Abstract
Plant specific SGS3-like proteins are composed of various combinations of an RNA-binding XS domain, a zinc-finger zf-XS domain, a coil–coil domain and a domain of unknown function called XH. In addition to being involved in de novo 2 (IDN2) and SGS3, the Arabidopsis genome encodes 12 uncharacterized SGS3-like proteins. Here, we show that a group of SGS3-like proteins act redundantly in RNA-directed DNA methylation (RdDM) pathway in Arabidopsis. Transcriptome co-expression analyses reveal significantly correlated expression of two SGS3-like proteins, factor of DNA methylation 1 (FDM1) and FDM2 with known genes required for RdDM. The fdm1 and fdm2 double mutations but not the fdm1 or fdm2 single mutations significantly impair DNA methylation at RdDM loci, release transcriptional gene silencing and dramatically reduce the abundance of siRNAs originated from high copy number repeats or transposons. Like IDN2 and SGS3, FDM1 binds dsRNAs with 5′ overhangs. Double mutant analyses also reveal that IDN2 and three uncharacterized SGS3-like proteins FDM3, FDM4 and FDM5 have overlapping function with FDM1 in RdDM. Five FDM proteins and IDN2 define a group of SGS3-like proteins that possess all four-signature motifs in Arabidopsis. Thus, our results demonstrate that this group of SGS3-like proteins is an important component of RdDM. This study further enhances our understanding of the SGS3 gene family and the RdDM pathway.
INTRODUCTION
In many eukaryotes, RNA-directed DNA methylation (RdDM) is often associated with transcriptional silencing (TGS) and is considered as an essential mechanism to maintain genome stability and to suppress the proliferation of transposable elements (1,2). A key component of RdDM is ∼20–24 nt small interfering RNA derived from transposon or repetitive sequences (rasiRNA) that associates with the argonaute (AGO) proteins to guide de novo cytosine methylation at its homolog loci (1,2). In Arabidopsis thaliana, the generation of 24-nt rasiRNAs depends on the RNA-dependent RNA polymerase 2 (RDR2), dicer-like 3 (DCL3), the SNF2-like chromatin-remodeling factor classy 1 (CLSY1) and the plant specific DNA-dependent RNA polymerase IV (Pol IV) (3–6). Pol IV associates with siRNA-generating loci and is thought to generate single-stranded RNAs (ssRNAs) from these loci, which are presumably converted into double-stranded RNAs (dsRNAs) by RDR2 and subsequently processed by DCL3 into 24 nt rasiRNA duplex (3–7). CLSY1 is required for the correct localization of Pol IV and RDR2 (8).
After generation, one strand of siRNA duplexes is loaded into AGO4, AGO6 or AGO9 (9–11). Presumably through base-pairing between siRNA and Pol V-dependent transcripts and/or physical interaction with NRPE1, which is the largest subunit of Pol V, AGO4 is guided to targets to recruit domains rearranged methyltransferase 2 (DRM2) to catalyze de novo cytosine DNA methylation at symmetric CG, CHG (H is adenine, thymine or cytosine) and asymmetric CHH context (12–14). It was recently shown that Pol II might recruit AGO4, Pol IV and Pol V to chromatin through its transcripts or transcription activity at intergenic low copy number loci (7). Additional RdDM components include suppressor of Ty insertion 5-like (SPT5L, also known as KTF1), defective in RNA-directed DNA methylation 1 (DRD1), defective in meristem silencing 3 (DMS3) and RNA-directed DNA methylation 1 (RDM1) (15–21). SPT5L interacts with both Pol V transcripts and AGO4 and is thought to act downstream of the RdDM pathway (16,18), whereas DRD1, DMS3 and RDM1 form a DDR complex that is required for the generation of Pol V-dependent transcripts (17,21).
The plant specific SGS3 gene family encodes proteins containing at least one of the following protein domains: XS, XH and zf-XS that were named after Arabidopsis SGS3 and its rice homolog X1 (22,23). Among these protein domains, the XS domain is an RNA-binding domain, zf-XS domain is a C2H2 type zinc finger domain and XH domain refers to X-homolog domain with unknown function (22). In addition to these protein domains, some of SGS3-like proteins also contain a coil–coil domain localized between the XS and XH domains (15,24). Arabidopsis encodes 14 SGS3-like proteins including SGS3 and involved in de novo 2 (IDN2, also called RDM12) (15,23,24). While SGS3 is an essential component of post transcriptional gene silencing (PTGS) required for the production of sense-transgene-induced siRNAs and trans-acting siRNAs (23,25), IDN2/RDM12 is for RdDM and required for TGS (15,24). Both SGS3 and IDN2 bind dsRNAs with a 5′ overhang (15,26). However, the functions of remaining 12 SGS3-like proteins are still unknown.
Here, we identify five SGS3 homologs, factor of DNA methylation (FDM) 1, 2, 3, 4 and 5, as important components of RdDM. Using a combination of transcriptome co-expression analysis and reverse genetics, we found that FDM1 and FDM2 display a highly correlated expression pattern with known components of RdDM. Both FDM1 and FDM2 act redundantly in DNA methylation, accumulation of Pol V-dependent rasiRNAs and silencing of RdDM loci. However, FDM1 and FDM2 are not required for the accumulation of Pol V- and Pol II-dependent scaffold transcripts. Furthermore, we show that IDN2 and three uncharacterized SGS3-like proteins FDM3, FDM4 and FDM5 have overlapping function with FDM1 in RdDM. FDM2 also have redundant function with IDN2 in RdDM. These findings broaden our knowledge of RdDM and the function of the SGS3 gene family.
MATERIALS AND METHODS
Plant materials
The T-DNA insertional mutants, fdm1-1 (SALK_075813) and fdm2-1 (SAIL_291_F01) and idn2-3 (Salk_152144) were obtained from the ABRC Stock Center (www.arabidopsis.org). The T-DNA insertions were identified through combination of gene-specific primers and T-DNA left border primer (primers FDM1RP, FDM1LP and LBa1 for fdm1-1; primers FDM2RP, FDM2LP and LB3 for fdm2-1; primers IDN2RP, IDN2LP and LBa1 for idn2-3; Supplementary Table S2). The fdm1-1 fdm2-1, fdm1-1 idn2-3, fdm2-1 idn2-3 mutants were constructed by crossing single mutants. nrpe1-1 (27), dcl3-1 (6) and the myc-AGO4 transgenic line were kindly gifts from Dr Xuemei Chen. Myc-AGO4 is in the Ler genetic background, whereas other mutants are in the Columbia genetic background.
Phylogenetic analyses
Protein sequences for 14 Arabidopsis SGS3-like proteins were obtained from the Arabidopsis website (http://www.arabidopsis.org). Full-length protein sequences of 14 SGS3-like proteins were aligned using CLUSTALW at The Biology Work Bench (http://workbench.sdsc.edu/). Phylogenetic analysis was done by the unrooted neighbor-joining method. To assess the degree of reliability for each branch on the tree, bootstrap confidence values of each node were calculated with 1000 replicates using PAUP 4.0 (http://paup.csit.fsu.edu/).
DNA methylation assays
Genomic DNA was extracted from flowers and digested overnight with different methylation-sensitive restriction enzyme (HaeIII, AvaII, HpaII and MspI) or 1 h with McrBC. Approximately 5% of the digested DNA was subsequently used for PCR analysis of AtSN1, IGN5, FWA SINE and siR02. The undigested genomic DNA was amplified simultaneously as loading controls. PCR conditions were: 94°C for 30 s, 54°C for 30 s, 72°C for 1 min, 32 cycles and 72°C for 10 min. For Southern blotting, 5 µg of genomic DNA treated with HaeIII, HpaII and MspI overnight was resolved in 1.2% agarose gel and transferred to Hybond-N+ membranes. 5S rDNA, MEA-ISR and AtMU1 Southern blotting were carried out as described (18,27,28). Primers used for DNA methylation analyses were listed in the Supplementary Table S2. The primer information was obtained from references (14,18,27,28).
RT–PCR analysis
Total RNA was extracted from flowers using Trizol reagent (Sigma). After DNase treatment, 2–5 µg of total RNA was used to synthesize cDNA with SuperScript III (Invitrogen) using oligo-dT or gene-specific primers. The diluted cDNA reaction mixture was used for RT–PCR of AtSN1, siR02 and 5s rRNA spacer as previously described (7,29). The constitutively expressed UBQ5 was used as an internal control. The cDNA reaction mixture without reverse transcriptase was used in PCR amplification to determine the absence of DNA contamination. Pol II- and Pol V-dependent transcripts were detected by RT–PCR according to (14). Primers used for RT–PCR analysis are listed in the Supplementary Table S2.
siRNA and miRNA detection
RNA isolation and hybridization were performed according to the method described by (30). siR1003, AtSN1, AtCopia2, SimpleHAT2, siR02, Cluster4 and TR2558 were detected using 5′-end-labeled (32P) antisense LNA oligonucleotides (7). Probe and primer sequences are listed in the Supplementary Table S2.
Immunolocalization
Leaves from 28-day-old plants were harvested and the immunolocalization experiments were performed as described (8,31).
RNA binding assay
The RNA and DNA binding assays were performed as previously described (32). GST and a truncated form of FDM1 (GST-FDM1ΔZH, amino acids 114–498) fused to GST were expressed in E. coli BL21 and purified as described by (33). The templates for RNA1, 2 and 3 were produced by PCR using primers RNA1F/1R, RNA2F/2R and RNA1F/3R, respectively. The template for RNA 1, 2 and 3 is the B region of the AtSN1 locus. Primers RNA1F, RNA2R and RNA3R contain the T7 promoter. The RNAs were synthesized by in vitro transcription with T7 RNA polymerase at the presence or absence of [α32P] UTP. RNA1 was used as ssRNAs in the binding assay. RNA1/RNA2 were annealed to generated dsRNAs with 5′ overhangs at both ends. RNA3 is a dsRNA with 3′ overhangs at both ends. Annealing was performed in the annealing buffer [10 mM Tris–HCl (pH 8.0), 20 mM NaCl, 1 mM EDTA (pH 8.0)] by incubating RNAs at 95°C for 5 min and then gradually cooling to room temperature. Sequences for primers are listed in the Supplementary Table S2.
RESULTS
At1g15910 and At4g00380 co-expressed with genes in the RdDM pathway
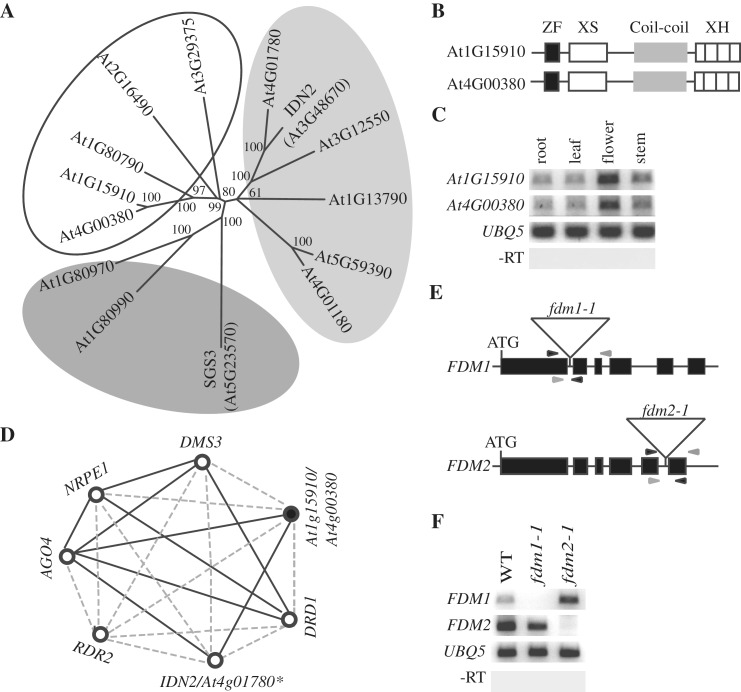
Phylogenic analyses using full-length protein sequences assigned 14 Arabidopsis SGS3 family members into three subfamilies (Figure 1A; 34). SGS3 from the first subgroup and IDN2 from the second subgroup have been shown to act in PTGS and TGS, respectively (15,23,24). However, no members from the third subgroup were studied. To extend our understanding of SGS3-like proteins, we selected At1g15910 and At4g00380 from subgroup 3 for functional characterization as they contain the zf-XS, XS, XH and coil–coil domains (Figure 1B). The protein sequences of At1g15910 and At4g00380 are highly similar (93% identities and 96% similarities; Supplementary Figure S1), indicating that they might have redundant function. This was supported by the similar expression pattern between At1g15910 and At4g00380 in leaves, flowers, stem and roots (Figure 1C). However, they displayed altered expression levels in leaves, flowers, stem and roots, suggesting that their expression may be developmentally regulated (Figure 1C).
Figure 1.
FDM1 and FDM2 are putative components of RdDM pathway. (A) Unrooted neighbor-joining phylogenies based on full-length amino acid sequences of 14 Arabidopsis SGS3 like proteins. Bootstrap values were given for branch node. Dark gray, subfamily 1; Light gray, subfamily 2; White, subfamily 3. (B) A scheme of protein structures of At1G15910 (FDM1) and At4G00380 (FDM2). Black box, the zf-XS domain; open box, the XS domain; Gray box, the coil–coil domain; hatched box, the XH domain. (C) RT–PCR analysis of At1G15910 and At4G00380 expression in root, leaf, flower and stem. Amplification of UBIQUITIN5 (At3g26650; UBQ5) with or without reverse transcription (RT) is shown as a control. (D) Correlation among several genes involved in RdDM pathway and FDM1/FDM2. Black circle, FDM1/FDM2; Open circle: genes involved in RdDM. solid black line, r > 0.9; dot line: 0.9 > r > 0.830. Asterisk: Because of cross hybridization of IDN2 andAt4g01780 in the microarray experiment, they were considered as a single gene during co-expression analysis. (E) Diagrams of T-DNA-insertion in fdm1-1 and fdm2-1, respectively. Black box, coding region; solid black line, intron; open triangle, T-DNA insertion site. Gray arrowheads, primer used for T-DNA genotyping; Black arrowheads, primer used for RT–PCR analysis. (F) RT–PCR analysis of FDM1 and FDM2 expression in fdm1-1, fdm2-1 and Col (wild-type: WT). Amplification of UBQ5 with or without RT is shown as a control.
To infer the functions of At1g15910 and At4g00380, we searched for their co-expression genes within the ATTED-II developmental expression data set using a co-expression analysis program at the RIKEN PRIMe website (35,36). This search was based on the hypothesis that genes involved in a particular biological process often share regulatory systems thus having a similar expression pattern (37). Because of cross-hybridization between At1g15910 and At4g00380 in the microarray experiments, they were considered as a single gene in the analysis. The results showed that At1g15910/At4g00380 had a very strong correlation with AGO4, NRPE1, DRD1, DMS3, IDN2 and RDR2 (correlation coefficiency r > 0.83; Figure 1D). These RdDM genes were coordinately expressed with At1g15910/At4g00380 in roots, embryos, siliques, leaves, stems and flowers (Supplementary Figure S2), according to the Arabidopsis eFP-Browser, which was developed to interpret gene expression data of Arabidopsis (38). At1g15910/ At4g00380 also had a considerably high correlation with DCL3, NRPD1 and DRM2 (0.68 < r < 0.83; Supplementary Table S1) as their expression was overlapped in various tissues and at different development stages (Supplementary Figure S2). Altogether, these results showed the correlation between At1g15910/At4g00380 and known genes involved the RdDM pathway, and therefore, suggested their potential role in RdDM. We named At1g15910 and At4g00380 factor of DNA methylation 1 (FDM1) and factor of DNA methylation 2 (FDM2), respectively, because we subsequently showed that they acted in RdDM (see below).
FDM1 and FDM2 have redundant and essential roles in RdDM
To examine the function of FDM1 and FDM2, two T-DNA insertion lines, SALK_075378 for FDM1 (39) and SAIL_291_F01 for FDM2 (40) were obtained from the Arabidopsis stock center (http://www.arabidopsis.org) and further characterized. As a first step, plants homozygous for SALK_075378 (named fdm1-1) and SAIL_291_F01 (named fdm2-1) were identified by PCR genotyping (Supplementary Figure S3). Sequence analysis of the flanking regions of the T-DNA revealed that fdm1-1 contained a T-DNA insertion in the first intron (949 bp downstream from the ATG site) of FDM1 and fdm2-1 harbored a T-DNA insertion in the fifth intron (2252 bp downstream from the ATG site) of FDM2 (Figure 1E). Using RT–PCR analysis, we failed to detect the transcripts of FDM1 and FDM2 in fdm1-1 and fdm2-1 (Figure 1F), respectively, indicating that they are potentially null alleles of FDM1 and FDM2. As FDM1 and FDM2 might have redundant functions, we constructed a fdm1-1 fdm2-1 double mutant by crossing the two respective single mutant lines. No obvious phenotypic abnormalities were observed in fdm1-1, fdm2-1 and fdm1-1 fdm2-1 (Supplementary Figure S4).
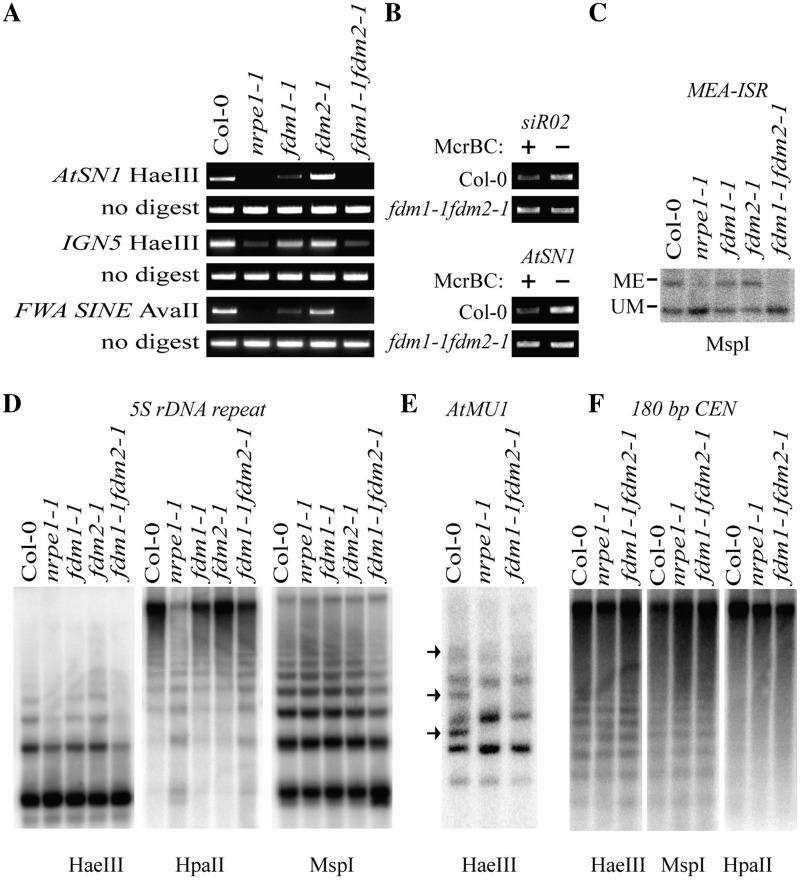
To evaluate whether FDM1 and FDM2 have roles in the RdDM pathway, we examined DNA methylation status at known RdDM-regulated retrotransposon such as AtSN1 and ING5 in fdm1-1, fdm2-1, fdm1-1 fdm2-1 and Arabidopsis ecotype Columbia (wild-type control; WT) plants by using methylation sensitive HaeIII restriction enzyme digestion followed by PCR that identifies CHH methylation. HaeIII cannot cleave ATSN1 and ING5 DNAs from WT due to DNA methylation at its cleavage site (14,41). A reduction in DNA methylation will cause AtSN1 and ING5 DNAs to be less resistant to HaeIII cleavage, resulting in reduced or undetectable PCR products (14,41). As shown in Figure 2A, fdm1-1 but not fdm2-1 showed a moderate reduction of DNA methylation at AtSN1 and ING5 loci relative to WT. A reduction of DNA methylation at short interspersed repetitive elements upstream of FWA gene (FWA SINE) in fdm1-1 but not in fdm2-1 was also detected by methylation sensitive AvaII enzyme digestion analysis (Figure 2A) (27). The reduction of DNA methylation in fdm1-1 but not fdm2-1 may be correlated with the reduction of FDM2 transcript abundance in fdm1-1 and increased FDM1 transcript levels in FDM2-1 (Figure 1F). Introducing the WT FDM1 genomic DNA into fdm1-1 fully recovered the DNA methylation levels at the AtSN1 locus (Supplementary Figure S5A), demonstrating that the reduction in DNA methylation in fdm1-1 is due to FDM1 loss-of-function. The restriction digestion patterns of AtSN1, ING5 and FWA SINE DNAs in fdm1-1 fdm2-1 were similar to nrpe1-1, indicating a strong loss of DNA methylation at these loci (Figure 2A). The reduction of DNA methylation at AtSN1 locus in fdm1-1 fdm2-1 was further confirmed by McrBC enzyme digestion followed by PCR (Figure 2B). The McrBC enzyme cuts methylated but not unmethylated DNA. A reduction in DNA methylation will result in increased PCR products after McrBC treatment. This assay also revealed a reduction in DNA methylation at the siR02 locus in fdm1-1 fdm2-1 (Figure 2B). We further examined the DNA methylation status of 5S rDNA, AtMU1 and MEA-ISR using the methylation-sensitive restriction enzyme HaeIII, HpaII (for CG and CHG methylation) and MspI (for CG methylation) followed by Southern blotting (18,27,28). A strong reduction in DNA methylation at 5S rDNA, AtMU1 and MEA-ISR loci comparable to nrpe1-1 was observed in fdm1-1 fdm2-1 but not in fdm1-1 and fdm2-1 (Figure 2C–E). Next, we examined the methylation status of the highly repetitive 180-bp centromeric repeat that is not an RdDM target (11). The DNA methylation at this locus showed no obvious alteration in fdm1-1 fdm2-1 and nrpe1-1 compared with WT (Figure 2F). This indicated that the function of FDM1 and FDM2 in DNA methylation is rasiRNA dependent. To confirm that the strong reduction of DNA methylation in fdm1-1 fdm2-1 is due to lack of both FDM1 and FDM2, we introduced the WT FDM1 or FDM2 genomic DNA into fdm1-1 fdm2-1. Two randomly chosen transgenic fdm1-1 fdm2-1 lines harboring the FDM1 transgene showed comparable DNA methylation levels at AtSN1 and ING5 with WT and fdm2-1, while two fdm1-1 fdm2-1 lines containing the FDM2 transgene have similar DNA methylation levels to fdm1-1 (Supplementary Figure S5B). These results demonstrated that FDM1 and FDM2 act redundantly in RdDM.
Figure 2.
FDM1 and FDM2 play redundant and essential roles in RdDM. (A) Reduced DNA methylation at AtSN1, IGN5 and FWA SINE in fdm1-1 fdm2-1. HaeIII-digested genomic DNAs from various genotypes were used for PCR amplification of AtSN1 and IGN5, whereas AvaII-treated genomic DNAs were used for the amplification of FWA SINE. Amplifications of undigested genomic DNA are used as loading controls. Col: WT plants. (B) Reduced DNA methylation at AtSN1 and siR02 loci in fdm1-1 fdm2-1. McrBC-digested and -undigested DNAs (control) were used for the amplification of AtSN1 and siR02. (C) Reduced DNA methylation at MEA-ISR in fdm1-1 fdm2-1. MspI-digested genomic DNAs from various genotypes were probed for MEA-ISR. Bands representing methylated (ME) or unmethylated (UM) DNA are indicated. (D) Reduced DNA methylation at 5S rDNA locus in fdm1-1 fdm2-1. HaeIII, HpaII or MspI-digested genomic DNAs from various genotypes were probed for 5S rDNA. (E) Reduced DNA methylation at AtMU1 locus in fdm1-1 fdm2-1. HaeIII-digested genomic DNAs were probed for AtMU1. The three undigested bands presented in Col (WT) but not in nrpe1-1 and fdm1-1 fdm2-1 were indicated by arrows. (F) Unaffected DNA methylation at 180 bp centromeric repeats in fdm1-1 fdm2-1. Following HpaII, MspI or HaeIII treatment, genomic DNAs from various genotypes were probed for 180-bp centromeric repeats.
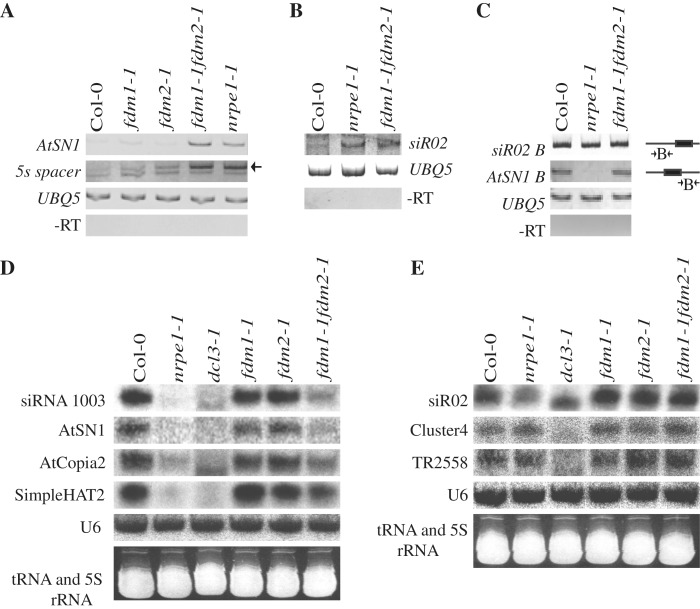
Next, we examined the expression levels of AtSN1, 5s rRNA spacer and siR02 in fdm1-1, fdm2-1 and fdm1-1 fdm2-1, nrpe1-1 and WT by RT–PCR. Their transcripts in fdm1-1 fdm2-1 but not in fdm1-1 and fdm2-1 were significantly increased to levels comparable to nrpe1-1 (Figure 3A and B). These results revealed that the reduction of DNA methylation in fdm1-1 fdm2-1 is correlated with derepression of RdDM target loci.
Figure 3.
FDM1 and FDM2 prompt the accumulation of type I rasiRNAs and are required for silencing of RdDM loci. (A and B) Enhanced transcription levels of AtSN1, 5S rRNA spacer and siR02 in fdm1-1 fdm2-1. Transcripts of RdDM targets were detected by RT–PCR. For 5S rRNA spacer transcripts, the band (∼210 bp) indicated by an arrow corresponds to the silenced transcripts in Col (WT). (C) Unaffected Pol II- and Pol V-dependent non-coding transcripts at flanking region of ATSN1 and siR02 in fdm1-1 fdm2-1. The transcripts were detected by strand-specific RT–PCR. The positions of amplified region by RT–PCR are indicated in the diagram on the right. Amplification of UBQ5 with or without RT is served as a control. (D) Reduced accumulation of type I rasiRNAs in fdm1-1 fdm2-1. (E) Unaffected accumulation of type II rasiRNAs in fdm1-1 fdm2-1. Various rasiRNAs were detected by northern blotting. The controls U6 rRNA blots and ethidium bromide-stained tRNAs were shown below the corresponding rasiRNA blots.
The levels of Pol V-dependent rasiRNAs are reduced in fdm1-1 fdm2-1
Based on their dependence on Pol V and Pol IV, rasiRNAs are classified into two types (27). The accumulation of type I rasiRNAs that are derived from highly repetitive DNA sequences, including AtSN1, siR1003 (from 5S rDNA), AtREP2, SimpleHAT2 and AtCopia2, depends on both Pol V and Pol IV, whereas the levels of type II rasiRNAs generated from low-copy number DNA repeats, such as siR02, Cluster4, TR2558, Cluster2 and soloLTR, require Pol IV but not Pol V (27).
We examined the accumulation of both type I rasiRNAs and type II rasiRNAs in fdm1-1, fdm2-1 and fdm1-1 fdm2-1 by northern blotting. The accumulation of both type I rasiRNAs (AtSN1, siRNA 1003, Atcopia and SimpleHAT2) and type II rasiRNAs (siR02, Cluster4, TR2558) was reduced in dcl3-1 but not in fdm1-1 and fdm2-1 relative to WT (Figure 3D and E). Like in nrpe1-1, the accumulation of type I but not type II rasiRNAs was significantly reduced in fdm1-1 fdm2-1 compared with WT (Figure 3D and E). These results suggested that FDM1 and FDM2 act redundantly to promote the accumulation of type I rasiRNAs but not type II rasiRNAs. We next tested whether FDM1 and FDM2 were involved in the accumulation of microRNAs (miRNAs). However, the levels of DCL1-dependent miR172 and miR173 in fdm1-1, fdm2-1 and fdm1-1 fdm2-1 were similar to those in WT (Supplementary Figure S6).
FDM1 and FDM2 are not required for the localization of NRPD1, RDR2, NRPE1 and AGO4 and for the accumulation of Pol V- or Pol II-dependent non-coding transcripts
To explore the role of FDM1 and FDM2 in RdDM, we examined the nuclear localization of NRPD1, RDR2, NRPE1 and AGO4 in fdm1-1 fdm2-1. As shown in Figure 4, in both WT and fdm1-1 fdm2-1 nuclei NRPD1 displayed punctate foci signals in the nucleoplasm. In contrast, as previously reported (31,42), RDR2, NRPE1 and AGO4 showed a round-shaped nucleolar signal in addition to puncta or diffuse signals outside the nucleolus both in WT and fdm1-1 fdm2-1 (Figure 4). Thus, the fdm1-1 and fdm2-1 double mutations have no effects on the localization of the RdDM players NRPD1, NRPE1, RDR2 and AGO4.
Figure 4.
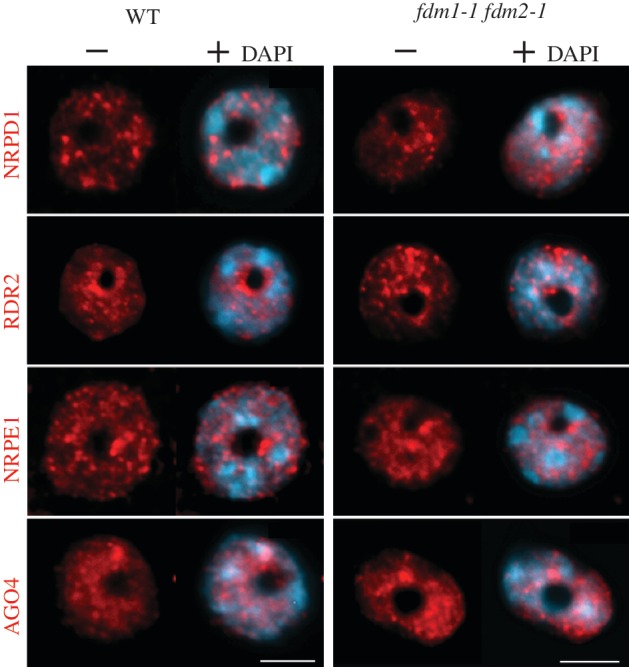
The fdm1-1 and fdm2-1 double mutations have no effects on RdDM proteins nuclear localization. NRPD1, RDR2, NRPE1 and AGO4. Peptide antibodies specifically recognizing native NRPD1, RDR2, NRPE1 or AGO4 (in red) were used to perform immunolocalization experiments in Arabidopsis leaf nuclei from ecotype Columbia (WT) and fdm1-1 fdm2-1 mutant line. DNA was counterstained with DAPI. Scale bar corresponds to 5 µm.
Next, we tested the requirement of FDM1 and FDM2 for the accumulation of Pol V- or Pol II-dependent non-coding transcripts that serve as scaffolds to recruit AGO4–siRNA complex to chromatin (7,43). RT–PCR analyses showed that the Pol V-dependent transcripts at AtSN1 locus (interval B) and Pol II-dependent transcripts at siR02 locus (interval B) were not affected in fdm1-1 fdm2-1 (Figure 3C).
FDM1 binds dsRNAs with 5′ overhangs
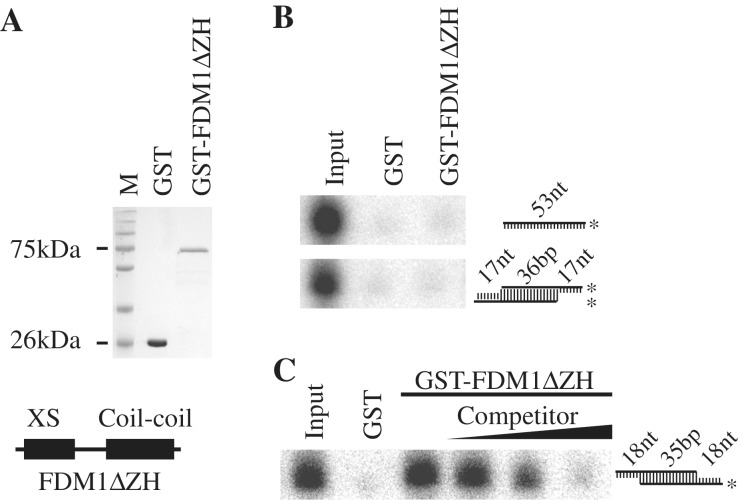
As the SGS3 and IDN2 have been shown to bind dsRNAs, we tested whether FDM1 is an RNA-binding protein using a GST-pull down assay. Because the truncated SGS3 and IDN2 proteins containing the XS and coil–coil domains are able to bind dsRNAs, we expressed a truncated version of FDM1 lacking the zinc finger and XH domain fused with GST tag at its N-terminus (GST-FDM1ΔZH) and a GST control protein in E. coli. The GST-FDM1ΔZH and GST proteins were purified with glutathione beads (Figure 5A). We prepared various radioactive-labeled RNA species including ssRNAs, dsRNAs with 3′ overhangs and dsRNAs with 5′ overhangs (Figure 5B and C). These probes were incubated with the glutathione beads containing GST-FDM1ΔZH or GST alone. GST-FDM1ΔZH retained radioactive 35 bp dsRNAs with 18 nt 5′ overhangs at each end but not 53 nt ssRNAs and a 36 bp dsRNA with 17 nt 3′ overhang at each end, whereas GST alone did not bind any RNA species (Figure 5B and C). Furthermore, addition of unlabeled dsRNAs of the same sequence efficiently reduced the binding of radioactive probe by GST-FDM1ΔZH (Figure 5C). These results demonstrated that FDM1 binds dsRNAs with 5′ overhangs.
Figure 5.
FDM1 binds dsRNAs with 5′ overhangs. (A) The two purified proteins used in the binding assay, GST and GST-FDM1ΔZH (truncated FDM1 containing XS and SMC domain) were resolved in SDS–PAGE gel and stained with coomassie Blue. The protein molecular weights are indicated on the right. (B and C) RNA-binding assays of FDM1 with various probes. The structure of various probes is shown on the right. Asterisk indicates radioactive labeled RNA strand. Approximately 50 µg of protein was used for the binding assay. For dsRNAs with 5′ overhang, 1×, 10× and 150× unlabeled RNAs of the same sequence were used for the competition assay.
RNA-mediated in vitro AGO4–FDM1 interaction
We next tested whether FDM1 interacts with AGO4 and RDR2 by in vitro protein pull-down assay in order to gain insight on the function of FDM1 in RdDM. A full-length FDM1 fused with a GST-tag at its N-termini was expressed in E. coli and purified with glutathione beads (Supplementary Figure S7). The glutathione beads conjugated with GST-FDM1 were incubated proteins extracts containing HA-RDR2 or MYC-AGO4. Western blot detected the enrichments of MYC-AGO4 but not HA-RDR2 in the GST-FDM1 complex (Supplementary Figure S7). In contrast, the control GST protein alone failed to pull down MYC-AGO4 (Supplementary Figure S7). Because both AGO4 and FDM1 are RNA binding proteins, we tested whether the interaction is RNA-mediated. RNase A treatment abolished AGO4–FDM1 interaction (Supplementary Figure S7).
FDM1 and FDM2 have overlapping functions with IDN2 in the RdDM pathway
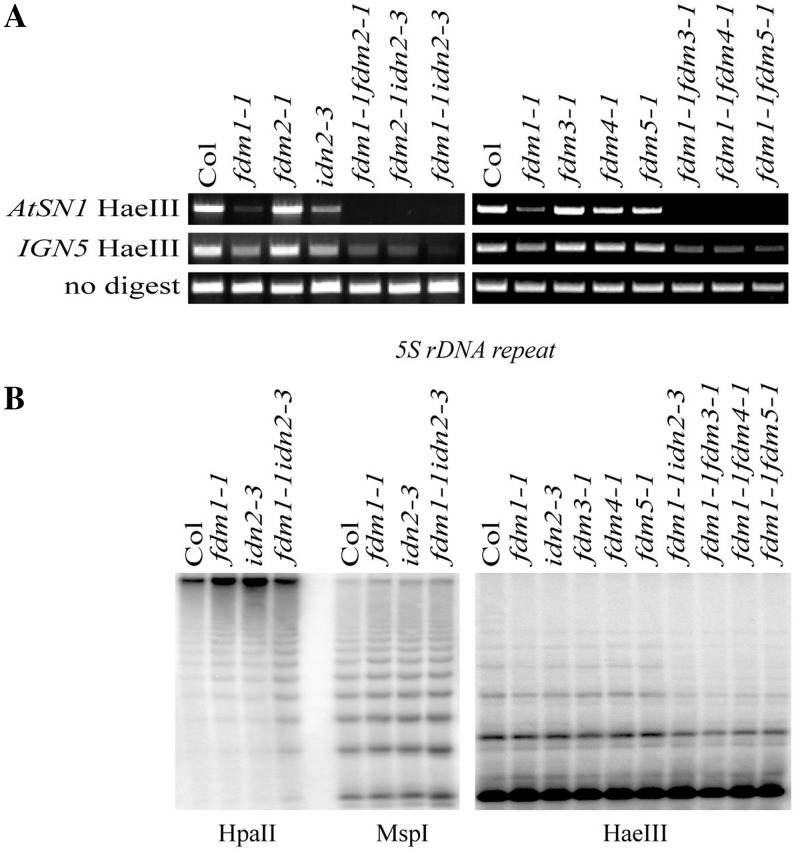
Because FDM1 and FDM2 protein sequences share considerable similarities with that of IDN2/RDM12 (∼60%) and all of them are involved in RdDM, we asked whether they have overlapping functions. We obtained a T-DNA insertion line Salk_152144 for IDN2/RDM12 from the Arabidopsis stock center and identified homozygous mutants by PCR genotyping (Supplementary Figure S8). We named this line idn2-3. The transcript levels of IDN2 were reduced in idn2-3 (Supplementary Figure S8C), resulting in a moderate reduction in DNA methylation at AtSN1 and ING5 loci (Figure 6A). We constructed two double mutants, fdm1-1 idn2-3 and fdm2-2 idn2-3 by crossing single mutants and analyzed DNA methylation status at AtSN1 and ING5 loci. Like fdm1-1 fdm2-1, fdm1-1 idn2-3 and fdm2-1 idn2-3 showed strong reduction in DNA methylation compared with each of single mutants (Figure 6A). It was noticed that the fdm1-1 idn2-3 showed a stronger reduction in DNA methylation at IGN5 locus than fdm1-1 fdm2-1 and fdm2-1 idn2-3. This result may be related to the reduced expression of FDM2 in the fdm1-1 genetic background (Figure 1F). fdm1-1 idn2-3 also displayed reduced DNA methylation at 5S rDNA locus relative to fdm1-1 and idn2-3 (Figure 6B).
Figure 6.
FDM1 has overlapping functions with IDN2, FDM3, FDM4 and FDM5. (A) DNA methylation levels at AtSN1 and IGN5 loci in various genotypes. HaeIII-digested genomic DNAs were used for PCR amplification of ATSN1 and ING5. Amplification of undigested DNAs was used as loading controls. (B) DNA methylation at 5S rDNA locus in various genotypes.
FDM3, FDM4 and FDM5 act redundantly with FDM1 in RdDM
IDN2/RDM12, FDM1 and FDM2 have three additional homologs At3G12550 (subfamily 2), At1g13790 (subfamily 2) and At1g80790 (subfamily 3) that contain all four-signature motifs of SGS3 protein family in Arabidopsis. We named these proteins FDM3, FDM4 and FDM5 respectively and tested whether they have functions in RdDM. Homozygous T-DNA insertion lines Salk_020841 for At3G12550 (fdm3-1), Salk_008738 (fdm4-1) for At1G13790 and Salk_052192 (fdm5-1) for At1G80790 were obtained from Arabidopsis center (Supplementary Figure S8). No transcripts for FDM3, FDM4 were detected in fdm3-1 and fdm4-1, respectively, whereas the abundance of FDM5 transcripts was reduced significantly in fdm5-1 (Supplementary Figure S8). The DNA methylation status of ATSN1, ING5 and 5S rDNA loci in fdm3-1, fdm4-1 and fdm5-1 showed no alteration relative to WT. We next tested whether FDM3, FDM4 and FDM5 have redundant functions with FDM1. In facts, the DNA methylation contents of ATSN1, ING5 and 5S rDNA loci are strongly reduced in fdm1-1 fdm3-1, fdm1-1 fdm4-1, fdm1-1 fdm5-1 compared with each of single mutants and WT. In fdm1-1 fdm3-1, fdm1-1 fdm4-1 and fdm1-1 fdm5-1 expressing the FDM3, FDM4 and FDM5 transgenes under the control of their native promoters, respectively, the DNA methylation content of ATSN1 and ING5 is comparable with that in fdm1-1, indicating that lack of FDM3, FDM4 or FDM5 is responsible for the enhanced DNA methylation defects in the double mutants (data not shown).
DISCUSSION
The SGS3-like genes encode a large uncharacterized protein family. In this study, through a combination of transcriptome co-expression analysis, reverse genetics and biochemical assays, we show that two SGS3-like proteins FDM1 and FDM2 from Arabidopsis are essential components of gene silencing triggered by small RNAs. FDM1 and FDM2 share high similarity and lack of both of them causes great reduction in DNA methylation levels and Pol V-dependent rasiRNA accumulation, resulting in release of TGS. These results demonstrate that FDM1 and FDM2 have essential and redundant roles in the RdDM pathway.
Co-expression analysis revealed that AGO4, NRPE1, DRD1, DMS3, IDN2/RDM12, FDM1/FDM2 DCL3 and RDR2 are highly correlated with each other (r > 0.76; Figure 1D and Supplementary Table S1). NRPD1 and DRM2 also display considerable correlation with these genes (r > 0.6 and r > 0.5; respectively; Supplementary Table S1). These results are supported by their coordinated high expression at DNA-replication active tissues such as inflorescence meristem, shoot meristem and developing embryo (Supplementary Figure S2), which agrees with their role in directing de novo DNA methylation (1,2). The correlation among genes involved in RdDM indicates that they may share a common regulatory system and tend to be co-expressed. Consequently, searching for co-expressed genes combined with reverse genetic analysis could be a powerful tool to identify novel genes that are involved in RdDM, especially those with functional redundancy.
How do FDM1 and FDM2 function in RdDM? They appear not to be required for the correct localization of NRPD1, RDR2, NRPE1 and AGO4, as these proteins have similar localizations in fdm1-1 fdm2-1 as in WT (Figure 4). Like IDN2 and SGS3 (15,26), FDM1 binds dsRNAs with 5′ overhangs (Figure 5). Given its sequence similarity and functional redundancy with FDM1, FDM2 most likely interacts with dsRNA with 5′ overhangs too. These observations suggest at least two hypotheses for FDM1 and FDM2 function, as indicated for IDN2/RDM12 (15,24). The first is that FDM1 and FDM2 may bind dsRNA produced by RDR2 to stabilize it, which may be required for rasiRNA biogenesis (24). The second is that FDM1 may interact with AGO4-bound dsRNAs generated by base pairing between rasiRNAs and target transcripts produced by Pol II or Pol V to stabilize rasiRNA-target interaction or recruit downstream components such as DRM2 to chromatin (15,24). fdm1-1 fdm2-1 displayed reduced DNA methylation levels of both types I and II rasiRNA generating loci (Figure 2) as well as reduced amount of type I rasiRNAs but not type II rasiRNAs (Figure 3). These molecular phenotypes of fdm1-1 fdm2-1 resemble those of nrpe1, ago4, rdm1 and drd1, indicating that like NRPE1, AGO4, DRD1 and RDM1, FDM1 and FDM2 may act downstream of ra-siRNA initiation in RdDM. In addition, FDM1 and FDM2 are not required for the accumulation of both Pol V-dependent and Pol II-dependent scaffold transcripts, indicating FDM1 and FDM2 may act downstream of Pol V and Pol II activities. Thus, we favor the suggestion that FDM1/FDM2 binds the rasiRNA-target duplex. In fact, an RNA-mediated AGO4–FDM1 association is observed, whereas an RDR2–FDM1 interaction is not detected (Supplementary Figure S7).
The Arabidopsis genome encodes 14 SGS3-like proteins (34) that can be assigned into three subfamilies. IDN2 and FDM1/FDM2 belong to subfamily 2 and 3, respectively (Figure 1A). However, their protein sequences are very similar (∼60% similarity), indicating that they may have closely related functions. This notion is strongly supported by the facts that fdm1-1 idn2-3 and fdm2-1 idn2-3 show much stronger reduction in DNA methylation than each of single mutants (Figure 5). Arabidopsis encodes six SGS3-like proteins from family 2 and family 3, including IDN2, FDM1 and FDM2, FDM3, FDM4 and FDM5, which contain all four-signature domains of SGS3-like proteins. The double mutant analyses reveal that FDM3, FDM4 and FDM5 have redundant roles with FDM1 in RdDM (Figure 6). Thus our study defines a group of SGS3-like proteins that play important roles in RdDM. Clearly, further work is required to determine their molecular role in RdDM.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figures 1–8 and Supplementary Methods.
FUNDING
National Science Foundation Grant MCB-1121193 (to B.Y.), an Enhancing Interdisciplinary Teams Grant from the University of Nebraska-Lincoln (to B.Y.) and Mallinckrodt Junior Foundation (to O.P. and P.C.N.). Funding for open access charge: Startup grant from the University of Nebraska-Lincoln (to B.Y.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Xuemei Chen from University of California-Riverside for nrpe1-1, dcl1-3 and myc-AGO4 seeds and Drs David Holding and Heriberto Cerutti from the University of Nebraska-Lincoln for critical reading of the manuscript.
REFERENCES
- 1.Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010;330:622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- 4.Kanno T, Huettel B, Mette MF, Aufsatz W, Jaligot E, Daxinger L, Kreil DP, Matzke M, Matzke AJ. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat. Genet. 2005;37:761–765. doi: 10.1038/ng1580. [DOI] [PubMed] [Google Scholar]
- 5.Onodera Y, Haag JR, Ream T, Nunes PC, Pontes O, Pikaard CS. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 6.Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng B, Wang Z, Li S, Yu B, Liu JY, Chen X. Intergenic transcription by RNA polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes Dev. 2009;23:2850–2860. doi: 10.1101/gad.1868009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith LM, Pontes O, Searle I, Yelina N, Yousafzai FK, Herr AJ, Pikaard CS, Baulcombe DC. An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell. 2007;19:1507–1521. doi: 10.1105/tpc.107.051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Havecker ER, Wallbridge LM, Hardcastle TJ, Bush MS, Kelly KA, Dunn RM, Schwach F, Doonan JH, Baulcombe DC. The Arabidopsis RNA-directed DNA methylation argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell. 2010;22:321–334. doi: 10.1105/tpc.109.072199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng X, Zhu J, Kapoor A, Zhu JK. Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO J. 2007;26:1691–1701. doi: 10.1038/sj.emboj.7601603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- 12.Cao X, Aufsatz W, Zilberman D, Mette MF, Huang MS, Matzke M, Jacobsen SE. Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr. Biol. 2003;13:2212–2217. doi: 10.1016/j.cub.2003.11.052. [DOI] [PubMed] [Google Scholar]
- 13.El-Shami M, Pontier D, Lahmy S, Braun L, Picart C, Vega D, Hakimi MA, Jacobsen SE, Cooke R, Lagrange T. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev. 2007;21:2539–2544. doi: 10.1101/gad.451207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ausin I, Mockler TC, Chory J, Jacobsen SE. IDN1 and IDN2 are required for de novo DNA methylation in Arabidopsis thaliana. Nat. Struct. Mol. Biol. 2009;16:1325–1327. doi: 10.1038/nsmb.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bies-Etheve N, Pontier D, Lahmy S, Picart C, Vega D, Cooke R, Lagrange T. RNA-directed DNA methylation requires an AGO4-interacting member of the SPT5 elongation factor family. EMBO Rep. 2009;10:649–654. doi: 10.1038/embor.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao Z, Liu HL, Daxinger L, Pontes O, He X, Qian W, Lin H, Xie M, Lorkovic ZJ, Zhang S, et al. An RNA polymerase II- and AGO4-associated protein acts in RNA-directed DNA methylation. Nature. 2010;465:106–109. doi: 10.1038/nature09025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He XJ, Hsu YF, Zhu S, Wierzbicki AT, Pontes O, Pikaard CS, Liu HL, Wang CS, Jin H, Zhu JK. An effector of RNA-directed DNA methylation in Arabidopsis is an ARGONAUTE 4- and RNA-binding protein. Cell. 2009;137:498–508. doi: 10.1016/j.cell.2009.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanno T, Mette MF, Kreil DP, Aufsatz W, Matzke M, Matzke AJ. Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr. Biol. 2004;14:801–805. doi: 10.1016/j.cub.2004.04.037. [DOI] [PubMed] [Google Scholar]
- 20.Kanno T, Bucher E, Daxinger L, Huettel B, Bohmdorfer G, Gregor W, Kreil DP, Matzke M, Matzke AJ. A structural-maintenance-of-chromosomes hinge domain-containing protein is required for RNA-directed DNA methylation. Nat. Genet. 2008;40:670–675. doi: 10.1038/ng.119. [DOI] [PubMed] [Google Scholar]
- 21.Law JA, Ausin I, Johnson LM, Vashisht AA, Zhu JK, Wohlschlegel JA, Jacobsen SE. A protein complex required for polymerase V transcripts and RNA- directed DNA methylation in Arabidopsis. Curr. Biol. 2010;20:951–956. doi: 10.1016/j.cub.2010.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bateman A. The SGS3 protein involved in PTGS finds a family. BMC Bioinformatics. 2002;3:21. doi: 10.1186/1471-2105-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N, et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- 24.Zheng Z, Xing Y, He XJ, Li W, Hu Y, Yadav SK, Oh J, Zhu JK. An SGS3-like protein functions in RNA-directed DNA methylation and transcriptional gene silencing in Arabidopsis. Plant J. 2010;62:92–99. doi: 10.1111/j.1365-313X.2010.04130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fukunaga R, Doudna JA. dsRNA with 5′ overhangs contributes to endogenous and antiviral RNA silencing pathways in plants. EMBO J. 2009;28:545–555. doi: 10.1038/emboj.2009.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pontier D, Yahubyan G, Vega D, Bulski A, Saez-Vasquez J, Hakimi MA, Lerbs-Mache S, Colot V, Lagrange T. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev. 2005;19:2030–2040. doi: 10.1101/gad.348405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao X, Jacobsen SE. Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc. Natl Acad. Sci. USA. 2002;99(Suppl. 4):16491–16498. doi: 10.1073/pnas.162371599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vaillant I, Schubert I, Tourmente S, Mathieu O. MOM1 mediates DNA-methylation-independent silencing of repetitive sequences in Arabidopsis. EMBO Rep. 2006;7:1273–1278. doi: 10.1038/sj.embor.7400791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pontes O, Li CF, Nunes PC, Haag J, Ream T, Vitins A, Jacobsen SE, Pikaard CS. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell. 2006;126:79–92. doi: 10.1016/j.cell.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 32.Jiao X, Trifillis P, Kiledjian M. Identification of target messenger RNA substrates for the murine deleted in azoospermia-like RNA-binding protein. Biol. Reprod. 2002;66:475–485. doi: 10.1095/biolreprod66.2.475. [DOI] [PubMed] [Google Scholar]
- 33.Yu B, Bi L, Zheng B, Ji L, Chevalier D, Agarwal M, Ramachandran V, Li W, Lagrange T, Walker JC, et al. The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc. Natl Acad. Sci. USA. 2008;105:10073–10078. doi: 10.1073/pnas.0804218105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qin Y, Ye H, Tang N, Xiong L. Systematic identification of X1-homologous genes reveals a family involved in stress responses in rice. Plant Mol. Biol. 2009;71:483–496. doi: 10.1007/s11103-009-9535-5. [DOI] [PubMed] [Google Scholar]
- 35.Akiyama K, Chikayama E, Yuasa H, Shimada Y, Tohge T, Shinozaki K, Hirai MY, Sakurai T, Kikuchi J, Saito K. PRIMe: a Web site that assembles tools for metabolomics and transcriptomics. In Silico Biol. 2008;8:339–345. [PubMed] [Google Scholar]
- 36.Obayashi T, Kinoshita K, Nakai K, Shibaoka M, Hayashi S, Saeki M, Shibata D, Saito K, Ohta H. ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res. 2007;35:D863–D869. doi: 10.1093/nar/gkl783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ihmels J, Levy R, Barkai N. Principles of transcriptional control in the metabolic network of Saccharomyces cerevisiae. Nat. Biotechnol. 2004;22:86–92. doi: 10.1038/nbt918. [DOI] [PubMed] [Google Scholar]
- 38.Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 40.McElver J, Tzafrir I, Aux G, Rogers R, Ashby C, Smith K, Thomas C, Schetter A, Zhou Q, Cushman MA, et al. Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics. 2001;159:1751–1763. doi: 10.1093/genetics/159.4.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamilton A, Voinnet O, Chappell L, Baulcombe D. Two classes of short interfering RNA in RNA silencing. EMBO J. 2002;21:4671–4679. doi: 10.1093/emboj/cdf464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li CF, Pontes O, El-Shami M, Henderson IR, Bernatavichute YV, Chan SW, Lagrange T, Pikaard CS, Jacobsen SE. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell. 2006;126:93–106. doi: 10.1016/j.cell.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 43.Wierzbicki AT, Ream TS, Haag JR, Pikaard CS. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat. Genet. 2009;41:630–634. doi: 10.1038/ng.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.