Abstract
Bacteria and bacteriophages have evolved DNA modification as a strategy to protect their genomes. Mom protein of bacteriophage Mu modifies the phage DNA, rendering it refractile to numerous restriction enzymes and in turn enabling the phage to successfully invade a variety of hosts. A strong fortification, a combined activity of the phage and host factors, prevents untimely expression of mom and associated toxic effects. Here, we identify the bacterial chromatin architectural protein Fis as an additional player in this crowded regulatory cascade. Both in vivo and in vitro studies described here indicate that Fis acts as a transcriptional repressor of mom promoter. Further, our data shows that Fis mediates its repressive effect by denying access to RNA polymerase at mom promoter. We propose that a combined repressive effect of Fis and previously characterized negative regulatory factors could be responsible to keep the gene silenced most of the time. We thus present a new facet of Fis function in Mu biology. In addition to bringing about overall downregulation of Mu genome, it also ensures silencing of the advantageous but potentially lethal mom gene.
INTRODUCTION
The temperate bacteriophage Mu infects Escherichia coli and several other enteric bacteria (1). One of the factors contributing to the wide host range of Mu is a novel post-replicative DNA modification function encoded by the phage gene mom (2–4). Sequence specific modification of adenine to N-6 acetamidoadenine by the gene product of mom renders the phage DNA resistant to several host encoded restriction/modification systems (5). Expression of mom, a late function in the lytic cycle of Mu, is cytotoxic to the host (6–8). An elaborate regulatory network has evolved to curb untimely mom expression (Supplementary Figure S1) (9). While Com (product of the overlapping com ORF within the mom operon) is essential to achieve translation of mom mRNA (8,10,11), a complex interplay between the host proteins Dam (DNA adenine methyltransferase) and OxyR governs transcription from mom promoter (Pmom) (12–14). In addition, owing to factors like suboptimal spacing between the poor −35 and −10 elements and intrinsic curvature conferred by a T stretch, Pmom is far from being a classical promoter and is dependent on transcription activator C (15–17). Furthermore, activation of the activator C itself is subject to availability of Mg+2 (18). Moreover, in the absence of C protein, RNA polymerase (RNAP) gets recruited at a divergent overlapping promoter momP2, thus posing competition to the already weak mom promoter (17,19).
The simultaneous existence of multiple mechanisms to fine-tune a single phage function might seem excessive. For instance, despite the existence of a multi-layered transcriptional control, a further check at the translational level appears needless. However, to restrain mom gene from exerting its cytotoxic effect, its expression must be precisely modulated. The currently known mom regulatory circuit is not only elaborate but also apparently failsafe (9). In this article, we report a hitherto unidentified mechanism operating to add to the fortification. While characterizing the role of the transactivator C, we observed a Pmom specific DNA binding activity in crude cell extracts of E. coli (Figure 1A, V. Ramesh, unpublished data). This observation prompted us to identify the host factor and investigate the possibility of it regulating the expression of mom. Here, we have elucidated the role of this new player in mom regulation.
Figure 1.
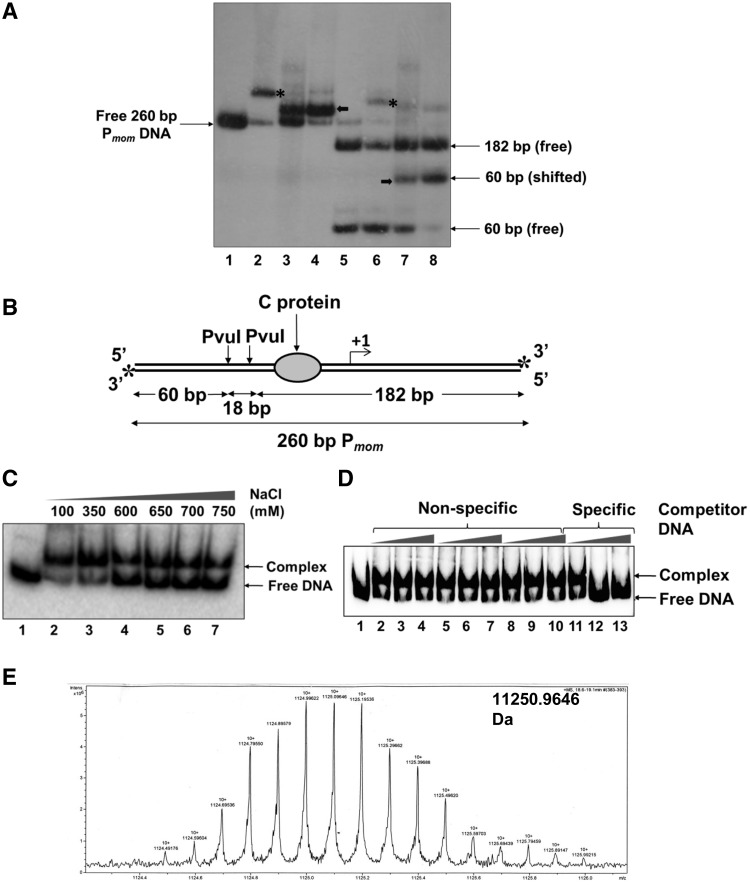
Identification of the host factor that binds specifically to Pmom. (A) Pmom specific activity in crude cell extracts of E. coli. End-labeled 260-bp Pmom fragment (lane 1) was incubated with 220 nM purified C (lane 2) and 1 µg of crude sonicated extract (lane 3) and partially purified cell extract (lane 4) of E. coli MG1655. Lane 5 onwards, PvuI digested Pmom (lane 5) was incubated with 220 nM purified C (lane 6) and 1 µg of crude sonicated extract (lane 7) and partially purified cell extract (lane 8) of E. coli MG1655. EMSAs with cell extracts were carried out in the presence of 200 ng poly (dI – dC). Mobility shift caused by binding of C is depicted by asterisks while that caused by the unknown host factor is depicted by bold arrows. (B) Pmom fragment used for EMSAs. A 260-bp fragment of Pmom obtained upon digestion of plasmid pUW4 with EcoRI and HindIII was radiolabeled at both the 3′-ends (denoted by asterisks) by Klenow polymerase-mediated end filling. Digestion of this fragment with PvuI yields three fragments of sizes 60, 18 and 182 bp of which only the terminal fragments (60 and 182 bp) carry the radiolabel. The larger fragment (182 bp) contains the C-binding site whereas the smaller fragment (60 bp) comes from the upstream region. (C) Effect of ionic strength on stability of the complex. 220-bp Pmom obtained upon digestion of pUW4 with EcoRI and BamHI (lane 1) was incubated with 1 µg of crude cell extract of E. coli MG1655 in the presence of increasing concentrations of NaCl as indicated. The host factor–DNA complex was stable even in 750 mM NaCl. (D) Effect of non-specific competitor DNA on complex formation. End labeled 220 bp Pmom fragment (lane 1) was incubated with 1 µg of crude cell extract of E. coli MG1655 in the presence of 0-, 50- and 100-fold molar excess of three different unlabeled non-specific competitor DNA fragments (lanes 2–10) and unlabeled Pmom fragment (lanes 11–13). Labeled Pmom DNA is competed by the specific unlabeled Pmom DNA but not by any of the three non-specific DNA fragments, showing that the complex results from specific binding of a molecule in the extract to Pmom. (E) LC/ESI-MS analysis of the active fraction. Crude cell extract of E. coli MG1655 was processed as described in ‘Materials and Methods’ section. Eluted fractions from SP Sepharose column that tested positive for Pmom specific DNA binding activity in EMSA were pooled, dialyzed and subjected to LC/ESI–MS for whole protein molecular weight detection. Analysis of the fraction revealed a molecular mass of 11.25 kDa.
MATERIALS AND METHODS
Bacterial strains and plasmids
The bacterial strains and plasmids used in this study are as described in Supplementary Table S1.
Electrophoretic Mobility Shift Assay
End labeled Pmom DNA was incubated with variable amounts of cell extracts or purified protein in TMEG100 buffer [20 mM Tris–HCl (pH 7.4), 5 mM MgCl2, 1 mM EDTA (pH 8.0), 10% glycerol, 100 mM NaCl, 5 mM β-mercaptoethanol] for 10 min on ice. Loading buffer [20 mM Tris–HCl (pH 7.5), 10% glycerol] containing 0.01% bromophenol blue and 0.01% xylene cyanol was added to the reaction and the samples were analyzed on 4 or 8% native polyacrylamide gels (30:0.8) in 0.5× TBE buffer [45 mM Tris–borate, 1 mM EDTA (pH 8.3)] at 4°C.
Purification of the host factor
Cultures of E. coli MG1655 were grown to log phase (OD600 ∼ 0.6) and harvested. About 5 g of cell pellet was resuspended in 15 ml lysis buffer [20 mM Tris–HCl (pH 7.4), 5 mM MgCl2, 1 mM EDTA (pH 8.0), 10% glycerol, 500 mM NaCl, 200 µg/ml lysozyme, 5 mM β-mercaptoethanol] and sonicated on ice. The extract was centrifuged at 100 000g for 2 h. An aliquot of the supernatant (crude cell extract) was used for Electrophoretic Mobility Shift Assays (EMSAs) and the rest was proceeded with as follows. Nucleic acids were precipitated using 1% (v/v) polyethyleneimine (pH 8.0). Hereafter, at each step of purification, fractions containing the host factor were selected based on their ability to bind Pmom in EMSA. Proteins in the extract were fractionated by a 0–65% saturated ammonium sulfate cut. The precipitated solution was spun down and the pellet was resuspended and dialyzed against TMEG100 buffer. The dialyzed fraction was mixed overnight with 5 ml pre-equilibrated phosphocellulose matrix (Whatman). After a wash with 10 ml TMEG100 buffer, elution was carried out with 5 ml TMEG1000 buffer [20 mM Tris–HCl (pH 7.4), 5 mM MgCl2, 1 mM EDTA (pH 8.0), 10% glycerol, 1 M NaCl]. The eluate was equilibrated in TMEG100 and passed through 1 ml SP sepharose column (GE Healthcare). Eluted fractions possessing Pmom specific DNA binding activity were pooled, dialyzed against 10 mM Tris–HCl (pH 7.4), lyophilized and subjected to micro LC ESI-MS (Bruker apex-ultra mass spectrometer) for whole protein molecular weight detection.
Proteins
C protein was overproduced from pVR7 in E. coli BL21(DE3) and purified as described earlier (20). RNAP was purified from E. coli K12 MG1655 as described (21). Fis was overproduced from the IPTG inducible lac promoter in pUHE25-2fis (22) in E. coli W3110 (23). The protocol described by Osuna et al. (24) was modified to include the cation exchangers phosphocellulose, sulfopropyl sepharose and heparin sepharose to purify Fis. Protein concentration was estimated by the method of Bradford (25) as well as by analyzing on SDS–PAGE gel. Restriction enzymes and DNA modifying enzymes were obtained from New England Biolabs.
In vitro transcription
Multiple round run-off transcription reactions were carried out using the linear 327 bp mom or tin7 promoter fragments (spanning the promoter from positions −136 to +79 relative to mom +1 start site; PCR amplified from pUW4mom and pUW4tin7 (20,26), respectively, using pUC forward and reverse primers). For generating transcription templates with specific promoter regions deleted, the full-length promoter fragment (described earlier) was digested with BstUI or PvuI followed by extraction from agarose gel. Fis, at indicated concentrations was incubated on ice with 40 nM template DNA in transcription buffer [40 mM Tris–HCl (pH 8.0), 5 mM magnesium acetate, 0.1 mM EDTA, 0.1 mM DTT, 100 mM KCl, 100 µg/ml BSA] for 5 min. C protein was included wherever indicated. RNAP (100 nM) was added to the reaction and closed complex was allowed to form for 10 min on ice. The reactions were then shifted to 37°C for 10 min to allow open complex formation. Transcription was initiated by adding 4 µCi [α32P]ATP and 0.3 mM NTP mix and the reactions were allowed to proceed at 37°C for 30 min. Reactions were terminated by the addition of 20 µl urea loading dye (8 M urea, 0.05% bromophenol blue, 0.05% xylene cyanol), heated to 65°C for 5 min and quenched on ice. Samples were loaded on 8% urea polyacrylamide gels and analyzed by phosphorimaging. [α-32P]ATP was purchased from Perkin–Elmer Life Sciences. NTPs and dNTPs were obtained from GE Healthcare.
Closed complex formation assays
5′ 32P labeled 78-bp Ptin7 promoter fragments (wild-type or with mutations in Fis binding sites) extending from positions −58 to +20 relative to +1 start site (Supplementary Figure S2) were incubated with indicated concentrations of Fis in transcription buffer for 10 min on ice. RNAP (20 nM) was added and further incubated for 10 min on ice. The reactions were analyzed on 5% (w/v) native polyacrylamide gels and visualized by phosphorimaging.
DNase I footprinting
Plasmid pUW4 (0.3 pmol) was incubated on ice with indicated concentrations of Fis for 30 min in footprinting buffer [10 mM Tris–HCl (pH 7.6), 2.5 mM MgCl2, 0.5 mM CaCl2]. DNase I (0.05 U, NEB) was added and incubated for 30 s at 25°C. Reactions were terminated by the addition of stop buffer [100 mM Tris–HCl (pH 7.5), 25 mM EDTA, 0.5% SDS]. Samples were deproteinized by successive extractions with phenol/chloroform/isoamylalcohol (25:24:1) and chloroform/isoamylalcohol (24:1). DNA was ethanol precipitated in the presence of glycogen as a carrier and primer extension was carried out as described by Gralla (27). Briefly, DNase I treated DNA samples were used as templates for extension with Klenow polymerase using end-labeled mom reverse primer (5′ GGAATCCGCCTTAAATAACA 3′), which anneals downstream of the +56 position with respect to the +1 transcription start site of mom. Samples were ethanol precipitated and resuspended in 4 µl of formamide loading dye. After denaturation at 95°C for 5 min, samples were electrophoresed on 8% urea polyacrylamide gels alongside dideoxy sequencing reactions and analyzed by phosphorimaging.
Construction of pLW4tin7 plasmids with mutations in Fis binding sites
The plasmid pLW4tin7 harbors Ptin7 extending from positions −136 to +79 (with respect to mom transcription start point) cloned in pNM480, resulting in a transcriptional fusion of Ptin7 with lacZ (19). Mutant plasmids pLW4tin7 FBS − 49 − 38, FBS − 49 − 38 + 3 and FBS + 3 were generated by the megaprimer method for site-directed mutagenesis using the plasmid pLW4tin7 as template. Sequences of oligonucleotides used for megaprimer generation are listed in Supplementary Table S2. The mutagenic megaprimers were then used in the Stratagene QuickChange™ side-directed mutagenesis method using Phusion high fidelity DNA polymerase (Finnzymes) to obtain mutant plasmids. Mutations introduced thus were confirmed by DNA sequencing.
β-Galactosidase assays
Plasmid borne reporter construct pLW4tin7 (explained in the previous section) and mutant constructs were introduced into E. coli CSH50 (28) and CSH50 fis::kan (29). For assessing the effect of activator C, cells were transformed with the C encoding plasmid pVN184 (19) in addition to the reporter construct. Overnight cultures were diluted 1:50 into fresh LB media containing appropriate antibiotics and incubated at 37°C until their OD600 readings were between 0.3 and 0.5. β-galactosidase activity was assayed as described (28). Assays were carried out in triplicate for each culture and the average values plotted.
RESULTS
Pmom-specific DNA binding activity in E. coli cell extracts
The finding that a host protein binds to Pmom came across serendipitously during the course of our earlier studies investigating the role of transactivator C. A 260-bp fragment encompassing Pmom was obtained by digesting pUW4 with EcoRI and HindIII (Figure 1A, lane 1 and Supplementary Figure S2). Both the strands of the fragment were radiolabeled at their 3′-ends by Klenow end-filling. In EMSA with E. coli cell extracts, it was observed that a host factor bound specifically to the fragment (Figure 1A, lanes 3 and 4). Notably, binding of the factor caused only a small shift in the mobility of Pmom as compared to that caused by the 33kDa transactivator C dimer (lane 2). We next assessed the binding region of the host factor. For this purpose, the Pmom fragment was digested with PvuI, yielding three fragments of sizes 60, 18 and 182 bp (Figure 1A, lane 5 and Figure 1B). Purified activator C bound only to the larger (182 bp) fragment, consistent with the location of the C binding site therein (Figure 1A, lane 6 and Figure 1B). On the other hand, crude cell extracts bound specifically to the 60-bp fragment indicating that the host factor binds to a site upstream of the C binding site (Figure 1A, lanes 7 and 8).
Identification of the host factor that binds specifically to Pmom
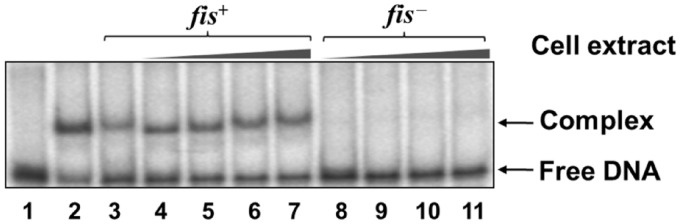
In order to identify this unknown factor, we resorted to isolate it from crude extracts of E. coli. We observed that the host factor-Pmom complex was stable at high ionic strength and competitor DNA concentrations (Figure 1C and D). In addition, its strong affinity to cation exchange resins was an indicator of its basic nature. We employed a protocol essentially encompassing cation exchange chromatography to isolate the host factor from log phase cultures of E. coli MG1655 (see ‘Materials and Methods’ section). Mass spectrometric analysis of the final fraction revealed a mass of 11.25 kDa, precisely the molecular weight of the nucleoid associated protein Fis (Figure 1E). To determine if the Pmom specific DNA binding was indeed due to Fis, we examined DNA binding activity of the cell extracts from isogenic fis− and fis+ strains JW3229-1 and BW25113, respectively (30). Specific retardation pattern, as had been observed first with E. coli MG1655 crude cell extracts was observed with fis+ strain but not with fis− strain (Figure 2, lanes 3–11). To further ascertain the identity of the host factor as Fis, we compared the DNA binding activity in crude cell extracts with that of purified Fis. Crude extracts were able to retard Pmom DNA to the same extent as purified Fis, indicating that the host factor that binds specifically to Pmom was indeed Fis (Figure 2, lanes 2 and 3).
Figure 2.
EMSA with cell extracts of wild-type (fis+) and fis− strains. The 60-bp fragment (from positions −136 to −85) of Pmom (lane 1) was incubated with 5 nM Fis (lane 2), 1 µg of crude extract of E. coli MG1655 (lane 3), and 1, 5, 10 and 15 µg of crude extracts of E. coli BW25113 (lanes 4–7, respectively) and isogenic fis− strain JW3229-1 (lanes 8–11, respectively). Specific shift as was observed with purified Fis could be seen with fis+ but not with fis− extracts. Binding buffer for all reactions contained 400 mM NaCl.
Effect of Fis on mom promoter activity
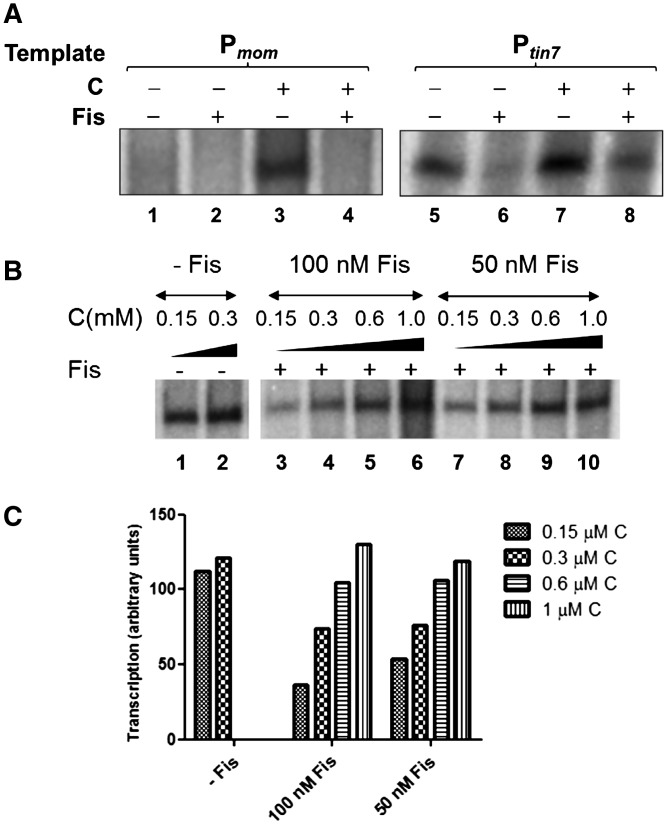
Fis, besides modulating chromatin architecture, is known to influence numerous promoters of E. coli and several other bacteria (31–33). We hypothesized that specific binding of Fis could affect the activity of mom promoter. To test this hypothesis, we conducted in vitro transcription with mom and tin7 promoters in the presence or absence of Fis. Activation of mom promoter is dependent on transactivator C. A spontaneous mutant of mom promoter—tin7 has a single base substitution at −14 position which converts it into an extended −10 promoter and thus activator independent (19). Fis repressed transcription from both Pmom and Ptin7 (Figure 3A) in a concentration dependent manner (Supplementary Figure S3). In these in vitro reactions, Fis repressed transcription even in the presence of C (Figure 3A). However, the repressive effect of Fis was reversed upon increasing the concentration of the activator C and transcription was restored to levels observed in the absence of Fis (Figure 3B and C). From these data, it is apparent that C can override Fis-mediated repression.
Figure 3.
Effect of Fis on mom promoter activity. (A) Effect of Fis on in vitro transcription from Pmom and Ptin7. Multiple round run-off transcription reactions were carried out using linear promoter fragments (−136 to +79) Pmom (lanes 1–4) and Ptin7 (lanes 5–8) in the presence or absence of 200 nM Fis. C (300 nM) was included as indicated. Transcription reactions were proceeded as described in ‘Materials and Methods’ section. At Ptin7, transcription is observed even in the absence of C. (B) Effect of C on Fis-mediated transcriptional repression. In vitro transcription reactions using Ptin7 template were carried out with increasing concentrations of C (as indicated) in the absence (lanes 1 and 2) or presence of Fis (100 nM in lanes 3–6 and 50 nM in lanes 7–10). (C) Densitometric analysis of the transcript bands from (B) was carried out and the values plotted.
Fis-mediated in vivo repression
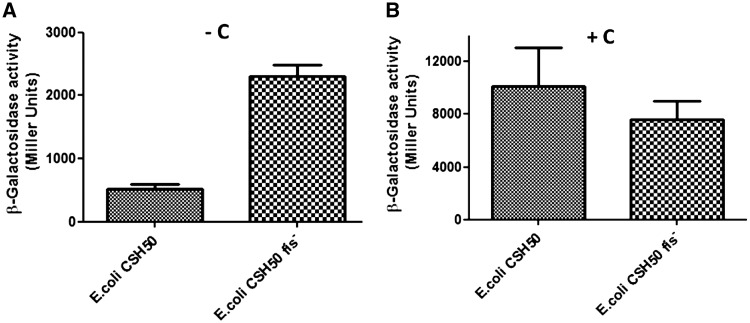
We next investigated the in vivo scenario. In order to assess the impact of Fis on transcription from Pmom, it was necessary to obviate the requirement for the positive regulator C. We therefore took advantage of the C-independent Ptin7 promoter. Plasmid borne reporter construct pLW4tin7 containing tin7–lacZ fusion was transformed into E. coli CSH50 wild-type and fis− cells and the β-galactosidase activity was measured. β-galactosidase levels in fis− cells were found to be significantly higher than those in the wild-type (fis+) strain (Figure 4A), consistent with the Fis-mediated transcriptional repression observed in vitro.
Figure 4.
Influence of Fis on promoter activity in vivo. (A) β-Galactosidase expression was measured in E. coli CSH50 wild-type and fis− strains containing Ptin7-lacZ fusion reporter construct. β-Galactosidase activity in wild-type cells was 4- to 5-fold lower than in the fis− strain. (B) β-Galactosidase activity measured in CSH50 wild-type and fis− strains containing the C encoding plasmid in addition to the plasmid borne Ptin7-lacZ fusion reporter construct.
To address the opposing effects of Fis and C in vivo and to see if C could overcome the repression imparted by Fis, wild-type and fis− cells were transformed with the C encoding plasmid and promoter activity assayed. No significant difference in the β-galactosidase levels was observed between the two strains (Figure 4B) in contrast to the scenario in the absence of C, shown in Figure 4A. This finding is in agreement with the in vitro results indicating that C can counter the repressive effect of Fis to activate transcription.
DNase I footprinting on Pmom shows multiple Fis binding sites
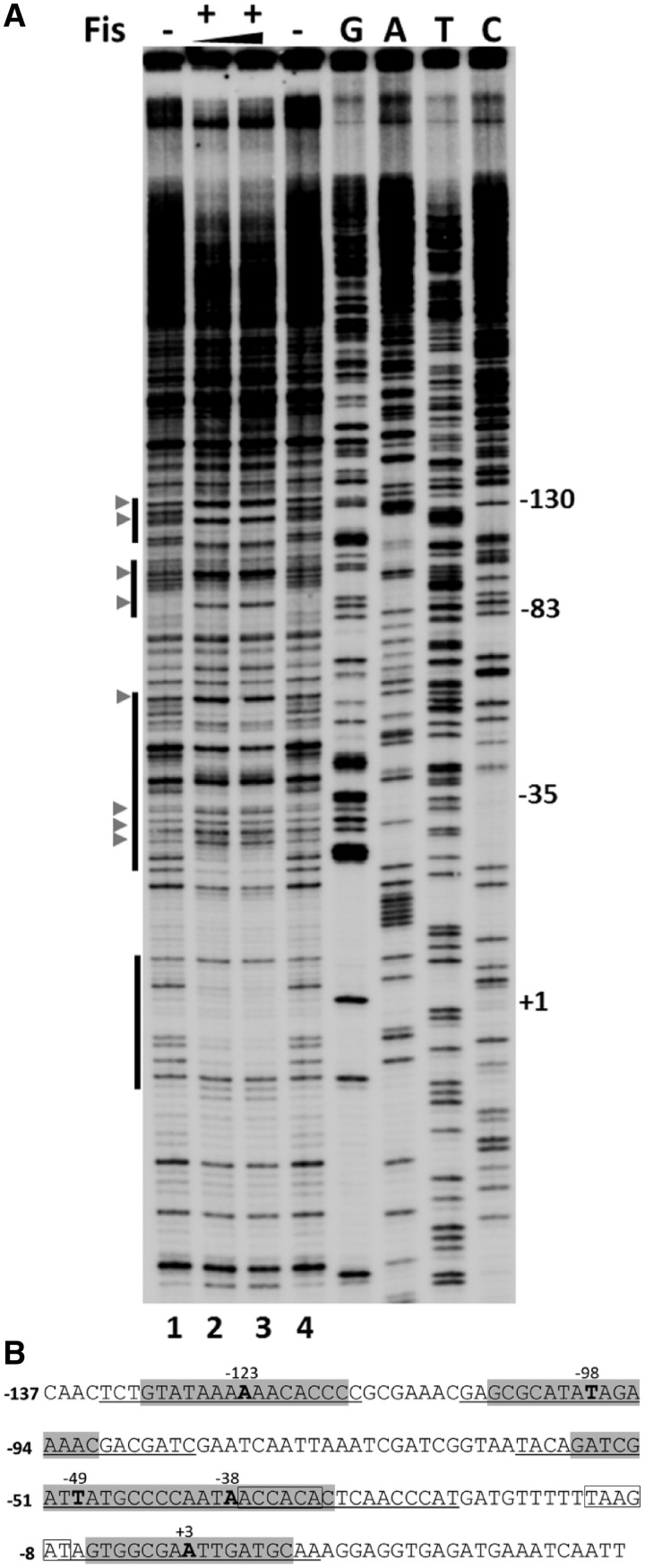
DNase I footprinting was employed to map the Fis binding sites located in Pmom (Figure 5). Fis protected a region between positions −7 to +12 in the immediate vicinity of the mom transcription start site as well as an extended region from −60 to −22. Further upstream, Fis protected two other regions between positions −107 to −84 and −133 to −115. Besides, addition of Fis also led to DNase I hypersensitivity around certain positions in the promoter, consistent with the properties of Fis to induce conformational changes in DNA (34,35). Inside the limits of each of the protected regions, a sequence similar to the consensus Fis binding site was detected (36–40). In the protected region spanning positions −60 to −22, two closely spaced sequences matching the Fis consensus site were detected. Totally, we identified five Fis binding sites in Pmom centered at positions −123, −98, −49, −38 and +3, respectively. EMSAs with the promoter fragments and purified Fis exhibited a ladder-like appearance, indicative of multiple Fis binding sites in Pmom, corroborating the footprinting data (Supplementary Figure S4 and Figure 6A, lanes 1–6).
Figure 5.
Footprinting analysis of Fis binding at Pmom. (A) DNase I footprinting analysis of Fis binding sites in Pmom. Plasmid pUW4 was incubated with Fis (288 and 384 nM in lanes 2 and 3, respectively) prior to digestion with DNase I. Primer extension was carried out with Klenow polymerase using mom reverse primer. The vertical bars on the left side indicate regions protected from DNase I in the presence of Fis. G, A, T and C refer to Sanger's dideoxy sequencing ladders of Pmom obtained using mom reverse primer. Numbers on the right denote nucleotide positions with respect to the +1 start site of Pmom. DNase I hypersensitive sites are depicted by arrowheads. (B) Sequence of mom promoter. Regions protected by Fis are underlined and sequences matching with the consensus Fis binding site are highlighted with the central nucleotide emboldened and its position indicated. The −35 and −10 promoter elements are boxed.
Figure 6.
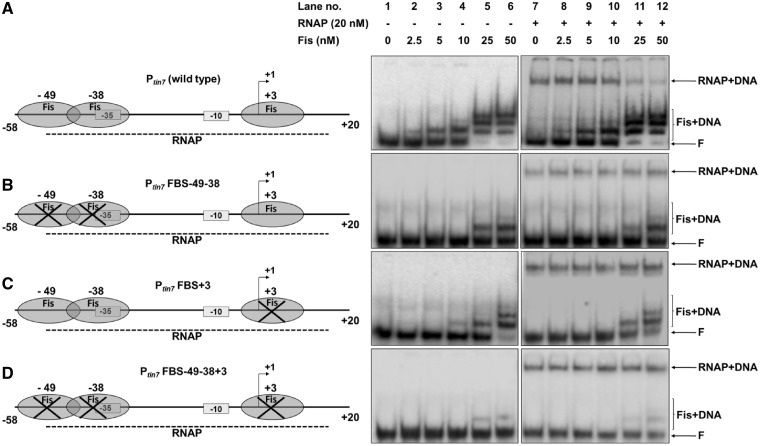
Effect of Fis on closed complex formation on various promoter fragments. (A) Ptin7 (−58 to +20 relative to +1 start point) (lane 1) was incubated with increasing concentrations of Fis as indicated (lanes 2–6). Subsequently, samples were incubated with 20 nM RNAP to allow closed complex formation (lanes 7–12). Mutant promoter fragments namely Ptin7FBS-49-38 (Fis binding sites at −49 and −38 mutated) (B), Ptin7FBS+3 (Fis binding site at +3 mutated) (C), Ptin7FBS-49-38+3 (all three Fis binding sites at −49, −38 and +3 mutated) (D) were also examined for the effect of Fis on closed complex formation at the respective mutant promoters. On the left are schematic depictions of occurrence or disruption of Fis binding sites in the respective fragments. F denotes free promoter DNA. DNA–protein complexes are indicated.
Fis excludes RNAP binding in vitro
The occurrence of multiple Fis binding sites spread over the mom regulatory region together with the observed inhibition of transcription suggested that indeed Fis binding may interfere with interaction of RNAP with the promoter. It was important at this point to examine if the transcriptional repression imparted by Fis diminished upon deletion/mutation of Fis binding sites. The Fis binding site at −123 (the site involved in Fis binding in cell extracts, see Figure 1) was targeted first for deletion. The 327-bp transcription template (encompassing −136 to +79 region of mom promoter) was digested with BstUI, removing the Fis binding site at −123 (Supplementary Figure S2). When this shortened promoter was used as a template for in vitro transcription, surprisingly, Fis continued to repress transcription (Supplementary Figure S5). A similar observation was made upon removal of the further downstream Fis binding site centered around −98 using PvuI, thus removing both the Fis binding sites located at −123 and −98 (Supplementary Figures S2 and S5). From these analyses, it appeared that the remaining promoter proximal Fis binding sites (centered at −49, −38 and +3) might be involved in transcriptional regulation of the promoter.
In parallel, we investigated the possibility of Fis directly interfering with RNAP binding. Indeed, one of the various ways by which Fis represses transcription is by competing with RNAP for binding to the promoter (41–44). Besides, the occurrence of three Fis binding sites (centered at −49, −38 and +3, respectively) overlapping the RNAP binding region substantiated the notion that Fis binding could pose steric hindrance to the polymerase and thus interfere with transcription initiation. To pursue this line of investigation further, we analyzed the effect of Fis on formation of RNAP closed complex on mom promoter, employing Ptin7, as it is amenable to closed complex analysis in the absence of C. Remarkably, at a 78-bp promoter fragment spanning positions −58 to +20 with respect to +1 start site, containing Fis binding sites at −49, −38 and +3, we observed that increasing concentrations of Fis progressively inhibited the binding of RNAP to the promoter, as evident from a diminishing closed complex (Figure 6A, lanes 7–12).Thus, Fis appears to downregulate the promoter activity by denying access to RNAP at the promoter. To test this further, we disrupted the above-mentioned Fis binding sites (singly and in combination) and examined the ability of Fis to compete with RNAP at the respective mutant promoter variants. At promoter fragments carrying mutations in Fis binding sites, Fis could no longer compete with RNAP, as evident by a stable closed complex even in the presence of high concentrations of Fis (Figure 6B–D). From these results, it appears that Fis binding at multiple sites in the promoter region is necessary for efficient repression. These competition experiments also showed that disruption of single Fis binding site markedly reduces the ability of Fis to prevent closed complex formation.
Disruption of Fis binding sites alleviates repression in vivo
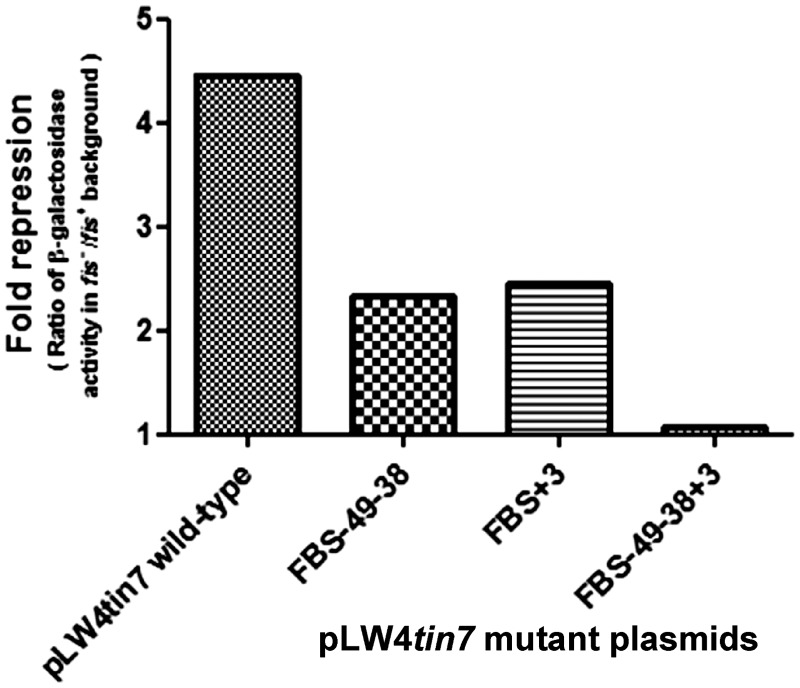
Having observed in vitro that disrupting Fis binding sites renders Fis incapable of competing with RNAP, we next tested the effect of disruption of these sites on the promoter activity in vivo. Complete alleviation of Fis-mediated repression is observed when all the three Fis binding sites were mutated whereas disruption of the site at +3 alone or those at −49 and −38 resulted in partial reduction of repression (Figure 7 and Supplementary Table S3). Thus, although both in vitro and in vivo lines of evidence highlight the importance of multiple Fis binding for maximal repression of transcription, the effect of disruption of the +3 site singly or those at −49 and −38 is not very pronounced in vivo. Perhaps this difference could be attributed to the fact that in addition to the repressive action of Fis, mom is subjected to a complex array of regulation with other factors also influencing the promoter activity (Supplementary Figure S1).
Figure 7.
Effect of disrupting Fis binding sites on Fis-mediated repression. pLW4tin7 and variants containing mutations in Fis binding sites were introduced into isogenic strains CSH50 fis− and CSH50 wild-type (fis+)and β-galactosidase activity was measured. Fold repression (the ratio of the β-galactosidase activity in fis− strain to that in fis+ strain) was plotted.
DISCUSSION
Fis was first discovered for its role in stimulating Gin catalyzed inversion of the G segment of phage Mu genome (45). Three Fis binding sites within the coding region of gin, the upstream neighbor of mom were identified in the course of the above investigation (46–48). These sites are located ∼550 bp upstream of the mom transcription start site and are distinct from those which we report in this study to be involved in regulating mom expression. Our study demonstrates that Fis represses transcription from mom promoter. However, it would be too presumptuous to attribute a primary regulatory role to Fis, given that mom expression is subject to a multitude of regulatory checkpoints. In addition, mom promoter presents a classic case of a weak, activator dependent promoter (15). Upstream of mom transcription start site is a busy area containing one or more binding sites for proteins Dam, OxyR, C, RNAP and Fis, subjecting the gene to a complex regulatory system (Supplementary Figure S1). Occlusion of mom promoter by Fis appears to prevent RNAP recruitment and thus transcription from Pmom. The present study provides evidence that Fis, a host nucleoid associated protein has been employed by the phage to achieve silencing of its potentially lethal but advantageous function. Deployment of nucleoid associated proteins as transcriptional silencers could be an effective strategy to maintain toxic genes like mom in the phage genome.
It is noteworthy that nutrient availability and growth phase of the bacterial culture have a pronounced effect on the intracellular concentrations of Fis. Fis levels markedly fluctuate from over 50 000–100 000 copies per cell in the early exponential phase or following a nutrient upshift to less than 100 copies per cell during the stationary phase and starvation (49). Earlier work has appreciated the role of Fis in Mu lysogeny. Studies employing the temperature sensitive mutant repressor cts62 have demonstrated that Fis aids the repressor in downregulating both early gene expression and transposition of phage Mu, thus contributing to maintain the phage in a lysogenic state (50). Furthermore, earlier observations have indicated that conditions of limited growth such as starvation and stationary phase (when Fis levels are low) could favor spontaneous induction of phage Mu (51,52). In such Fis depleted situations when the lytic cycle sets in, Fis-mediated repression of mom can be anticipated to diminish. In the lysogenic phase, however, there is a requirement to keep the genome silenced and abundance of Fis could effectively ensure silencing of mom. In other words, regulation of mom expression by Fis is likely to be mediated by a change in the protein abundance. Fis is known to suppress transcription from guaB, crp, fis, gyrA promoters by directly competing with RNAP (41–44). Pre-binding of Fis at these promoters impedes RNAP recruitment on account of steric hindrance. We suggest that a similar phenomenon of promoter occlusion by Fis could be responsible for silencing mom expression (Figure 8). Although it appears that binding of Fis at multiple sites in mom promoter has a cumulative repressive effect, each site on its own can also exert a partial negative effect on transcription from this promoter (Figure 7).
Figure 8.
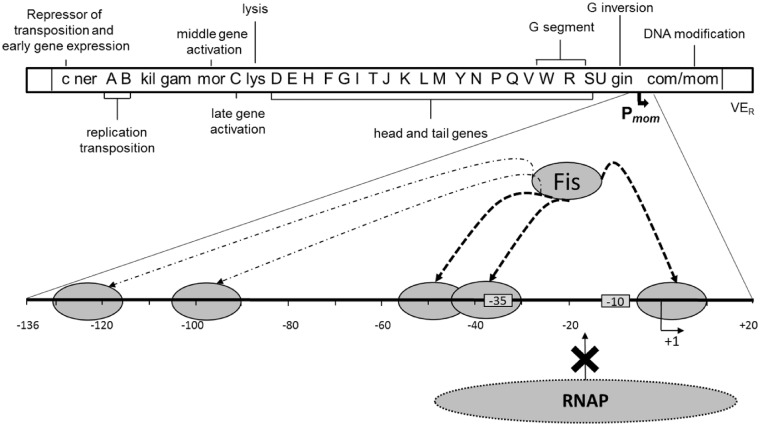
Model showing Fis-mediated silencing of mom. The rightmost gene in the Mu genome is mom. Pmom, the regulatory region of mom has been enlarged. The −35 and −10 promoter elements of Pmom are boxed. The dashed arrows indicate binding of Fis (small oval) to multiple sites in the promoter (as shown by DNase I footprinting). RNAP (large oval) cannot bind to Pmom because Fis is occluding the RNAP binding region. The Fis binding sites implicated in interfering with RNAP binding and thus with transcription are indicated by thickened arrows. Effect of Fis on other regions of the genome (see ‘Discussion’ section) is not shown.
An orchestrated cascade operating at transcriptional and translational levels regulates mom expression (Supplementary Figure S1). Most importantly, transcription of mom is contingent on C protein, a well-established activator of Pmom that is expressed only in the middle phase of the lytic cycle (53). C facilitates RNAP recruitment by realigning the out of phase −35 and −10 promoter elements (54). At a further step, C enhances promoter clearance and overcomes the influence of negative factors responsible for silencing of mom (55). Multistep activation of Pmom by C ensures mom expression at the late lytic phase. This irreversible genetic switch would have to overcome Fis-mediated repression. Since the binding region of C and two of the Fis sites overlap around the −35 region of the promoter, it is likely that under conditions of Fis abundance and in the lysogenic state, Fis would predominantly occupy the region. During the lytic cycle, low concentrations of C might not be sufficient to displace Fis bound to the promoter (Figure 3A). However, higher concentrations of the activator effectively promote RNAP recruitment and activity, evident from the significant stimulation of transcription brought about by increased concentrations of C even in the presence of inhibitory concentrations of Fis (Figure 3B and C). RNAP recruited at the promoter would also compete out Fis bound to the downstream site. Thus, the activator and the polymerase seem to join hands to overcome the repression imparted by Fis.
Silencing of Pmom by Fis is important in yet another context. Promoter-up mutations are not uncommon in nature. Spontaneous emergence of promoter-up mutations in Pmom during lysogeny would result in premature mom expression and death of the host. The role of Fis in silencing mom becomes especially important during the lysogenic state to avoid the lethal effects of untimely expression arising due to promoter mutations. The ability of Fis to repress transcription from Ptin7, an activator independent and spontaneous promoter-up mutant of Pmom emphasizes its importance as a silencer of mom, capable of eliminating any leaky expression from the promoter. Thus, Fis appears to play a dual role in Mu lysogeny; not only does it contribute to overall silencing of gene expression and transposition but also represses a toxic gene of Mu. We propose that Fis-mediated repression adds an additional layer to the already elaborate mom regulatory circuit. After all, the selective advantage imparted by the anti-restriction function of mom can outweigh the risk associated with its untimely expression by silencing of this cytotoxic gene till the late lytic phase.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3 and Supplementary Figures 1–5.
FUNDING
Funding for open access charge: Department of Science and Technology, Government of India (grant and J.C. Bose Fellowship to V.N.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to V. Ramesh whose initial observations lead to this study. We owe special thanks to Dr Georgi Muskhelishvili and Prof. Charles Dorman for providing the Fis knockout and overexpressing strains. We thank Soumitra Ghosh for his help in Fis protein purification, Sharmila for technical assistance and other colleagues for helpful discussions. S.K. is a recipient of junior research fellowship from Council of Scientific and Industrial Research, India.
REFERENCES
- 1.Taylor AL. Bacteriophage-induced mutation in Escherichia coli. Proc. Natl Acad. Sci. USA. 1963;50:1043–1051. doi: 10.1073/pnas.50.6.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Toussaint A. The DNA modification function of temperate phage Mu-1. Virology. 1976;70:17–27. doi: 10.1016/0042-6822(76)90232-4. [DOI] [PubMed] [Google Scholar]
- 3.Hattman S. Unusual modification of bacteriophage Mu DNA. J. Virol. 1979;32:468–475. doi: 10.1128/jvi.32.2.468-475.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swinton D, Hattman S, Crain PF, Cheng CS, Smith DL, McCloskey JA. Purification and characterization of the unusual deoxynucleoside, alpha-N-(9-beta-D-2′-deoxyribofuranosylpurin-6-yl)glycinamide, specified by the phage Mu modification function. Proc. Natl Acad. Sci. USA. 1983;80:7400–7404. doi: 10.1073/pnas.80.24.7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kahmann R. Methylation regulates the expression of a DNA-modification function encoded by bacteriophage Mu. Cold Spring Harb. Symp. Quant. Biol. 1983;47(Pt 2):639–646. doi: 10.1101/sqb.1983.047.01.075. [DOI] [PubMed] [Google Scholar]
- 6.Hattman S, Ives J. S1 nuclease mapping of the phage Mu mom gene promoter: a model for the regulation of mom expression. Gene. 1984;29:185–198. doi: 10.1016/0378-1119(84)90179-3. [DOI] [PubMed] [Google Scholar]
- 7.Kahmann R. The mom gene of bacteriophage Mu. Curr. Top. Microbiol. Immunol. 1984;108:29–47. doi: 10.1007/978-3-642-69370-0_4. [DOI] [PubMed] [Google Scholar]
- 8.Kahmann R, Seiler A, Wulczyn FG, Pfaff E. The mom gene of bacteriophage Mu: a unique regulatory scheme to control a lethal function. Gene. 1985;39:61–70. doi: 10.1016/0378-1119(85)90108-8. [DOI] [PubMed] [Google Scholar]
- 9.Hattman S. Unusual transcriptional and translational regulation of the bacteriophage Mu mom operon. Pharmacol. Ther. 1999;84:367–388. doi: 10.1016/s0163-7258(99)00042-x. [DOI] [PubMed] [Google Scholar]
- 10.Hattman S, Ives J, Wall L, Maric S. The bacteriophage Mu com gene appears to specify a translation factor required for mom gene expression. Gene. 1987;55:345–351. doi: 10.1016/0378-1119(87)90295-2. [DOI] [PubMed] [Google Scholar]
- 11.Wulczyn FG, Kahmann R. Post-transcriptional regulation of the bacteriophage Mu mom gene by the com gene product. Gene. 1987;51:139–147. doi: 10.1016/0378-1119(87)90302-7. [DOI] [PubMed] [Google Scholar]
- 12.Bolker M, Kahmann R. The Escherichia coli regulatory protein OxyR discriminates between methylated and unmethylated states of the phage Mu mom promoter. EMBO J. 1989;8:2403–2410. doi: 10.1002/j.1460-2075.1989.tb08370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hattman S, Sun W. Escherichia coli OxyR modulation of bacteriophage Mu mom expression in dam+ cells can be attributed to its ability to bind hemimethylated Pmom promoter DNA. Nucleic Acids Res. 1997;25:4385–4388. doi: 10.1093/nar/25.21.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hattman S, Goradia M, Monaghan C, Bukhari AI. Regulation of the DNA-modification function of bacteriophage Mu. Cold Spring Harb. Symp. Quant. Biol. 1983;47(Pt 2):647–653. doi: 10.1101/sqb.1983.047.01.076. [DOI] [PubMed] [Google Scholar]
- 15.Gindlesperger TL, Hattman S. In vitro transcriptional activation of the phage Mu mom promoter by C protein. J. Bacteriol. 1994;176:2885–2891. doi: 10.1128/jb.176.10.2885-2891.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun W, Hattman S. Bidirectional transcription in the mom promoter region of bacteriophage Mu. J. Mol. Biol. 1998;284:885–892. doi: 10.1006/jmbi.1998.2228. [DOI] [PubMed] [Google Scholar]
- 17.Cabot TL. Transcriptional activation of the bacteriophage Mu mom promoter by C protein. Ph.D. Thesis. 1995 University of Rochester, Rochester, NY. [Google Scholar]
- 18.Swapna G, Saravanan M, Nagaraja V. Conformational changes triggered by Mg2+ mediate transactivator function. Biochemistry. 2009;48:2347–2354. doi: 10.1021/bi8022448. [DOI] [PubMed] [Google Scholar]
- 19.Balke V, Nagaraja V, Gindlesperger T, Hattman S. Functionally distinct RNA polymerase binding sites in the phage Mu mom promoter region. Nucleic Acids Res. 1992;20:2777–2784. doi: 10.1093/nar/20.11.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramesh V, De A, Nagaraja V. Engineering hyperexpression of bacteriophage Mu C protein by removal of secondary structure at the translation initiation region. Protein Eng. 1994;7:1053–1057. doi: 10.1093/protein/7.8.1053. [DOI] [PubMed] [Google Scholar]
- 21.Kumar KP, Chatterji D. An improved method for the purification of DNA-dependent RNA polymerase from Escherichia coli. J. Biochem. Biophys. Methods. 1988;15: 235–240. doi: 10.1016/0165-022x(88)90010-3. [DOI] [PubMed] [Google Scholar]
- 22.Deufel A, Hermann T, Kahmann R, Muskhelishvili G. Stimulation of DNA inversion by FIS: evidence for enhancer-independent contacts with the Gin-gix complex. Nucleic Acids Res. 1997;25:3832–3839. doi: 10.1093/nar/25.19.3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kohara Y, Akiyama K, Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987;50:495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- 24.Osuna R, Finkel SE, Johnson RC. Identification of two functional regions in Fis: the N-terminus is required to promote Hin-mediated DNA inversion but not lambda excision. EMBO J. 1991;10:1593–1603. doi: 10.1002/j.1460-2075.1991.tb07680.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 26.Basak S, Olsen L, Hattman S, Nagaraja V. Intrinsic DNA distortion of the bacteriophage Mu momP1 promoter is a negative regulator of its transcription. A novel mode of regulation of toxic gene expression. J. Biol. Chem. 2001;276:19836–19844. doi: 10.1074/jbc.M011790200. [DOI] [PubMed] [Google Scholar]
- 27.Gralla JD. Rapid “footprinting” on supercoiled DNA. Proc. Natl Acad. Sci. USA. 1985;82:3078–3081. doi: 10.1073/pnas.82.10.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 29.Koch C, Vandekerckhove J, Kahmann R. Escherichia coli host factor for site-specific DNA inversion: cloning and characterization of the fis gene. Proc. Natl Acad. Sci. USA. 1988;85:4237–4241. doi: 10.1073/pnas.85.12.4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100050. 2006 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bradley MD, Beach MB, de Koning AP, Pratt TS, Osuna R. Effects of Fis on Escherichia coli gene expression during different growth stages. Microbiology. 2007;153:2922–2940. doi: 10.1099/mic.0.2007/008565-0. [DOI] [PubMed] [Google Scholar]
- 32.Browning DF, Grainger DC, Busby SJ. Effects of nucleoid-associated proteins on bacterial chromosome structure and gene expression. Curr. Opin. Microbiol. 2010;13:773–780. doi: 10.1016/j.mib.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 33.Dorman CJ, Deighan P. Regulation of gene expression by histone-like proteins in bacteria. Curr. Opin. Genet. Dev. 2003;13:179–184. doi: 10.1016/s0959-437x(03)00025-x. [DOI] [PubMed] [Google Scholar]
- 34.Stella S, Cascio D, Johnson RC. The shape of the DNA minor groove directs binding by the DNA-bending protein Fis. Genes Dev. 2010;24:814–826. doi: 10.1101/gad.1900610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan CQ, Finkel SE, Cramton SE, Feng JA, Sigman DS, Johnson RC. Variable structures of Fis-DNA complexes determined by flanking DNA-protein contacts. J. Mol. Biol. 1996;264:675–695. doi: 10.1006/jmbi.1996.0669. [DOI] [PubMed] [Google Scholar]
- 36.Hubner P, Arber W. Mutational analysis of a prokaryotic recombinational enhancer element with two functions. EMBO J. 1989;8:577–585. doi: 10.1002/j.1460-2075.1989.tb03412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finkel SE, Johnson RC. The Fis protein: it's not just for DNA inversion anymore. Mol. Microbiol. 1992;6:3257–3265. doi: 10.1111/j.1365-2958.1992.tb02193.x. [DOI] [PubMed] [Google Scholar]
- 38.Hengen PN, Bartram SL, Stewart LE, Schneider TD. Information analysis of Fis binding sites. Nucleic Acids Res. 1997;25:4994–5002. doi: 10.1093/nar/25.24.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shao Y, Feldman-Cohen LS, Osuna R. Functional characterization of the Escherichia coli Fis-DNA binding sequence. J. Mol. Biol. 2008;376:771–785. doi: 10.1016/j.jmb.2007.11.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cho BK, Knight EM, Barrett CL, Palsson BO. Genome-wide analysis of Fis binding in Escherichia coli indicates a causative role for A-/AT-tracts. Genome Res. 2008;18:900–910. doi: 10.1101/gr.070276.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Husnain SI, Thomas MS. Downregulation of the Escherichia coli guaB promoter by FIS. Microbiology. 2008;154:1729–1738. doi: 10.1099/mic.0.2008/016774-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ninnemann O, Koch C, Kahmann R. The E. coli fis promoter is subject to stringent control and autoregulation. EMBO J. 1992;11:1075–1083. doi: 10.1002/j.1460-2075.1992.tb05146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gonzalez-Gil G, Kahmann R, Muskhelishvili G. Regulation of crp transcription by oscillation between distinct nucleoprotein complexes. EMBO J. 1998;17:2877–2885. doi: 10.1093/emboj/17.10.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneider R, Travers A, Kutateladze T, Muskhelishvili G. A DNA architectural protein couples cellular physiology and DNA topology in Escherichia coli. Mol. Microbiol. 1999;34:953–964. doi: 10.1046/j.1365-2958.1999.01656.x. [DOI] [PubMed] [Google Scholar]
- 45.Koch C, Kahmann R. Purification and properties of the Escherichia coli host factor required for inversion of the G segment in bacteriophage Mu. J. Biol. Chem. 1986;261:15673–15678. [PubMed] [Google Scholar]
- 46.Kahmann R, Rudt F, Koch C, Mertens G. G inversion in bacteriophage Mu DNA is stimulated by a site within the invertase gene and a host factor. Cell. 1985;41:771–780. doi: 10.1016/s0092-8674(85)80058-1. [DOI] [PubMed] [Google Scholar]
- 47.Koch C, Ninnemann O, Fuss H, Kahmann R. The N-terminal part of the E. coli DNA binding protein FIS is essential for stimulating site-specific DNA inversion but is not required for specific DNA binding. Nucleic Acids Res. 1991;19:5915–5922. doi: 10.1093/nar/19.21.5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kahmann R, Mertens G, Klippel A, Bräuer B, Rudt F, Koch C. The Mechanism of G Inversion. New York: Alan R. Liss; 1987. [Google Scholar]
- 49.Ball CA, Osuna R, Ferguson KC, Johnson RC. Dramatic changes in Fis levels upon nutrient upshift in Escherichia coli. J. Bacteriol. 1992;174:8043–8056. doi: 10.1128/jb.174.24.8043-8056.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Betermier M, Poquet I, Alazard R, Chandler M. Involvement of Escherichia coli FIS protein in maintenance of bacteriophage Mu lysogeny by the repressor: control of early transcription and inhibition of transposition. J. Bacteriol. 1993;175:3798–3811. doi: 10.1128/jb.175.12.3798-3811.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Higgins NP. Death and transfiguration among bacteria. Trends Biochem. Sci. 1992;17:207–211. doi: 10.1016/0968-0004(92)90376-k. [DOI] [PubMed] [Google Scholar]
- 52.Shapiro JA, Higgins NP. Differential activity of a transposable element in Escherichia coli colonies. J. Bacteriol. 1989;171:5975–5986. doi: 10.1128/jb.171.11.5975-5986.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stoddard SF, Howe MM. Localization and regulation of bacteriophage Mu promoters. J. Bacteriol. 1989;171:3440–3448. doi: 10.1128/jb.171.6.3440-3448.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Basak S, Nagaraja V. DNA unwinding mechanism for the transcriptional activation of momP1 promoter by the transactivator protein C of bacteriophage Mu. J. Biol. Chem. 2001;276:46941–46945. doi: 10.1074/jbc.M107476200. [DOI] [PubMed] [Google Scholar]
- 55.Chakraborty A, Nagaraja V. Dual role for transactivator protein C in activation of mom promoter of bacteriophage Mu. J. Biol. Chem. 2006;281:8511–8517. doi: 10.1074/jbc.M512906200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.