restriction enzymes, and ligated using T4 DNA ligase (NEB) into the binary vector pBinAR (25).
The sequence encoding the P5SM RNA element from O. sativa was derived from the genomic fragment of Os02g0116000 previously cloned into the TOPO vector (23). To generate the eGFP-OsP5SM construct, overlap extension PCR was performed to insert OsP5SM in place of the original P5SM from A. thaliana. To generate eGFP-HyP5SM, the purine-rich insertion was introduced directly into the primers used to amplify eGFP-OsP5SM as two DNA fragments, then the two overlapping pieces were extended by PCR. After sequence confirmation of these constructs in the TOPO vector, they were cloned into pBinAR as described above.
Leaf-based fluorescence assay
In vivo reporter fluorescence assays were performed by Agrobacterium-mediated leaf infiltration in Nicotiana benthamiana as previously described (19). Briefly, each leaf half was infiltrated with a 1:1:1 mixture of Agrobacterium transformed with pBinAR plasmids carrying the reporter construct, inducer (AtL5 or OsL5) or the control (LUC), and DsRed2 as a normalization standard. For assaying the fLUC reporter, untransformed Agrobacterium was used as the control instead. In vivo fluorescence was measured 3 days post-infiltration using the Typhoon laser-based scanning system (GE Healthcare) using excitation and emission wavelength settings of 488/520 nm for eGFP and 532/580 nm for DsRed2. The ratios of eGFP/DsRed2 fluorescence readings were taken for each leaf half in order to normalize differences in transformation efficiency. On average, ∼15% difference was observed between DsRed2 fluorescence for leaf halves from the same leaf sample.
To calculate relative fold induction by AtL5 or OsL5, the eGFP/DsRed2 ratio for the leaf half coexpressing the inducer was divided by the other half coexpressing the control. The average relative fold induction was calculated from these values for several leaf samples, and the number of independent leaf samples analyzed (n) and standard deviations are as indicated. No more than two leaf samples were analyzed from the same plant. Autofluorescence (defined as fluorescence measured for uninfiltrated, or ‘blank’, leaves) was not measured, and so was not subtracted from the eGFP fluorescence readings for all experiments except for the one showing relative fluorescence of HyP5SM without DsRed2. For this experiment, autofluorescence was measured and subtracted from the eGFP fluorescence readings of both leaf halves, and fold induction for each individual leaf was calculated by taking the ratio of the half coexpressing OsL5 versus the other coexpressing LUC (control).
RNA isolation and RT–PCR analyses
Total RNA was isolated from ∼100 mg N. benthamiana leaf tissue collected 2 days post-infiltration using the Universal RNA Purification Kit (CHIMERX) according to the manufacturer's instructions. Leaf tissue samples were immediately frozen in liquid nitrogen after being placed in microtubes, then pulverized using a bead mill (TissueLyser, Qiagen) prior to RNA extraction. The samples were kept frozen during the milling process by being placed in an adapter rack that was pre-chilled at −80°C. The entire procedure was performed as quickly as possible, and the integrity of the RNA after isolation was checked by analysis on an agarose gel.
cDNA was generated from 500 ng RNA samples that had been treated with RQ1 DNase (Promega) using iSCRIPT reverse transcriptase (Bio-Rad) and oligo d(T) primers following the manufacturer's instructions. Spliced products were amplified from cDNA samples by PCR with Taq polymerase (NEB) using target-specific primers (Supplementary Table S1). Spliced products were visualized by agarose gel electrophoresis and confirmed by sequencing after gel purification and TOPO cloning (see Supplementary Figure S6 for sequence maps of spliced products). RT–qPCR analysis was performed on the Bio-Rad CFX96 instrument using spliced product-specific primers (Supplementary Table S1) and SsoFast EvaGreen Supermix (Bio-Rad). The specificity of spliced product amplification was confirmed by melting curve analysis and visualization of PCR products on an agarose gel. Control samples in which no reverse transcriptase was added to the reaction also were analyzed after PCR to confirm the absence of DNA contamination. Transcript abundances for different samples were calculated using standard curves determined for each target. Primer efficiencies were determined to be within the range of 100 ± 5%. Relative transcript amounts were normalized to the relative amount of DsRed2 transcript and the sample with the control LUC was set to 1. For biological replicates, no more than two leaf samples were taken from the same plant.
Luciferase assay
Firefly luciferase (fLUC) enzyme activity was measured using the Luciferase Assay System (Promega) according to the manufacturer's instructions. N. benthamiana leaf tissue samples were collected and analyzed 3 days post-infiltration. Tissue lysate samples were normalized to total protein content determined by using the detergent-compatible (DC) protein assay (Bio-Rad), and relative luminescence was measured in triplicate using a GloMax 96 Microplate Luminometer (Promega). Fold induction was determined relative to samples without coexpression of inducer.
Bioinformatics
The TAIR9 release of 33 201 annotated protein coding sequences for A. thaliana was inputted into a MySQL database table. A database query was made for each pair of amino acids (corresponding to the bordering codons) within the first half of protein coding sequences. Next, a database query was made counting the number of protein entries not containing any of the amino acid pairs within the first half of the coding sequence. This number was subtracted from and divided by the total number of protein coding sequences to calculate the proteome coverage percentage. Similar queries were used to calculate the percentage of protein entries containing an E, K or Q amino acid within the first half of the coding sequence.
RESULTS
A natural splicing cassette bearing a suicide exon can be employed to regulate other genes
We and others have previously reported the discovery of the plant 5S rRNA mimic (P5SM), an RNA element residing within a highly conserved, alternatively spliced suicide exon that controls expression of transcription factor IIIA (TFIIIA) in land plants (23,26). We have shown that TFIIIA expression is activated by skipping of the suicide exon, which is induced by ribosomal protein L5 binding to the P5SM RNA element (23). Our initial design for a splicing cassette conservatively included not only the P5SM exon (175 nt in length) and flanking intronic regions (150 and 98 nt in length) from the A. thaliana TFIIIA gene, but also the two bordering codons, which encode amino acids Glu (E) and Arg (R), respectively (Figure 1A). This design was intended to balance preserving the relative splice site strengths, in case they were important for alternative splicing activity, with minimizing the gene context requirements.
Using the overlap extension PCR method (24), the above splicing cassette, called P5SME/R, was inserted in place of the codons for E-96 and R-97 within the coding sequence for enhanced green fluorescent protein to generate the reporter construct eGFP-P5SME/R (Figure 1A). Thus, outside of the inserted cassette, the only change to the coding sequence of eGFP was a silent mutation of codon 97 from CGC to AGA (both encode Arg). This codon was changed in order to match the sequence surrounding the 3′-splice site (ss) in TFIIIA.
The function of the splicing cassette in this non-native gene context was compared to that for the control construct, TFIIIA-eGFP, in which the cassette remains in the native TFIIIA context with eGFP fused to the C-terminal end (Figure 1A). RT–PCR analysis of eGFP-P5SME/R detected only the expected exon-skipped (SP-I) and exon-retained (SP-II) spliced products (Figure 1B). The SP-I transcript was identical to the original eGFP coding sequence, except for the silent mutation, and its levels were increased ∼1.5-fold by coexpression of A. thaliana L5 (AtL5), similar to previous observations for TFIIIA-eGFP (Figure 1C) (23). The decrease in SP-II does not match the increase in SP-I, most likely because the abundance of the exon-retained spliced product is governed by NMD; this agrees with previous observations that the SP-II of TFIIIA is degraded (27).
Consistent with the RT–PCR analysis, increases in protein fluorescence with AtL5 induction were comparable between eGFP-P5SME/R and TFIIIA-eGFP (Figure 1D). The representative leaf scans for each construct (Figure 1E) show similar AtL5 induction of eGFP fluorescence, as well as some background fluorescence induced by endogenous L5. These results support that the splicing cassette is fully functional within a foreign context—the eGFP coding sequence—with conservation of only the immediate bordering codons.
We further inserted P5SME/R into several A. thaliana protein coding sequences, in place of E/R sequences naturally present within the first half of each ORF. In each of these constructs, splicing fidelity and regulation appear to be maintained, although some splicing intermediates were observed that correspond to unspliced and partially spliced pre-mRNA (Supplementary Figure S1). It is unclear whether these intermediates build up to detectable levels due to slower spliceosome processing or RNA degradation, but we later determined that they are not observed when the splicing cassette is inserted between other amino acid pairs within these coding sequences.
Determination of the minimal gene context requirements for the P5SM-splicing cassette expands its general utility
Expanding the repertoire of gene contexts beyond the wild-type E/R sequence would enable more facile design of splicing-regulated transgenes. Thus, to define the minimal context required for cassette function, we analyzed the effects of systematically mutating the bordering codons. Mutations upstream of the 5′-ss at positions −1 to −3 and downstream of the 3′-ss at positions +1 to +3 were performed on the TFIIIA-eGFP reporter (Figure 2A). Since the amino acid changes are not in the eGFP sequence, the changes in fluorescence reflect the activity of the splicing cassette.
Figure 2.
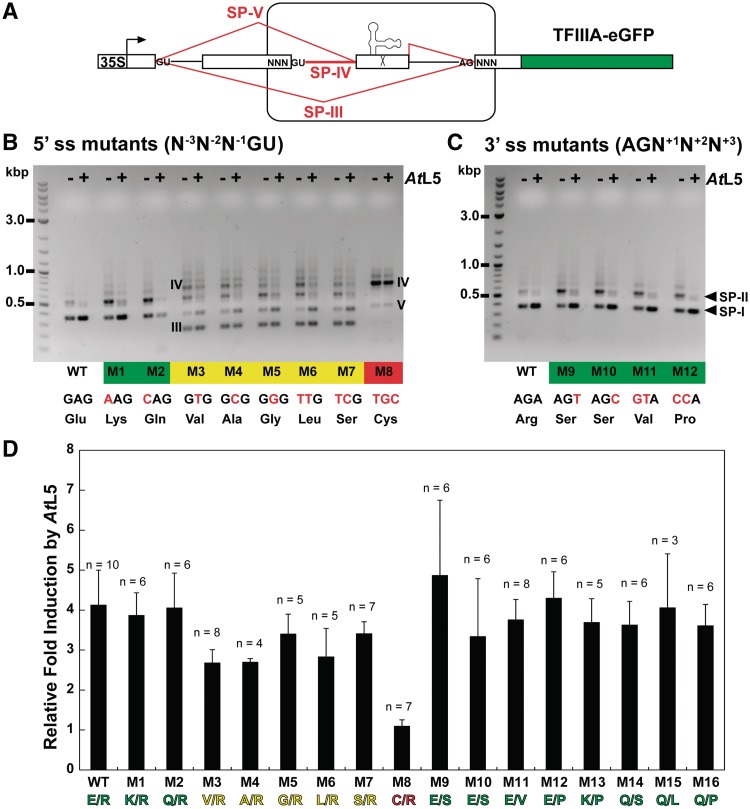
Splicing activity of the cassette tolerates many, but not all, mutations to the bordering codons. (A) Schematic representation of the TFIIIA-eGFP gene construct showing the location of the mutated bordering codons (labeled NNN). The eGFP sequence is shown in green. Aberrant spliced products (red lines) are described in the text and the sequences are shown in Supplementary Figure S6. (B, C) Comparison of spliced products detected by RT–PCR for select 5′-ss and 3′-ss mutations tested within the TFIIIA-eGFP context (also see Supplementary Figure S2). Mutants were coexpressed with AtL5 (+) or LUC as a control (−). Spliced products are labeled and the sequences are shown in Supplementary Figure S6. The mutants are categorized as fully functional (green), semi-functional (yellow) or non-functional (red). Red nucleotides indicate the mutations made to the bordering codon, and the corresponding encoded amino acid is listed below. (D) Fold induction of protein expression quantified by eGFP fluorescence for select TFIIIA-eGFP mutants (also see Supplementary Figure S2). Induction with AtL5 was measured by comparison to induction with LUC as a control. For each mutant construct, the two amino acids encoded by the 5′- and 3′-bordering codons are indicated.
The effect of mutations to the bordering codons on splicing was analyzed by RT–PCR and on gene expression was quantified by fluorescence leaf scanning (Figure 2B–D and Supplementary Figure S2). The results are consistent with the reported conservation pattern for nucleotides proximal to annotated 5′-ss in A. thaliana (28), which we took to reflect the extent of influence on splice site strength. Position −3 had not been found to be highly conserved, and accordingly, the M1 and M2 constructs appear to retain full splicing activity. These mutants were characterized as splicing to SP-I and SP-II only, responding to AtL5 by an increase in SP-I, and exhibiting a similar fluorescence induction to WT. The U substitution at position −3 was not tested because it would generate a premature termination codon within the coding sequence.
Mutations at the more highly conserved positions −2 and −1 lead to partial or full loss of splicing fidelity and regulation. M3-M7 constructs harboring various mutations at position −2 were characterized as semi-functional. These mutants exhibit aberrant splicing but still some splicing to SP-I and SP-II, which leads to overall lower fluorescence induction relative to WT. Sequence analysis of the observed aberrant spliced products, SP-III and SP-IV, showed that they are generated by improper usage of an upstream 5′-ss or intron retention, respectively (Figure 2A). This is consistent with the idea that these mutations weaken recognition of the 5′-ss. The M8 construct, which deliberately substitutes the least frequently observed nucleotides at positions −1 through −3, is fully non-functional. This mutant displays no induction of protein expression and only aberrant spliced products (SP-IV and SP-V), consistent with an impaired 5′-ss.
In contrast, alterations near the 3′-ss do not appear to affect splicing activity, as evidenced by the M9-M12 constructs. Even the M12 construct, in which positions +1 and +2 are substituted with the least frequently observed nucleotides, maintains full splicing activity. We did not rigorously test mutations to position +3 because no nucleotide conservation was observed at this position (28). In addition, M13-M17 constructs confirm that double mutations to the bordering codons are tolerated as well as the corresponding single mutations (Figure 2D and Supplementary Figure S2).
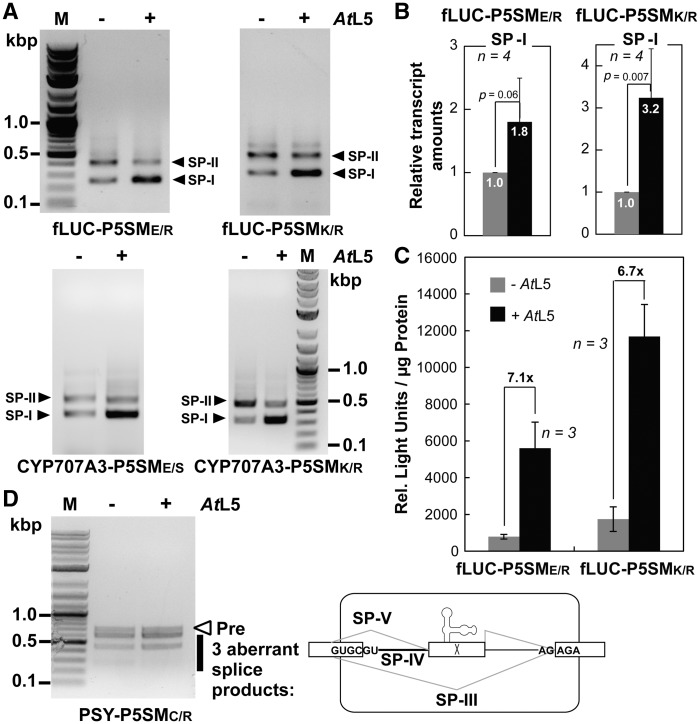
Taken together, these mutagenesis results suggest that full splicing activity for the splicing cassette requires a 5′ context that extends to positions −1 and −2, but that there is no apparent requirement for the 3′ context past the canonical 3′-ss. In order to validate these surprisingly minimal exonic sequence requirements, we further analyzed the activity of the splicing cassette in ORF contexts corresponding to several of the tested mutations. The predicted functional contexts E/R, K/R and E/S were tested in the ORFs for fLUC or abscissic acid 8′-hydroxylase (CYP707A3) as a representative plant gene. In good agreement with the results for WT, M1 and M9 constructs, the splicing cassette in these ORFs displayed full splicing activity and (analyzed by enzyme activity for fLUC only) increased protein expression in response to AtL5 (Figure 3A–C). The predicted non-functional context C/R was also tested in the ORF for phytoene synthase (PSY). Similar to the result for the M8 construct, the splicing cassette in this ORF is deregulated and gives related aberrant spliced products (Figure 3D).
Figure 3.
The cassette functions predictably in different gene contexts. (A) RT–PCR detection of spliced products for fLUC and abscissic acid 8’-hydroxylase (CYP707A3) harboring the P5SM cassette inserted within the E/R, K/R or E/S sequence contexts, upon induction with AtL5 (+) or LUC as a control (−). (B) Relative amounts of SP-I transcript for fLUC constructs determined by RT–qPCR. Data are averages with number of biological replicates (n) and standard deviations shown. The p-value was determined using the paired t-test. (C) Luciferase activity in relative light units of fLUC constructs normalized to total protein upon induction with AtL5 (+) or control (−). Fold induction by AtL5 is shown at the top of each bar. (D) RT–PCR detection of spliced products for PSY harboring the P5SM cassette inserted within the C/R sequence context upon induction with AtL5 (+) or LUC as a control (−). A schematic representation of the identified aberrant spliced products (right) shows cryptic splicing from a proximal GU in the coding sequence (sequences are shown in Supplementary Figure S6).
These data provide strong evidence that the mutagenesis results can reliably predict splicing activity for the P5SM cassette in other ORF contexts. If predicted functional contexts are limited to the 12 amino acid pairs that have been validated as giving full splicing activity (Figure 2 and Supplementary Figure S2), bioinformatics analysis suggests that the splicing cassette can be inserted within the first half of 93% of annotated ORFs in the A. thaliana genome. However, our results also show that the 3′-ss context appears not to influence cassette activity (Figure 2). The minimal exonic context for full maintenance of splicing fidelity and regulation appears to be an AG immediately upstream of the 5′-ss. An in-frame NAG codon would encode any of the amino acids Glu, Lys or Gln. If we consider all ORFs that contain one of these three amino acids within the first half of their coding regions, the genome coverage estimate rises to 99.7%.
Rational adaptation of a species-divergent P5SM element gives robust, orthogonal gene activation
The natural P5SM cassette appears to maintain splicing fidelity and regulation within diverse coding sequences. However, one potential drawback is that basal activation from endogenous L5 contributes to background fluorescence and leads to relatively modest (∼4–7-fold) levels of gene induction by AtL5 (Figure 1D and 3C). To eliminate basal activation, one might consider knocking out the endogenous protein, but since L5 is a component of the ribosomal machinery, this is expected to be highly detrimental. Also, it is preferable to avoid requiring a specific genotype for use of the splicing cassette.
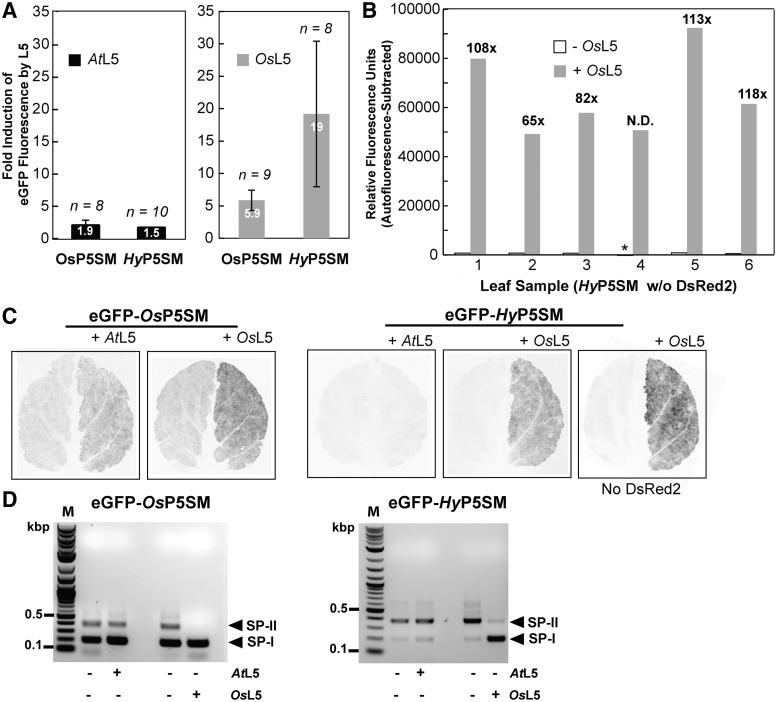
Alternatively, we reasoned that the P5SM RNA element from a more divergent plant species might be less responsive to the endogenous L5 as compared to the A. thaliana P5SM RNA element. Both A. thaliana and N. benthamiana, the model species in which the transient expression assays (see ‘Materials and Methods’ section) have been performed, are dicots. Thus, we replaced the P5SM RNA element in the reporter construct eGFP-P5SME/R with one derived from the monocot O. sativa, which was termed OsP5SM. We found that splicing activity is maintained for the new construct eGFP-OsP5SME/R, which also exhibits a higher level of induction by OsL5 than AtL5 (Figure 4A). This construct was expected to give lower background fluorescence due to reduced activation by the endogenous L5. Instead, higher background fluorescence is observed (Figure 4C, left side of leaves), corresponding to an increased ratio of SP-I to SP-II in the uninduced sample (Figure 4D).
Figure 4.
Rational design of the P5SM RNA element leads to generation of an orthogonal splicing cassette. (A) Fold induction of protein expression quantified by eGFP fluorescence for reporters harboring the OsP5SM or HyP5SM RNA elements within the splicing cassette. Induction with AtL5 or OsL5 was each measured in comparison to induction with LUC as a control. (B) Fluorescence of eGFP-HyP5SM without coinfiltration with DsRed2 and with subtraction of background autofluorescence for individual leaf samples. Fold induction by OsL5 relative to induction with LUC as a control is shown. Background subtraction gives a negative fluorescence value (asterisk) for leaf 4 in the absence of OsL5 induction, so the fold induction could not be determined for this sample. (C) Representative whole leaf scans of eGFP fluorescence for reporters harboring OsP5SM or HyP5SM. The right leaf halves coexpressed AtL5 or OsL5 and the left halves coexpressed LUC as a control. All samples except for the one labeled ‘No DsRed2’ also coexpressed DsRed2 on both leaf halves. (D) RT–PCR detection of spliced products for eGFP-OsP5SM and eGFP-HyP5SM upon induction with AtL5, OsL5, or LUC as a control.
This unexpected result can be rationalized in light of the proposed regulatory mechanism for how the P5SM-L5 interaction influences alternative splicing (23). Previous data have suggested that L5 protein binding to the P5SM RNA element acts to displace an exon-defining splice factor from the L2 loop, which results in exon skipping. The L2 loop of the A. thaliana P5SM RNA element has an extended purine-rich sequence that is shortened in the rice element (Supplementary Figure S3). It had been shown that substitution of this purine-rich sequence with the sequence UC leads to constitutive exon skipping (23). Thus, we hypothesized that swapping in the OsP5SM RNA element not only disrupted binding to endogenous L5 but also to the putative splice factor.
Loss of exon definition due to a reduction in splice factor binding or another mechanism involving the L2 loop could explain the increase in SP-I and high background fluorescence. To address this possibility, we converted the sequence of the OsP5SM L2 loop to match the purine-rich A. thaliana sequence and tested a construct containing the hybrid P5SM sequence, eGFP-HyP5SM (Supplementary Figure S3). This construct exhibits strong induction (∼19-fold) by OsL5, almost no induction (∼1.5-fold) by AtL5, and very low background fluorescence (Figure 4A and C). With such low background fluorescence, slight variations in this value resulted in a large standard deviation for fold induction between individual leaf samples.
Furthermore, it was determined that DsRed2, the fluorescent protein used as a normalization standard in all of the experiments (see ‘Materials and Methods’ section), contributed to the residual background fluorescence. In the absence of DsRed2, little to no background above autofluorescence was observed for eGFP-HyP5SM in the absence of OsL5 induction (Supplementary Figure S4). With the exclusion of DsRed2 and with subtraction of autofluorescence, both of which had not been done in previous experiments, it was revealed that the activation of gene expression is very strong (∼97-fold on average) (Figure 4B). Correspondingly, western blot analysis with a GFP-specific antibody to measure protein levels detected the expressed protein only in OsL5-induced samples (Supplementary Figure S5). Consistent with the protein fluorescence and immunoblot data, RT–PCR analysis revealed an almost complete switch in splicing upon induction by OsL5 but not AtL5 (Figure 4D).
DISCUSSION
Our construct designs and the associated bioinformatics estimates for genome coverage include two simplifying assumptions which impose conservative restrictions on gene context. First, we artificially limit the site of cassette insertion to within the first half of ORFs, so that even in the absence of NMD mechanisms, the exon-retained spliced product would not generate functional protein. However, premature termination codons have been shown to activate NMD as long as they are upstream of an exon junction by some distance. In N. benthamiana, introduction of the well-characterized Ls intron within the 3′-UTR of an otherwise stable mRNA triggers NMD when the stop codon is 99-nt but not 28-nt upstream of the location of intron placement (12). This observation is consistent with the general rule established in mammals, which is that NMD is usually triggered if a stop codon is ∼50–55 nt upstream of an exon junction (29). The premature termination codon in the P5SM suicide exon is 121 nt upstream of the exon junction, and the exon-retained spliced product of A. thaliana TFIIIA is subject to NMD (27). Regardless of gene context, the splicing cassette maintains the suicide exon at the same distance upstream of the exon junction, so its location probably does not need to be within the first half of the ORF to trigger NMD. However, cassette insertion at a roughly central location will help ensure that each ORF section is of sufficient length to be recognized as an exon.
Second, we only consider in-frame cassette insertion sites which allow us to generate proper splice site contexts via synonymous codon substitutions. It is very likely that the conserved AG sequence upstream of the 5′-ss does not have to be in-frame with the ORF, although this remains to be tested. Our observation that mutations in the extended 3′-ss context have no effect is consistent with the spliceosome cycle as elucidated in mammalian systems, in which the 5′-ss and branchpoint are first recognized (30). The branchpoint region of the 3′-intron can be readily identified, as it is 27-nt upstream of the 3′-ss and conforms to the plant consensus, CURAY (31). Thus, it appears that maintenance of the distance between the 3′-ss and the strong branchpoint signal is sufficient for proper splice site recognition. Taken together, it is possible that the P5SM splicing cassette will regulate the expression of any gene with the dinucleotide AG in its coding sequence, which effectively would mean that any gene can be regulated in a traceless fashion.
The splicing cassette was reliably engineered into three different ORFs, eGFP, fLUC and CYP707A3. The targeted ORFs did not originally possess introns at the site of cassette insertion, but in each case the suicide exon was spliced with no observable loss of fidelity. All of the predicted functional contexts we have tested maintain activation of alternative splicing by L5, demonstrating that the splicing cassette retains the necessary cis regulatory elements. A ∼2-fold difference in expression level was observed between the E/R and K/R contexts in the fLUC gene, regardless of the presence or absence of AtL5 (Figure 3C). Thus, there can be a slight sequence context effect that is independent of alternative splicing regulation, so we recommend testing two or three different contexts in the same ORF for optimal function. This strategy is practical because most coding sequences contain multiple candidate sites for cassette insertion, and overlap extension PCR employing different primers can be used to generate all of the desired constructs at the same time.
Essentially no fluorescence was measured for the eGFP-HyP5SM reporter in the absence of OsL5 induction (Figure 4B), even though the strong, constitutive CaMV 35S promoter was used to drive transcription and the experiment was performed in wild-type plants. This result shows that conditional splicing does not require the use of weak promoters or specialized genotypes. Furthermore, this result demonstrates that a plant-derived regulatory element could be employed in plants with almost no leaky expression. Basal activation by endogenous L5 was eliminated through rational improvements to the P5SM RNA element that were devised from our understanding of its role in alternative splicing. The splicing cassette harboring the HyP5SM RNA element was selectively activated by OsL5 over AtL5 (Figure 4A). Restoration of the purine-rich L2 loop was required to promote default exon retention, which is consistent with the observation that purine-rich motifs act as exonic splicing enhancers in A. thaliana (32). The ribosomal protein L5 for N. benthamiana has not been fully sequenced, so a direct assay has not been performed, but the little to no background induction observed in wild-type N. benthamiana supports that NbL5 also does not recognize the HyP5SM RNA element.
It is not immediately obvious which nucleotide changes to the RNA element are responsible for the discrimination in binding of OsL5 versus AtL5. There are very few differences in sequence between the original and hybrid P5SM in the regions that are homologous to 5S rRNA (the L2/P2 and P3c/L3 regions, Supplementary Figure S3). More differences are observed in the P3a/b stem, which in general exhibits higher sequence variability between representatives (23). Structural information about the P5SM-L5 complex will be needed to ascertain the molecular interactions which dictate recognition of the RNA element by the L5 protein from O. sativa but not from A. thaliana.
Because L5 is not a general splice factor and instead interacts specifically with the P5SM element, the activation of transgene expression by alternative splicing is highly robust and selective. Coexpression of OsL5 increases the levels of the reporter protein ∼97-fold (Figure 4B) but has no effect on the levels of another fluorescent protein, DsRed2, used as a normalization standard in all other experiments (Supplementary Figure S4). This result is particularly impressive given that only a single copy of the P5SM RNA element is present in the splicing cassette. The induction of protein expression is comparable to that obtained using a conditional promoter harboring six copies of the GAL4 upstream activating sequence upon dexamethasone treatment (33).
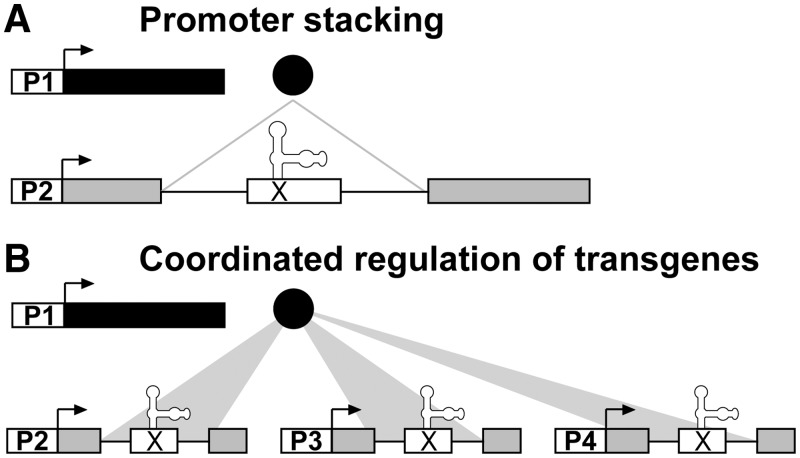
In conclusion, we have adapted a natural splicing cassette to serve as a portable regulatory element that robustly controls gene expression via alternative splicing. Gene regulation by the suicide exon is effectively traceless, as induced exon skipping affords the original ORF with at most one or two synonymous codon substitutions. This, along with the minimal requirements for sequence context, makes the regulation of any gene of interest quite facile using the P5SM splicing cassette. Thus, we have shown that conditional splicing can be a general and effective mechanism for transgene regulation. It is envisioned that the ability to combine DNA- and RNA-level regulation will enable novel strategies for controlling the expression of single genes with multiple promoters and for coordinating the expression of genes without the use of homologous promoters (Figure 5). We are currently pursuing these approaches to advance the genetic engineering of plant species for both basic research and biotechnology, and we expect that these strategies also will have application towards gene regulation in other eukaryotic organisms.
Figure 5.
Strategies for transgene regulation that combine conditional promoters and splicing cassettes. (A) Concept for promoter stacking with the P5SM splicing cassette. Expression of a single gene is regulated directly by its own promoter (P2) and indirectly by the promoter driving expression of the inducer (P1). Introduction of additional splicing cassettes would enable stacking of more than two promoters. (B) Concept for coordinated regulation of transgenes with the P5SM splicing cassette. A single conditional promoter (P1) drives expression of the inducer, which can target multiple suicide exons. The DNA sequences of the splicing cassettes can be non-homologous, as the recognition element is an RNA structure, to avoid gene silencing. Different constitutive promoters (P2–P4) are used for the individual transgenes, as expression is instead coordinated by conditional alternative splicing.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures S1–S6, Supplementary Methods and Supplementary Table S1.
FUNDING
Career Award at the Scientific Interface from the Burroughs Wellcome Fund (to M.C.H.). Funding for open access charge: Burroughs Wellcome Fund
Conflict of interest statement. The authors have filed a provisional application for a patent on the HyP5SM splicing cassette.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Patrice Kurnath, Thomas Kleist and Mary Lin for technical support.
REFERENCES
- 1.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lopez AJ. Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu. Rev. Genet. 1998;32:279–305. doi: 10.1146/annurev.genet.32.1.279. [DOI] [PubMed] [Google Scholar]
- 3.Terzi LC, Simpson GG. Regulation of flowering time by RNA processing. Curr. Top. Microbiol. 2008;326:201–218. doi: 10.1007/978-3-540-76776-3_11. [DOI] [PubMed] [Google Scholar]
- 4.Ali GS, Reddy AS. Regulation of alternative splicing of pre-mRNAs by stresses. Curr. Top. Microbiol. 2008;326:257–275. doi: 10.1007/978-3-540-76776-3_14. [DOI] [PubMed] [Google Scholar]
- 5.Gassmann W. Alternative splicing in plant defense. Curr. Top. Microbiol. 2008;326:219–233. doi: 10.1007/978-3-540-76776-3_12. [DOI] [PubMed] [Google Scholar]
- 6.Kojima S, Shingle DL, Green CB. Post-transcriptional control of circadian rhythms. J. Cell Sci. 2011;124:311–320. doi: 10.1242/jcs.065771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell MA, Haas BJ, Hamilton JP, Mount SM, Buell CR. Comprehensive analysis of alternative splicing in rice and comparative analyses with Arabidopsis. BMC Genomics. 2006;7:327. doi: 10.1186/1471-2164-7-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang BB, Brendel V. Genomewide comparative analysis of alternative splicing in plants. Proc. Natl Acad. Sci. USA. 2006;103:7175–7180. doi: 10.1073/pnas.0602039103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Padidam M. Chemically regulated gene expression in plants. Curr. Opin. Plant Biol. 2003;6:169–177. doi: 10.1016/s1369-5266(03)00005-0. [DOI] [PubMed] [Google Scholar]
- 10.Singer SD, Cox KD, Liu Z. Enhancer-promoter interference and its prevention in transgenic plants. Plant Cell Rep. 2011;30:723–731. doi: 10.1007/s00299-010-0977-7. [DOI] [PubMed] [Google Scholar]
- 11.Halpin C. Gene stacking in transgenic plants–the challenge for 21st century plant biotechnology. Plant Biotechnol. J. 2005;3:141–155. doi: 10.1111/j.1467-7652.2004.00113.x. [DOI] [PubMed] [Google Scholar]
- 12.Kertesz S, Kerenyi Z, Merai Z, Bartos I, Palfy T, Barta E, Silhavy D. Both introns and long 3'-UTRs operate as cis-acting elements to trigger nonsense-mediated decay in plants. Nucleic Acids Res. 2006;34:6147–6157. doi: 10.1093/nar/gkl737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kerenyi Z, Merai Z, Hiripi L, Benkovics A, Gyula P, Lacomme C, Barta E, Nagy F, Silhavy D. Inter-kingdom conservation of mechanism of nonsense-mediated mRNA decay. EMBO J. 2008;27:1585–1595. doi: 10.1038/emboj.2008.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weigand JE, Suess B. Tetracycline aptamer-controlled regulation of pre-mRNA splicing in yeast. Nucleic Acids Res. 2007;35:4179–4185. doi: 10.1093/nar/gkm425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Culler SJ, Hoff KG, Smolke CD. Reprogramming cellular behavior with RNA controllers responsive to endogenous proteins. Science. 2010;330:1251–1255. doi: 10.1126/science.1192128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim DS, Gusti V, Dery KJ, Gaur RK. Ligand-induced sequestering of branchpoint sequence allows conditional control of splicing. BMC Mol. Biol. 2008;9:23. doi: 10.1186/1471-2199-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooper TA. Use of minigene systems to dissect alternative splicing elements. Methods. 2005;37:331–340. doi: 10.1016/j.ymeth.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 18.Croft MT, Moulin M, Webb ME, Smith AG. Thiamine biosynthesis in algae is regulated by riboswitches. Proc. Natl Acad. Sci. USA. 2007;104:20770–20775. doi: 10.1073/pnas.0705786105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wachter A, Tunc-Ozdemir M, Grove BC, Green PJ, Shintani DK, Breaker RR. Riboswitch control of gene expression in plants by splicing and alternative 3' end processing of mRNAs. Plant Cell. 2007;19:3437–3450. doi: 10.1105/tpc.107.053645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bocobza S, Adato A, Mandel T, Shapira M, Nudler E, Aharoni A. Riboswitch-dependent gene regulation and its evolution in the plant kingdom. Gene Dev. 2007;21:2874–2879. doi: 10.1101/gad.443907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sudarsan N, Barrick JE, Breaker RR. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA. 2003;9:644–647. doi: 10.1261/rna.5090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammond MC, Wachter A, Breaker RR. A plant 5S ribosomal RNA mimic regulates alternative splicing of transcription factor IIIA pre-mRNAs. Nat. Struct. Mol. Biol. 2009;16:541–549. doi: 10.1038/nsmb.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horton RM, Cai ZL, Ho SN, Pease LR. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- 25.Hofgen R, Willmitzer L. Biochemical and genetic-analysis of different patatin isoforms expressed in various organs of potato (Solanum-Tuberosum) Plant Sci. 1990;66:221–230. [Google Scholar]
- 26.Fu Y, Bannach O, Chen H, Teune JH, Schmitz A, Steger G, Xiong L, Barbazuk WB. Alternative splicing of anciently exonized 5S rRNA regulates plant transcription factor TFIIIA. Genome Res. 2009;19:913–921. doi: 10.1101/gr.086876.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoine M, Ohto MA, Onai K, Mita S, Nakamura K. The lba1 mutation of UPF1 RNA helicase involved in nonsense-mediated mRNA decay causes pleiotropic phenotypic changes and altered sugar signalling in Arabidopsis. Plant J. 2006;47:49–62. doi: 10.1111/j.1365-313X.2006.02771.x. [DOI] [PubMed] [Google Scholar]
- 28.Schwartz SH, Silva J, Burstein D, Pupko T, Eyras E, Ast G. Large-scale comparative analysis of splicing signals and their corresponding splicing factors in eukaryotes. Genome Res. 2008;18:88–103. doi: 10.1101/gr.6818908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maquat LE, Tarn WY, Isken O. The pioneer round of translation: features and functions. Cell. 2010;142:368–374. doi: 10.1016/j.cell.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burge CB, Tuschl T, Sharp PA. Splicing of precursors to mRNAs by the spliceosomes. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. 2nd edn. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1999. pp. 525–560. [Google Scholar]
- 31.Schuler MA. Splice site requirements and switches in plants. Curr. Top. Microbiol. 2008;326:39–59. doi: 10.1007/978-3-540-76776-3_3. [DOI] [PubMed] [Google Scholar]
- 32.Egoavil C, Marton HA, Baynton CE, McCullough AJ, Schuler MA. Structural analysis of elements contributing to 5' splice site selection in plant pre-mRNA transcripts. Plant J. 1997;12:971–980. doi: 10.1046/j.1365-313x.1997.12050971.x. [DOI] [PubMed] [Google Scholar]
- 33.Aoyama T, Chua NH. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 1997;11:605–612. doi: 10.1046/j.1365-313x.1997.11030605.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.