Abstract
The ribosomal stalk is formed by four acidic phosphoproteins in Saccharomyces cerevisiae, P1α, P1β, P2α and P2β, which form two heterodimers, P1α/P2β and P1β/P2α, that preferentially bind to sites A and B of the P0 protein, respectively. Using mutant strains carrying only one of the four possible P1/P2 combinations, we found a specific phenotype associated to each P1/P2 pair, indicating that not all acidic P proteins play the same role. The absence of one P1/P2 heterodimer reduced the rate of cell growth by varying degrees, depending on the proteins missing. Synthesis of the 60S ribosomal subunit also decreased, particularly in strains carrying the unusual P1α–P2α or P1β–P2β heterodimers, although the distinct P1/P2 dimers are bound with similar affinity to the mutant ribosome. While in wild-type strains the B site bound P1β/P2α in a highly specific manner and the A site bound the four P proteins similarly, both the A and B binding sites efficiently bound practically any P1/P2 pair in mutant strains expressing truncated P0 proteins. The reported results support that while most ribosomes contain a P1α/P2β–P0–P1β/P2α structure in normal conditions, the stalk assembly mechanism can generate alternative compositions, which have been previously detected in the cell.
INTRODUCTION
The stalk is a functional domain of the large ribosomal subunit that is directly involved in the interaction and GTPase activity of several soluble factors during translation (1). In eukaryotes, the stalk is formed by a central 32-kDa protein, P0, which interacts through its N-terminal domain (NTD) with the highly conserved GTPase-associated region (GAR) of the large rRNA subunit, and with two heterodimers of the acidic 12-kDa P1 and P2 proteins, ultimately forming a P0–(P1/P2)2 pentamer. Some lower eukaryotic species possess more than one P1 or P2 protein forms, such as Saccharomyces cerevisiae that contains two: P1α/P1β and P2α/P2β (2,3). The last 13 amino acids of the C-terminal domain (CTD) in five components of the pentamer, which mediate stalk activity during protein synthesis, are identical and are almost totally conserved in all eukaryotic species. The presence of a functional CTD in P0 is a key differential feature of the eukaryotic stalk, rendering the acidic proteins P1 and P2 non-essential for translation (4). In contrast, the prokaryotic P1/P2 counterpart, the L7/L12 protein, is required for protein synthesis (5), as the L10 protein lacks the corresponding functional CTD.
The eukaryotic stalk is a dynamic structure, whereby there is considerable exchange of P1/P2 proteins between the ribosome and a cytoplasmic pool of free proteins (6–8), and ribosomes either totally or partially lacking these acidic proteins exist in the cell (9). As stalk stability is essential for its proper functioning during translation, the aforementioned exchange implies that the stalk structure undergoes cyclical conformational changes that affect the interaction of P0 with the P1/P2 heterodimers, thereby facilitating the release of the acidic proteins.
The CTD of the stalk acidic proteins exhibits a significant mobility, which has hindered the elucidation of the crystal structure of this ribosomal domain. Lately, a crystal structure of the prokaryotic stalk core was generated by removing most of the mobile region of the acidic proteins (10,11). Moreover, homology modelling has provided some insight into the structure of a large portion of the eukaryotic stalk (12,13), although direct experimental data remain elusive. Nonetheless, notable progress has been made in characterizing the interactions between eukaryotic stalk components, due to the use of a range of biochemical and biophysical approaches (14–25). By analysing the P0–P1/P2 interactions, P1/P2 heterodimers were seen to bind to specific contiguous sites in P0 (20,26,27). In wild-type S. cerevisiae, it has been reported that the first site (site A) lies between amino acids 199–230 in P0 and it binds the P1α/P2β heterodimers, while the P1β/P2α dimer mainly associates with site B lying between amino acids 231–258 (20). The size of the P0 and P1/P2 regions involved in these interactions (20,28), together with the information from homology modelling studies (12,13) and low resolution biophysical analyses of the complex (22), strongly suggest that the association of the acidic proteins with the core stalk protein is notably more complex in eukaryotes than in bacteria (10) or archaea (11). This complexity increases the potential of the stalk structure to undergo induced cyclical conformational changes, which probably mediate P1/P2 exchange. Moreover, small changes at the NTD can drastically affect the affinity of the acidic proteins for P0 (13).
While the need for two significantly different types of acidic proteins in eukaryotes (P1 and P2) remains unexplained, experimental evidence suggests that they might play distinct roles (29). The presence of multiple forms of P1 and P2 in some lower organisms remains equally perplexing, although they may represent an evolutionary response to environmental challenges to which higher organisms are not exposed. The four acidic proteins in S. cerevisiae appear to play distinct roles in standard laboratory conditions, and initial studies using stalk deletion mutants suggested that proteins P1α and P2β are more important for cell growth than P1β or P2α (30). A recently described stalk assembly model attributed a leading role to the P1α/P2β pair, proposed to be the first heterodimer to bind to site A of P0. The P1β/P2α pair would play a subsidiary function, subsequently binding to site B (20,24). The model implies a clear specificity of the P0 sites A and B for the corresponding P1/P2 couples, and suggests the formation of stable heterodimers prior to binding, at least in the case of P1α/P2β. However, evidence obtained from yeast strains lacking some of the stalk components suggests alternative combinations. Saccharomyces cerevisiae strains D47 and D56, which express only the ‘non-canonical’ P1α/P2α and P1β/P2β pairs, respectively, contain ribosomes with a functional stalk that carry the corresponding non-standard heterodimers (30). This observation suggests that the 12-kDa proteins can be assembled in the yeast stalk in different ways, depending on cell conditions, raising a number of questions regarding the specificity of the interactions between the distinct stalk components.
We performed a comparative detailed analysis of S. cerevisiae strains that individually express each of the four possible P1/P2 pairs, and quantified the structural and functional peculiarities of the corresponding ribosomal stalks, with a view to obtaining a better understanding of the function and assembly of this essential ribosomal domain.
MATERIALS AND METHODS
Organisms and growth conditions
Escherichia coli DH5α, grown in LB medium at 37°C, was used to propagate and maintain the plasmids. Bacterial transformations were performed as previously described (31). The S. cerevisiae strains used are listed in Supplementary Table S1, and they were grown at 30°C in YPD or SC medium with the appropriate metabolic requirements until mid-log phase. Yeast transformation was carried out using the lithium acetate method (32).
Plasmids
A series of plasmids encoding truncated P0 proteins that lack the P1/P2 binding sites A and B were constructed (Supplementary Table S2). The nucleotides encoding either site A (amino acids 198–230), site B (amino acids 230–258) or both sites together (amino acids 198–258) were removed from the RPP0 gene encoded in the plasmid BS-P0 (33). The constructs were generated by an overlapping PCR strategy (Quickchange II site-directed mutagenesis kit: Stratagene) using BS-P0 as the template and the appropriate oligonucleotide primers (Supplementary Table S3). The RPP0 gene was removed from BS-P0 as a BamHI–XhoI DNA fragment of 2.8–2.6 kbp, depending on the specific mutations incorporated, and inserted into either the BamHI–SalI sites of pFL36(LEU2) or the EcoRI–SalI sites of pFL37(HIS3), pFL38(URA3) and pFL39(TRP1) (34), depending on the genetic markers available in the strain to be transformed.
Cell fractionation and ribosome preparation
Total cell extracts and high-salt washed ribosomes from S. cerevisiae were prepared as summarized in the Supplementary Methods (28).
Analysis of polysomes and ribosomal subunits on sucrose gradients
Polysome profiles were prepared following standard methods, as summarized in the Supplementary Methods. In the ribosome dissociation experiments, the KCl concentration in cytoplasmic extracts was increased to 0.5 M (high salt-washed), and, the ribosomal subunits were resolved by centrifugation in a SW40Ti rotor at 39 000 rpm for 3 h 45 min at 4°C on a 10–30% (w/v) sucrose gradient in 15 mM Tris–HCl pH 7.4, 500 mM KCl and 5 mM MgCl2.
Protein analysis
Ribosomal proteins were analysed by 15% SDS–PAGE or by isoelectrofocusing in 5% polyacrylamide gels over a pH range of 2.0–5.0 (35). The proteins were detected by Western blot using specific monoclonal antibodies against yeast stalk P proteins (36), or by silver staining. Proteins from total cell extracts were resolved by 2D polyacrylamide gel electrophoresis as described previously (37).
RESULTS
Phenotypic analysis of S. cerevisiae strains that contain a single P1/P2 heterodimer in the ribosomal stalk
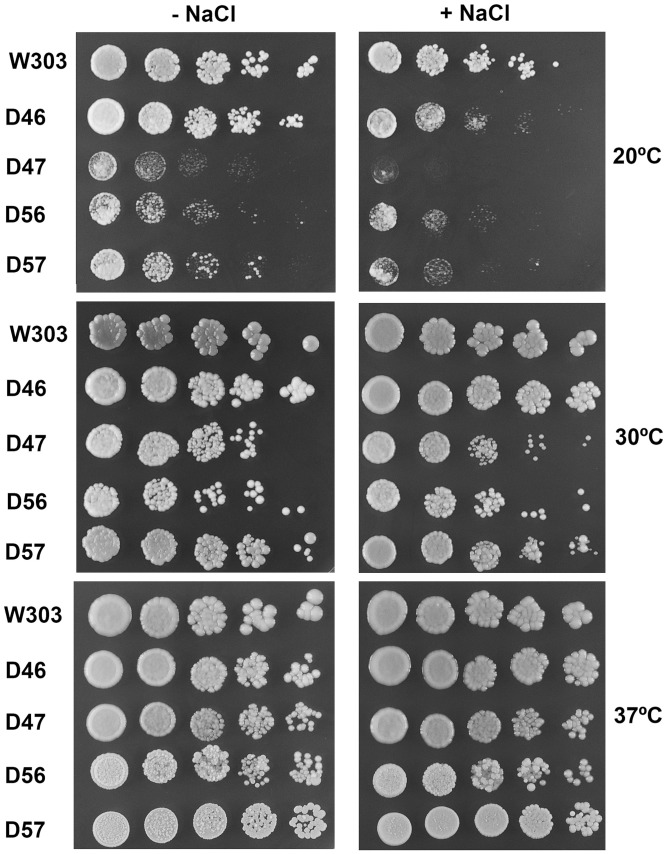
Wild-type S. cerevisiae contains four acidic P proteins, P1α, P1β, P2α and P2β, which form two distinct P1/P2 heterodimers. Mutant strains carrying only one of the four possible P1/P2 pairs have been previously obtained (30), the strains D46, D47, D56 and D57 carrying the acidic protein couples P1α/P2β, P1β/P2β, P1α/P2α and P1β/P2α, respectively. The absence of one of the P1/P2 pairs does not appear to be particularly deleterious to the cell, although the effect varies depending on the specific proteins missing. Thus, in liquid rich medium at 30°C, the absence of P1β and P2α (strain D46) results in a 30% increase in doubling time, which increases to 70% in the other three double-disrupted strains (30). In addition, we analysed the response of these four mutants to temperature and osmotic stress, growing the strains on rich medium agar plates at 20, 30 and 37°C, in the presence or absence of 0.3 M NaCl (Figure 1). The growth effects were less noticeable on agar plates at 30°C, and they were almost undetectable at 37°C. However, the differences in growth rate were strongly accentuated at 20°C. Particularly strains D47 and D56, which contain the non-canonical heterodimers P1β/P2β and P1α/P2α, respectively, displayed a clear cold-sensitive phenotype, which was enhanced in the presence of NaCl.
Figure 1.
Response of S. cerevisiae stalk mutants to temperature and osmotic stress. Serial dilutions of cells from the four double mutants (D46, D47, D56 and D57), and of the parental W303 strain, were grown for 4 days on YEPD agar plates in the presence or absence of 0.3 M NaCl at 20, 30 or 37°C.
Protein synthesis inhibitor sensitivity of stalk mutants
The response of the strains to sordarin and cycloheximide, two well-known eukaryotic translation inhibitors, was investigated by growing the yeast in rich liquid medium containing increasing drug concentrations (Supplementary Data). Both positive and negative changes in cell sensitivity (IC50) to the drugs were detected depending on the proteins missing (Table 1). A significant decrease in IC50 was observed in the D56 and D57 strains treated with sordarin, while the drug sensitivity of the D46 and D47 strains did not change. In contrast, the response to cycloheximide was only significantly altered in strain D47, which contains the P1β/P2β dimer, resulting in an ∼4-fold increase in the IC50.
Table 1.
Sensitivity of ribosomal stalk mutants to translation inhibitors
| Strain | Stalk proteins present | Cycloheximidea | Sordarina |
|---|---|---|---|
| IC50 µg/ml | IC50 µg/ml | ||
| W303 | P1α, P1β, P2α, P2β | 0.12 (1.00) | 0.33 (1.00) |
| D46 | P1α, P2β | 0.10 (0.83) | 0.34 (1.02) |
| D47 | P1β, P2β | 0.40 (3.40) | 0.32 (0.97) |
| D56 | P1α, P2α | 0.14 (1.16) | 0.19 (0.57) |
| D57 | P1β, P2α | 0.10 (0.82) | 0.10 (0.30) |
aIC50 values relative to W303 in brackets.
Effect of stalk composition on subunit association and polysome formation
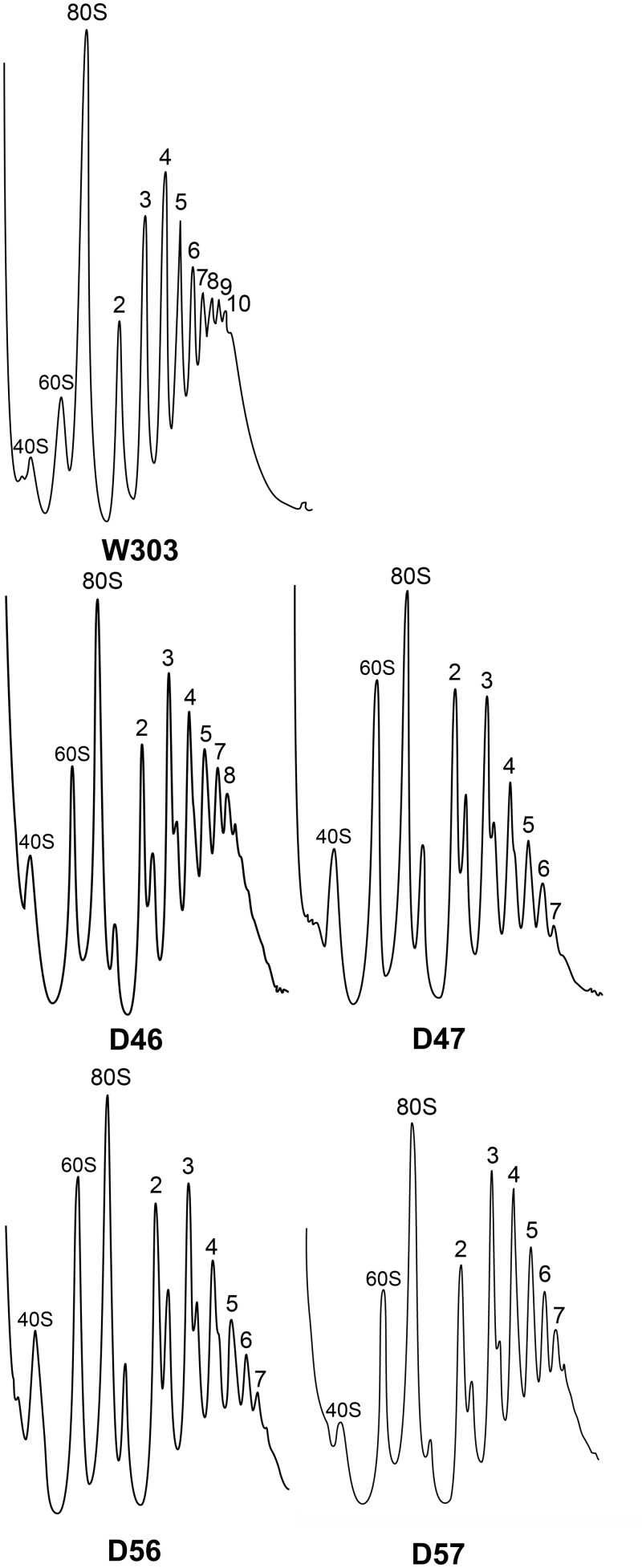
Cell extracts from the parental strain and from the four stalk mutants were resolved on sucrose gradients (Figure 2). Both the number of polysome peaks and the total amount of polysomes were lower in the mutants, although these effects were less pronounced in the D46 strain that exhibits a faster growth rate, and they were more evident in the slower growing mutants.
Figure 2.
Total cell extracts from the strains indicated grown to mid-logarithmic phase (OD600 = ∼0.5), were resolved in 10–50% sucrose gradients under the conditions described in ‘Materials and Methods’ section, and the A260 of the fractions was measured. The position of the 40S subunits, 60S subunits, 80S monosomes and polysomes containing increasing numbers of ribosomes is indicated.
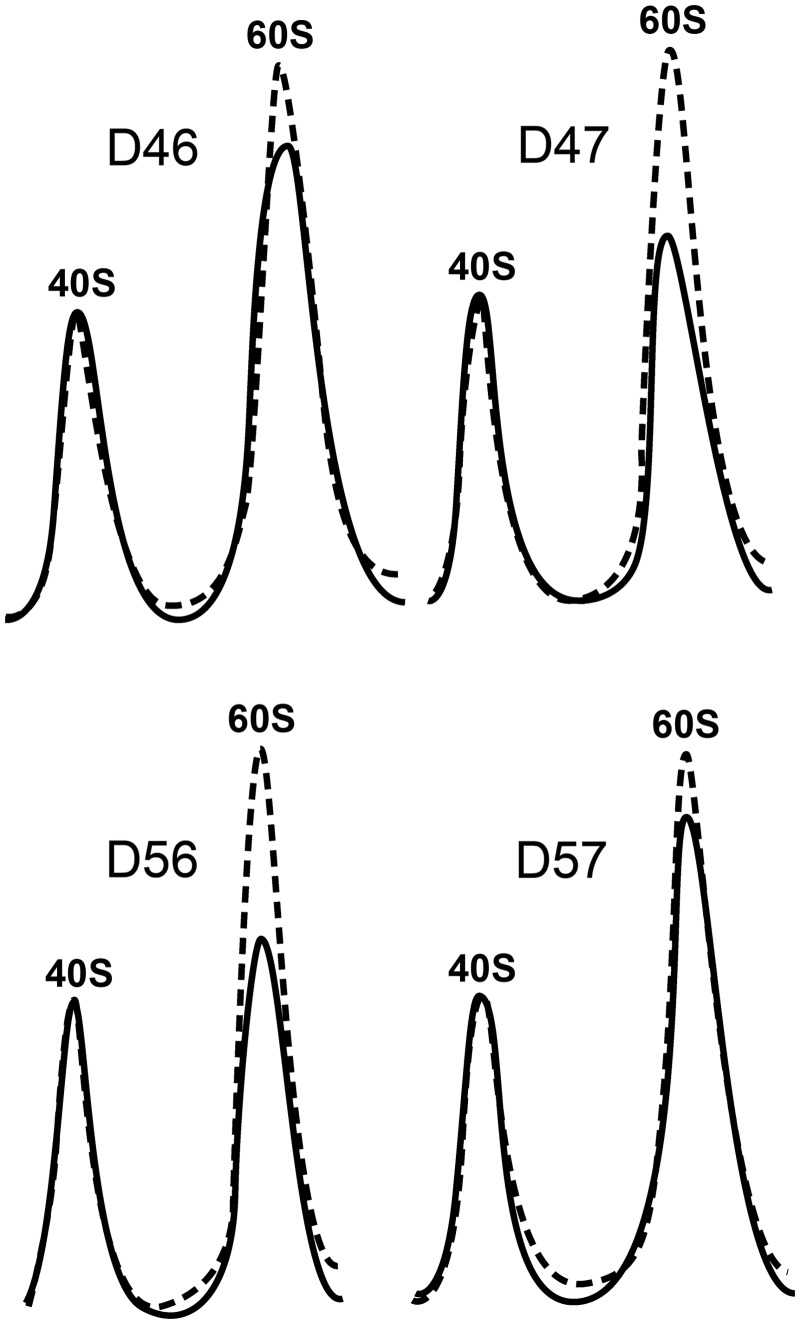
The presence of halfmers and an increase in the free subunit peaks was detected in the four mutant extracts, particularly in strains carrying non-canonical heterodimers (i.e. D47 and D56). The presence of halfmers in the polysome profiles may reflect a decrease in free 60S subunits in the cell, usually due to defective ribosome synthesis, or a decrease in the interactions between the ribosomal subunits that leads to the release of a fraction of the large subunits from polysomes during centrifugation. Calculating the overall 40S/60S ratio of the cell can help identify which of these two mechanisms is implicated. Thus, the total amount of each subunit was estimated by analysing cell extracts under ribosomal dissociation conditions, revealing a reduction in the amount of 60S subunits in all the mutant strains and particularly, in D47 and D56 (Figure 3).
Figure 3.
Total amount of ribosomal subunits in cell extracts resolved by sucrose gradients in dissociating conditions. The A260 profile of the mutant extracts (continuous lines) is superimposed upon that of the parental W303 strain (discontinuous line). The position of the 40S and 60S subunits is indicated.
Protein expression in ribosomal stalk mutants
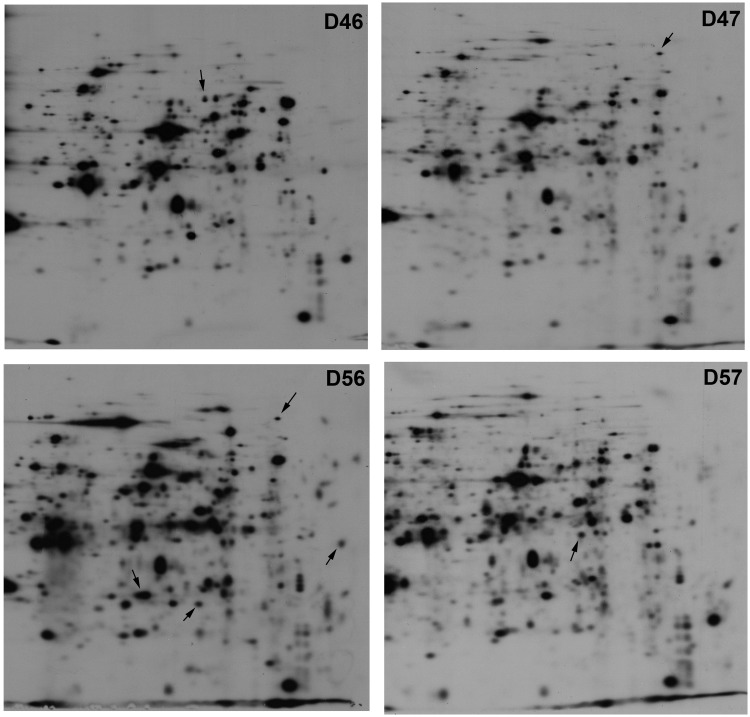
The overall protein expression in each of the mutant strains was determined by 2D gel electrophoresis (Figure 4), revealing clear differences between the four strains. The relative intensity of many spots differed between mutants, and more significantly, several spots were observed in some mutant strains but not others. Hence, it would appear that the composition of the ribosomal stalk determines, either directly or indirectly, the efficiency of translation of some mRNAs, as previously shown in a mutant strain lacking P1/P2 proteins in the ribosome (4).
Figure 4.
2D electrophoresis of total extracts from mutant strains D46, D47, D56 and D57. Some of the differential spots that appear in one sample but not in others are indicated.
Analysis of ribosomal stalks in S. cerevisiae double mutants
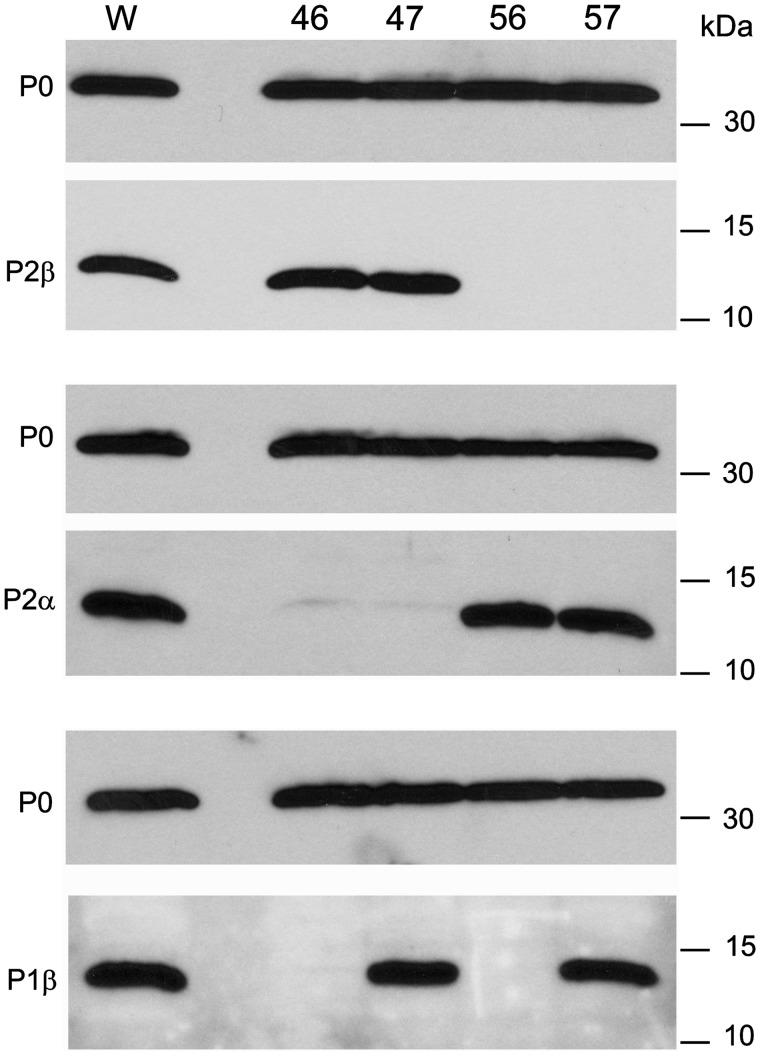
In each of the four mutant strains the acidic proteins present in the ribosomes purified by a standard high-salt washing protocol were estimated in immunoblots probed with specific monoclonal antibodies against proteins P0, P1β, P2α and P2β. The ratio of each protein to P0 was comparable in the ribosomes of each mutant strain and the parental W303 strain (Figure 5). Although an equivalent monoclonal antibody to P1α was not available, the results from its partner protein P2β strongly indicated that this protein was also present in comparable proportions in all strains that express it.
Figure 5.
Analysis of stalk proteins in purified ribosomes from mutant strains. Ribosomes purified from the indicated mutant strains and the parental W303 (W) strain were resolved by 15% SDS–PAGE and the indicated proteins were detected in immunoblots probed with specific monoclonal antibodies.
These results indicate that the four possible P1/P2 heterodimers, including P1α/P2α and P1β/P2β, exhibit the same degree of ribosomal binding, suggesting that the specificity of P0 binding sites is not particularly stringent under the experimental conditions used.
Ribosomal stalk components in strains expressing truncated P0 proteins that lack P1/P2 binding sites
To better understand how non-canonical P1/P2 heterodimers are assembled in stalks, plasmids were constructed encoding different P0-truncated derivatives, similar to those described previously (20). Accordingly, the protein P0ΔA lacks site A (amino acids 198–230), protein P0ΔB lacks site B (amino acids 230–258) and the P0ΔAB protein lacks both sites (amino acids 198–258). The three truncated proteins, as well as the native P0, were expressed in conditional P0 null strains (W303dGP0, D46dGP0, D47dGP0, D56dGP0 and D57dGP0) that only express the plasmid-encoded protein when grown in glucose medium (33). Total extracts from the transformed strains grown in glucose were resolved by SDS–PAGE and the proteins were detected in immunoblots probed with antibodies specific to the CTD of eukaryotic stalk proteins. There was an expected decrease in the size of the truncated P0 proteins (Supplementary Figure S1) and interestingly, the P1/P2 proteins were absent from the total extracts of double-disrupted mutants expressing the P0ΔAB form. Moreover, these proteins were notably reduced in the W303dGP0/P0ΔAB strain.
The effect of expressing a truncated P0 on the growth rate of double-disrupted mutants was estimated by serial dilution tests using rich medium agar plates. The presence of either P0ΔA or P0ΔB had no significant effect on the growth of strains D46 and D57, when compared with the strains expressing the wild-type P0 (Supplementary Figure S2).
Deletion of P0 acidic protein binding sites: effects on stalk composition
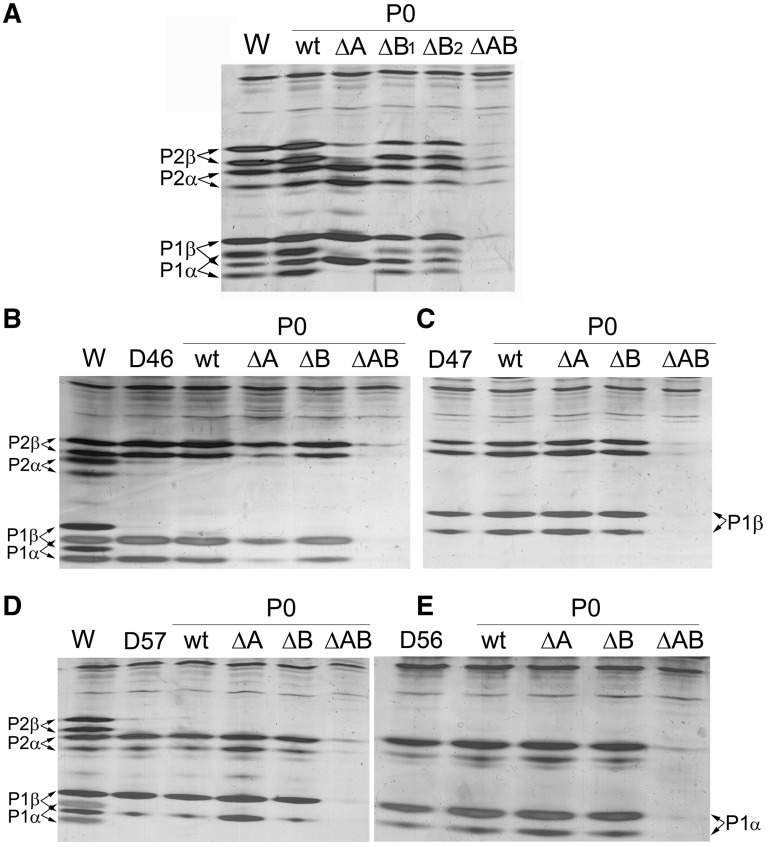
Ribosomes from the transformed strains, purified and high-salt washed according to standard protocols, were analysed by isoelectrofocusing in a pH range of 2.0–5.0 to resolve the different P1/P2 forms. The absence of site A in the parental W303dGP0/P0ΔA strain yielded ribosomes that almost exclusively contained P1β and P2α proteins (Figure 6A). In contrast, the four acidic proteins were present in comparable amounts in ribosomes from W303dGP0/P0ΔB, which only carries site A. This unexpectedly low specificity of site A was confirmed in cells from two different transformation experiments (ΔP0ΔB1 and ΔP0ΔB2 in Figure 6A). As expected, elimination of both binding sites resulted in the absence of acidic proteins in ribosome from W303dGP0/P0ΔAB.
Figure 6.
Ribosomal stalk composition of mutant yeast strains carrying truncated P0 proteins. Ribosomes were purified from the wild-type W303 strain (A), from the D46 (B) and D57 (D) mutants containing canonical heterodimers, and from the D47 (C) and D56 (E) mutants containing non-canonical heterodimers, expressing either wild-type P0 (P0wt) or truncated P0 proteins ΔA, ΔB and ΔAB (see Supplementary Figure S1). The ribosomes were resolved by isoelectrofocusing in a pH range of 2.0–5.0 and the proteins were detected by silver staining. As a control, purified ribosomes from the respective non-transformed strain were included in the corresponding gels. In gel A, ribosomes from two clones from two different transformations of W303dGP0 with protein P0-B (ΔB1 and ΔB2) were included. The upper and lower bands correspond to the phosphorylated and non-phosphorylated forms of each protein.
When the truncated proteins were expressed in strain D46dGP0 (that contains P1α and P2β alone), both proteins were detected in normal amounts in ribosomes containing site A (D46dGP0/P0ΔB), as was expected. However, both proteins were also notably present in D46dGP0/P0ΔA ribosomes that only contained site B (Figure 6B). Similarly, in D57 the amount of P1β and P2α was greater when the P0 carried only site B (D57dGP0/P0ΔA), although both proteins also bound to site A in D57dGP0/P0ΔB ribosomes (Figure 6D). In both these strains, the expression of P0ΔAB eliminates the acidic proteins from the cell (Supplementary Figure S1) and consequently from the ribosome (Figure 6B and D).
In strains D47 and D56 (which express the non-canonical heterodimers P1β/P2 and P1α/P2α, respectively), comparable amounts of acidic proteins were bound to the ribosomes regardless of the P0 protein present, except in the case of P0ΔAB that totally prevented the accumulation of all P1/P2 proteins in the cell (Figure 6C and E).
DISCUSSION
The stalk is essential for proper ribosomal function, and stalk defective ribosomes have a partially reduced translational efficiency (4). It is, therefore, not surprising that alteration in the stalk composition affects protein synthesis and consequently cell growth in eukaryotes. These effects vary depending on the proteins missing from the stalk. Thus, the absence of proteins P1β and P2α in strain D46 provoked a slight reduction in growth as compared to wild-type controls. This reduction was much more pronounced in D57, which lacks P1α and P2β, and particularly in D47 and D56, which carry the non-canonical pairs P1β/P2β and P1α/P2α, respectively. The growth effect was minimal at 30°C and almost undetectable at 37°C, although it was drastic at 20°C indicating a clear cold-sensitive phenotype. These results emphasize the different roles of the four acidic P proteins in stalk activity.
The changes in cell growth were largely paralleled by the polysome profiles of mutant extracts. Thus, the polysome area and the number of polysome peaks were greater in extracts from D46, the fastest growing mutant. Halfmers were also evident in these profiles and there was an increase in free ribosomal subunits in mutant extracts, particularly in D47, D56 and D57. Moreover, estimation of the total amount of 40S and 60S in extracts under dissociating conditions indicates a specific reduction in large subunits in the presence of a defective ribosomal stalk. This effect was more notable in D47 and D56, which express the non-canonical heterodimers P1β/P2β and P1α/P2α, respectively. Although it cannot be fully ruled out, it is unlikely that this reduction is due to the targeting of mature subunits carrying an altered stalk for degradation, as these subunits are functional and are efficiently incorporated into polysomes. The diminished expression of the 60S subunit is more likely to be due to alterations in the assembly pathway induced by the unusual P1/P2 expression in the mutants, which induces the generation of a fraction of defective pre-ribosomal particles that are degraded before maturing (38). The cold-sensitive phenotype of these mutants is in fact a typical feature of cells with a deregulated ribosomal assembly process (39). Thus, the P1/P2 proteins would appear to play a role in the 60S assembly process.
The functional differences displayed by the stalk mutants result in an alteration of the overall pattern of protein expressed. The total absence of P1/P2 proteins was shown to affect the translation of specific mRNAs, leading to the proposal that these proteins can regulate translation acting as modulators of ribosomal function (2,4). The present results indicate that ribosome carrying unusual ribosomal stalk compositions can also participate in the proposed regulatory process.
The sensitivity to two well-known inhibitors of translation elongation, cycloheximide and sordarin, was also influenced in a distinct manner by the stalk alterations of each mutant strain. While some mutants remained practically unaffected, others exhibited significant alterations in their IC50 values. Indeed, D47 was more resistant to cycloheximide while D56 and D57 were significantly more sensitive to sordarin than the parental strains. Given the poor understanding of the molecular mechanism of cycloheximide inhibition at the molecular level, the role of different ribosomal stalk components in drug activity remains unclear. In contrast, the well-documented mode of action of sordarin may help us interpret our results. GTP hydrolysis provokes conformational changes in the elongation factor, which generate a signal required for the completion of translocation. EF2-bound sordarin blocks the transmission of this signal, as revealed by crystal structure analysis of EF2-bound drug (40) and Cryo-EM analysis of the 80S–EF2–sordarin complex (41). Mutations in the ribosomal stalk protein P0 can induce resistance to sordarin without blocking drug binding, which takes place far from the stalk (42,43). These mutations probably restore signal transmission in the presence of the drug, thereby implicating P0 in this process. Our results indicate that P1α and particularly P2β are relevant to this process, but not P1β and P2α, as mutants lacking the P2 protein (D56) or both proteins (D57) were notably more sensitive to sordarin.
The specificity observed for the P1/P2 binding sites of the P0 protein was particularly interesting. First, the non-canonical couples P1α/P2α and P1β/P2β were detected in purified ribosomes from strains D56 and D47 at comparable proportions to the canonical pairs in strains D46 and D57 (Figure 5), suggesting that site specificity is not particularly stringent regarding the heterodimer composition. This conclusion was confirmed by expressing a truncated P0 lacking one P1/P2 binding site in the double mutants expressing only one of the four possible P1/P2 pairs. In all cases, the P1 and P2 proteins expressed bound to the ribosome in roughly stoichiometric amounts, irrespective of the P0 binding site. Taken together, these results suggest that the P0 sites A and B can bind any P1/P2 combination with comparable efficiency. However, the response of the deletion of the P0 binding sites observed in W303 strain, which contains the four acidic proteins, was different. In this case, site A exhibits little specificity and it binds the four proteins in similar proportions when site B is deleted, while site B shows strong specificity for P1β/P2α when site A is deleted. This conclusion is confirmed by the reduced levels of P1α/P2β observed in strain D46ΔA in which only site B is present.
These results do not support the stalk assembly pathway involving binding at site A prior to site B previously proposed (24). As this model was based on data from fully assembled ribosomes, it is difficult to deduce directly how the stalk was actually assembled. Moreover, the results indicated a stabilization of the structure upon formation of the stalk pentamer (24), implying that significant changes occur in the preceding complexes during the assembly process. The differential specificity of P0 sites reported here suggests an alternative assembly pathway (Supplementary Figure S3). We propose that stalk assembly begins with the binding of P1β/P2α to site B. Positive coupling of both binding sites then facilitates the subsequent interaction of P1α/P2β at site A, which is also likely to be promoted by the increased local concentration of these heterodimers resulting from the prior binding of P1β/P2α at site B. The occupation of both sites stabilizes the assembled P1α/P2β–P0–P1β/P2α pentamer, as reported previously (24). However, the weak specificity of site A also permits a small proportion of P1β/P2α heterodimers to bind to this site and form a P0–(P1β/P2α)2 complex (see Supplementary Figure S3). Accordingly, experimental evidence demonstrated that in the cell, ribosomes may carry unusual P1/P2 heterodimer combinations. The formation of P protein homodimers has been described in ribosome cross-linking studies (29), demonstrating that two copies of the same heterodimer may exist in the stalk. Moreover, fluorescence correlation spectroscopy using strains expressing GFP-labelled stalk components has shown that cells may contain a small fraction of ribosomes carrying unexpected amounts of P proteins incompatible with the standard stalk composition (44).
Our current understanding of stalk assembly is limited. Where and when P1/P2 proteins bind to P0, and the possible role of helper proteins in this process, remain unclear (45–47). As such, any proposed assembly model should be considered as a working hypothesis that awaits further experimental support. Nevertheless, the data available indicate that in the yeast wild-type ribosome, P1α/P2β dimers are predominantly found at site A and P1β/P2α at site B, although a fraction of particles may contain alternative P1/P2 heterodimer distributions. Thus, stalk assembly mechanism appears to be able to generate a variable level of stalk heterogeneity in the ribosomal population. This heterogeneity is likely to be important for the proposed regulatory functions of the stalk (2) and it is probably controlled by the cell, which regulates the expression of different P1 and P2 proteins, thereby affecting the proportion of distinct P1/P2 pairs in the cytoplasm (48). In connection with this issue, the drastic decrease in P1/P2 protein accumulation in the presence of P0ΔAB (Supplementary Figure S1) indicates a possible involvement of P0 in the control of P1/P2 expression, which require further investigation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–3, Supplementary Methods and Supplementary References [49–51].
FUNDING
Spanish Ministry of Science and Innovation (MICINN) (grant BFU2009-09738 to J.P.G.B.); Fundación Ramón Areces (Institutional Grant to Centro de Biología Molecular Severo Ochoa). Funding for open access charge: Grant from the Spanish Ministry of Science and Innovation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank M.C. Fernández Moyano for expert technical assistance.
REFERENCES
- 1.Mandava CS, Peisker K, Ederth J, Kumar R, Ge X, Szaflarski W, Sanyal S. Bacterial ribosome requires multiple L12 dimers for efficient initiation and elongation of protein synthesis involving IF2 and EF-G. Nucleic Acids Res. 2012;40:2054–2064. doi: 10.1093/nar/gkr1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballesta JPG, Remacha M. The large ribosomal subunit stalk as a regulatory element of the eukaryotic translational machinery. Prog. Nucleic Acid Res. Mol. Biol. 1996;55:157–193. doi: 10.1016/s0079-6603(08)60193-2. [DOI] [PubMed] [Google Scholar]
- 3.Tchorzewski M. The acidic ribosomal P proteins. Int. J. Biochem. Cell Biol. 2002;34:911–915. doi: 10.1016/s1357-2725(02)00012-2. [DOI] [PubMed] [Google Scholar]
- 4.Remacha M, Jimenez-Diaz A, Bermejo B, Rodriguez-Gabriel MA, Guarinos E, Ballesta JPG. Ribosomal acidic phosphoproteins P1 and P2 are not required for cell viability but regulate the pattern of protein expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:4754–4762. doi: 10.1128/mcb.15.9.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang C, Mandava CS, Sanyal S. The ribosomal stalk plays a key role in IF2-mediated association of the ribosomal subunits. J. Mol. Biol. 2010;399:145–153. doi: 10.1016/j.jmb.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 6.Zinker S, Warner JR. The ribosomal proteins of Saccharomyces cerevisiae. Phosphorylated and exchangeable proteins. J. Biol. Chem. 1976;251:1799–1807. [PubMed] [Google Scholar]
- 7.Tsurugi K, Ogata K. Evidence for the exchangeability of acidic ribosomal proteins on cytoplasmic ribosomes in regenerating rat liver. J. Biochem. 1985;98:1427–1431. doi: 10.1093/oxfordjournals.jbchem.a135410. [DOI] [PubMed] [Google Scholar]
- 8.Scharf K-D, Nover L. Control of ribosome biosynthesis in plant cell cultures under heat shock conditions. II. Ribosomal proteins. Biochim. Biophys. Acta. 1987;909:44–57. doi: 10.1111/j.1432-1033.1986.tb09971.x. [DOI] [PubMed] [Google Scholar]
- 9.Guarinos E, Santos C, Sanchez A, Qiu DY, Remacha M, Ballesta JP. Tag-mediated fractionation of yeast ribosome populations proves the monomeric organization of the eukaryotic ribosomal stalk structure. Mol. Microbiol. 2003;50:703–712. doi: 10.1046/j.1365-2958.2003.03733.x. [DOI] [PubMed] [Google Scholar]
- 10.Diaconu M, Kothe U, Schlunzen F, Fischer N, Harms JM, Tonevitsky AG, Stark H, Rodnina MV, Wahl MC. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell. 2005;121:991–1004. doi: 10.1016/j.cell.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 11.Naganuma T, Nomura N, Yao M, Mochizuki M, Uchiumi T, Tanaka I. Structural basis for translation factor recruitment to the eukaryotic/archaeal ribosomes. J. Biol. Chem. 2010;285:4747–4756. doi: 10.1074/jbc.M109.068098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee KM, Yu CW, Chan DS, Chiu TY, Zhu G, Sze KH, Shaw PC, Wong KB. Solution structure of the dimerization domain of ribosomal protein P2 provides insights for the structural organization of eukaryotic stalk. Nucleic Acids Res. 2010;38:5206–5216. doi: 10.1093/nar/gkq231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camargo H, Nusspaumer G, Abia D, Briceno V, Remacha M, Ballesta JP. The amino terminal end determines the stability and assembling capacity of eukaryotic ribosomal stalk proteins P1 and P2. Nucleic Acids Res. 2011;39:3735–3743. doi: 10.1093/nar/gkq1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jose MP, Santana-Roman H, Remacha M, Ballesta JPG, Zinker S. The eukaryotic phosphoproteins interact with the ribosome through their amino terminal domain. Biochemistry. 1995;34:7941–7948. doi: 10.1021/bi00024a019. [DOI] [PubMed] [Google Scholar]
- 15.Uchiumi T, Kominami R. Binding of mammalian ribosomal protein complex P0-P1-P2 and protein L12 to the GTPase-associated domain of 28 S ribosomal RNA and effect on the accessibility to anti-28 S RNA autoantibody. J. Biol. Chem. 1997;272:3302–3308. doi: 10.1074/jbc.272.6.3302. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalo P, Lavergne JP, Reboud JP. Pivotal role of the P1 n-terminal domain in the assembly of the mammalian ribosomal stalk and in the proteosynthetic activity. J. Biol. Chem. 2001;276:19762–19769. doi: 10.1074/jbc.M101398200. [DOI] [PubMed] [Google Scholar]
- 17.Lalioti VS, Perez-Fernández J, Remacha M, Ballesta JPG. Characterization of interaction sites in the Saccharomyces cerevisiae ribosomal stalk components. Mol. Microbiol. 2002;46:719–729. doi: 10.1046/j.1365-2958.2002.03179.x. [DOI] [PubMed] [Google Scholar]
- 18.Shimizu T, Nakagaki M, Nishi Y, Kobayashi Y, Hachimori A, Uchiumi T. Interaction among silkworm ribosomal proteins P1, P2 and P0 required for functional protein binding to the GTPase-associated domain of 28S rRNA. Nucleic Acids Res. 2002;30:2620–2627. doi: 10.1093/nar/gkf379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tchorzewski M, Krokowski D, Boguszewska A, Liljas A, Grankowski N. Structural characterization of yeast acidic ribosomal P proteins forming the P1A-P2B heterocomplex. Biochemistry. 2003;42:3399–3408. doi: 10.1021/bi0206006. [DOI] [PubMed] [Google Scholar]
- 20.Krokowski D, Boguszewska A, Abramczyk D, Liljas A, Tchorzewski M, Grankowski N. Yeast ribosomal P0 protein has two separate binding sites for P1/P2 proteins. Mol. Microbiol. 2006;60:386–400. doi: 10.1111/j.1365-2958.2006.05117.x. [DOI] [PubMed] [Google Scholar]
- 21.Santos C, Ballesta JP. Characterization of the 26S rRNA-binding domain in Saccharomyces cerevisiae ribosomal stalk phosphoprotein P0. Mol. Microbiol. 2005;58:217–226. doi: 10.1111/j.1365-2958.2005.04816.x. [DOI] [PubMed] [Google Scholar]
- 22.Grela P, Helgstrand M, Krokowski D, Boguszewska A, Svergun D, Liljas A, Bernado P, Grankowski N, Akke M, Tchorzewski M. Structural characterization of the ribosomal P1A-P2B protein dimer by small-angle X-ray scattering and NMR spectroscopy. Biochemistry. 2007;46:1988–1998. doi: 10.1021/bi0616450. [DOI] [PubMed] [Google Scholar]
- 23.Naganuma T, Shiogama K, Uchiumi T. The N-terminal regions of eukaryotic acidic phosphoproteins P1 and P2 are crucial for heterodimerization and assembly into the ribosomal GTPase-associated center. Genes Cells. 2007;12:501–510. doi: 10.1111/j.1365-2443.2007.01067.x. [DOI] [PubMed] [Google Scholar]
- 24.Grela P, Krokowski D, Gordiyenko Y, Krowarsch D, Robinson CV, Otlewski J, Grankowski N, Tchorzewski M. Biophysical properties of the eukaryotic ribosomal stalk. Biochemistry. 2010;49:924–933. doi: 10.1021/bi901811s. [DOI] [PubMed] [Google Scholar]
- 25.Francisco-Velilla R, Remacha M. In vivo formation of a stable pentameric (P2alpha/P1beta)-P0-(P1alpha/P2beta) ribosomal stalk complex in Saccharomyces cerevisiae. Yeast. 2010;27:693–704. doi: 10.1002/yea.1765. [DOI] [PubMed] [Google Scholar]
- 26.Hagiya A, Naganuma T, Maki Y, Ohta J, Tohkairin Y, Shimizu T, Nomura T, Hachimori A, Uchiumi T. A mode of assembly of P0, P1, and P2 proteins at the GTPase-associated center in animal ribosome: in vitro analyses with P0 truncation mutants. J. Biol. Chem. 2005;280:39193–39199. doi: 10.1074/jbc.M506050200. [DOI] [PubMed] [Google Scholar]
- 27.Perez-Fernandez J, Remacha M, Ballesta JP. The acidic protein binding site is partially hidden in the free Saccharomyces cerevisiae ribosomal stalk protein P0. Biochemistry. 2005;44:5532–5540. doi: 10.1021/bi047332r. [DOI] [PubMed] [Google Scholar]
- 28.Briceno V, Camargo H, Remacha M, Santos C, Ballesta JP. Structural and functional characterization of the amino terminal domain of the yeast ribosomal stalk P1 and P2 proteins. Int. J. Biochem. Cell Biol. 2008;41:1315–1322. doi: 10.1016/j.biocel.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 29.Qiu D, Parada P, Marcos AG, Cardenas D, Remacha M, Ballesta JP. Different roles of P1 and P2 Saccharomyces cerevisiae ribosomal stalk proteins revealed by cross-linking. Mol. Microbiol. 2006;62:1191–1202. doi: 10.1111/j.1365-2958.2006.05445.x. [DOI] [PubMed] [Google Scholar]
- 30.Remacha M, Santos C, Bermejo B, Naranda T, Ballesta JPG. Stable binding of the eukaryotic acidic phosphoproteins to the ribosome is not an absolute requirement for in vivo protein synthesis. J. Biol. Chem. 1992;267:12061–12067. [PubMed] [Google Scholar]
- 31.Hanahan D. Techniques for transformation of E. coli. In: Glover DM, editor. DNA Cloning: A Practical Approach. Oxford: IRL Press; 1985. pp. 109–136. [Google Scholar]
- 32.Gietz R, Woods R. High efficiency transformation with lithium acetate. In: Johnston JR, editor. Molecular Genetics of Yeast, A practical Approach. Oxford: IRL Press; 1994. pp. 121–134. [Google Scholar]
- 33.Santos C, Ballesta JP. Ribosomal protein P0, contrary to phosphoproteins P1 and P2, is required for ribosome activity and Saccharomyces cerevisiae viability. J. Biol. Chem. 1994;269:15689–15696. [PubMed] [Google Scholar]
- 34.Bonneaud N, Ozier-Kalogeropoulos O, Li G, Labouesse M, Minvielle-Sebastia L, Lacroute F. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast. 1991;7:609–615. doi: 10.1002/yea.320070609. [DOI] [PubMed] [Google Scholar]
- 35.Zambrano R, Briones E, Remacha M, Ballesta JPG. Phosphorylation of the acidic ribosomal P proteins in Saccharomyces cerevisiae. A reappraisal. Biochemistry. 1997;36:14439–14446. doi: 10.1021/bi971494o. [DOI] [PubMed] [Google Scholar]
- 36.Vilella MD, Remacha M, Ortiz BL, Mendez E, Ballesta JPG. Characterization of the yeast acidic ribosomal phosphoproteins using monoclonal antibodies. Proteins L44/L45 and L44′ have different functional roles. Eur. J. Biochem. 1991;196:407–414. doi: 10.1111/j.1432-1033.1991.tb15831.x. [DOI] [PubMed] [Google Scholar]
- 37.Santaren JF. Towards establishing a protein database of Drosophila. Electrophoresis. 1990;11:254–267. doi: 10.1002/elps.1150110309. [DOI] [PubMed] [Google Scholar]
- 38.Lafontaine DL. A ‘garbage can’ for ribosomes: how eukaryotes degrade their ribosomes. Trends Biochem Sci. 2010;35:267–277. doi: 10.1016/j.tibs.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 39.Woolford JJ. The structure and biogenesis of yeast ribosomes. Adv. Genet. 1991;29:63–118. doi: 10.1016/s0065-2660(08)60107-8. [DOI] [PubMed] [Google Scholar]
- 40.Jorgensen R, Yates SP, Teal DJ, Nilsson J, Prentice GA, Merrill AR, Andersen GR. Crystal structure of ADP-ribosylated ribosomal translocase from Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:45919–45925. doi: 10.1074/jbc.M406218200. [DOI] [PubMed] [Google Scholar]
- 41.Spahn CM, Gomez-Lorenzo MG, Grassucci RA, Jorgensen R, Andersen GR, Beckmann R, Penczek PA, Ballesta JP, Frank J. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004;23:1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gómez-Lorenzo MG, Garcia-Bustos JF. Ribosomal P-protein stalk function is targeted by sordarin antifungals. J. Biol. Chem. 1998;273:25041–25044. doi: 10.1074/jbc.273.39.25041. [DOI] [PubMed] [Google Scholar]
- 43.Justice MC, Ku T, Hsu MJ, Carniol K, Schmatz D, Nielsen J. Mutations in ribosomal protein L10e confer resistance to the fungal-specific eukaryotic elongation factor 2 inhibitor sordarin. J. Biol. Chem. 1999;274:4869–4875. doi: 10.1074/jbc.274.8.4869. [DOI] [PubMed] [Google Scholar]
- 44.Garcia-Marcos A, Sanchez SA, Parada P, Eid JS, Jameson DM, Remacha M, Gratton E, Ballesta JP. Yeast ribosomal stalk heterogeneity in vivo shown by two-photon FCG and molecular brightness analysis. Biophys. J. 2008;94:2884–2890. doi: 10.1529/biophysj.107.121822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodriguez-Mateos M, Garcia-Gomez JJ, Francisco-Velilla R, Remacha M, de la Cruz J, Ballesta JP. Role and dynamics of the ribosomal protein P0 and its related trans-acting factor Mrt4 during ribosome assembly in Saccharomyces cerevisiae. Nucleic Acids Res. 2009;37:7519–7532. doi: 10.1093/nar/gkp806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lo KY, Li Z, Wang F, Marcotte EM, Johnson AW. Ribosome stalk assembly requires the dual-specificity phosphatase C for the exchange of Mrt4 with P0. J. Cell. Biol. 2009;186:849–862. doi: 10.1083/jcb.200904110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kemmler S, Occhipinti L, Veisu M, Panse VG. Yvh1 is required for a late maturation step in the 60S biogenesis pathway. J. Cell. Biol. 2009;186:863–880. doi: 10.1083/jcb.200904111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bermejo B, Remacha M, Ortiz-Reyes B, Santos C, Ballesta JP. Effect of acidic ribosomal phosphoprotein mRNA 5’-untranslated region on gene expression and protein accumulation. J. Biol. Chem. 1994;269:3968–3975. [PubMed] [Google Scholar]
- 49.Rothstein RJ. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 50.Rodriguez-Gabriel MA, Remacha M, Ballesta JPG. The RNA interacting domain but not the protein interacting domain is highly conserved in ribosomal protein P0. J. Biol. Chem. 2000;275:2130–2136. doi: 10.1074/jbc.275.3.2130. [DOI] [PubMed] [Google Scholar]
- 51.Sanchez-Madrid F, Reyes R, Conde P, Ballesta JPG. Acidic ribosomal proteins from eukaryotic cells. Effect on ribosomal functions. Eur. J. Biochem. 1979;98:409–416. doi: 10.1111/j.1432-1033.1979.tb13200.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.